Abstract
The mechanisms and rates by which genotypic and phenotypic variation is generated in opportunistic, eukaryotic pathogens during growth in hosts are not well understood. We evaluated genomewide genetic and phenotypic evolution in Candida albicans, an opportunistic fungal pathogen of humans, during passage through a mouse host (in vivo) and during propagation in liquid culture (in vitro). We found slower population growth and higher rates of chromosome-level genetic variation in populations passaged in vivo relative to those grown in vitro. Interestingly, the distribution of long-range loss of heterozygosity (LOH) and chromosome rearrangement events across the genome differed for the two growth environments, while rates of short-range LOH were comparable for in vivo and in vitro populations. Further, for the in vivo populations, there was a positive correlation of cells demonstrating genetic alterations and variation in colony growth and morphology. For in vitro populations, no variation in growth phenotypes was detected. Together, our results demonstrate that passage through a living host leads to slower growth and higher rates of genomic and phenotypic variation compared to in vitro populations. Results suggest that the dynamics of population growth and genomewide rearrangement contribute to the maintenance of a commensal and opportunistic life history of C. albicans.
OPPORTUNISTIC pathogens such as Candida albicans often reside in the host as benign, commensal organisms until the immune system is weakened. In patients undergoing organ transplants or chemotherapy, or when indigenous competitors are eliminated upon antibiotic treatment, opportunistic pathogens may gain access to vulnerable tissues, causing death in ≤50% of infected patients (Wilson et al. 2002). Consequently, it is important to understand the genetic mechanisms underlying the survival and adaptation of opportunistic pathogens to growth in host environments (Margolis and Levin 2007). Here, we used a genomewide array of single nucleotide polymorphisms (SNPs) to characterize the rates of genetic and phenotypic evolution accompanying the growth of C. albicans in contact with a mammalian host and compared these to rates of evolution during in vitro growth.
Genome evolution during interactions with hosts varies considerably across different microbial pathogens. The specific genome rearrangements leading to phase change and antigenic switching that allow pathogens to evade host immune responses are well described for only a few pathogens such as trypanosomes (Borst and Rudenko 1994) and Plasmodium (Kyes et al. 2001). Obligate intracellular symbiotic microbes, such as Buchnera (Moran 1996) and Pneumocystis (Strobel and Arnold 2004), propagate asexually and often carry a minimal but stable genome, making them wholly dependent on life within their hosts (Wren 2000). Although both opportunistic and obligate pathogens commonly propagate by asexual means, these organisms often maintain large genomes and generate substantial genomic and phenotypic variation via genome rearrangements (Victoir and Dujardin 2002; Kline et al. 2003) and heritable silencing at telomeres (Cross et al. 1998; Borst 2002; Gupta 2005). Given that many commensal and apparently harmless symbionts may become invasive pathogens in immunocompromised hosts, the mechanisms underlying the maintenance of genetic variation and of the commensal state bear investigation (Levin et al. 2000).
As the most common commensal fungus of the human microbial flora, C. albicans provides a model for the study of opportunistic pathogens because it reproduces primarily asexually and demonstrates a high degree of genetic and genomic variability among isolates (Cowen et al. 1999; Iwaguchi et al. 2000; Joly et al. 2002; Pujol et al. 2002; Legrand et al. 2004). The complete genome sequence revealed high levels of heterozygosity (∼4%) across the 16-Mb diploid genome (Jones et al. 2004; van het Hoog et al. 2007), and population-level variation has been demonstrated in clinical populations from different continents, regions, hospitals, and families (Forche et al. 1999; Pujol et al. 2002; Bougnoux et al. 2006). However, the genome and population processes underlying observed variation in host populations is not well understood. Appreciable rates of mitotic recombination estimated at specific genome regions (Lephart et al. 2005; Lephart and Magee 2006) and in repetitive regions (Zhang et al. 2003) have been evaluated primarily from in vitro cultures. Chromosomal variation as well as point mutations accumulate rapidly in populations evolving resistance to azole antifungal drugs (Selmecki et al. 2006; Coste et al. 2007), and in a few cases, the evolution of a pathogen within the same individual has been studied over the time course of antifungal drug treatment (Lopez-Ribot et al. 1998; Marr et al. 1998; Coste et al. 2007; Selmecki et al. 2008). Together, clinical studies reveal the accumulation of variation in host-associated populations, but the evolutionary relationship among isolates is not clear, and the number of isolates obtained during the course of infection are insufficient to allow a comprehensive view of population dynamics.
With the goal of understanding mechanisms by which genetic and phenotypic variation arise as a pathogen propagates in its host, we tracked genomewide dynamics in C. albicans populations during passage through a susceptible host (in vivo) and compared results to populations propagated in liquid culture (in vitro). We first asked if population growth rates differ when cells are grown in a mammalian host relative to when they are grown in liquid culture. We then compared the rates and types of short- and long-range loss of heterozygosity (LOH) events that arose during in vivo relative to in vitro propagation. Finally, we determined the rates and types of phenotypic variation in colony growth that arose during in vivo and in vitro propagation. To conduct the analyses, we exploited the counterselectable marker GAL1, measured recombination as LOH using genomewide SNPs, and evaluated changes in chromosome copy number using competitive genome hybridization (CGH) (Forche et al. 2005; Selmecki et al. 2005). We found fivefold lower population growth rates and distinctly different genome dynamics arising in response to growth in vivo compared to growth in vitro. Furthermore, we found that variation in C. albicans colony size and morphology arose during in vivo propagation only and was positively associated with short-range and chromosome-level recombination events. Taken together, our results suggest that passage through a mammalian host is accompanied by slow population growth and elevated levels of genetic and phenotypic variation relative to the rates of variation observed with propagation in the laboratory.
MATERIALS AND METHODS
Strains and media used in this study:
To study recombination events across the entire C. albicans genome, we used a system with two components: a heterozygous counterselectable GAL1 marker that permitted selection of isolates in which LOH at GAL1 had occurred and 123 SNP loci positioned ∼100 kb apart across most of the genome (Table 1; supporting information, Table S1). C. albicans strain AF7 is a derivative of sequenced strain SC5314 in which one copy of GAL1 was replaced with URA3 to generate the GAL1/gal1 heterozygous locus (GAL1/Δgal1∷URA3) (Forche et al. 2003). Gal+ strains are sensitive to 2-deoxygalactose (2DGS) because metabolism of 2DG results in a toxic product (Platt 1984) but grow on media with galactose as the sole carbon source. Gal− cells lack a functional GAL1 locus and are 2DG resistant (2DGR), but do not grow on media with galactose as the sole carbon source.
TABLE 1.
The number and chromosomal location of heterozygous SNP loci analyzed in reference strains SC5314 and AF7
| Chromosome | Strain SC5314 | Strain AF7 |
|---|---|---|
| R | 19 | 19 |
| 1 | 23 | 13 |
| 2 | 19 | 19 |
| 3 | 11 | 10 |
| 4 | 21 | 20 |
| 5 | 12 | 12 |
| 6 | 11 | 11 |
| 7 | 7 | 6 |
| Total | 123 | 110 |
SNPs heterozygous in AF7 were analyzed for all experimental strains.
Colonies of C. albicans were routinely grown on the nonselective YEPD medium (1% yeast extract, 1% yeast peptone, 2% glucose; 1.5% agar for plate cultures). To distinguish between Gal− (2DGR) and Gal+ (2DGS) phenotypes, strains are plated onto a synthetic 2-deoxygalactose medium (2DG; 0.67% yeast nitrogen base without amino acids, 0.1% 2-deoxygalactose, 1.5% agar) and counterselected on synthetic galactose medium (0.67% yeast nitrogen base without amino acids, 2% galactose, 1.5% agar).
