This Special Issue of Annals of Botany celebrates the career of Professor Michael David Bennett (known to all as Mike; Fig. 1). It includes 14 papers covering a diverse range of current genomics research topics, which were solicited to reflect the breadth of his research interests. The Special Issue arises from the meeting entitled ‘Plant Genome Horizons – Vistas and Visions’ held at the Royal Botanic Gardens, Kew (16–17 April, 2007) and attended by more than 80 colleagues, ex-students and friends representing all stages of his career from 1963 to 2006.
Fig. 1.

Professor Mike Bennett.
This article gives an overview of some of Mike's most notable work carried out at various stages of his career. It also provides a link by placing his work in the context of the research outlined in the following papers.
In his entry in the Kew Science Directory (see Box 1), Mike summarized his research as being ‘focused on holistic genomics to improve understanding of the organisation, behaviour and evolution of genomes and their chromosomes’, and although this description was written to cover his work at Kew, it actually describes his career from his early days at Aberystwyth and Cambridge, through to his time at Kew to retirement.
Box 1 Entry for Mike Bennett in the Kew Science Directory (www.kew.org/science/directory/)
‘Research ranged widely over plant karyogenomics since 1963. Work on nuclear DNA and angiosperms has focused on: (1) genome size evolution, (2) reproductive cell biology; (3) higher order genome organisation, and (4) biosystematics. Work at Kew aims to describe and improve understanding of the extent, nature and significance of variation in nuclear DNA amount (DNA C-value and genome size) as fundamental biodiversity characters in flowering plants and other embryophyta. It continues to explore their adaptive significance for plant distribution and behaviour, and to develop the nucleotype hypothesis as a unifying concept, testing its predictive value. The work has produced the “Plant DNA C-values database”, greatly improving its taxonomic, geographic and phytoformic representation. Other interests are: polyploidy and wide hybrids, and work to develop and use chromosome painting techniques as powerful tools in plant biosystematic research.’
Mike began his scientific career as an undergraduate at the Department of Agricultural Botany, University College of Wales, Aberystwyth from 1962–1965. During this time he caught the eye of Professor Huw Rees and went on to study for a PhD under his direction. Part of this involved investigating the work of Pierce (1937), who had reported how chromosomes of Viola conspersa varied in size by over 300 % depending on the amount of phosphate in the culture solution. In his first paper (Bennett and Rees, 1967), published in Nature, Mike showed that in rye (Secale cereale) there was no change in genome size despite changes in chromosome volume of 50 % depending on the phosphate level. Further studies on Allium cepa (Bennett and Rees, 1969) revealed similar results, and studies on Vicia faba showed that chromosomes prepared from the main root were 2–3 times larger than those in small lateral roots, although the DNA content remained constant (Fig. 2; Bennett, 1970). Mike's PhD thesis was entitled ‘Experimental control of chromosome structure and behaviour’, and the degree was awarded in 1968 (Bennett, 1968).
Fig. 2.
