Abstract
Background and Aims
The genus Sinojackia consists of eight species, all endemic to China. All species of Sinojackia are endangered or threatened owing to poor recruitment within populations. Information on molecular phylogenetics is critical for developing successful conservation strategies for this genus.
Methods
Combined DNA sequence data from the nuclear ribosomal internal transcribed spacer regions and plastid psbA–trnH intergenic spacer and microsatellite data were used to infer a phylogeny of the genus.
Key Results
Parsimony analysis of the combined sequence data and multivariate analysis based on fruit characters indicated that Sinojackia dolichocarpa is monophyletic and genetically well separated from the other Sinojackia species, thus supporting its rank at the generic level as Changiostyrax. Phylogenetic relationships within Sinojackia sensu stricto are unresolved from the combined sequence data. A UPGMA dendrogram based on seven microsatellite loci of 96 individual plants yielded a first-diverging cluster of all individuals of S. microcarpa. The remaining species form another cluster without any definitive patterns corresponding to current species circumscriptions, suggesting either extensive hybridization or incipient speciation.
Conclusions
The results suggest that there are too many species recognized within Sinojackia sensu stricto, but this must be further assessed with comprehensive morphological and taxonomic revisionary work. The implications of the phylogenetic data for conservation are discussed.
Key words: Changiostyrax, conservation, phylogeny, Sinojackia, Styracaceae
INTRODUCTION
Sinojackia is a Chinese endemic genus of Styracaceae comprising eight species (Yao et al., 2007a). The first report of this genus was based on a specimen from Jiangsu Province, in eastern China, which was recognized as S. xylocarpa Hu (Hu, 1928). Since then, Hu (1930) described S. rehderiana Hu from Jiangxi, and Merrill (1937) transferred Pterostyrax henryi Dummer to Sinojackia as S. henryi (Dummer) Merr. Subsequently, Luo (1992) described S. sarcocarpa L. Q. Luo from Sichuan, and Chen discovered S. microcarpa C. T. Chen & G. Y. Li and S. oblongicarpa C. T. Chen & T. R. Cao from Zhejiang and Hunan, respectively (Chen and Li, 1997; Chen, 1998). The genus has a widespread but disjunct distribution in China, occurring from Zhejiang Province in eastern China to Sichuan Province in south-western China. Most species of the genus have potential horticultural value (Chen and Chen, 1996). All species of Sinojackia are endangered or threatened as a result of small population size and lack of regeneration within populations (Fu, 1992; Wang and Xie, 2004; Yao et al., 2005). It is therefore important to conduct molecular phylogenetic studies to provide effective conservation measures for this endangered genus.
Chen (1995) segregated Sinojackia dolichocarpa as the new monotypic genus Changiostyrax. This was done because S. dolichocarpa differs significantly from Sinojackia in characters of the trunk, flowers and fruit. Hwang and Grimes (1996), however, indicated that the systematic position of S. dolichocarpa was indefinite, and the taxonomic position of this species remains in dispute (Cao et al., 2006). The other seven species of Sinojackia are morphologically similar, and delimitation relies substantially on fruit morphology. During surveys of the morphological features of Sinojackia, however, a high degree of variation in fruit morphology was observed in S. xylocarpa within and among populations sampled from Jiangsu and Henan provinces (X. Yao et al., unpubl. res.). Such variation also occurs in S. huangmeiensis, S. sarcocarpa, S. oblongicarpa and S. rehderiana (Luo, 2005; Yao et al., 2005). Therefore, primary reliance on fruit characters as indicators of phylogenetic relationships and species circumscription in Sinojackia appears to be problematic.
A recent phylogenetic study of Styracaceae based on morphological and molecular data (Fritsch et al., 2001) supported the distinctness of S. dolichocarpa from the two other Sinojackia species sampled. The sampling of Sinojackia is expanded herein to all known species except for S. henryi to assess phylogenetic relationships in the group. Numerous studies have documented the utility of the internal transcribed spacer region (ITS, comprising the ITS-1 and ITS-2 spacers and the 5·8S gene) of nuclear ribosomal DNA for resolving relationships among closely related species (Baldwin et al., 1995; Soltis and Soltis, 1998). The ITS region was therefore used to sample the nuclear genome of Sinojackia. We also attempted to base the phylogenetic reconstruction on five regions of the plastid genome: the trnS–trnG, atpB–rbcL and psbA–trnH intergenic spacers (IGS), the trnL intron and the rps4 gene. Microsatellite [=simple sequence repeat (SSR)] loci were also used as an independent approach to provide phylogenetic resolution not achieved with the use of DNA sequence data. The objectives of the present study were to (1) reconstruct species relationships within Sinojackia, (2) assess the taxonomic rank and position of S. dolichocarpa (≡Changiostyrax dolichocarpus), (3) assess the distinctness of the other species within the genus and (4) address conservation concerns about Sinojackia in the context of the results.