In vivo populations:
In a previous study, 106 cells of the parent strain AF7 (GAL1/Δgal1∷URA3) were injected into the tail vein of 13 outbred ICR male mice (22–25 g; Harlan, Indianapolis) (Forche et al. 2003). Mice were observed and, when moribund at 5–7 days, anesthetized using isofluorane and euthanized, and both kidneys were removed. Kidneys were combined, homogenized with 1 ml of water, and dilutions at 1:1000 of the kidney homogenate were plated onto YEPD medium to obtain total colony counts. The same homogenate was diluted 1:10 and plated onto 2DG medium to obtain Gal− colony counts at 3 days. At 3 days, Gal− colonies arising by mutation do not grow to an observable size but colonies are apparent for a control Δgal1/Δgal1 strain. (Forche et al. 2003).
In vitro populations:
To generate in vitro populations, strain AF7 was streaked out on synthetic galactose medium and grown 2 days at 30° to obtain single colonies, which were removed and transferred to each of 20, 5 ml YEPD liquid cultures (Beckerman et al. 2001; Spell and Jinks-Robertson 2004; Mookerjee and Sia 2006). These were grown for 16 hr in a roller tube incubator at 30°. The cultures were spun down, washed once with sterile water, and resuspended in 1 ml of sterile water. The resuspended cells were then plated onto YEPD plates at 10−7 dilutions and grown at 30° for 2 days to obtain the total cell count and plated on 2DG plates at 10−3 dilution and grown at 30° to obtain a count of Gal− colonies at 3 days.
Diagnostic PCR to determine GAL1 status:
Diagnostic PCRs were carried out using primers flanking the GAL1 locus to determine if Gal− phenotypes (2DGR) obtained in the above experiments were due to loss of the remaining GAL1 copy or due to mutation in the GAL1 ORF. Primers gal1-detF, ura3-detR, and 2020 were used for upstream and primers 2278, 2279, and 2280 were used for downstream diagnostic PCR (Table S1). Total genomic DNA extractions were carried out as described previously (Beckerman et al. 2001). PCRs were carried out in a total volume of 25 μl with 10 mm Tris–HCl (pH 8.0); 50 mm KCl; 3 mm MgCl2; 100 μm each dATP, dCTP, dGTP, and dTTP; 2.5 units Taq polymerase (rTaq, TAKARA); either 5 or 10 μmol of each primer (see Table S1); and 30 ng of genomic DNA under the following conditions: initial denaturation for 3 min at 94°, 30 cycles of a denaturation step for 1 min at 94°, a primer annealing step for 30 sec at 54°, and an extension step for 1 min at 72°. The final extension step was 5 min at 72°. PCR fragments were size fractionated on agarose gels [1% in 1× TBE (0.89 m Tris; 0.02 m EDTA–NA2H2O; 0.89 m boric acid)] and compared to the size of products from positive control strains [SC5314 (GAL1/GAL1) and AF7 (GAL1/gal1Δ)] and the negative control strain [AF27 (Δgal1/Δ gal1)] (Forche et al. 2003).
Population growth rate:
For in vivo experimental populations, we used two methods to obtain estimates of net population growth rate and the number of cell divisions. First, we obtained and analyzed published results from a careful time-course experiment tracking C. albicans population growth in vivo (MacCallum and Odds 2004). Our study used strain genotypes and inoculation cell numbers comparable to those used in that study (MacCallum and Odds: SC5314 at 5 × 104 cells/g body weight of BALB/c mice; our study: AF7 at 4 × 104 cells/g body weight of ICR mice), both with tail-vein infection. Strain AF7 is derived from SC5314, is prototrophic, shows no growth rate differences, and is not attenuated in virulence in comparison to SC5314 (Forche et al. 2003). Numbers of colony-forming units (CFUs) in kidney tissues over time were log transformed and regressed against time (minutes) in Microsoft Excel 2004, version 11.3.7. Obtaining an increasing function, the slope of the regression of ln(cell number) vs. time for the period of 2–72 hr after inoculation provided an estimate of the intrinsic population growth rate, r. The number of cells generated per parent cell during time, t, of increasing growth was calculated as ert, and the number of doublings (cell divisions) was calculated by solving for x = log2(ert). A second estimate of population growth in vivo was made using total colony counts from homogenized kidney tissue and by estimating the number of doublings (cell divisions) required to produce the observed increases in cell numbers: x = log2(ert). We assumed a bottleneck population size as a fraction of the final population size (MacCallum and Odds (2004) to estimate starting population sizes in the kidney.
For in vitro experimental populations, cell numbers at the start and end of population growth were counted as CFUs on YEPD plates and population growth rates were estimated as described above for in vivo experiments.
Rates of LOH at GAL1:
For both the in vivo and in vitro experiments, we used Lea and Coulson's (1949) method of the median to estimate the rate at which the heterozygous GAL1/Δgal1 (2DGS) locus is converted to the homozygous state, Δgal1/Δgal1 (2DGR). The method uses the variance among independent cultures for the proportion of 2DGR cells generated relative to the total number of cells generated over the same time to estimate the rates of the event per generation (Table S2). The method accounts for differences of in vitro and in vivo growth rates and permits direct comparison of LOH rates at GAL1.
SNP microarray analysis:
We expanded a previously described SNP microarray (Forche et al. 2005), adding 98 SNP loci to cover a total of 123 SNP loci (Table 1). The SNP loci are positioned ∼100 kb apart across the eight C. albicans chromosomes except for regions of low polymorphism in the fully sequenced genome of strain SC5314 (Chr3 right arm, Chr7 left arm, and telomere distal regions of ChrR (van het Hoog et al. 2007). Design of allele-specific oligonucleotides, probe generation, slide preparation and hybridization, and data analysis was performed as described earlier (Forche et al. 2005). Four multiplex PCR reactions with 28–34 primer pairs/reaction (Table S1) were carried out for each C. albicans isolate, using 60 ng of genomic DNA and a Qiagen Multiplex PCR kit following the manufacturer's instructions (Qiagen, Valencia, CA).
The SNP genotypes were determined using the allelic fraction (AF), which describes the relative intensity of the control probe (SC5314: labeled with Cy5) compared to the experimental probe (test strain: labeled with Cy3) in competitive hybridizations to the microarray according to the protocol detailed in Forche et al. (2005). The AF values are normalized to give an expected value of 0.5 for heterozygous loci. The distribution of AF values for homozygous and heterozygous loci and the cutoff values to score homozygous states were as described previously (Forche et al. 2005). For each individual SNP locus, AF values between 0.40 and 0.60, inclusive, were scored as heterozygous, AF values <0.40 were scored as allele 1 homozygous, and AF values >0.60 were scored as allele 2 homozygous.
We used SNP genotypes and information on their physical linkage to infer chromosome-level rearrangement and altered ploidy. Individual SNP loci were scored as potentially homozygous, heterozygous, or trisomic by AF values as follows: 0–0.342 (homozygous for a1); 0.343–0.422 (trisomy; two copies of a1 and one copy of a2); 0.423–0.573 (heterozygous; a1/a2); 0.574–0.657 (trisomy; 2 copies of a2 and 1 copy of a1); and 0.658–1.0 (homozygous for a2) (data not shown). Where AF values suggested altered ploidy at linked SNP loci over large segments or entire chromosomes, we confirmed ploidy levels using CGH (Selmecki et al. 2005; Legrand et al. 2008) as described below.
Genomic distribution of short-range LOH events:
For in vivo and in vitro populations, we counted the number of short-range LOH events occurring on each chromosome and compared the observed number of LOH events to that expected for a random distribution across the genome within each population. We tested the significance of higher or lower numbers of observed events compared to that expected using a binomial test (Sokal and Rolf 1981) with an expected value set at the average number. Because the number of events detected is very low relative to the number of loci evaluated and because the binomial test lacks power, we also conducted a permutation test. Here, we randomly permuted loci (columns in Table S3) across all chromosomes and divided columns into “chromosomes” with the same number of loci as in the empirical data set. Repeating permutations, we generated 1000 random distribution data sets. Significance was evaluated as the number of times that the observed number of events occurred in the permuted data sets, divided by 1000, the total number of randomized data sets.
Comparative genome hybridization:
Comparative genome hybridization uses a microarray format with probes for all of the ∼6800 C. albicans ORFs and detects changes in gene copy number using hybridization signal intensity (Selmecki et al. 2005). CGH was carried out as described previously (Selmecki et al. 2005) using strains SC5314 and AF7 as heterozygous, disomic reference strains. An updated version of the Ch_map program, provided by Sven Bergmann (Selmecki et al. 2005), was used to visualize combined SNP and CGH results.