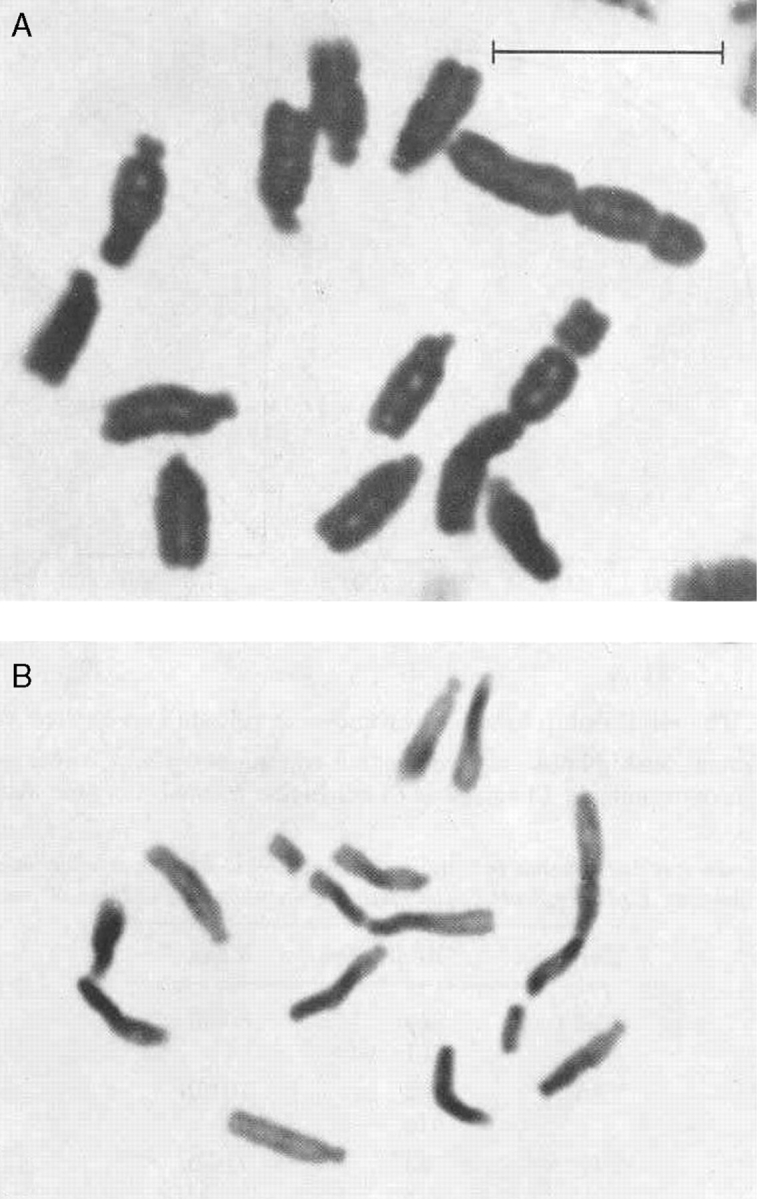
Metaphase chromosomes of Vicia faba prepared from (A) the main root and (B) lateral roots. The difference in volume between the two is three-fold although no difference in DNA amount was found (from Bennett, 1970). Scale bar = 10 µm.
From Aberystwyth, Mike went on to work with Sir Ralph Riley at the Plant Breeding Institute, Cambridge (PBI) looking at the mechanisms and timing of meiosis in cereals including diploid rye and barley (Hordeum vulgare), hexaploid wheat (Triticum aestivum) and hexaploid and octoploid triticale [×Triticale (Triticum × Secale)]. As part of this work he estimated the duration of both male and female meiosis and demonstrated that it was correlated with genome size at the diploid level (Bennett, 1971). He went on to study how factors such as polyploidy, temperature, environment and individual chromosomes could affect the timing of meiosis and discussed some of the possible wider implications for plants (Bennett, 1977b). Arising from this he became interested in the consequences of genome size variation on developmental processes and life cycles of plants, leading him to coin the term ‘nucleotype’ to define those conditions of the DNA that affect the phenotype independently of the encoded information in the DNA. Since then the term has been extensively used in the field of genome size research. Indeed, in two papers in this Special Issue, the consequences of genome size variation at the cellular (Francis et al., 2008) and phenotypic level (Knight and Beaulieu, 2008) are discussed and reviewed, demonstrating the far-reaching consequences that variation in DNA amount can have on an organism.
His interest in nucleotypic effects of genome size variation led him to collate widely scattered genome size data into a usable database. In 1976 the first of eight lists was published (Bennett and Smith, 1976); this was, of course, in hard copy only. With the rapid development of computer and database technologies, the electronic Plant DNA C-values database (www.kew.org/genomesize/homepage.html) went live in 1997, and subsequent updates have been invaluable to the genome size community, enabling broad analyses of the data. Mike, together with colleagues, has used the database to analyse large-scale patterns of genome size evolution across angiosperms (Leitch et al., 1998; Soltis et al., 2003) and land plants (Leitch et al., 2005), the relationship between genome size and weediness (Bennett et al., 1998) and seed weight (Beaulieu et al., 2007), and patterns of genome size change following polyploidy (Leitch and Bennett, 2004). Others have also made use of the database (e.g. Jasienski and Bazzaz, 1995; Knight and Ackerly, 2002; Vinogradov, 2003; O'Meara et al., 2006; Ross-Ibarra, 2007). Two additional analyses are included in this volume (Francis et al., 2008; Jones et al., 2008).