MATERIALS AND METHODS
Plant material
Sources of plant material used in this study are listed in Table 1. Bruinsmia styracoides, Halesia carolina, H. diptera, H. macgregori, Melliodendron xylocarpum, Pterostyrax corymbosus, P. hispidus, P. psilophyllus and Rehderodendron macrocarpum were included in the analysis, with B. styracoides used as outgroup in accordance with the results of Fritsch et al. (2001). Seven taxa representing all Sinojackia species except S. henryi were included in the phylogenetic analysis. No wild populations or individuals of S. henryi have been found since the type specimen was first collected in 1937, suggesting that this species may be extinct in the wild (Yao et al., 2005). A pilot study with single individuals of seven Sinojackia species and P. psilophyllus was initially performed to search for the most variable DNA regions. rps4, atpB–rbcL, trnS–trnG, psbA–trnH and trnL were surveyed (Table 2). ITS sequences were obtained from GenBank for several additional taxa, including S. xylocarpa, S. rehderiana, S. dolichocarpa, B. styracoides, H. carolina, H. diptera, H. macgregori, M. xylocarpum, P. corymbosus, P. hispidus, P. psilophyllus and R. macrocarpum (Table 1). All species of Sinojackia excluding S. henryi have been conserved ex situ in the Wuhan Botanical Garden, the Chinese Academy of Sciences (WBG, CAS). The complete data sets are available upon request from the first author. Ten individuals of S. dolichocarpa, 13 of S. oblongicarpa, ten of S. sarcocarpa, 16 of S. huangmeiensis, 17 of S. rehderiana, 15 of S. microcarpa and 15 of S. xylocarpa from a single population of each species (Table 1) were used for analysis of seven microsatellite loci developed by Yao et al. (2006).
Table 1.
List of taxa examined in this study and accession numbers for sequences deposited in GenBank
| Taxa | Locality | Voucher specimen | psbA–trnH | ITS | rps4 | trnS–trnG | trnL | atpB–rbcL |
|---|---|---|---|---|---|---|---|---|
| Sinojackia dolichocarpa (C. J. Qi) C. T. Chen | Gaoqiaohe, Shimen, Hunan, China | Chen T, 940004 (IBSC) | DQ317992 | AF396439 | DQ318000 | DQ318012 | AF396165 | DQ317984 |
| Sinojackia huangmeiensis J. W. Ge & X. H. Yao | Xiaxin, Huangmei, Hubei, China | Yao X.H., 04003(HIB) | DQ317989 | DQ318017 | DQ318797 | DQ318009 | DQ318005 | DQ317981 |
| Sinojackia microcarpa T. Chen & G. Y. Li | Meicheng, Jiande, Zhejiang, China | Chen T, 9511041 (IBSC) | DQ317991 | DQ318016 | DQ318799 | DQ318011 | DQ318004 | DQ317983 |
| Sinojackia oblongicarpa T. Chen & T. R. Chao | Hejiatian, Huaihua, Hunan, China | Chen T 9511046 (IBSC) | DQ317993 | DQ318018 | DQ318001 | DQ318013 | DQ318003 | DQ317985 |
| Sinojackia rehderiana Hu | Hangzhou Botanical Garden, Zhengjiang, China | C.T. Chen & P.W. Fritsch 9704076 (CAS) | DQ317990 | AF396450 | DQ318798 | DQ318010 | AF396189 | DQ317982 |
| Sinojackia sarcocarpa L. Q. Luo | Wuyoushan, Leshan, Sichuan, China | Luo LQ, 1573(IBSC) | DQ317987 | DQ318015 | DQ318795 | DQ318007 | DQ318006 | DQ317979 |
| Sinojackia xylocarpa Hu | C.R. Parks residence, North Carolina, USA | P.W. Fritsch 1362 (RSA) | DQ317994 | AF396451 | DQ318002 | DQ318014 | AF396191 | DQ317986 |
| Bruinsmia styracoides Boerl. & Koord. | Sabah, Malaysia | C.H. Cannon 529 (DUKE) | EU336947 | AF396438 | ||||
| Halesia diptera J. Ellis | University of California Botanical Garden, Berkeley, CA, USA. | P.W. Fritsch 1482 (CAS) | EU336948 | AF396441 | ||||
| Halesia carolina L. | University of California Botanical Garden, Berkeley, CA, USA | P.W. Fritsch 1481 (CAS) | EU336949 | AF396440 | ||||
| Halesia macgregori Chun | Nanyue Arboretum, Hunan, China | C.T. Chen & P.W. Fritsch 9704096 (CAS) | EU336950 | AF396442 | ||||
| Melliodendron xylocarpum Hand.-Mazz. | Strybing Arboretum, San Francisco, CA, USA | P.W. Fritsch s.n. (CAS) | EU336951 | AF396444 | ||||
| Pterostyrax corymbosus Siebold & Zucc. | Hangzhou Botanical Garden, Zhejiang, China | C.T. Chen & P.W. Fritsch 9704067 (CAS) | EU336952 | AF396445 | ||||
| Pterostyrax hispidus Diels ex Perkins | University of California Botanical Garden, Berkeley, CA, USA | P.W. Fritsch 1483 (CAS) | EU336953 | AF396446 | ||||
| Pterostyrax psilophyllus Diels ex Perkins | Hangzhou Botanical Garden, Zhengjiang, China | C.T. Chen & P.W. Fritsch 9704070 (CAS) | DQ317988 | AF396447 | DQ318796 | DQ318008 | AF396183 | DQ317980 |
| Rehderodendron macrocarpum Hu | Washington Park Arboretum, Seattle, WA, USA | P.W. Fritsch 1359 (RSA) | EU336954 | AF396449 |
DUKE = Duke University; CAS = California Academy of Sciences; HIB = Herbarium of Wuhan Botanical Garden, Wuhan, China; IBSC = South China Botanical Garden Herbarium, Guangzhou, China; RSA= Herbarium of Rancho Santa Ana Botanic Garden.
Table 2.