Colony growth phenotype:
Colonies obtained from experimental populations were streaked for single colonies on YEPD plates, and colony morphology was scored as follows: the parental colony phenotype (PT) that is indistinguishable from that of AF7, the small colony phenotype (Sm), and the wrinkly colony phenotype (Wr). In addition, isolates were grown for 2 days at 23°, 32°, and 37° in duplicate YEPD plates. Strains that formed wrinkled colonies at all three temperatures were designated as “wrinkling not regulated by temperature” (Wr-N-R). Strains that formed smooth yeast colonies at 23° and formed filamentous, wrinkled colonies at 32° and 37° were designated as “wrinkling that is temperature regulated” (Wr-T-R). The frequency of each phenotype in experimental populations was recorded, and plates were photographed.
RESULTS
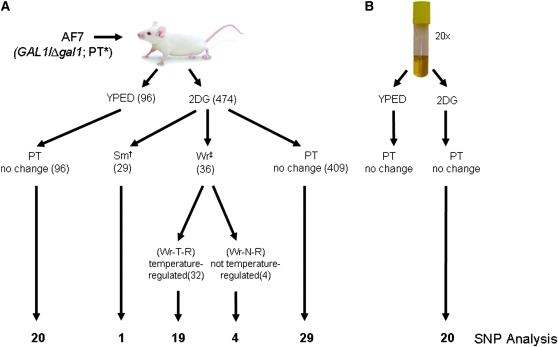
Our overall goal was to estimate and compare rates at which genetic and phenotypic variation is generated in C. albicans populations propagated in vivo and in vitro. From the in vivo experimental populations, 96 single-colony isolates (strains) were obtained from nonselective YEPD plates (Gal+, 2DGS) and 474 strains were isolated from 2DG plates (Gal−, 2DGR) for further study. From the in vitro experiments, 20 2DGR strains were isolated to serve as a control population (Figure 1). In the results reported below, GAL1 status of single colony isolates was determined by plating on selective media and by diagnostic PCR (data not shown; see materials and methods).
Figure 1.—
Propagation of C. albicans populations for in vivo and in vitro experiments, media used for isolation (YEPD, 2DG), and occurrence of altered colony phenotypes. Numbers of strains isolated for study are shown in parentheses, and numbers of isolates analyzed using the SNP microarray are shown at the bottom. (A) In vivo experiments. Colony growth phenotype indicated as PT*, parental type; SM†, small colony; and Wr‡, wrinkly colony. Wr-T-R, “wrinkly, temperature regulated”; Wr-N-R, “wrinkly, not regulated by temperature.” (B) In vitro analysis. Twenty strains isolated for SNP analyses from 2DG plates. PT colony growth phenotype observed for all isolates.
In vivo populations:
We estimated the total CFUs at 8.0 × 104–1.7 × 106 cells/g kidney across kidneys of 13 infected mice (Forche et al. 2004). Because kidney tissues were thoroughly homogenized, we assumed that each CFU represented a single nucleate hyphal or yeast cell. We estimated the proportion of cells that had undergone LOH at the GAL1 locus (2DGR) at 8.7 × 10−5–1.7 × 10−2/cell for all mice. Among 474 2DGR isolates, 65 (13.7%) exhibited colony phenotypes different from that of the parent AF7, and the remainder demonstrated AF7 PTs. The altered colony phenotypes included Sm colonies due to slow growth and Wr colonies due to filamentous cell growth compared to AF7 (Figure 1); these are fully characterized below. In contrast, none of the 96 colonies isolated on YEPD medium demonstrated altered colony growth phenotypes.
In vitro populations:
The proportion of cells that had undergone LOH at the GAL1 locus during in vitro propagation was 3.2 × 10−5–1.1 × 10−4/cell across the 20 independent YEPD cultures. No altered growth phenotypes were detected among ∼7500 colonies grown on YEPD or on 2DG plates after in vitro propagation (Figure 1).
Population growth rates in vivo:
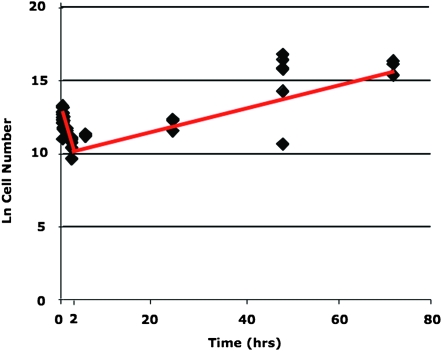
We employed two different approaches to estimate growth rates for in vivo populations, and these yielded similar results. We report net population growth rates because rates of birth and death cannot be separately estimated. First, we estimated the population growth rate and the number of cell divisions using the published results of a careful time-course study (MacCallum and Odds 2004). These data show that most C. albicans cells are cleared from the blood and that population numbers in the kidney greatly decline during the first 2 hr after inoculation (MacCallum and Odds 2004). The estimated population bottleneck size was 0.45% of the final population size at 2 hr. After 2 hr, populations in the kidneys demonstrated log growth, and the slope of the regression of ln(cell number) vs. time for the period of 2–72 hr after inoculation (Figure 2) provides an estimate of the intrinsic population growth rate, r, at 1.3 × 10−3 (r2 = 0.75; SE ± 0.0002). The number of cells generated during the time t = 70 hr of increasing growth is ert = 3.2 × 102 cells/cell/g kidney. On average, log2 (320) = 8.3 cell divisions/parent cell in the population is required to generate 320 cells.
Figure 2.—
Graph of log-phase population growth during in vivo propagation, ln (cell number) over time in hours. From the data of MacCallum and Odds (2004).
Second, we assumed a bottleneck population size at 0.45% of the final population size for our experimental in vivo populations and then calculated the average net increase in cell numbers required to obtain final population sizes of 2.2 × 102 cells/cell/g kidney. The estimated number of cell divisions is log2 (220) = 7.8 cell divisions/parent cell in the population, a value similar to that obtained above.
Rates of cell division are slower in vivo than in vitro:
The average rate of cell division for in vivo populations estimated from data of MacCallum and Odds (2004) is 8.3 cell divisions over 70 hr, or 0.12 generations/hr. From our experimental in vivo data, we estimated a rate of 7.8 cell divisions over 5 days, or 0.065 generations/hr, which is even slower. For in vitro populations, we used direct counts of total CFUs before and after growth in liquid culture to estimate an average 9.9 cell divisions/parent cell and an average rate of cell division over the 16-hr experiment at 0.62 divisions/hr. Thus, net population growth rates were 5- to 10-fold slower in vivo than in vitro.
Rates of recombination at GAL1 are higher in vivo than in vitro:
For both the in vivo and in vitro populations, we used Lea and Coulson's (1949) method of the median to estimate the rate at which the heterozygous GAL1/Δgal1 locus is converted to the homozygous state, Δgal1/Δgal1 (Table 2; Table S2). For the in vivo populations, we estimated the total CFUs at 8.0 × 104–1.7 × 106 cells/g kidney across the 13 mouse populations. We estimated the proportion of cells that had undergone LOH at the GAL1 locus to generate 2DGR cells at 8.7 × 10−5–1.7 × 10−2/cell across the 13 mouse populations and rates of LOH at GAL1 at 1.7 × 10−4 (±2.12 × 10−4 SD) events/generation.
TABLE 2.
Number, type, and rates per generation (±SD) of genome rearrangement in strains derived from in vitro and in vivo experiments
|
In vivo experiment
|
In vitro experiment
|
|||
|---|---|---|---|---|
| Type of events | No. of events | Ratea | No. of events | Ratea |
| GAL1 | 1.7 × 10−4 | 6.0 × 10−6 | ||
| (±2.2 × 10−4) | (±2.0 × 10−6) | |||
| SNP LOH | 16 | 3.3 × 10−4 | 7 | 3.5 × 10−4 |
| (±5.8 × 10−4) | (±2.4 × 10−5) | |||
| BIR | 1 | 0 | 0 | |
| Whole chromosomes | 5 | 0 | 0 | |
| All chromosomes | 6 | 1.2 × 10−3 | 0 | 0 |
| (±4.2 × 10−3) | ||||
Standard deviation was calculated across loci (short-range LOH) or across strains (chromosome-level rates).