Jones and his colleagues review the results of an analysis looking at the relationship between genome size and occurrence of B-chromosomes in angiosperms (Palestis et al., 2004; Trivers et al., 2004; Levin et al., 2005). They show that the presence of B-chromosomes is positively correlated with genome size (Jones et al., 2008). In addition, Jones et al. provide an overview of B-chromosome research in the century since they were first discovered in 1907.
Mike's interest in the nucleotypic relationship between genome size and cell cycle time led him to team up with Francis to investigate possible relationships between replicon size, rates of DNA replication, duration of S-phase and genome size (e.g. Francis et al., 1985; Kidd et al., 1987). In this Special Issue, Francis et al. (2008) present results of a new analysis from ‘data mining’ of the literature. In total 110 measurements of cell-cycle time for species with genomes sizes ranging 290-fold were combined to generate the largest cell-cycle time survey to date. This revealed a strong positive relationship between genome size and cell-cycle time independent of ploidy level and life cycle type. The relationship held whether all species were analysed together or if monocots and eudicot subsamples were analysed separately, although eudicots appeared to be characterized by a narrower range of cell-cycle times than monocots of equivalent DNA amount. Further, mean cell-cycle duration was seen to be significantly shorter in annuals than in perennials. The relationship was non-linear, with a striking increase in cell-cycle time in perennial monocots with C-values greater than approx. 25 pg. Although more data are needed to confirm this trend, the apparent threshold at approx. 25 pg is intriguing since a similar DNA amount was noted by Mike to be important in determining life cycle strategies in angiosperms: species with 1C values greater than approx. 25 pg were observed to be obligate perennials (Bennett, 1972; Bennett, 1987). Potential explanations for the observed correlations are discussed in the light of molecular studies. They point to the possibility that as DNA amount increases the amount of heterochromatin also increases, accompanied by conformational changes in the chromosome structure. This in turn may reduce the frequency of DNA replication origins and results in a longer DNA synthesis phase and hence longer cell-cycle time. While intriguing, these explanations do not explain the sudden and dramatic increase in cell cycle time in species with C-values greater than approx. 25 pg, and further work is clearly needed.
Throughout his career, genome size has remained one of Mike's major interests, woven through many other areas of research, and he has contributed to the field immensely. His thorough understanding of the principles of Feulgen staining (one of the main techniques used to estimate genome size) led him to make suggestions for best practice as early as 1976 (Bennett and Smith, 1976). With the advent of flow cytometry in the 1980s as an additional technique for genome size estimation, Mike pursued this with the same attention to detail. In particular, the potential for inhibitory cytosolic compounds to interfere with the quantitative binding of fluorochromes (essential for accurate genome size estimation) became a focus for some of his more recent research, leading to his paper published in this issue (Bennett et al., 2008) investigating the effects of anthocyanin on genome size estimation in Euphorbia pulcherrima. Overall, his contribution to this area of genome size research is reflected in the paper presented by Greilhuber (2008) who provides an overview of nuclear DNA content measurements in plants with a particular focus on technical difficulties.
Polyploidy has been another recurring theme in Mike's research career. Initially, this was focused on the impact of ploidy on the duration of meiosis in cereals (e.g. Bennett and Smith, 1972; Finch and Bennett, 1972; Bennett and Kaltsikes, 1973), showing that the duration of meiosis in polyploids was shorter than for diploids with corresponding genomes sizes (Bennett, 1977b). Since then his interest has expanded into many areas including studies of chromosome origin, behaviour and spatial organisation in the polyploid nucleus (e.g. Bennett, 1984b; 2004). These studies have been considerably enhanced by the development and application of genomic in situ hybridization (GISH; Le et al., 1989; Schwarzacher et al., 1989), which enables different genomes within a polyploid nucleus to be distinguished (Bennett et al., 1992; Bennett, 2004). Indeed, Mike's group was the first to use GISH to analyse the genomic structure of a wild allopolyploid, Milium montianum (Fig. 3; Bennett et al., 1992).
Fig. 3.