Oligonucleotide primers used to amplify the rps4, trnS–trnG, atpB–rbcL, psbA–trnH, trnL intron and ITS with amplification direction and reference
| Primer | Sequence | Direction | Reference |
|---|---|---|---|
| rps4 | |||
| rps5 | 5′-ATGTCCCGTTATCGAGGACCT-3′ | Forward | Souza-Chies et al. (1997) |
| trnS | 5′-TACCGAGGGTTCGAATC-3′ | Reverse | Souza-Chies et al. (1997) |
| trnS–trnG | |||
| trnS (GCU) | 5′-GCCGCTTTAGTCCACTCAGC-3′ | Forward | Hamilton (1999) |
| trnG (UCC) | 5′-GAACGAATCACACTTTTACCAC-3′ | Reverse | Hamilton (1999) |
| atpB–rbcL | |||
| atpB-1 | 5′-ACATCKARTACKGGACCAATAA-3′ | Forward | Chiang et al. (1998) |
| rbcL-1 | 5′-AACACCAGCTTTRAATCCAA-3′ | Reverse | Chiang et al. (1998) |
| psbA–trnH | |||
| trnHR | 5′-CGCGCATGGTGGATTCACAAATC-3′ | Forward | Tate (2002) |
| psbAF | 5′-GTTATGCATGAACGTAATGCTC-3′ | Reverse | Sang et al. (1997) |
| trnL | |||
| trnL-c | 5′-CGAAATCGGTAGACGCTACG-3′ | Forward | Taberlet et al. (1991) |
| trnL-d | 5′-GGGGATAGAGGGACTTGA AC-3′ | Reverse | Taberlet et al. (1991) |
| ITS | |||
| ITS4 | 5′-TCCTCCGCTTATTGATATGC-3′ | Forward | Swensen et al. (1998) |
| ITS5p | 5′-GGAAGGAGAAGTCGTAACAAGG-3′ | Reverse | Swensen et al. (1998) |
Morphological analysis
Styracaceae taxonomy follows Hwang and Grimes (1996), Chen and Li (1997), Fritsch et al. (2001) and Yao et al. (2007a). Five individuals from each of the seven species of Sinojackia and one individual from each of the species of the five genera of Styracaceae that were used in the analysis based on DNA sequences plus individuals of three other genera (Alniphyllum, Huodendron and Styrax) were included in the morphological analysis. Sixteen fruit characters considered useful in delimiting genera of Stryacaceae were included in the analysis (Appendix). These characters were derived from the generic descriptions in Hwang and Grimes (1996) and the data from Fritsch et al. (2001). The matrix of morphological characters was subjected to principal coordinates analysis (PCoA) in NTSYS-pc (Rohlf, 2000) based on a distance matrix derived from the similarity coefficient of Nei (1972).
Genomic DNA extraction, PCR amplification, sequencing and microsatellite typing
Total DNA was extracted from fresh leaf material by using the 2-CTAB method (Doyle and Doyle, 1987) as modified to include two initial phenol/chloroform/isoamyl alcohol (25 : 24 : 1) extractions and followed by a chloroform/isoamyl alcohol (24 : 1) extraction. The primers for all PCR amplification reactions are listed in Table 2. Reaction volumes were 25 µL and contained 1·5 U AmpliTaq DNA polymerase, Replitherm buffer, 2·0 mmol L−1 MgCl2, 1 mmol L−1 dNTP, 0·2 µmol L−1 primer and 25–60 ng sample DNA. PCR was performed in a PTC-200 thermocycler (BioRad, Hercules, California, USA). PCR amplifications for the five regions of the plastid genome were carried out at an initial denaturation of 94 °C for 4 min, followed by 35 cycles of 94 °C of 45 s, 50 °C for 1 min and 72 °C for 1 min, finally followed by an extension of 7 min at 72 °C. Amplification of the ITS region was conducted with 35 cycles of 94 °C for 45 s, 50 °C for 1 min and 72 °C for 1 min 30 s, followed by a final extension of 7 min at 72 °C. PCR products were purified by using the E.Z.N.A® Gel Extraction Kit (Omega Bio-Tek). All cleaned PCR products except those of the ITS region were cloned into a pMD-18-T vector (Takara). Ligation, transformation and plating were carried out by following the recommendations of the manufacturer. One clone of each species was obtained and plasmid DNA preparations were carried out by following the protocols for precipitation with Watson's plasmid mini-columns. Because Fritsch et al. (2001) reported that many ITS cloned sequences in Styracaceae may represent pseudogenes, the ITS region was directly sequenced following the methods of Fritsch et al. (2001). There are only a few point mutations in the 5·8S subunit of all of our Sinojackia samples, indicating that the sequences probably represent functional copies of the ITS region. Purified PCR products and plasmid DNA preparations were sequenced with the ABI BigDye™ Terminators Cycle Sequencing Kit (Applied Biosystems) in an ABI Prism 3100 automated sequencer.
Microsatellite typing was performed in accordance with the methods described in Yao et al. (2006) with the loci Sx11, Sx15, Sx40, Sx101, Sx112, Sx116 and Sx154. The identification of alleles was based on the disposition of the fragments in relation to a 10-bp marker ladder on a high-resolution polyacrylamide gel with silver staining.