GAL1 LOH rates were calculated as events per generation, SNP LOH rates were calculated as events per locus per generation, and chromosomal rates were calculated as events per chromosome per generation.
For the 20 in vitro cultures, we estimated the proportion of cells that had undergone LOH at the GAL1 locus at 3.2 × 10−5–1.1 × 10−4/cell across cultures and the rates of LOH at 6.0 × 10−6 (±2.0 × 10−6 SD) events/generation. Thus, the rate of recombination at GAL1 is ∼28-fold greater during in vivo growth than during in vitro growth (Table 2).
Rates of LOH at individual SNP loci are similar in vivo and in vitro:
For in vivo-propagated populations, we determined whole-genome SNP genotypes for 73 strains that were chosen to represent the range of observed 2DG and colony morphology phenotypes. We analyzed SNP genotypes for 20 2DGS and 53 2DGR strains, and among these 2DGR strains, we analyzed SNPs for 24 strains with altered colony phenotypes (Sm and Wr) and 29 strains with PT phenotypes. For the in vitro populations, we analyzed 20 2DGR isolates (Figure 1). In the course of these genotypic analyses, we found that 13 of the 123 SNP loci were homozygous in the AF7 parent strain; 10 on Chr1, and one each on Chr3, Chr4, and Chr7. These were excluded from further analyses, giving a total of 110 SNP loci analyzed for the 93 strains from both experiments (Table 1). Diagnostic PCR confirmed that all 2DGR strains lacked both copies of GAL1 and that all 2DGS strains retained at least one copy of GAL1 (data not shown).
After first determining that the LOH events located in genomes of different isolates obtained from the same mouse represented independent events, we counted 16 short-range LOH events over the 7442 individual SNP loci for which unambiguous data were obtained (Table S3); Of these, 15 altered only a single SNP locus and one longer recombination event spanned three contiguous SNP loci on Chr4 (Table 3). Assuming that each cell represents an average of 7.8–8.3 mitotic cell divisions, the estimated rate is 2.5 × 10−4 short-range LOH events/locus/generation (±5.2 × 10−4 SD). To determine the sensitivity of the estimate to the population evaluated, we analyzed results for only the 2DGR strains where 15 events were observed. The calculated rate is slightly higher at 3.3 × 10−4 events/locus/generation (±5.8 × 10−4 SD). Both estimates are comparable to the rate of LOH at GAL1 during in vivo growth that we obtained above (1.7 × 10−4 events/generation).
TABLE 3.
Short-range LOH and chromosome-level events observed for strains from in vivo and in vitro experiments
| Event | Chromosome | Description | Host ID | Strain |
|---|---|---|---|---|
| In vivo experiments | ||||
| Chr | R | Whole-chromosome LOH | E63(3)a | AF617 |
| Chr | R | Whole-chromosome LOH | A22(15) | AF3976 |
| Chr | R | Whole-chromosome trisomy | A22(15) | AF3977 |
| Chr | 2 | BIR,b 11 contiguous loci | B22(8) | AF21 |
| Chr | 2 | Whole-chromosome LOH | A22(15) | AF3990 |
| Chr | 2 | Whole-chromosome trisomy | B63(9) | AF540 |
| LOH | R | Single LOH at 1694/2254 | A22(15) | AF656 |
| LOH | R | Single LOH at 1645/2358 | A63(8) | AF121 |
| LOH | 1 | Single LOH at 2347/2406 | A53(4) | AF42 |
| LOH | 1 | Single LOH at 1916/2198 | A53(4) | AF51 |
| LOH | 1 | Single LOH at 2347/2406 | B63(9) | AF160 |
| LOH | 1 | Single LOH at CPH1 | B63(9) | AF540 |
| LOH | 1 | Single LOH at 2080/2297 | B63(9) | AF169 |
| LOH | 2 | Single LOH at 1760/2185 | C63(5) | AF194 |
| LOH | 3 | Single LOH at 2095/2215 | A63(8) | AF122 |
| LOH | 3 | Single LOH at 1629/1863 | B63(9) | AF157 |
| LOH | 4 | LOH (three continuous loci) | C63(5) | AF558 |
| LOH | 4 | Single LOH at 2101/2204 | C63(5) | AF558 |
| LOH | 6 | Single LOH at 1372/2355 | B53(5) | AF72 |
| LOH | 6 | Single LOH at 1372/2355 | B63(9) | AF148 |
| LOH | 7 | Single LOH at 6.1345 | B22(8) | AF22 |
| LOH | Unknown | Single LOH at B5B7 | A22(15) | AF3990 |
| In vitro experiments | ||||
| LOH | 2 | Single LOH at 1775/2077 | — | AF7-8 |
| LOH | 2 | Single LOH at 1179/2238 | — | AF7-14 |
| LOH | 2 | Single LOH at 2184/2321 | — | AF7-16 |
| LOH | 3 | Single LOH at 1629/1863 | — | AF7-3 |
| LOH | 3 | Single LOH at 2195/2207 | — | AF7-7 |
| LOH | 3 | Single LOH at 1629/1863 | — | AF7-17 |
| LOH | 4 | Single LOH at 2004/2043 | — | AF7-12 |
Number of strains analyzed per mouse in parentheses.
BIR, break-induced replication.
For the in vitro-propagated populations, we detected seven LOH events at single SNP loci and no events composed of multiple SNP loci over the 2012 individual SNP loci for which unambiguous data were obtained. Assuming that each cell represents an average of 9.9 mitotic cell divisions, we obtained a rate of 3.5 × 10−4 short-range LOH events/locus/generation (±2.4 × 10−5 SD) (Table 2). We conclude that the average rates of short-range LOH due to recombination and independent of those occurring at GAL1 are not significantly different for in vivo and in vitro populations.
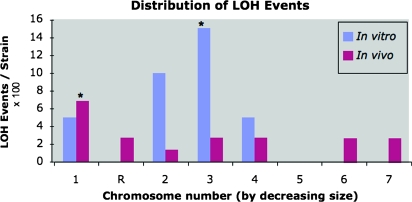
Genomewide distribution of short-range LOH events:
We next evaluated separately the distribution of short-range LOH events across chromosomes for in vivo and in vitro propagated populations (Figure 3). Among the in vivo-propagated strains, LOH events were distributed across all chromosomes except for Chr5 and Chr5. Comparing the number of LOH events/strain for each chromosome to that expected for each chromosome if LOH events were randomly distributed, the number of events on Chr1 was significantly greater than expected for a random distribution (P < 0.05; binomial test or permutation test; see materials and methods). The larger number of LOH events on Chr1 parallels the higher rate of LOH at GAL1, also located on Chr1.
Figure 3.—
Chromosomal distribution of short-range LOH events involving one to three SNP loci, analyzed separately for in vivo and in vitro data sets. Shown is the number of rearrangement events per strain for each chromosome (×100) ordered from largest (Chr1) to smallest (Chr7) chromosome. An asterisk indicates the chromosomes for which greater than expected (average) numbers of LOH events were observed (P < 0.05) within the in vivo or in vitro data sets.
For the in vitro-propagated populations, short-range LOH events were less evenly distributed across chromosomes than for in vivo strains. LOH occurred primarily on the larger chromosomes, Chr1, Chr2, Chr3, and Chr4, although no events were detected on the second largest chromosome (ChrR) or on the smaller chromosomes (Chr5, Chr6, and Chr7) (Figure 3). The number of events on Chr3 was significantly greater than expected for a random distribution (binomial test or permutation test; see materials and methods).
