Abstract
Background
In studies looking at individual polyploid species, the most common patterns of genomic change are that either genome size in the polyploid is additive (i.e. the sum of parental genome donors) or there is evidence of genome downsizing. Reports showing an increase in genome size are rare. In a large-scale analysis of 3008 species, genome downsizing was shown to be a widespread biological response to polyploidy. Polyploidy in the genus Nicotiana (Solanaceae) is common with approx. 40 % of the approx. 75 species being allotetraploid. Recent advances in understanding phylogenetic relationships of Nicotiana species and dating polyploid formation enable a temporal dimension to be added to the analysis of genome size evolution in these polyploids.
Methods
Genome sizes were measured in 18 species of Nicotiana (nine diploids and nine polyploids) ranging in age from <200 000 years to approx. 4·5 Myr old, to determine the direction and extent of genome size change following polyploidy. These data were combined with data from genomic in situ hybridization and increasing amounts of information on sequence composition in Nicotiana to provide insights into the molecular basis of genome size changes.
Key Results and Conclusions
By comparing the expected genome size of the polyploid (based on summing the genome size of species identified as either a parent or most closely related to the diploid progenitors) with the observed genome size, four polyploids showed genome downsizing and five showed increases. There was no discernable pattern in the direction of genome size change with age of polyploids, although with increasing age the amount of genome size change increased. In older polyploids (approx. 4·5 million years old) the increase in genome size was associated with loss of detectable genomic in situ hybridization signal, whereas some hybridization signal was still detected in species exhibiting genome downsizing. The possible significance of these results is discussed.
Key words: Genome downsizing, genome size, Nicotiana, polyploidy, sequence elimination, Solanaceae
INTRODUCTION
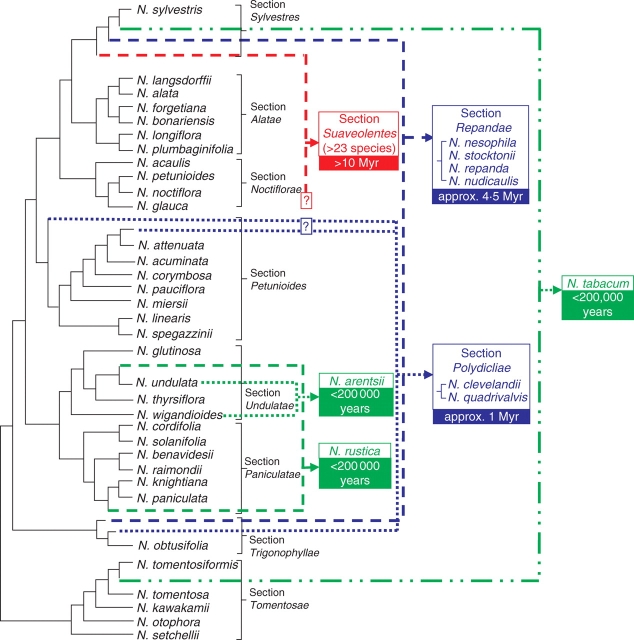
The genus Nicotiana, comprising approx. 75 species, displays a range of genomic changes including gene conversion, tandem and dispersed sequence evolution, intergenomic translocations, dysploidy, polyploidy, etc. (Kenton et al., 1993; Matzke et al., 2004; Melayah et al., 2004; Dadejova et al., 2007; Lim et al., 2007; Kovarik et al., 2008). Given the increasingly robust phylogenetic framework now available (Chase et al., 2003; Clarkson et al., 2004, 2005), Nicotiana is thus ideally suited to study patterns of genome evolution.
Polyploidy is common in the genus, with approx. 40 % of species being allotetraploid. They comprise (a) N. tabacum (section Nicotianae), (b) N. rustica (section Rusticae), (c) N. arentsii (section Undulatae), (d) N. clevelandii and N. quadrivalvis (section Polydicliae), (e) N. nudicaulis, N. repanda, N. nesophila and N. stocktonii (section Repandae) and (f) all approx. 23 species in section Suaveolentes. All polyploids, except some of those in section Suaveolentes, are 2n = 4x = 48, representing a doubling of the diploid chromosome number for the genus (2n = 2x = 24). In section Suaveolentes, polyploid evolution has been accompanied by changes in chromosome number, probably through dysploid reductions via chromosome deletions or fusions (2n ranges from 32 to 48).
Based on cytological and floral morphology, combined with sequence data from plastid and nuclear genes (Goodspeed, 1954; Aoki and Ito, 2000; Chase et al., 2003; Clarkson et al., 2004), the likely parentage of nearly all allotetraploid species has now been determined, together with estimates of their ages based on combining molecular clock analysis and calibration with the ages of oceanic volcanic islands (Clarkson et al., 2005; Kovarik et al., 2008) (Fig. 1). These data show that the polyploid species range considerably in age. The youngest (N. tabacum, N. rustica and N. arentsii) are each estimated to have arisen <200 000 years ago, followed by the two species in section Polydicliae, which are approx. 1 million years (Myr) old, and the five species in section Repandae, which are approx. 4·5 Myr old (Clarkson et al., 2005). The oldest polyploids, in section Suaveolentes, are considered to have originated from a single polyploid event >10 Myr ago, followed by speciation to produce the approx. 23 species known today.
Fig. 1.
Summary of phylogenetic relationships of Nicotiana species with proposed origins of polyploids. Data used in analyses include plastid and internal transcribed spacer loci. Figure modified and adapted from Knapp et al. (2004) using more recent phylogenetic information taken from glutamine synthase (J. J. Clarkson et al., unpubl. res.). Uncertainty concerning one of the parental genome donors for sections Polydicliae and Suaveolentes is indicated by question marks.