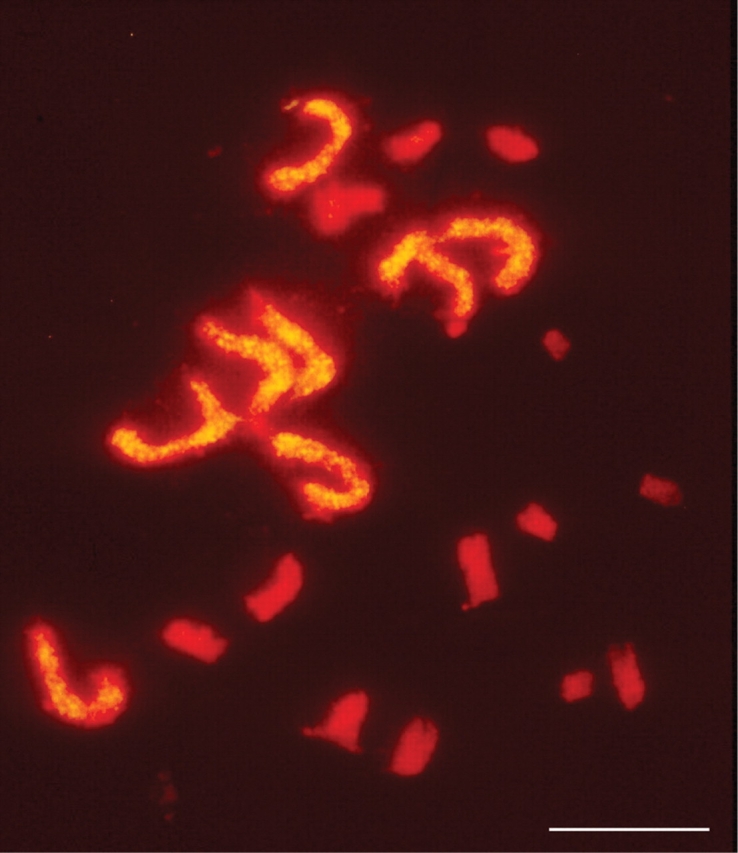
Use of genomic in situ hybridization to distinguish different genomes in the allopolyploid Milium montianum (2n = 22; Poaceae). Scale bar = 10 µm. From Bennett et al. (1992).
The effect of polyploidy on genome size evolution has also been studied by Mike. A large-scale analysis of DNA amounts in 2185 diploid and 823 polyploids showed that loss of DNA following polyploidy was a widespread, although not universal, phenomenon (Leitch and Bennett, 2004). This result was supported by previous studies analysing genome size evolution in individual polyploid species (reviewed in Leitch and Bennett, 2004), and more recent data coming from genome sequencing that shows extensive loss of DNA in polyploids such as maize (Zea mays; Messing et al., 2004) and rice (Oryza sativa; Wang et al., 2005; Bruggmann et al., 2006). This Special Issue includes a paper by Leitch et al. (2008), who report recent research on genome size evolution in the genus Nicotiana. Here, genome size changes were documented in nine allopolyploids, ranging in age from <200 000 years to approx. 4·5 million years. Although genome downsizing occurred in four polyploids, in the remaining five increases in genome size were apparent, with the size of the increase being positively correlated with increasing age of the polyploid species. Assuming that the apparent changes in the polyploids are not artefacts caused by the incorrect choice of putative parental species used for comparison or by subsequent genome size changes in the parental diploids following polyploid formation, then the results provide new insights into how genome size may evolve following polyploidy.
At the molecular level, research into both naturally occurring and synthetic polyploids has shown that combining different genomes in the same nucleus can have dramatic effects, leading to changes at the level of the genome, transcriptome, proteome and metabolome (reviewed by Chen, 2007). Mike's interest in the molecular evolution of polyploids is reflected in three papers in this Special Issue.