Comparing the results for in vivo and in vitro populations, it is apparent that the distribution of short-range LOH events differs (Figure 3). For either the in vitro or the in vivo populations, results show no significant correlation between the number of LOH events and either chromosome size or number of SNP loci analyzed per chromosome (data not shown). For both populations, permutation tests revealed that low numbers of events on some chromosomes could have occurred by chance. These results suggest that the observed differences in the distribution of short-range LOH events across the genome are driven more by the growth environment or the chromosome structure than by the size of the chromosome.
Chromosome-level changes were observed for in vivo, but for not in vitro, populations:
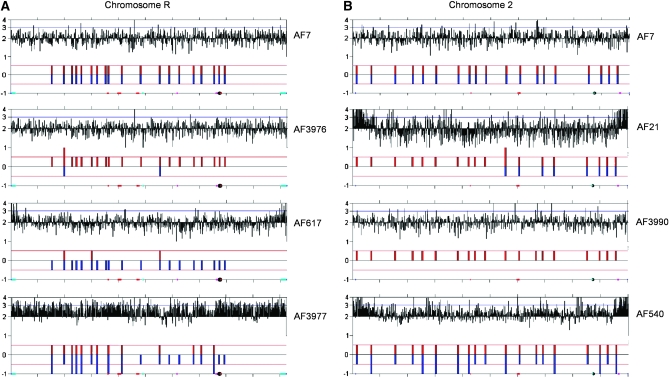
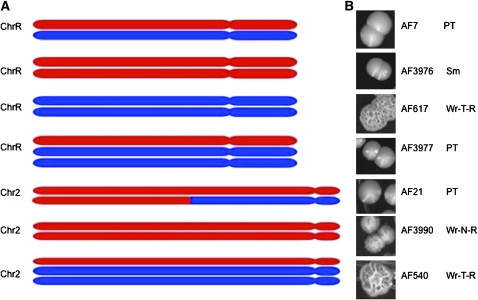
We inferred long-range chromosome-level rearrangement events when AF values for SNP loci and CGH results demonstrated changes spanning all or most of a chromosome (Selmecki et al. 2005). Importantly, we detected six long-range events in isolates from in vivo populations (AF617, AF3976, AF3977 for ChrR; AF21, AF540, AF3990 for Chr2) but no long-range events in isolates from in vitro populations. The six long-range events that we detected involved two chromosomes, ChrR and Chr2 (Tables 2 and 3).
For ChrR, we recovered two strains with disomic, homozygous ChrR and one strain carrying a trisomic ChrR. Homozygous disomy for ChrR was inferred for strains AF617 and AF3976 because the AF values at most individual SNP loci were close to either 0 or 1, indicating that the entire ChrR was homozygous (Table S3). CGH analysis for these same strains demonstrated that ChrR was present in two copies (Figure 4), suggesting that one ChrR homolog was lost and the second homolog reduplicated. Interestingly, AF values were ∼0 for strain AF617 and ∼1 for AF3976 at some SNPs (e.g., locus1381/2345); they were the opposite at other SNPs: ratios were ∼1 for strain AF617 and ∼0 for AF3976 (e.g., locus 1694/2254) (Table S3). We infer that strains AF617 and AF3976 are homozygous for different ChrR haplotypes (phases of alleles). We defined ChrR haplotype 1 as the set of SNP alleles found in strain AF3976 and ChrR haplotype 2 as the set of alleles found in strain AF617. Because strains AF617 and AF3976 were obtained from different mice, they must represent independent chromosome loss and reduplication, or nondisjunction, events.
Figure 4.—
Correspondence of CGH and SNP array results showing chromosome copy number for (A) ChrR and (B) Chr2. Results are shown for parental strain AF7 and those strains demonstrating chromosomal-level rearrangement. Data are plotted as a function of chromosome position using a modified version of Ch_map (Selmecki et al. 2005). Strong signals at telomeres for CGH results are due to incomplete digestion of genomic DNA.
Trisomy was inferred for ChrR of strain AF3977 because most SNP loci demonstrated AF values at either ∼0.3 or ∼0.7, which is intermediate between that expected for heterozygous loci (∼0.5) and that expected for homozygous loci (∼0 or 1) (Table S3). CGH analyses confirmed that ChrR is trisomic (Figure 4A). Following the haplotype designations above, strain AF3977 likely carries two copies of haplotype 2 and one copy of haplotype 1 at ChrR. Nondisjunction could explain the ChrR complement for strains disomic AF3976 and trisomic AF3977 because these strains were isolated from the same mouse.
For Chr2, we recovered three different strains demonstrating chromosome-level events (Figure 4B, Table 3). Because these were recovered from populations in three different mice, we assume that they represent independent events. In strain AF3990, the AF values for most SNP loci were close to either 0 or 1, revealing that Chr2 was homozygous. CGH analysis determined that Chr2 was disomic. In strain AF540, AF values were intermediate between expected homozygous and heterozygous values, suggesting that markers on Chr2 were trisomic, a result confirmed by CGH analysis. In the third strain, AF21, only SNP loci on the left arm of Chr2 were homozygous, and CGH showed that Chr2 was disomic, leading to the conclusion that break-induced replication (BIR) or a single crossover (Kraus et al. 2001) generated the left arm of Chr2 (Figure 5). We designated haplotype 1 as the set of alleles found homozygous on Chr2 in strain AF3990. This same set of alleles was homozygous for most of the left arm of Chr2 in strain AF21. Haplotype 2 was defined as the set of alleles with AF values of ∼0.7 on trisomic Chr2 in strain AF540 (Table S3).
Figure 5.—
Chromosome rearrangement is associated with altered colony phenotypes. (A) Chromosome diagrams for ChrR and Chr2. Haplotype 1 is indicated in red and haplotype 2 is indicated in blue. (B) Colony morphology, strain identification, and colony growth phenotype associated with chromosomal genotypes.
In summary, all six chromosome-level recombination events were recovered from in vivo propagated strains, and these involved only two chromosomes, ChrR and Chr2 (Tables 2 and 3). Among these six long-range LOH events, we found two cases of whole-chromosome trisomy (one for ChrR and one for Chr2), three cases of chromosome homolog loss with duplication of the remaining homolog by reduplication or nondisjunction, and one case in which most of one chromosome arm likely underwent BIR or a single crossover (Kraus et al. 2001) (Tables 2 and 3). The in vivo rate of long-range chromosome-level events was estimated at 1.2 × 10−3 events/chromosome/cell division (±4.3 × 10−3 SD) among the 584 chromosomes evaluated, an order of magnitude higher than the rate of short-range LOH events in this same population. Chromosome-level events were not detected among the 160 chromosomes evaluated from in vitro-propagated isolates (Tables 2 and 3).
LOH at GAL1 is associated with recovery of additional rearrangement events:
We asked whether LOH at GAL1, detected by 2DG selection, was associated with recovery of genome rearrangement events occurring independently of those at the GAL1 locus. Among the in vivo-propagated strains, we performed SNP analysis on 53 2DGR and 20 2DGS strains. Among the 2DGR isolates, we found 14 short-range LOH events covering a single SNP locus, one event covering three SNPs, and six chromosome-level events described above. In the 20 2DGS strains, we detected only a single SNP LOH event, on ChrR, in strain AF656 (Table 3). From the in vitro-propagated populations, 20 2DGR strains demonstrated seven different LOH events at single SNP loci and no chromosome-level events.
An association test showed that genome rearrangement events were significantly associated with 2DGR strains (Gstat = 6.23, d.f. = 1, P = 0.01). From the data available, we cannot determine if events at GAL1 and other SNP loci or chromosomes occurred in the same cell cycle or at different times during cell division in the host. Nonetheless, the 2DG selection for strains exhibiting LOH at GAL1 yields strains that have undergone additional LOH events at other loci and chromosomes.