Nicotiana polyploids not only vary in age but also in the relatedness of the parental species contributing genomes to the polyploid nucleus. For example, N. arentsii is an intrasectional polyploid, the hybrids and diploid progenitors all belonging to section Undulatae. In N. rustica (section Rusticae) the parental species are in closely related sections (Paniculatae and Undulatae) (Clarkson et al., 2004; Kovarik et al., 2008). In contrast, the diploid species giving rise to N. tabacum and polyploids in section Repandae, Polydicliae and Suaveolentes are from distantly related sections (Figs 1 and 2).
Fig. 2.
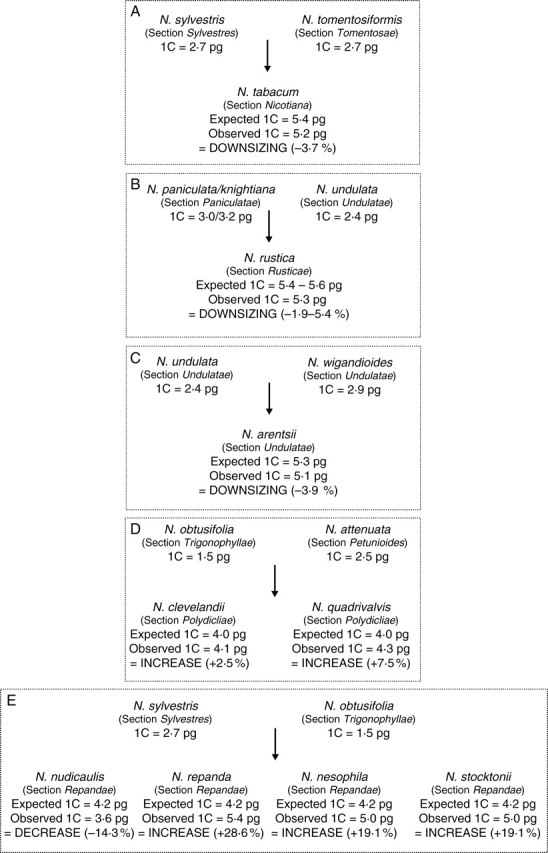
Direction of genome size evolution in Nicotiana polyploids varying in age from <200 000 years (A–C), approx. 1 Myr (D) to approx. 4·5 Myr (E) old. The observed versus expected 1C DNA amounts in nine polyploids were based on comparison between genome sizes determined for diploid and polyploid species using genome size data taken from Table 1. Putative diploid parental genome donors used for comparison of the various polyploids are based on a range of molecular and cytogenetic results (see text).
Insights into contrasting patterns of molecular evolution in Nicotiana polyploids have been gained through the study of natural and synthetic species. These have involved analysing both specific DNA sequences [e.g. ribosomal DNA (rDNA), non-coding tandem repeats and retrotransposons] and global genome organization using fluorescent in situ hybridization (FISH) [including genomic in situ hybridization (GISH); Kovarik et al., 1996, 2004; Chase et al., 2003; Lim et al., 2004, 2005, 2006b, 2007; Clarkson et al., 2005; Skalicka et al., 2005; Petit et al., 2007]. Such studies have shown that allopolyploidy in Nicotiana has been accompanied by numerous genetic changes including (depending on the polyploid species in question) rDNA homogenization and loss of loci (Matyasek et al., 2003; Kovarik et al., 2004; Clarkson et al., 2005), intergenomic translocations (Kenton et al., 1993; Chase et al., 2003; Lim et al., 2004) and changes in copy number and organization of both tandem and dispersed repeats (Melayah et al., 2004; Petit et al., 2007). By combining these data with the known ages of the polyploids, a temporal perspective on polyploidy evolution has been obtained. Lim et al. (2007) showed that during early evolution of Nicotiana polyploids (i.e. those <200 000 years old) intergenomic translocations and rearrangement and loss of repeated DNA sequences may take place. Over the next 1–2 Myr, considerable exchange of repeats between the parental genomes becomes apparent, and by 5 Myr there can be near complete genomic turnover including evolution of new repeats not present in the parental species.
Studies of genome size evolution in polyploids in a diverse range of angiosperms have been conducted at a number of levels. For specific polyploid species, there are many reports showing no change or a decrease in DNA amount relative to the proposed progenitor species (see review in Leitch and Bennett, 2004); those showing an increase are much rarer but include a few naturally occurring species of Hordeum (Jakob et al., 2004) and artificially induced dihaploids of Nicotiana (Dhillon et al., 1983). At the other end of the spectrum, large-scale analyses combining available genome size data for 3008 angiosperms have led to the proposal that genome downsizing is a widespread biological response to polyploidization leading to diploidization of the polyploid genome (Leitch and Bennett, 2004). Such conclusions have been supported by comparative molecular studies. For example, nearly 80 % of the duplicated genes have been lost from rice (Oryza sativa) since the polyploid event that gave rise to it approx. 70 Myr ago (Wang et al., 2005), and >50 % of the duplicated genes in maize (Zea mays) have been lost since its polyploid origin approx. 3–11 Myr ago (Messing et al., 2004). In addition, comparative analyses of DNA sequences surrounding the AdhA locus in diploid and polyploid Gossypium genomes suggest that polyploidization may lead to increased rates of illegitimate recombination resulting in a greater loss of DNA from polyploids than related diploids (Grover et al., 2007b). Rapid loss of DNA (based both on genome size measurements or loss of AFLP and/or RFLP bands or specific sequences) has also been reported following synthesis of various artificial hybrids and polyploids of Brassica (Song et al., 1995), Nicotiana (Skalicka et al., 2003, 2005; Petit et al., 2007) and Aegilops and Triticum (Ozkan et al., 2001, 2003; Kashkush et al., 2002; Levy and Feldman, 2004).