Phylogenetic analysis
Sequences were initially aligned using the CLUSTAL W algorithm (Thompson et al., 1994) and then edited manually. Because the informative characters for trnL, rps4, trnS–trnG and atpB–rbcL were too few for construction of a phylogenetic tree, these regions were excluded from subsequent analyses (Table 3). Phylogenetic trees were thus based on psbA–trnH and ITS data only. Optimal trees were inferred using maximum parsimony (MP) as implemented in PAUP* version 4·0b10 (Swofford, 2003). For all analyses, characters were equally weighted, gaps were treated as missing data, parsimony-uninformative characters were excluded and multistate characters were treated as uncertainties. Analyses were performed with the following options implemented: heuristic search mode used 1000 random-addition sequence replicates, tree bisection–reconnection (TBR) branch-swapping, MULTrees option on, steepest descent off and branches collapsed when maximum length was zero. Bootstrap analyses were conducted using full heuristic searches, 1000 bootstrap replicates and ten random-addition starting sequences with all trees saved, removing taxa with identical sequences. Separate and combined phylogenetic analyses of the Sinojackia matrix were performed. The incongruence length difference (ILD) test was conducted to determine whether the plastid DNA and nrDNA data partitions differed significantly from random partitions of the combined data (Farris et al., 1994). This was implemented as the partition homogeneity test in PAUP* by using 100 replicates and 1000 random-addition starting sequences. Maximum likelihood (ML; with PAUP*) and Bayesian (MrBayes 2·01, Huelsenbeck and Ronquist, 2001) analyses were also conducted for all three data sets using substitution models estimated with Modeltest v. 3·06 (Posada and Crandall, 1998). Because the results from these analyses were congruent with the results of the MP analyses, they are not presented here.
Table 3.
Sequence characteristics of the different DNA regions
| Aligned length | Sequence length range | Variable characters | Informative characters | Mean G + C content (mol %) | |
|---|---|---|---|---|---|
| rps4 | 902 | 888–896 | 48 | 1 | 36·0 |
| trnS–trnG | 743 | 712–728 | 84 | 4 | 38·1 |
| atpB–rbcL | 885 | 843–879 | 42 | 2 | 30·1 |
| psbA–trnH | 550 | 177–529 | 80 | 47 | 29·4 |
| trnL | 509 | 506–509 | 6 | 2 | 33·0 |
| ITS | 682 | 633–658 | 139 | 67 | 64·5 |
The unweighted pair group mean analysis (UPGMA) cluster method was used to generate a dendrogram of all individual plants based on Nei's genetic distance values (Nei, 1972). The analysis was performed by using the SAHN and TREE programs provided with the software NTSYS pc2·0 (Rohlf, 2000).
RESULTS
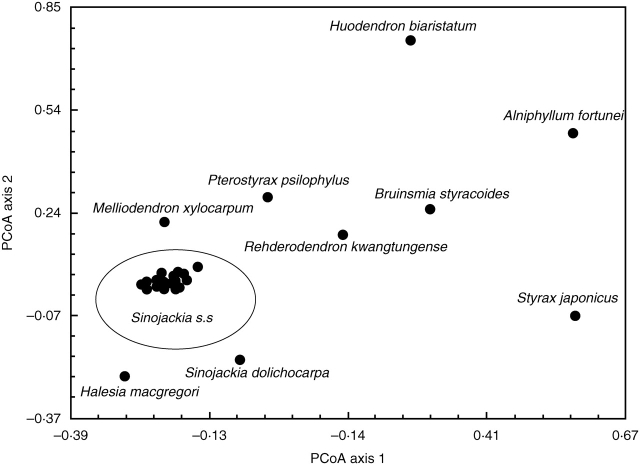
The multivariate analysis indicated a clear morphological separation of Sinojackia dolichocarpa from other species of the genus and from the other genera of Styracaceae sampled, and little morphological variation among the individuals of S. dolichocarpa. The PCoA revealed that all individuals of the six species of Sinojackia other than S. dolichocarpa clustered closely together and interspecific differentiation was not apparent (Fig. 1).
Fig. 1.
Plot of the first two axes from a principal co-ordinates analysis of individuals of Sinojackia Hu and eight other genera of Styracaceae based on the fruit characters.
The length of the psbA–trnH sequences ranged from 177 to 529 bp. Alignment of nine taxa for psbA–trnH yielded 550 nucleotide sites, of which 84 were potentially parsimony-informative. The G + C content ranged from 26·7 to 34·9 % with a mean of 29·4 %. The aligned length of the ITS region comprised 682 characters for nine taxa. The data set contained 69 potentially parsimony-informative characters. The G + C content ranged from 63·5 to 67 % with a mean of 64·5 %.
The rps4 gene contained one potentially parsimony-informative and 47 uninformative nucleotide substitutions. The trnS–trnG region contained four potentially parsimony-informative and 83 uninformative nucleotide substitutions. The atpB–rbcL and trnL regions had two potentially parsimony-informative characters each. The topology of the strict consensus tree with the data from trnL, rps4, atpB–rbcL and trnS–trnG included did not differ from that based on ITS and psbA–trnH alone. The former regions contained little phylogenetic signal and were therefore excluded from the final analysis (Table 3). The ILD test indicated that the ITS and psbA–trnH data sets were not significantly incongruent (P = 0·12).
Parsimony analyses based on the combined ITS + psbA–trnH data yielded 4127 most-parsimonious trees (MPTs) with a length of 358, consistency index of 0·76 and a retention index of 0·85 (Fig. 2). The strict consensus trees of the MPTs based on either ITS or psbA–trnH alone (data not shown) were similar to that based on the combined data as regards the relationships among the species of Sinojackia. In the combined strict consensus, S. dolichocarpa was placed as part of a large polytomy that also included most genera of Styracaceae. The other species of Sinojackia (Sinojackia sensu stricto) formed a clade with strong bootstrap support (100 %). Phylogenetic relationships within Sinojackia sensu stricto were unresolved (bootstrap support <50 %).
Fig. 2.