Altered colony phenotypes arose during in vivo but not in vitro propagation:
We next determined the rate at which altered colony growth phenotypes arose and the relationship between these phenotypes and genomewide genetic changes. For the in vivo populations, when grown at 32°, 65 of the 474 2DGR isolates (13.7%) exhibited colony growth phenotypes different from that of parent strain AF7 (“PT” in Figure 5). Of these 65 strains with altered colony phenotype, 29 formed smaller colonies (Sm) than did AF7, and 36 exhibited filamentous Wr colony morphologies with varying degrees of wrinkling relative to strain AF7. The Sm phenotype was due to slower cell growth, as determined by growth-rate measurements in liquid culture for two of these Sm strains (data not shown). Of the 36 Wr strains, 32 formed smooth colonies at 23°, but formed filamentous, wrinkled colonies at 32° and 37°, the Wr-T-R phenotypes. Four of the 36 Wr strains formed wrinkled colonies at all three temperatures, the Wr-N-R phenotypes (Figure 5, Table 4).
TABLE 4.
Association of genotypic and phenotypic change
| Colony growth phenotypea
|
|||||
|---|---|---|---|---|---|
| Parental
|
Nonparental
|
||||
| Genome change | PT | Sm | Wr-N-R | Wr-T-R | Totalb |
| No change | 42 | 0 | 0 | 10 | 52 |
| Chromosome and BIRc | 2 | 1 | 1 | 2 | 6 |
| LOH | 5 | 0 | 4 | 7 | 16 |
| Total | 49 | 1 | 5 | 19 | 74 |
Goodness-of-fit statistic: 26.45, d.f. = 10, P < 0.005.
PT, parental colony phenotype; Sm, small colony phenotype; Wr-N-R, not regulated by temperature; Wr-T-R, colony phenotype dependent on temperature.
One strain exhibited two LOH events, giving a total of 74 events in 73 strains.
BIR, break-induced replication.
In striking contrast to the in vivo populations, we did not observe altered colony phenotypes among >7000 colonies observed during the in vitro experiment. Although filamentous colony phenotypes are detected occasionally during routine maintenance of SC5314 and its derivative strains, these events are rarely observed with growth at 32°. We discount the explanation that 2DG media is mutagenic and leads to increased rates of LOH and phenotypic variation because the in vitro-propagated cells that were subjected to 2DG media did not show elevated rates of LOH or phenotypic variation. Thus, the higher rate of altered colony phenotypes observed in strains from in vivo propagation and selected on 2DG is attributable to different growth conditions in vivo compared to those in vitro, and not to the 2DG selection itself.
Genome rearrangement is associated with altered colony phenotypes:
We examined the relationship of altered colony phenotypes and genome rearrangement within the data set for in vivo populations. Using the goodness-of-fit test, there is a significant association between altered colony phenotypes (Sm, Wr) and 2DGR phenotypes relative to 2DGS phenotypes (Gstat = 19.45, d.f. = 3, P = 0.01).
We then evaluated the relationship of altered colony phenotypes (Sm, Wr) and those short-range and chromosome-level LOH events at loci other than GAL1. We found that altered phenotypes were significantly and positively associated with LOH events while the PT smooth colony phenotype was more often associated with the absence of detected LOH events than would be expected by chance (Table 4). Examining the specific events and strains, we found that four of the five strains in which LOH occurred along an entire chromosome also exhibited an altered colony phenotype. For ChrR, homozygous haplotype 1 (strain AF3976) was associated with the Sm phenotype while homozygous haplotype 2 (strain AF617) was associated with the Wr-T-R phenotype (Figure 5). Interestingly, trisomy of ChrR with an extra copy of haplotype 2 (strain AF3977) is not associated with an altered colony phenotype. These results suggest that the loss of specific ChrR alleles leads to altered colony phenotypes, while a change in the ratios of alleles from 1:1 to 2:1 on ChrR does not have the same effect.
We also detected changes in colony phenotype associated with chromosome-level changes on Chr2. Homozygous haplotype 1 is associated with the Wr-N-R colony phenotype (strain AF3990), and trisomy of Chr2 with two copies of haplotype 2 is associated with the Wr-T-R colony phenotype (strain AF540) (Figure 5). In contrast to ChrR results, we found that both the loss of a haplotype or an imbalance of allele ratios on Chr2 gave rise to altered colony phenotypes. Finally, conversion of the left half of Chr2 to homozygous haplotype 1 was not associated with a change in colony phenotype (strain AF21), which suggests that the Wr-T-R phenotype in strain AF3990 is due to homozygous haplotype 1 loci on the right half of Chr2. Together, results show varying effects of trisomy and hemizygosity at ChrR and Chr2 on growth phenotype.
Synthesizing results for LOH at GAL1, rearrangement events independent of GAL1, and altered colony phenotypes, we find that altered colony phenotypes are observed more frequently in strains that have undergone at least one LOH event at GAL1 but that the underlying cause of altered colony phenotypes is additional, GAL1-independent LOH events. A large proportion of isolates with altered colony phenotypes also had undergone chromosomal changes via short-range LOH events (11/17, or 65%) or chromosome-level events (4/6, or 80%) (Table 4). Overall, our results show that the choice of isolates with altered colony phenotypes increased the detection of genome changes and are consistent with findings that LOH at many different loci influences filamentous growth in C. albicans (Uhl et al. 2003).
DISCUSSION
Opportunistic pathogens often live as commensal associates of plants and animals, becoming dangerous infectious agents when host defenses are weakened or new virulence types arise by mutation (Margolis and Levin 2007). Since many of these microbial pathogens propagate asexually, understanding the mechanisms by which opportunistic pathogen populations respond to host environments is essential to developing effective disease control and antifungal therapy and to understanding the fundamental evolutionary question of how adaptation occurs in asexual lineages. Here, we report differing types and distribution of recombination events across the genome for in vivo- vs. in vitro-propagated populations and that altered colony growth phenotypes arose only within the in vivo populations. Our results demonstrate that C. albicans populations generate considerably more phenotypic and genetic variation during growth in a living host than in the relatively benign environment of in vitro culture.
Net population growth rates are 5- to 10-fold lower in vivo than in vitro. Immediately after C. albicans is introduced into the mouse host, population sizes dramatically decline in the bloodstream and kidneys and subsequently expand exponentially in the kidney, causing renal failure and death (Odds et al. 2000; MacCallum and Odds 2004). Because log growth is density independent, the results suggest that the slower growth trajectory characteristic of in vivo populations is less likely a result of direct competition among C. albicans cells and more likely a result of higher cell mortality due to the host immune response or slower cell division due to nutrient limitations (Barelle et al. 2006; Bernardis et al. 2007). The linear growth rate of pseudohyphal and hyphal cells (Hausauer et al. 2005) may also contribute to slower population growth in the in vivo environment. Slower growth in vivo is not surprising; the result is important because relative rates of genetic change can be evaluated and because of the empirical insight provided into the population dynamics of an important pathogen.
We found that average rates of LOH at individual SNP loci across the genome are quite similar for in vivo and in vitro populations (∼10−4 events/generation). Yet the distribution of events differed with greater numbers of short-range LOH events on Chr1 for in vivo populations and on Chr3 for in vitro-propagated populations than expected for a random distribution across all SNP loci and chromosomes. Higher rates of LOH at the GAL1 locus on Chr1 were observed during in vivo growth than during in vitro growth. Loss of the URA3 gene with LOH at the engineered GAL1 locus could lead to lower virulence (Staab and Sundstrom 2003), but such an effect should decrease, rather than increase, the recovery of Gal− strains. Previous results show little evidence that URA3 copy number affected survival in our experimental system (Forche et al. 2003). The results show that the growth environment strongly affects the genomic distribution of short-range LOH events and that, in either environment, LOH events are unevenly distributed across the genome.
Both the rates and the distribution of chromosome-level events across the genome differed from the rates and distribution of short-range LOH events. Most striking, chromosome-level events were observed only for in vivo populations, and in those populations, only for ChrR and Chr2. Our results are supported by those of Diogo et al. (2009) for a limited number of C. albicans strains isolated from the digestive tract of healthy individuals where chromosome-level events (trisomy, nondisjunction) were prevalent. We conclude that conditions during growth in a mammalian host affect chromosome nondisjunction more strongly than they affect mitotic crossover or other recombination processes. While the type of rearrangements differs from the mutational spectrum observed in bacterial pathogens during growth in stressful conditions (Victoir and Dujardin 2002; Tenaillon et al. 2004; Ponder et al. 2005), common mechanisms involving recombination and repair may yet be identified.