Given contrasting ages of Nicotiana polyploids (<200 000 years to >10 Myr), differences in the extent of genome divergence in diploid progenitors and the diversity of molecular changes that have been documented to occur following polyploidization, it seems timely to examine patterns of genome size evolution in Nicotiana polyploids of different ages. Although there have been several previous studies reporting genome sizes for species of Nicotiana, the most extensive has been by Narayan (1987) who listed C-values for 51 Nicotiana taxa, including all but two of the species studied here. However, in his discussion, genome size evolution was only assessed in the three ‘young’ polyploids (N. tabacum, N. rustica and N. arentsii), and there were considerable discrepancies between some of his genome size estimates compared with those previously published by himself (Narayan and Rees, 1974), other authors (e.g. Ingle et al., 1975; Galbraith et al., 1983) and here (see Discussion). Thus, genome sizes were estimated in nine diploid and nine polyploid species. Unfortunately, taxonomic relationships and parental genome donors of polyploid Suaveolentes species are not yet sufficiently understood to be investigated in this way.
METHODS AND MATERIALS
Plant material
Table 1 lists the 18 species analysed in the current work together with their origin.
Table 1.
List of Nicotiana species studied together with chromosome number, 1C and 4C DNA amount, calibration standard and method of estimating genome size
| Species | Accession number (where available) and source of material* | 2n | 4C DNA (± s.d.) (pg) | 1C DNA (± s.e.) (pg) | Standard used† | Method used‡ |
|---|---|---|---|---|---|---|
| Section Nicotiana | ||||||
| N. tabacum L. | Nee et al. 51789a | 48 | 20·70 (1·34) | 5·2 (0·060) | Pisum | Fe |
| Section Petunioides | ||||||
| N. attenuata Torrey ex S.Watson | TW13b | 24 | 9·93 (0·05) | 2·5 (0·002) | Pisum | FC |
| Section Paniculatae | ||||||
| N. knightiana Goodsp. | CPGe | 24 | 12·64 (0·06) | 3·2 (0·002) | Pisum | FC |
| N. paniculata L. | TW99b | 24 | 11·78 (0·08) | 3·0 (0·007) | Pisum | FC |
| Section Polydicliae | ||||||
| N. clevelandii A.Gray | TW30b | 48 | 16·56 (0·21) | 4·1 (0·018) | Pisum | FC |
| N. quadrivalvis Pursh. | TW18b | 48 | 17·01 (0·06) | 4·3 (0·005) | Hordeum | FC |
| Section Repandae | ||||||
| N. repanda Willd. | TW110b | 48 | 21·76 (0·35) | 5·4 (0·005) | Zea | FC |
| N. nesophila I.M.Johnston | 964750097d | 48 | 20·13 (0·07) | 5·0 (0·005) | Pisum | FC |
| N. nudicaulis S.Watson | 964750051d | 48 | 14·22 (0·07) | 3·6 (0·008) | Pisum | FC |
| N. stocktonii Brandegee | 974750101d | 48 | 19·99 (0·06) | 5·0 (0·005) | Pisum | FC |
| Section Rusticae | ||||||
| N. rustica L. | NIC 616147c | 48 | 21·19 (0·08) | 5·3 (0·040) | Pisum | FC |
| Section Sylvestres | ||||||
| N. sylvestris Speg. & Comes | TW127b | 24 | 10·78 (0·10) | 2·7 (0·002) | Pisum | FC |
| Section Tomentosae | ||||||
| N. tomentosiformis Goodsp. | Nee et al. 51771a | 24 | 10·97 (0·19) | 2·7 (0·020) | Pisum | FC |
| Section Trigonophyllae | ||||||
| N. obtusifolia M.Martens & Galeotti | 894750176d | 24 | 6·18 (0·03) | 1·5 (0·002) | Solanum | FC |
| Section Undulatae | ||||||
| N. arentsii Goodsp. | NIC 445/82c | 48 | 20·22 (0·15) | 5·1 (0·060) | Pisum | FC |
| N. glutinosa L. | Wood 11732a | 24 | 8·94 (0·10) | 2·2 (0·010) | Solanum | FC |
| N. undulata Ruiz & Pav. | RBG, Kewf | 24 | 9·66 (0·04) | 2·4 (0·006) | Pisum | FC |
| N. wigandioides Koch & Fintelm. | Nee et al. 51764a | 24 | 11·38 (0·50) | 2·9 (0·020) | Pisum | Fe |
* Source of material: a = New York Botanic Garden, New York, USA; b = USDA, North Carolina State University, Raleigh, NC, USA; c = IPK, Gatersleben, Germany; d = Botanical and Experimental Garden, Radboud University Nijmegen,The Netherlands; e = Chelsea Physic Garden, London, UK; f = Royal Botanic Gardens, Kew, UK.
† Species and 4C values used for calibration standards are as follows: Pisum = Pisum sativum cv. Minerva Maple, 4C = 17·52 pg; Solanum = Solanum lycopersicum, 4C = 4·00 pg; Hordeum = Hordeum vulgare cv. Sultan, 4C = 22·24 pg; Zea = Zea mays CE-777, 4C = 11·34 pg.
‡ FC = flow cytometry; Fe = Feulgen microdensitometry.
Genome size estimation
Two methods were used to estimate genome size in Nicotiana species: flow cytometry and Feulgen microdensitometry. Both methods have been shown to produce comparable results (Doležel et al., 1998).
Feulgen microdensitometry using a Vickers M85a microdensitometer followed the methods described in Hanson et al. (2001), and flow cytometry was conducted using a Partec CyFlow or PAII as described in Hanson et al. (2005). The calibration standard and method used for each species are listed in Table 1. Expected genome sizes of the polyploids were determined by adding the genome sizes of the putative diploid progenitor species.