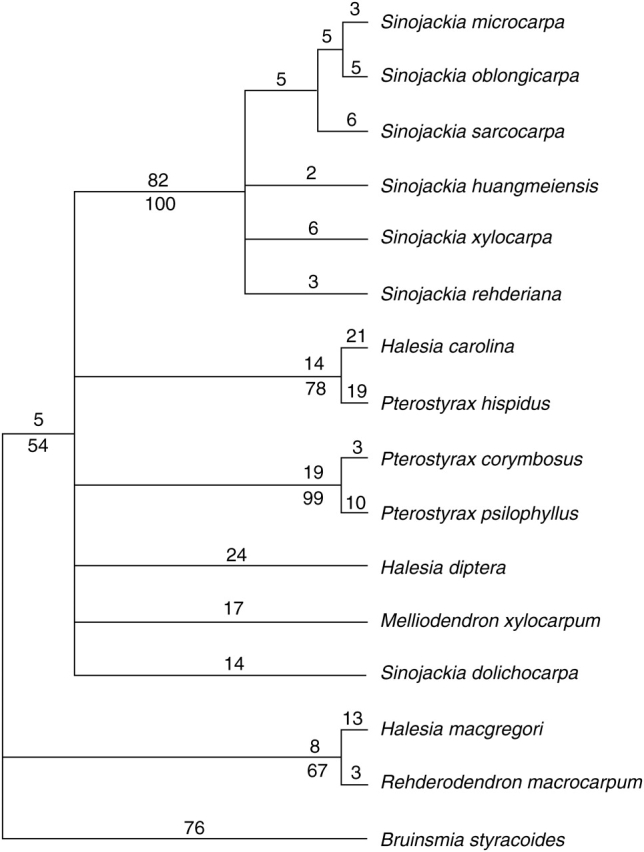
Strict consensus tree of 4127 most-parsimonious trees of 358 steps (CI = 0·76, RI = 0·85) for Sinojackia based on the combined ITS and psbA–trnH data set. Numerals above each branch are branch lengths. Bootstrap percentage (>50 %) are given below branches.
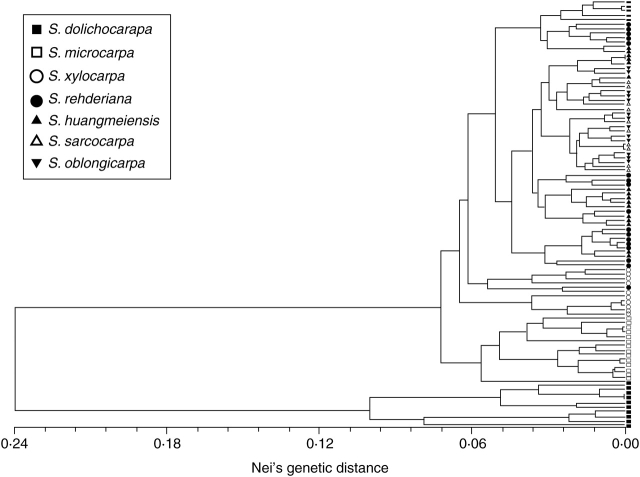
The UPGMA dendrogram based on microsatellite data of the 96 individual plants of Sinojackia yielded a first-diverging cluster containing all individuals of S. dolichocarpa (Fig. 3). All individuals of S. microcarpa formed the next cluster, but were much closer to the remaining individuals of Sinojackia sensu stricto than to those of S. dolichocarpa. The individuals of the remaining Sinojackia species were intermixed with no clearly discernible patterns.
Fig. 3.
The UPGMA dendrogram for 96 individuals of Sinojackia based on Nei's (1972) genetic distances of seven microsatellite loci.
DISCUSSION
Phylogenetic relationships of Sinojackia
The phylogenetic analyses based on both the combined sequence and microsatellite data, as well as the PCoA analysis of fruit characters, confirm the findings of Fritsch et al. (2001) that S. dolichocarpa is a distinct lineage from the other members of Sinojackia. Thus, our data strongly support Chen's (1995) transfer of S. dolichocarpa to the monotypic genus Changiostyrax. Chen delimited Changiostyrax from Sinojackia sensu stricto using the following unique combination of characteristics: trunk without thorns (versus with thorns), vegetative buds without scales (versus with scales), inflorescences ebracteate (versus bracteate), flowers 4-merous (versus 5- or 6-merous), stamens 8, equal in length (versus 10 or 12, unequal in length), anther connectives not prolonged (versus prolonged), ovary semi-inferior (versus fully inferior) and fruit terminated by a long rostrum (versus a short rostrum). The sequence data and multivariate analysis based on fruit characters provide strong support for the monophyly of Sinojackia sensu stricto (Figs 1 and 2), consistent with the results of Fritsch et al. (2001) based on a more limited sample.
The consensus tree based on combined ITS and psbA–trnH sequence data exhibited no resolution among the species of Sinojackia sensu stricto. This is consistent with the observation that species of Sinojackia sensu stricto show low levels of interspecific morphological divergence, i.e. only in flower size, petal shape, and fruit size and shape (Yao et al., 2007a). The UPGMA analysis based on microsatellite loci indicated a first-diverging position for S. microcarpa, whereas all other species of Sinojackia sensu stricto exhibited no distinct grouping patterns. Compared with the other species of Sinojackia sensu stricto, S. microcarpa possessed little intra-specific morphological variation in floral and fruit characters (Yao et al., 2007a). In combination with our microsatellite data, this suggests that, other than S. microcarpa, the entities within Sinojackia sensu stricto are undergoing either extensive hybridization or incipient speciation.