The rates of chromosome-level recombination that we observed (∼10−3 events/generation) are comparable to those reported for rates in other fungi at ∼10−3 events/kb/generation (Awadalla 2003). Although high rates of chromosomal rearrangement have been observed at specific chromosomal loci of other pathogens (Henderson et al. 1999; Victoir and Dujardin 2002; Kline et al. 2003; Uhl et al. 2003; Zhang et al. 2003), few quantitative estimates of genomewide change are available to explain the variation apparent in clinical populations (Graeser et al. 1996; Fries and Casadevall 1998; Iwaguchi et al. 2000; Forche et al. 2005). The rates determined in our study are certainly sufficient to generate heritable variation in C. albicans populations during a single passage through the host.
The variation in colony phenotypes generated during in vivo growth is of most direct consequence to C. albicans fitness in the host. We estimated the recovery of altered colony phenotypes at ∼10−6/generation with a large fraction of these also regulated by temperature (Wr-T-R) and the greatest trait expression at 37°. This finding is consistent with genetic studies that found that many genes, when hemizygous, affect filamentous growth in vitro (Uhl et al. 2003) and suggests that the number of genes that affect the quality of filamentous growth is larger than the number of genes that affect the temperature regulation of filamentous growth. We found a positive association between altered colony phenotypes and short-range and chromosome-level LOH events and infer that choosing phenotypic variants for analysis increased the overall rate of the recovery of rearrangement events. Within in vitro-propagated strains, we did not observe altered colony growth phenotypes, which is consistent with the very low recovery of chromosomal rearrangement or altered colony growth phenotypes observed for in vitro growth (reviewed in Rustchenko 2007). Our observation of the appreciable phenotypic variation arising during a single passage through a mouse host is striking and is of sufficient magnitude to strongly affect the evolution of C. albicans populations within the short lifetime of the affected host.
Recombination events as well as the altered colony phenotypes were positively associated with the recovery of 2DGR strains. We considered the possibility that selective growth on 2DG, like growth on 5-fluoroorotic acid (Boeke et al. 1987), might be mutagenic. However, no altered colony phenotypes were recovered among the many 2DGR strains derived from in vitro experiments, and controls show that cells were subject to 2DG selection for too little time to generate and recover colonies arising from GAL1 mutation. Further, rates at GAL1 (10−6/generation) in vitro are comparable to those reported for Saccharomyces cerevisiae at the URA3 locus at ∼5 × 10−6 events/generation (calculated from Hiraoka et al. 2000). The GAL1 locus resides on Chr1, suggesting that the higher rate of LOH at GAL1 for in vivo populations results from a greater rate of recovering short-range LOH events there. We conclude that 2DG selection is not mutagenic; rather, it provides a portal through which genome rearrangement events and associated phenotypic variation can be efficiently recovered and studied.
Together, our results show that relatively few mitotic generations occurring during a single passage through a mouse host generate measurable genomewide and phenotypic variation within populations. Our results shed light on the role of recombination in the evolution of asexual organisms (Dunham et al. 2002), especially opportunistic pathogens. The evolution of genetic factors that increase recombination rates will be favored under environmental uncertainty (Lenormand and Otto 2000; Otto 2002), and previous studies with S. cerevisiae demonstrate an advantage to recombining populations during infectious growth (Grimberg and Zeyl 2005). We propose that the commensal state of opportunistic pathogens such as C. albicans is maintained by the dynamic fluctuations of populations (Levin et al. 2000; Levin and Anita 2001). In this model, population bottlenecks decrease the efficiency of selection in removing deleterious phenotypes that arise during infectious growth. Even though population sizes may be quite large at later infection stages, populations will be dominated by those mutations arising early in passage through the host. In contrast, traits under selection during commensal growth will likely differ from those beneficial to invasive growth, and larger population sizes allow efficient selection. Either the retention of deleterious alleles by drift when population sizes are small, or clonal interference among genotypes carrying beneficial mutations (Kao and Sherlock 2008; Campos and Wahl 2009) when populations are large, will slow evolution of virulence and host adaptation. If so, further experiments should find that colony growth and virulence phenotypes generated during host passage are more variable than expected under strong directional selection for increased virulence and instead constitute “sink” populations that are unable to compete in the commensal environment or survive past host death (Romani et al. 2003; Sokurenko et al. 2006). As in drug resistance evolution (Anderson 2005), it will be important to evaluate fitness associated with differing growth conditions and varying rates of genome rearrangement. Results presented here portray the evolution of a genome strongly responsive to growth environment as well as suggest an important role of recurrent bottlenecks and population expansion during host passage and reinfection common to clinical settings.
Acknowledgments
We greatly appreciate Frank Odds for providing original data. The Minnesota Super Computing Institute provided computational resources and support. We thank Mark McClellan for production of whole-genome microarrays used for the competitive genome hybridization (CGH) portion of this study and Brett Couch, Ruth Shaw, Peter Tiffin, and Mike Travisano for helpful comments on evolutionary analyses and interpretation. Research was supported by National Institutes of Health (NIH) grant AI46351 to G.M. and P.T.M., NIH/National Institute of Allergy and Infectious Diseases grant RO1-AI062427 to A.F. and J.B., and a Microbial and Plant Genomics Institute Integrative fellowship to A.S. Funding for CGH microarray production was provided in collaboration with Mira Edgerton (DE10641-S).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103325/DC1.
References
- Anderson, J. B., 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. 3 547–556. [DOI] [PubMed] [Google Scholar]
- Awadalla, P., 2003. The evolutionary genomics of pathogen recombination. Nat. Rev. 4 50–60. [DOI] [PubMed] [Google Scholar]
- Barelle, C. J., C. L. Priest, D. M. MacCallum, N. A. R. Gow, F. C. Odds et al., 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman, J., H. Chibana, J. Turner and P. T. Magee, 2001. Single-copy IMH3 allele is sufficient to confer resistance to mycophenolic acid in Candida albicans and to mediate transformation of clinical Candida species. Infect. Immun. 69 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardis, F. D., H. Liu, R. O'Mahony, R. L. Valle, S. Bartollino et al., 2007. Human domain antibodies against virulence traits of Candida albicans inhibit fungus adherence to vaginal epithelium and protect against experimental vaginal candidiasis. J. Infect. Dis. 196 149–157. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., J. Trueheart, G. Natsoulis and G. Fink, 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154 164–175. [DOI] [PubMed] [Google Scholar]
- Borst, P., 2002. Antigenic variation and allelic exclusion. Cell 109 5–8. [DOI] [PubMed] [Google Scholar]
- Borst, P., and G. Rudenko, 1994. Antigenic variation in African trypanosomes. Science 264 1872–1873. [DOI] [PubMed] [Google Scholar]
- Bougnoux, M. E., D. Diogo, N. Francois, B. Sendid, S. Veirmeire et al., 2006. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J. Clin. Microbiol. 44 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, P. R., and L. M. Wahl, 2009. The effects of population bottlenecks on clonal interference, and the adaptation effective population size. Evolution 63 950–958. [DOI] [PubMed] [Google Scholar]
- Coste, A., A. Selmecki, A. Forche, D. Diogo, M. E. Bougnoux et al., 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6 1889–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen, L. E., C. Sirjusingh, R. C. Summerbell, S. Walmsley, S. Richardson et al., 1999. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 43 2930–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, G. A. M., E. Wirtz and M. Navarro, 1998. Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 91 77–91. [DOI] [PubMed] [Google Scholar]
- Diogo, D., C. Bouchier, C. d'Enfert and M. E. Bougnoux, 2009. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet. Biol. 46 159–168. [DOI] [PubMed] [Google Scholar]