GISH
GISH was carried out as described in Clarkson et al. (2005). Briefly, slides were pretreated with RNase A (100 µg mL–1, 1 h) and pepsin (0·25 µg mL–1, 5 min), followed by denaturation in 70 % formamide in 2× SSC (0·3 m sodium chloride, 0·03 m sodium citrate) at 70 °C for 2 min. The hybridization mixture included 8 µg mL–1 digoxigenin-labelled N. sylvestris DNA and 8 µg mL–1 biotin-labelled N. obtusifolia DNA. In situ hybridization was carried out overnight at 37 °C. Post-hybridization washes included formamide [20 % (v/v) in 0·1× SSC, 42 °C] giving an estimated hybridization stringency of 80–85 %. Sites of probe hybridization were detected with fluorescein-conjugated anti-digoxigenin IgG (Roche Biochemicals) (20 µg mL–1) and Cy3-conjugated avidin (Amersham Biosciences; 5 µg mL–1). Chromosomes were counterstained with DAPI (4',6-diamidino-2-phenylindole; 2 µg mL–1 in 4× SSC,) and mounted in Vectashield medium (Vector Laboratories). Metaphases were photographed on a Leica DMRA2 epifluorescence microscope with an Orca ER camera. Images were processed for colour balance, contrast and brightness uniformly.
RESULTS
Genome size estimates in Nicotiana
Table 1 lists genome size estimates obtained for the species analysed. Figure 2 shows the genome size of the diploid species considered to be most closely related to the species that gave rise to the polyploids, together with the expected versus the observed genome size estimates for these polyploids. Genome downsizing was observed in four of the polyploids studied and genome size increases were observed in the remaining five polyploids.
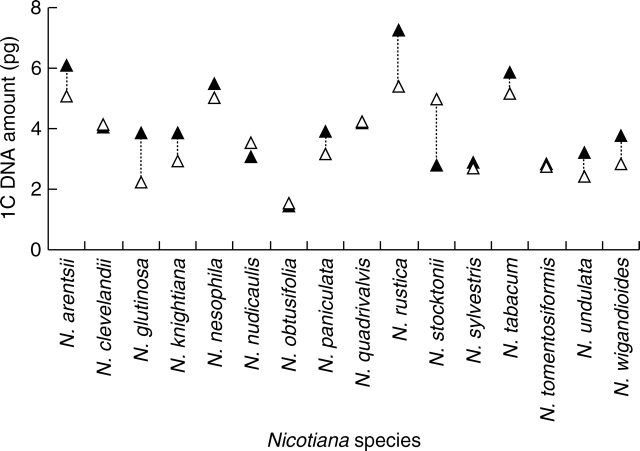
Comparisons between genome size estimates obtained in the present work for 16 species with those reported by Narayan (1987) are shown diagrammatically in Fig. 3. For 11 species Narayan's estimates were larger than ours whereas in the remaining five species our estimates were greater than Narayan's.
Fig. 3.
Comparison between 1C DNA amounts reported by Narayan (1987) (closed triangles) and those given in Table 1 of this paper (open triangles).
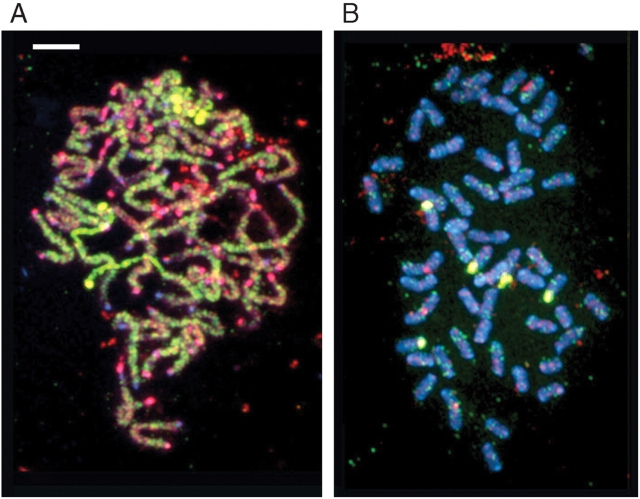
GISH in section Repandae
When GISH was applied to chromosome preparations of the polyploid species N. nudicaulis and N. nesophila (both in section Repandae) the labelling patterns were quite different (Fig. 4). In N. nudicaulis both probes (i.e. genomic DNA from N. sylvestris and N. obtusifolia) hybridized weakly across all chromosomes, although some chromosomes were more strongly labelled with N. sylvestris probe (green signal) and others were more strongly labelled with the N. obtusifolia probe (red signal). Given the amount of cross-hybridization it was not possible to distinguish the genomic origin of each chromosome in N. nudicaulis.
Fig. 4.
GISH to (A) Nicotiana nudicaulis and (B) N. nesophila root-tip metaphase probed with N. sylvestris genomic DNA (digoxigenin labelled, FITC detected, yellow/green) and N. obtusifolia (biotin labelled, Cy3 detected, red). Chromosomes were counterstained with DAPI (blue). Scale bar = 10 µm.
In contrast, hybridization of both probes to chromosomes of N. nesophila was weak, with only scattered signal detected from each probe. The only exception to this was the strong yellow hybridization signals corresponding to the location of ribosomal DNA loci.