The differences in geographical range, elevation and flowering period of species of Sinojackia sensu stricto suggest that opportunities for hybridization or gene introgression are currently limited. Most species have limited ranges, with populations of small size that occur in isolated patches scattered throughout the low-elevation mountain ranges of east and central China. It has been hypothesized, however, that Sinojackia species may be remnants of a formerly more continuous and widespread series of populations in China that has been disrupted by human activity (Yao et al., 2005). Interspecific hybridization, with or without subsequent introgression from one species into another, occurs commonly in natural populations of many groups of plants (Rieseberg and Carney, 1998). Potential natural hybridization within Sinojackia sensu stricto is supported by an artificial reciprocal cross between S. xylocarpa and S. rehderiana, the progeny of which readily set seeds (Ye et al., 2006). Moreover, parentage analysis based on microsatellites indicates that nearly 5 % of candidate fathers of progeny of S. xylocarpa are from S. rehderiana in Wuhan Botanical Garden, which is consistent with a pattern of natural hybridization (J. J. Zhang et al., pers. comm.).
Regardless of whether the complex microsatellite pattern within and among species of Sinojackia sensu stricto observed in the present study is due to extensive hybridization or incipient speciation, the present data suggest that the ultimate morphological basis for recognizing species in Sinojackia sensu stricto should now be revisited, especially when considered in combination with observations of high morphological variation within and similarity among several of the species, as previously observed (Luo, 2005; Yao et al., 2005). This has already occurred in the case of S. oblongicarpa, in which variation in flower and fruit dimensions lies within that found in S. sarcocarpa and has prompted the treatment of the former as a synonym of the latter (Luo, 2005). On the basis of our microsatellite data alone, all species of Sinojackia sensu stricto excluding S. microcarpa could be reduced to synonyms of S. xylocarpa, the first described species of the group. Whether this is ultimately the best alternative, however, should await further careful morphological and anatomical studies focusing on variation within and among populations as part of a comprehensive taxonomic revision of the group.
The use of microsatellites in phylogenetic studies of Sinojackia
The intergenic spacers of atpB–rbcL, trnS–trnG and psbA–trnH have been shown to be variable in studies of plant population genetic diversity (e.g. Hamilton, 1999; Wang et al., 2004). The low DNA sequence variation observed among the species of Sinojackia sensu stricto for these plastid DNA regions, however, was not useful in resolving relationships. The nearly exclusive use of DNA sequence data from the ITS region to resolve relationships at low taxonomic levels reflects the paucity of alternatives from the nuclear genome for this purpose (Baldwin et al., 1995; Soltis and Soltis, 1998; Manos et al., 1999; Kyndt et al., 2005). Despite its more rapid rate of evolution than plastid DNA, ITS often exhibits a lack of intrageneric variation (e.g. Leclerc et al., 1998; Schilling et al., 1998; Blattner et al., 2001; Zhang et al., 2001; Després et al., 2003), and there are presumably ITS studies that remain unpublished due to lack of resolution. Like the plastid DNA spacers, the ITS region was not useful in resolving relationships among species of Sinojackia sensu stricto.
Phylogenetic inference at low taxonomic levels is often limited in plants by the lack of suitable DNA regions, such as mitochondrial DNA in animals (Després et al., 2003). In plants, microsatellites have been widely used in cultivar identification, genomic mapping and population genetics (Jarne and Lagoda, 1996; Zheng et al., 2004; Somers et al., 2004). They have not, however, been widely used in phylogenetic inference due to rapid evolution and potential for homoplasy (Jarne and Lagoda, 1996), but these problems should be alleviated through maximizing the sample size for each species and using suitable genetic distance measures (Nei et al., 1983; Goldstein and Pollock, 1997). Microsatellites have the potential to resolve interspecific phylogenetic relationships, especially for closely related species for which ITS or plastid DNA variation is low or absent (Goldstein and Pollock, 1997). The recent development of relatively rapidly evolving microsatellite loci for S. xylocarpa and their successful amplification here in congeners including S. dolichocarpa provided a source of phylogenetic information within the Sinojackia sensu stricto clade.
Conservation implications of the data for Sinojackia and Changiostyrax
An attribute shared by nearly all members within Sinojackia sensu stricto and Changiostyrax is their endangerment due to continued habitat destruction and harvesting for fuel wood (Yao et al., 2005, 2007b). The use of phylogenetic analysis to help determine conservation priorities has been advocated for cases in which resources are scarce and not all taxa can receive equal protection (Vane-Wright et al., 1991; Linder, 1995; Faith, 1996; Hopper et al., 1999). This type of analysis provides an indication of evolutionary relationships and is not necessarily what would be predicted from morphological characters alone (Hopper et al., 1999). For example, differences in the placement of S. dolichocarpa (Changiostyrax) in the cladograms of Qi (1981) and Chen (1995) would clearly lead to quite different conservation priorities; i.e. when S. dolichocarpa is transferred to the monotypic genus Changiostyrax, the conservation status of this species is reinforced. Only C. dolichocarpus and S. xylocarpa are currently considered vulnerable by the International Union for the Conservation of Nature (IUCN, http://www.iucnredlist.org), but the other species of Sinojackia sensu stricto have suffered serious population declines in recent years (Wang and Xie, 2004; Yao et al., 2005); whether they are considered one or several species will have a marked effect on assessment of their conservation status. In consideration of the levels of molecular variation found within and among populations, the method advocated by Ennos et al. (2005) in conserving the evolutionary processes that generate taxonomic biodiversity may be the best way to conserve the Chinese endemic genus Sinojackia.