- Dunham, M. J., H. Badrane, T. Ferea, J. Adams and P. O. Brown, 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A., G. Schonian, Y. Graser, R. Vilgalys and T. G. Mitchell, 1999. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet. Biol. 28 107–125. [DOI] [PubMed] [Google Scholar]
- Forche, A., G. May, J. Beckerman, S. Kauffman, J. Becker et al., 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39 38–50. [DOI] [PubMed] [Google Scholar]
- Forche, A., P. T. Magee, B. B. Magee and G. May, 2004. Genome-wide single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell 3 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A., G. May and P. T. Magee, 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans during infection. Eukaryot. Cell 4 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries, B. C., and A. Casadevall, 1998. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J. Infect. Dis. 178 1761–1766. [DOI] [PubMed] [Google Scholar]
- Graeser, Y., M. Volovsek, J. Arrington, G. Schoenian, W. Presber et al., 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. USA 93 12473–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg, B., and C. Zeyl, 2005. The effects of sex and mutation rate of adaptation in test tubes and to mouse hosts by Saccharomyces cerevisiae. Evolution 59 431–438. [PubMed] [Google Scholar]
- Gupta, S., 2005. Parasite immune escape: new views into host-parasite interactions. Curr. Opin. Microbiol. 8 428–433. [DOI] [PubMed] [Google Scholar]
- Hausauer, D. L., M. Gerami-Nejad, C. Kistler-Anderson and C. A. Gale, 2005. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot. Cell 4 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, I. R., P. Owen and J. P. Nataro, 1999. Molecular switches: the ON and OFF of bacterial phase variation. Mol. Microbiol. 33 919–932. [DOI] [PubMed] [Google Scholar]
- Hiraoka, M., K. Watanabe, K. Umezu and H. Maki, 2000. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics 156 1531–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaguchi, S. I., T. Kanbe, T. Tohne, P. T. Magee and T. Suzuki, 2000. High-frequency occurrence of chromosome translocation in a mutant strain of Candida albicans by a suppressor mutation of ploidy shift. Yeast 16 411–422. [DOI] [PubMed] [Google Scholar]
- Joly, S., C. Pujol and D. R. Soll, 2002. Microevolutionary changes and chromosomal translocations are more frequent at RPS loci in Candida dubliniensis than in Candida albicans 1. Infect. Genet. Evol. 2 19–37. [DOI] [PubMed] [Google Scholar]
- Jones, T., N. Federspiel, H. Chibana, J. Dungan, S. Kalman et al., 2004. The diploid genome of Candida albicans. Proc. Natl. Acad. Sci. USA 101 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, K. C., and G. Sherlock, 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, K. A., E. V. Sechman, E. P. Skaar and H. S. Seifert, 2003. Recombination, repair and replication in the pathogenic Neisserieae: the 3 r's of molecular genetics of two human-specific bacterial pathogens. Mol. Microbiol. 20 3–13. [DOI] [PubMed] [Google Scholar]
- Kraus, E., W. Y. Leung and J. E. Haber, 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes, S., P. Horrocks and C. Newbold, 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55 673–707. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49 264–285. [DOI] [PubMed] [Google Scholar]
- Legrand, M., P. Lepart, A. Forche, F.-M. C. Mueller, T. J. Walsh et al., 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangments and tetraploid formation. Mol. Microbiol. 52 1451–1462. [DOI] [PubMed] [Google Scholar]
- Legrand, M., A. Forche, A. Selmecki, C. Chan, D. T. Kirkpatrick et al., 2008. Haplotype mapping of a diploid non-meiotic organism using existing and induced anueploides. PLoS Genet. 4 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand, T., and S. P. Otto, 2000. The evolution of recombination in a heterogenous environment. Genetics 156 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lephart, P. R., and P. T. Magee, 2006. Effect of the major repeat sequence on mitotic recombination in Candida albicans. Genetics 174 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lephart, P. R., H. Chibana and P. T. Magee, 2005. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot. Cell 4 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, B. R., and R. Anita, 2001. Why we don't get sick: the within-host population dynamics of bacterial infections. Science 292 1112–1114. [DOI] [PubMed] [Google Scholar]
- Levin, B. R., V. Perrot and N. Walker, 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White et al., 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42 2932–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum, D. M., and F. C. Odds, 2004. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses 48 151–161. [DOI] [PubMed] [Google Scholar]
- Margolis, E., and B. R. Levin, 2007. Within-host evolution for the invasiveness of commensal bacteria: an experimental study of bacteremias resulting from Haemophilus influenzae nasal carriage. J Infect. Dis. 196 1068–1075. [DOI] [PubMed] [Google Scholar]
- Marr, K. A., C. N. Lyons, T. R. Rustad, R. A. Bowden and T. C. White, 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42 2484–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookerjee, S. A., and E. A. Sia, 2006. Overlapping contributions of Msh1p and putative recombination proteins Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting events in the mitochondrial genome of Saccharomyces cerevisiae. Mutat. Res. 595 91–106. [DOI] [PubMed] [Google Scholar]
- Moran, N. A., 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds, F. C., L. V. Nuffel and N. A. Gow, 2000. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology 146 1881–1889. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3 252–261. [DOI] [PubMed] [Google Scholar]
- Platt, T., 1984. Toxicity of 2-deoxygalactose to Saccharomyces cerevisiae cells constitutively synthesizing galactose-metabolizing enzymes. Mol. Cell. Biol. 4 994–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder, R. G., N. C. Fonville and S. M. Rosenberg, 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19 791–804. [DOI] [PubMed] [Google Scholar]
- Pujol, C., M. Pfaller and D. R. Soll, 2002. Ca3 fingerprinting of Candida albicans bloodstream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 40 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani, L., F. Bistoni and P. Puccetti, 2003. Adaptation of Candida albicans to the host environment: the role of morphogenesis in virulence and survival in mammalian hosts. Curr. Opin. Microbiol. 6 338–343. [DOI] [PubMed] [Google Scholar]
- Rustchenko, E., 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7 2–11. [DOI] [PubMed] [Google Scholar]
- Selmecki, A., S. Bergmann and J. Berman, 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55 1553–1565. [DOI] [PubMed] [Google Scholar]
- Selmecki, A., A. Forche and J. Berman, 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki, A., M. Gerami-Nejad, C. Paulson, A. Forche and J. Berman, 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68 624–641. [DOI] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rolf, 1981. Biometry: The Principles and Practice of Statistics in Biological Research. W. H. Freeman, New York.
- Sokurenko, E. V., R. Gomulkiewicz and D. E. Dykhuizen, 2006. Source-sink dynamics of virulence evolution. Nat. Rev. 4 548–555. [DOI] [PubMed] [Google Scholar]
- Spell, R. M., and S. Jinks-Robertson, 2004. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae, pp. 3–12 in Genetic Recombination: Review and Protocols, edited by A. S. Waldman. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Staab, J. F., and P. Sundstrom, 2003. URA3 as a selectable marker for disruption and virulence assessment in Candida albicans genes. Trends Microbiol. 11 69–73. [DOI] [PubMed] [Google Scholar]
- Strobel, G. L., and J. Arnold, 2004. Essential eukaryotic core. Evolution 58 441–446. [PubMed] [Google Scholar]
- Tenaillon, O., E. Denamur and I. Matic, 2004. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 12 264–270. [DOI] [PubMed] [Google Scholar]
- Uhl, M. A., M. Biery, N. Craig and A. D. Johnson, 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 22 2668–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog, M., T. J. Rast, M. Martchenko, S. Grindle, D. Dignard et al., 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8 R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoir, K., and J.-C. Dujardin, 2002. How to succeed in parasitic life without sex? Asking Leishmania. Trends in Parasitology 18 81–85. [DOI] [PubMed] [Google Scholar]
- Wilson, L. S., C. M. Reyes, M. Stolpman, J. Speckman, K. Allen et al., 2002. The direct cost and incidence of systemic fungal infections. Value Health 5 26–34. [DOI] [PubMed] [Google Scholar]
- Wren, B. W., 2000. Microbial genome analysis: insights into virulence, host adaptation and evolution. Nat. Rev. Genet. 1 30–39. [DOI] [PubMed] [Google Scholar]
- Zhang, N., A. L. Harrrex, B. R. Holland, L. E. Fenton, R. D. Cannon et al., 2003. Sixty alleles of the ALS7 open reading frame in Candida albicans: ALS7 is a hypermutable contingency locus. Genome Res. 13 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]