DISCUSSION
Comparisons with previous genome size estimates
The most extensive survey of genome sizes in species of Nicotiana prior to the present one was that of Narayan (1987) who estimated DNA amounts in 51 species. However, as illustrated in Fig. 3, there are considerable discrepancies between some of his values and those estimated here (Table 1). The most notable are those for N. rustica for which Narayan's estimate is nearly one-third larger than ours (1C = 7·2 versus 5·3 pg), and N. stocktonii for which our estimate is nearly double that of Narayan's (1C = 5·0 versus 2·8 pg). Further discrepancies are noted between Narayan's estimates and those published by other workers. For example, whereas Galbraith et al. (1983) and we (Table 1) reported 1C values of 2·1 and 2·2 pg, respectively, for N. glutinosa, Narayan gave an estimate of 1C = 3·9 pg. The reasons for these discrepancies remain unknown, but the observation that the differences are not always in the same direction (i.e. sometimes Narayan's estimates are larger or smaller than ours and previous workers) suggests that there is not a simple, methodological basis behind the differences. Instead the discrepancies may be due to taxonomic issues (e.g. incorrect identification).
Evolution of genome size in Nicotiana polyploids
It is noted that the observations and discussions presented below assume that (a) the diploid species used are indeed closely related to the actual progenitors that gave rise to the polyploids, and that (b) since polyploid formation the genome sizes of these diploids have remained largely unchanged. Any violation of these two assumptions will affect interpretation of the results, but at present it is the best approximation available. Choice of diploid progenitor is based on extensive molecular and morphological analyses of the majority of species in the genus and is thus as comprehensive as possible. The possibility that the incorrect species have been selected is low but cannot be ruled out completely, particularly for the older polyploids. The second assumption is impossible to confirm as it requires estimating genome sizes in the diploid progenitor and polyploid species at the time of polyploidization – thousands to millions of years ago (depending on the polyploid). With these caveats, the following results and discussion on the evolution of genome size in polyploids of Nicotiana are presented.
Polyploids <200 000 years old
Three independently produced allotetraploids of Nicotiana (all 2n = 4x = 48), N. tabacum, N. rustica and N. arentsii, are estimated to be approx. 200 000 years old or less. Based on flower and chromosome morphology (Goodspeed, 1954) and plastid and nuclear DNA sequence data (Aoki and Ito, 2000; Chase et al., 2003) the parental genomes of these polyploids are considered to be derived from ancestors of the extant species shown in Fig. 2A–C. All three polyploids appear to have undergone a limited amount of genome downsizing, losing between 3·7 % and 5·4 % over approx. 200 000 years. These results agree with the lower numbers of certain repeats reported in these polyploids compared with their diploid progenitors. For example, in N. tabacum there are lower numbers of several repeat sequences, including NTRS, A1/A2 (Lim et al., 2004), a pararetroviral repeat sequence NtoEPRV (Gregor et al., 2004) and various retrotransposons (Melayah et al., 2004; Petit et al., 2007), than in the diploids. In N. rustica, an examination of satellite repeats showed they too differed in abundance and distribution relative to the diploid progenitor species (Lim et al., 2005). One repeat (NUNSSP) was similar in organization and copy number to the diploid progenitors, whereas a comparison of a second repeat (NPAMBO) revealed minor changes in its chromosomal distribution but copy number was reduced by at least 10-fold compared with N. paniculata (the maternal donor species used for comparison). Such sequence elimination probably occurred since N. rustica evolved and could have contributed to the observed 2–5% reduction in DNA amount (Fig. 2B). Alternatively these sequences may have increased in the diploids after polyploid formation.
In all three polyploids the number of 35S rDNA loci is additive, yet within this framework considerable sequence elimination of individual copies has taken place with only 30 %, 50 % and 80 % of the expected copy numbers (based on estimates from the diploid species) observed in N. tabacum, N. arentsii and N. rustica, respectively. In N. tabacum and N. rustica, most of the remaining repeats from the maternal genome donor have been replaced, via gene conversion, with those originating from the paternal genome donor (Kovarik et al., 2004, 2008). Similar observations have been reported for N. arentsii, although here the direction of homogenization is different, with the 35S rDNA repeats being homogenized to the maternal repeat type (Kovarik et al., 2004, 2008).
Despite these observations, extensive sequence elimination is not apparently a universal response throughout the genomes of these young Nicotiana polyploids. In all three, the number of loci and copies of the 5S rDNA sequences are the sum of the diploid progenitors (i.e. additive), with no evidence for sequence loss or homogenization (Fulnecek et al., 2002). Further, the copy number of a satellite repeat isolated from N. paniculata (NPAMBE) was greater in N. rustica, although the number of extra copies varied 1·7-fold between the seven accessions analysed (Lim et al., 2005).
Polyploids approx. 1 million years old
The two polyploid species comprising section Polydicliae, N. quadrivalvis and N. clevelandii, are estimated to have formed approx. 1 Myr ago. Sequencing of both nuclear and plastid DNA indicates that they most likely arose from two different polyploidization events but involved the same diploid parents; an ancestor of N. obtusifolia (section Trigonophyllae) as the maternal genome donor and a progenitor of the lineage that later gave rise to N. attenuata (section Petunioides) as the paternal genome donor (Chase et al., 2003; Clarkson et al., 2004; Knapp et al., 2004; Qu et al., 2004). Analysis of genome size shows that in contrast to the younger polyploids discussed above, these two polyploids have undergone an increase in genome size. In N. clevelandii, genome upsizing is small – approx. 2·5 % – but in N. quadrivalvis it is more substantial (+7·5 %; Fig. 2D). NB These changes are based on using the genome size of N. attenuata as the species most closely related to the paternal genome donor of Polydicliae. However, given the lingering controversy concerning the choice of paternal genome donor (i.e. whether an ancestor of N. attenuata or an ancestor of all species in section Petunioides, Chase et al., 2003), an additional estimate of the observed versus expected genome size in Polydicliae was determined by taking the mean of six of the eight species recognized in this section (Knapp et al., 2004) as the genome size for the paternal genome donor (mean 1C = 2·57 pg; I. J. Leitch; L. Hanson, unpubl. res.). Using this approach differences in genome size were still detected for N. clevelandii and N. quadrivalvis but were smaller than just using the value for N. attenuata (+0·5 % and +6 %, respectively).