ACKNOWLEDGEMENTS
We are grateful to Dr Tao Chen and Mr Fanzhang Du for assistance in collecting samples. This work was supported in part by the Natural Scientific Foundation of China (30370153) and WZ No.050809 of the Conservation Genetics Laboratory, Wuhan Botanical Garden, Chinese Academy of Sciences.
APPENDIX
List of fruit characters and their states used in the morphological analysis of Styracaceae (refer to Fritsch et al., 2001).
Fruit dehiscent (0); fruit indehiscent (1).
Fruit unwinged (0); fruit two-winged (1); fruit four-winged (2).
Fruit wall not ribbed (0); fruit wall ribbed (1).
Fruit glabrous (0); fruit stellate-pubescent (1).
Fruit wrinkled after drying (0); fruit plump after drying (1).
Fruit ovoid (0); fruit ellipsoid (1).
Fruit beak long (0); fruit beak short (1); fruit without beak (2).
Fruit beak conical at apex (0); fruit beak acuminate at apex (1).
Fruit diameter <1 cm diameter (0); fruit 1–1·2 cm diameter (1); fruit >1·2 cm diameter (2).
Mesocarp absent (0); mesocarp present (1).
Endocarp surface longitudinally ridged (0); endocarp surface not longitudinally ridged (1).
Endocarp lacunae absent (0); endocarp lacunae present (1).
Seed coat indurate (0); seed coat fragile (1).
Seed to carpel ratio <1 (0); seed to carpel ratio 1–2 (1); seed to carpel ratio >2 (2).
Seed coat not appendaged (0); seed coat appendaged (1).
Seed surface areolate (0); seed surface fibrous (1); seed surface papillose (2).
LITERATURE CITED
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- Blattner FR, Weising K, Bänfer G, Maschwitz U, Fiala B. Molecular analysis of phylogenetic relationships among myrmecophytic Macaranga species (Euphorbiaceae) Molecular Phylogenetics and Evolution. 2001;19:331–344. doi: 10.1006/mpev.2001.0941. [DOI] [PubMed] [Google Scholar]
- Cao PJ, Yao QF, Ding BY, Zeng HY, Zhong YX, Fu CX, Jin XF. Genetic diversity of Sinojackia dolichocarpa (Styracaceae), a species endangered and endemic to China, detected by inter-simple sequence repeat (ISSR) Biochemical Systematics and Ecology. 2006;34:231–239. [Google Scholar]
- Chen T. Changiostyrax, a new genus of Styracaceae from China. Guihaia. 1995;15:289–292. [Google Scholar]
- Chen T. A new species of Sinojackia Hu (Styracaceae) from Hunan, south China. Edinburgh Journal of Botany. 1998;55:235–238. [Google Scholar]
- Chen T, Chen ZY. The geographical distribution of Styracaceae. Bulletin of Botanical Research. 1996;16:57–66. [Google Scholar]
- Chen T, Li GY. A new species of Sinojackia Hu (Styracaceae) from Zhejiang, east China. Novon. 1997;7:350–352. [Google Scholar]
- Chiang TY, Schaal BA, Peng CI. Universal primers for amplification and sequencing a non-coding spacer between the atpB and rbcL genes of chloroplast DNA. Botanical Bulletin of Academia Sinica. 1998;39:245–250. [Google Scholar]
- Després L, Gielly L, Redoutet B, Taberlet P. Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Molecular Phylogenetics and Evolution. 2003;27:185–196. doi: 10.1016/s1055-7903(02)00445-1. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Ennos RA, French GC, Hollingsworth PM. Conserving taxonomic complexity. Trends in Ecology and Evolution. 2005;20:164–168. doi: 10.1016/j.tree.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation priorities and phylogenetic pattern. Conservation Biology. 1996;10:1286–1289. [Google Scholar]
- Farris J S, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- Fritsch PW, Morton CM, Chen T, Meldrum C. Phylogeny and biogeography of the Styracaceae. International Journal of Plant Science. 2001;162(Suppl.):S95–S116. [Google Scholar]
- Fu LK. China plant red data book—rare and endangered plants. Beijing: Science Press; 1992. [Google Scholar]
- Goldstein DB, Pollock DD. Launching microsatellites: a review of mutation processes and method of phylogenetic inference. Journal of Heredity. 1997;88:335–342. doi: 10.1093/oxfordjournals.jhered.a023114. [DOI] [PubMed] [Google Scholar]
- Hamilton MB. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology. 1999;8:521–523. [PubMed] [Google Scholar]
- Hopper SD, Fay MF, Rossetto M, Chase MW. A molecular phylogenetic analysis of the bloodroot and kangaroo paw family, Haemodoraceae: taxonomic, biogeographic and conservation implications. Botanical Journal of the Linnean Society. 1999;131:285–299. [Google Scholar]
- Hu HH. Sinojackia, a new genus of Styracaceae from southeastern China. Journal of the Arnold Arboretum. 1928;9:130–131. [Google Scholar]
- Hu HH. Notulae systematicae ad floram sinensem, II. Journal of the Arnold Arboretum. 1930;11:224–228. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hwang SM, Grimes J. In: Sinojackia. Wu CY, Raven P, editors. vol. 15. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; 1996. pp. 268–270. [Google Scholar]