Currently there is little information on specific molecular changes that have accompanied evolution of species in section Polydicliae, although a reduction in genome size might be expected based on the chromosomal distribution of 35S and 5S rDNA sites using FISH. Kovarik et al. (2008) showed a loss of loci; both polyploids had just three 35S and one 5S loci compared with the expected five 35S and two 5S loci based on the numbers of loci observed in N. attenuata and N. obtusifolia (Kovarik et al., 2008). In addition, Wu et al. (2006) analysed low-copy genes encoding a family of trypsin-proteinase inhibitors. From cDNA, intron and promoter sequence analysis and Southern blotting, they deduced that only the maternally inherited genes of N. obtusifolia were retained in both N. clevelandii and N. quadrivalvis whereas those of N. attenuata were deleted. Thus there is apparent incongruence between the sequence data showing mostly uniparental eliminations and the increased genome sizes. An explanation may stem from the GISH results on N. quadrivalvis by Lim et al. (2007) showing (a) intergenomic mixing of DNA between the two parental genomes and (b) the invasion of N. attenuata subtelomeric repeat sequences onto N. obtusifolia chromosomes followed by their replacement. Adding the genome size data to these observations suggests that genome evolution in these polyploids has been accompanied by increases in the number of existing repeats. Perhaps increase in genome size arises from copy and paste mechanisms that also blur the distinction between the two parental genomes, as revealed by GISH (Lim et al., 2007). Further work is needed to characterize the nature and number of a representative sample of repeats in these polyploids.
Intergenomic sequence invasions involving both rDNA (Wendel et al., 1995) and certain classes of transposable elements (Zhao et al., 1998) have been observed in Gossypium polyploids that are estimated to be between 1 and 2 Myr old and thus similar in age to species in section Polydicliae. However, whether these rates of intergenomic sequence invasions in polyploid nuclei are ubiquitous or widespread in other polyploid groups of these ages needs to be determined.
Polyploids approx. 4·5 million years old
Section Repandae comprises four polyploid species, N. nudicaulis, N. repanda, N. stocktonii and N. nesophila (Knapp et al., 2004). Based on DNA sequence data from both the internal transcribed spacer of nuclear rDNA (Chase et al., 2003) and plastid DNA (Clarkson et al., 2004) the group has been shown to be monophyletic with N. nudicaulis sister to and distinct (both morphologically and genetically) from the remaining three species (which differ minimally). Clarkson et al. (2005) suggested that the original allopolyploidization event occurred approx. 4·5 Myr ago. This was followed by subsequent speciation with N. nudicaulis diverging from the rest approx. 2–3 Myr ago and N. stocktonii and N. nesophila diverging from N. repanda more recently, approx. 1 Myr ago. Sequence data from glutamine synthase (Clarkson et al., 2005) and plastid loci (Clarkson et al., 2004) indicate that the maternal genome donor of these four polyploids was an ancestor of section Sylvestres, which today comprises a single species N. sylvestris (1C = 2·7 pg). The paternal genome donor is an ancestor of section Trigonophyllae, which also now comprises a single extant species N. obtusifolia (1C = 1·5 pg). If genome size evolution in the four polyploids were additive then one would expect each to have a 1C value of 4·2 pg. However, this is not the case (Table 1, Fig. 2). Instead, the following were observed: (a) genome decreases in N. nudicaulis (1C = 3·6 pg) with a loss of approx. 14·3 % of DNA compared with the diploid progenitors; (b) genome upsizing of 28·6 % in N. repanda (1C = 5·4pg) and 19·1 % in N. nesophila and N. stocktonii (both with 1C = 5·0 pg).
There are considerable differences in the GISH-labelling patterns between N. nudicaulis (Fig. 4A) and N. nesophila (Fig. 4B). GISH to N. nudicaulis gives substantial labelling of the chromosomes, with some chromosomes that predominantly label with N. sylvestris genomic DNA and others predominantly with N. obtusifolia genomic DNA. However, due to cross hybridization of the probes, it is not possible to distinguish the parental origin of most chromosomes of the complement. In contrast, GISH to N. nesophila generates little signal at all, with only rDNA (large yellow signal in Fig. 4B) and some minor signals from both GISH probes. Lim et al. (2007) described the loss of signal as ‘near-complete genome turnover’ (see also Grover et al., 2007a).
Comparing GISH signals with genome size data is informative. The presence of some GISH signal to N. nudicaulis is associated with genome downsizing, whereas the near absence of GISH signal to N. nesophila is associated with genome size increases. Although mobility of repeats and homogenization mechanisms would act to reduce GISH discrimination between the two parental chromosome sets, it would not almost completely eliminate GISH signal, as seen in N. nesophila. Genome downsizing is thought to involve illegitimate recombination leading to the production of small indels and unequal intrastrand homologous recombination in, for example, long terminal repeat retroelements (e.g. Devos et al., 2002; Vitte and Panaud, 2003; Bennetzen et al., 2005). If these mechanisms are occurring in N. nudicaulis then the overall similarity of the repeat elements must be retained sufficiently to enable GISH to work partially. In contrast, near absence of GISH signal on N. nesophila is associated with genome upsizing. Mechanisms responsible for this phenomenon include amplification, transposition and insertion of retroelements (Vitte and Bennetzen, 2006) and evolution and amplification of satellite repeats (Lim et al., 2000, 2006a). Presumably these mechanisms, including evolution of the new satellite repeat NNE in N. nesophila, are occurring so rapidly that almost the entire repetitive portion of the genome has ‘turned over’, and little of the original character of the chromosomes remains (Lim et al., 2007). Thus, these results suggest that genome downsizing leads to a less dramatic alteration of genome characteristics than genome upsizing. It is likely that evolution and amplification of new sequences in association with genome upsizing replace many of the original sequences that were in the ancestral polyploid genome.
CONCLUDING REMARKS
Genomic responses to polyploidy in Nicotiana are complex, variable and determined by many factors, with age and genomic similarity of the parental genome donors potentially playing a role. Age is important in determining the extent of DNA sequence divergence encountered in polyploids, with genome turnover becoming extensive after approx. 4·5 Myr (Lim et al., 2007). Genomic relatedness of the parental genome donors may also be important in determining the extent of genome evolution in the polyploid genome. In Nicotiana the only polyploid shown to have undergone unequivocal intergenomic translocations is N. tabacum, which combines genomes from phylogenetically widely separated sections (Fig. 1; Kenton et al., 1993; Lim et al., 2004). The next most highly diverged genomes brought together are found in section Repandae, although genomic turnover and homogenization that has taken place since the four polyploids formed prevents GISH from effectively identifying any ancestral intergenomic translocations. No intergenomic translocations have been found in any of the other polyploids examined (Chase et al., 2003; Lim et al., 2004, 2007). These observations are similar to those of Song et al. (1995), who noted that, in synthetic polyploids of Brassica, the most extensive genomic changes occurred in polyploids with the most widely diverged parental genomes.
In terms of genome size evolution, no trends with increasing age of the polyploids or increasing distance between diploid progenitors of the polyploids were discernable, with four Nicotiana polyploids showing decreases and five showing increases. Even different polyploids originating from the same parental genome donors (i.e. section Repandae) responded differently with one species (N. nudicaulis) exhibiting decreases and the other three (N. nesophila, N. stocktonii and N. repanda) showing increases. The only noticeable trend was an increase in the extent of DNA amount change with increasing age of the polyploids. Thus, whereas ‘young’ polyploids (<200 000 years old) showed only small losses of DNA (2–5 %), after approx. 4·5 Myr of evolution DNA amount changes ranged from 14 % to 29 % (Fig. 2). The potential relationship between genome size increases and turnover (replacement of ancestral repeats with new repeats over time) and genome decreases with maintenance of ancestral repeats will need to be explored.
Studying this relationship in other systems may, however, be difficult because the only other reliable examples of genome increases in naturally occurring polyploids are found in a few species of Hordeum (Jakob et al., 2004). The scarcity of genome size increases in polyploids in general suggests strong selection against this process. Perhaps the reason is that a newly formed polyploid already has a large genome compared with its diploid parental competitors so selection favours early polyploids which have undergone genome size decreases rather than increases. Nevertheless, the identification of genome size increases in five Nicotiana polyploids suggests that here either the additional DNA does confer some competitive advantage or, perhaps more likely, it has no function but there is no strong selection against it.
ACKNOWLEDGEMENTS
We thank NERC and the Grant Agency of the Czech Republic (521/07/0116) for support.
LITERATURE CITED
- Aoki S, Ito M. Molecular phylogeny of Nicotiana (Solanaceae) based on the nucleotide sequence of the matK gene. Plant Biology. 2000;2:316–324. [Google Scholar]
- Bennetzen J, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, et al. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae) Annals of Botany. 2003;92:107–127. doi: 10.1093/aob/mcg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Molecular Phylogenetics and Evolution. 2004;33:75–90. doi: 10.1016/j.ympev.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytologist. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Dadejova M, Lim KY, Souckova-Skalicka K, Matyasek R, Grandbastien MA, Leitch A, et al. Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids. New Phytologist. 2007;174:658–668. doi: 10.1111/j.1469-8137.2007.02034.x. [DOI] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon SS, Wernsman EA, Miksche JP. Evaluation of nuclear DNA content and heterochromatin changes in anther-derived dihaploids of tobacco (Nicotiana tabacum) cv. Coker 139. Canadian Journal of Genetics and Cytology. 1983;25:169–173. [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, Meister A, Lysak MA, Nardi L, et al. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany. 1998;82(Suppl. A):17–26. [Google Scholar]
- Fulnecek J, Lim KY, Leitch AR, Kovarik A, Matyasek R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity. 2002;88:19–25. doi: 10.1038/sj.hdy.6800001. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Goodspeed TH. The genus Nicotiana. Waltham, MA: Cronica Botanica Co; 1954. [Google Scholar]
- Gregor W, Mette MF, Staginnus C, Matzke MA, Matzke AJM. A distinct endogenous pararetrovirus family in Nicotiana tomentosiformis, a diploid progenitor of polyploid tobacco. Plant Physiology. 2004;134:1191–1199. doi: 10.1104/pp.103.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C, Hawkins JS, Wendel JF. Tobacco genomes quickly go up in smoke. New Phytologist. 2007;a 175:599–602. doi: 10.1111/j.1469-8137.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- Grover CE, Kim H, Wing RA, Paterson AH, Wendel JF. Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium) The Plant Journal. 2007;b 50:995–1006. doi: 10.1111/j.1365-313X.2007.03102.x. [DOI] [PubMed] [Google Scholar]
- Hanson L, McMahon KA, Johnson MAT, Bennett MD. First nuclear DNA C-values for 25 angiosperm families. Annals of Botany. 2001;87:251–258. doi: 10.1006/anbo.2000.1325. [DOI] [PubMed] [Google Scholar]