- Jarne P, Lagoda PJL. Microsatellites, from molecules to populations and back. Trends in Ecology and Evolution. 1996;11:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Droogenbroeck BV, Romeijn-Peeters E, Romero-Motochi JP, Scheldeman X, Goetghebeur P, et al. Molecular phylogeny and evolution of Caricaceae based on rDNA internal transcribed spacers and chloroplast sequence data. Molecular Phylogenetics and Evolution. 2005;37:442–459. doi: 10.1016/j.ympev.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Leclerc MC, Barriel V, Lecointre G, de Reviers B. Low divergence in rDNA ITS sequences among five species of Fucus (Phaeophyceae) suggests a very recent radiation. Journal of Molecular Evolution. 1998;46:115–120. doi: 10.1007/pl00006278. [DOI] [PubMed] [Google Scholar]
- Linder HP. Setting conservation priorities: the importance of endemism and phylogeny in the Southern African orchid genus Herschelia. Conservation Biology. 1995;9:585–595. [Google Scholar]
- Luo LQ. A new species of Sinojackia from Sichuan. Acta Scientiarum Naturalium Universitatis Sunyatseni. 1992;31:78–79. [Google Scholar]
- Luo LQ. A new synonym in the genus Sinojackia (Styracaceae) Acta Phylotaxonomica Sinica. 2005;43:561–564. [Google Scholar]
- Merrill ED. Miscellanea sinensia. Sunyatsenia. 1937;3:246–262. [Google Scholar]
- Manos O, Doyle JJ, Nixon KC. Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae) Molecular Phylogenetics and Evolution. 1999;12:333–349. doi: 10.1006/mpev.1999.0614. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. The American Naturalist. 1972;106:283–292. [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Qi CJ. A new species of Styracaceae from Hunan. Acta Phytotaxonomica Sinica. 1981;19:526–528. [Google Scholar]
- Rieseberg LH, Carney SE. Plant hybridization. Tansley Review, 102. New Phytologist. 1998;140:599–624. doi: 10.1046/j.1469-8137.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. NTSYS-pc, numerical taxonomy and multivariate analysis system. New York: Exeter Software; 2000. Version 2·0. [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution and biogeography of Paeonia (Paeoniaceae) American Journal of Botany. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Schilling EE, Linder CR, Noyes RD, Rieseberg LH. Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal spacer region sequence data. Systematic Botany. 1998;23:177–187. [Google Scholar]
- Soltis DE, Soltis PS. Choosing an approach and an appropriate gene for phylogenetic analysis. In: Soltis DE, Doyle JJ, editors. Molecular systematics of plants: II, DNA sequencing. Boston: Kluwer Academic Publishers; 1998. pp. 1–42. [Google Scholar]
- Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B. Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Systematics and Evolution. 1997;204:109–123. [Google Scholar]
- Swensen SM, Luthi JN, Rieseberg LH. Datiscaceae revisited: monophyly and the sequence of breeding system evolution. Systematic Botany. 1998;233:157–169. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (* and other methods) Sunderland, MA: Sinauer Associates; 2003. Version 4b 10. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tate JA. The University of Texas at Austin; 2002. Systematics and evolution of Tarasa (Malvaceae): an enigmatic Andean polyploid genus. PhD thesis. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane-Wright RI, Humphries CJ, Williams PH. What to protect?—Systematics and the agony of choice. Biological Conservation. 1991;55:235–254. [Google Scholar]
- Wang S, Xie Y. China species red list. Beijing: Higher Education Press; 2004. [Google Scholar]
- Wang T, Su YJ, Li XY, Zheng B, Chen GP, Zeng QL. Genetic structure and variation in the relict populations of Alsophila spinulosa from southern China based on RAPD markers and cpDNA atpB–rbcL sequence data. Hereditas. 2004;140:8–17. doi: 10.1111/j.1601-5223.2004.01659.x. [DOI] [PubMed] [Google Scholar]
- Yao XH, Ye QG, Kang M, Huang HW. Geographic distribution and current status of the endangered genera Sinojackia and Changiostyrax. Biodiversity Science. 2005;13:339–346. [Google Scholar]
- Yao XH, Ye QG, Kang M, Zhou JF, Xu YQ, Wang Y, Huang HW. Characterization of microsatellite markers in the endangered Sinojackia xylocarpa (Styracaceae) and cross-species amplification in closely related taxa. Molecular Ecology Notes. 2006;6:133–136. [Google Scholar]
- Yao XH, Ye QG, Ge JW, Kang M, Huang HW. A new species of Sinojackia (Styracaceae) from Hubei, central China. Novon. 2007;a 17:138–140. [Google Scholar]
- Yao XH, Ye QG, Kang M, Huang HW. Microsatellite analysis reveals interpopulation differentiation and gene flow in the endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China. New Phytologist. 2007;b 176:472–480. doi: 10.1111/j.1469-8137.2007.02175.x. [DOI] [PubMed] [Google Scholar]
- Ye QG, Yao XH, Zhang SJ, Kang M, Huang HW. Potential risk of hybridization in ex situ collections of two endangered species of Sinojackia Hu (Styracaceae) Journal of Integrative Plant Biology. 2006;48:867–872. [Google Scholar]
- Zhang LB, Comes HP, Kadereit JW. Phylogeny and Quaternary history of the European montane/alpine endemic Soldanella (Primulaceae) based on ITS and AFLP variation. American Journal of Botany. 2001;88:2331–2345. [PubMed] [Google Scholar]
- Zheng Y, Li Z, Huang H, Wang Y. Molecular characterization of kiwifruit (Actinidia) cultivars and selections using SSR markers. Journal of the American Society for Horticultural Science. 2004;129:374–382. [Google Scholar]