- Hanson L, Boyd A, Johnson MAT, Bennett MD. First nuclear DNA C-values for 18 eudicot families. Annals of Botany. 2005;96:1315–1320. doi: 10.1093/aob/mci283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle J, Timmis JN, Sinclair J. Relationship between satellite deoxyribonucleic acid, ribosomal ribonucleic acid gene redundancy, and genome size in plants. Plant Physiology. 1975;55:496–501. doi: 10.1104/pp.55.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob SS, Meister A, Blattner FR. Considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Molecular Biology and Evolution. 2004;21:860–869. doi: 10.1093/molbev/msh092. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenton A, Parokonny AS, Gleba YY, Bennett MD. Characterization of the Nicotiana tabacum L. Molecular & General Genetics. 1993;240:159–169. doi: 10.1007/BF00277053. genome by molecular cytogenetics. [DOI] [PubMed] [Google Scholar]
- Knapp S, Chase MW, Clarkson JJ. Nomenclatural changes and a new sectional classification in Nicotiana (Solanaceae) Taxon. 2004;53:73–82. [Google Scholar]
- Kovarik A, Fajkus J, Koukalova B, Bezdek M. Species-specific evolution of telomeric and rDNA repeats in the tobacco composite genome. Theoretical and Applied Genetics. 1996;92:1108–1111. doi: 10.1007/BF00224057. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Matyasek R, Lim KY, Skalicka K, Koukalova B, Knapp S, et al. Concerted evolution of 18-5·8-26S rDNA repeats in Nicotiana allotetraploids. Biological Journal of the Linnean Society. 2004;82:615–625. [Google Scholar]
- Kovarik A, Dadejova M, Lim KY, Chase MW, Clarkson JJ, Knapp S, et al. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Annals of Botany. 2008;101:815–823. doi: 10.1093/aob/mcn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Levy AA, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society. 2004;82:607–613. [Google Scholar]
- Lim KY, Matyasek R, Lichtenstein CP, Leitch AR. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma. 2000;109:245–258. doi: 10.1007/s004120000074. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biological Journal of the Linnean Society. 2004;82:599–606. [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Fulnecek J, Leitch AR. Cytogenetic and Genome Research. 2005;109 doi: 10.1159/000082413. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Chase MW, Knapp S, McCarthy E, et al. Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. The Plant Journal. 2006;a 48:907–919. doi: 10.1111/j.1365-313X.2006.02930.x. [DOI] [PubMed] [Google Scholar]
- Lim KY, Souckova-Skalicka K, Sarasan V, Clarkson JJ, Chase MW, Kovarik A, et al. A genetic appraisal of a new synthetic Nicotiana tabacum (Solanaceae) and the Kostoff synthetic tobacco. American Journal of Botany. 2006;b 93:875–883. doi: 10.3732/ajb.93.6.875. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson J, Grandbastien MA, et al. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Matyasek R, Lim KY, Kovarik A, Leitch AR. Ribosomal DNA evolution and gene conversion in Nicotiana rustica. Heredity. 2003;91:268–275. doi: 10.1038/sj.hdy.6800333. [DOI] [PubMed] [Google Scholar]
- Matzke M, Gregor W, Mette MF, Aufsatz W, Kanno T, Jakowitsch J, et al. Endogenous pararetroviruses of allotetraploid Nicotiana tabacum and its diploid progenitors, N. sylvestris and N. tomentosiformis. Biological Journal of the Linnean Society. 2004;82:627–638. [Google Scholar]
- Melayah D, Lim KY, Bonnivard E, Chalhoub B, De Borne FD, Mhiri C, et al. Distribution of the Tnt1 retrotransposon family in the amphidiploid tobacco (Nicotiana tabacum) and its wild Nicotiana relatives. Biological Journal of the Linnean Society. 2004;82:639–649. [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, et al. Sequence composition and genome organization of maize. Proceedings of the National Academy of Sciences of the USA. 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan RKJ. Nuclear DNA changes, genome differentiation and evolution in Nicotiana (Solanaceae) Plant Systematics and Evolution. 1987;157:161–180. [Google Scholar]
- Narayan RKJ, Rees H. Nuclear DNA, heterochromatin and phylogeny of Nicotiana amphidiploids. Chromosoma. 1974;47:75–83. [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. The Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H, Tuna M, Arumuganathan K. Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. Journal of Heredity. 2003;94:260–264. doi: 10.1093/jhered/esg053. [DOI] [PubMed] [Google Scholar]
- Petit M, Lim K, Julio E, Poncet C, Dorlhac de Borne Fo, Kovarik A, et al. Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (Nicotiana tabacum) Molecular Genetics and Genomics. 2007;278:1–15. doi: 10.1007/s00438-007-0226-0. [DOI] [PubMed] [Google Scholar]
- Qu N, Schittko U, Baldwin IT. Consistency of Nicotiana attenuata's herbivore- and jasmonate-induced transcriptional responses in the allotetraploid species Nicotiana quadrivalvis and Nicotiana clevelandii. Plant Physiology. 2004;135:539–548. doi: 10.1104/pp.103.037036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Koukalova B, Leitch AR, Kovarik A. Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. American Journal of Botany. 2003;90:988–996. doi: 10.3732/ajb.90.7.988. [DOI] [PubMed] [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytologist. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Song KM, Lu P, Tang KL, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proceedings of the National Academy of Sciences of the USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proceedings of the National Academy of Sciences of the USA. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Panaud O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Molecular Biology and Evolution. 2003;20:528–540. doi: 10.1093/molbev/msg055. [DOI] [PubMed] [Google Scholar]
- Wang XY, Shi XL, Hao BL, Ge S, Luo JC. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytologist. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proceedings of the National Academy of Sciences of the USA. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Baldwin IT. Evolution of protein inhibitor defenses in North American allopolyploid species of Nicotiana. Planta. 2006;224:750–760. doi: 10.1007/s00425-006-0256-6. [DOI] [PubMed] [Google Scholar]
- Zhao XP, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, et al. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research. 1998;8:479–492. doi: 10.1101/gr.8.5.479. [DOI] [PubMed] [Google Scholar]