The first of these concerns the evolution of ribosomal DNA (rDNA) sequences in allopolyploid Nicotiana species by Kovarik et al. (2008). Here, the links between chromosomal organization, rDNA sequence homogenization, concerted evolution and nucleolar dominance are discussed within a temporal and comparative framework by taking advantage of Nicotiana polyploids formed over widely different time-frames (thousands to millions of years). The results suggest that establishment of nucleolar dominance through epigenetic silencing (even as early as in the F1 hybrid) plays a significant role in influencing the subsequent evolution of rDNA sequences. Active rDNA sequences appear vulnerable to homogenization leading to concerted evolution at such loci. In contrast, inactive, epigenetically silenced rDNA sequences do not appear to undergo homogenization. Because selection cannot act on such silenced genes, they accumulate mutations and are eventually eliminated from the genome. Based on the time-frames involved, results of this study suggest that sequence elimination is detectable in polyploids ≥1 million years.
Ma and Gustafson (2008) report dynamic and genome-specific changes occurring after the formation of synthetic hexaploid and octoploid triticale (polyploids containing both wheat and rye genomes). More changes were apparent in the rye genome than in the wheat genome (based on analysis of AFLP fragments), a result that is congruent with some of Mike's earlier work with Gustafson on chromosomal evolution in triticale in which spontaneous changes in the telomeric rye C-bands were reported (Gustafson and Bennett, 1982; Gustafson et al., 1983). Genome-specific changes as observed in triticale have also been reported in some other polyploids (e.g. Song et al., 1995; Zhao et al., 1998; Ozkan et al., 2001; Skalicka et al., 2005).
Understanding endopolyploidy, particularly the development of endosperm (which is initially triploid, although later higher polytriploid levels are found), is important given that the endosperm of a range of species is a major source of food. However, the origin, significance and development of endosperm is still poorly understood (Bennett, 2004). Mike has contributed to this area of research through his detailed studies of the timing and cytological changes associated with endosperm development in cereals, such as wheat (Bennett et al., 1973) and rye (Bennett et al., 1975), and of how this is altered in triticale (Bennett, 1974; 1977a; Gustafson and Bennett, 1982). In particular, he noted a relationship between the presence of late-replicating sub-telomeric heterochromatin on particular rye chromosomes and grain shriveling, and suggested that this heterochromatin needed to be eliminated for successful triticale breeding (Gustafson and Bennett, 1982).
In an attempt to understand endosperm development more fully, one area of research has focused on unravelling the role that genomic interactions play, given the inbuilt genomic imbalance in the endosperm (combining one paternal and two maternal genomes). To this end, molecular tools have been applied using, for example, reporter lines and gene expression approaches to examine the consequences of creating endosperms with maternal and paternal genomic excesses and hence the role that gene dosage, genomic imprinting and the combining of two sets of chromatin-remodeling machinery play in the development of a functional endosperm. In this Special Issue, Pennington et al. (2008) use such approaches together with morphological analysis to examine how maize endosperm development is influenced by maternal and paternal genomic excesses, uncovering the importance of parental genomic imprinting (in which alleles of a particular gene are differentially expressed, depending on whether they originated from the male or female parent) among other factors, in normal endosperm development.
At Cambridge, Mike worked primarily with plant breeders and much of the focus of his research was to understand better the biology of crop plants, especially in Poaceae. In addition to his work on the importance of organized endosperm development in triticale, (mentioned above) he carried out detailed analyses of meiosis and gametophytic development to shed light on factors affecting seed development in cereals (Bennett et al., 1975). His interest in crops is reflected in this Special Issue by two papers. King et al. (2008) describe the development of microsatellite markers for perennial ryegrass (Lolium perenne; an important forage and amenity grass worldwide) and their potential application in gene isolation, analysis of genes involved in the control of target traits and marker-assisted selection in breeding programmes.
Together with a knowledge of the genetics of cultivated forms of crop plants, plant breeders also depend on an understanding of the origins of these forms and of diversity in progenitors and wild relatives (e.g. Vaughan et al., 2007). An example of this is illustrated in the paper by Saeidi et al. (2008) in this Special Issue in which genetic diversity of Aegilops tauschii (2n = 2x = 14), the D-genome progenitor of bread wheat, is examined in Iran using IRAPs (inter-retrotransposon amplified polymorphisms), which detect retrotransposon insertional polymorphisms (Kalendar and Schulman, 2006; Schulman, 2007). These data are used to provide insights into the patterns of genomic diversity, evolutionary relationships and phylogeography of the species.
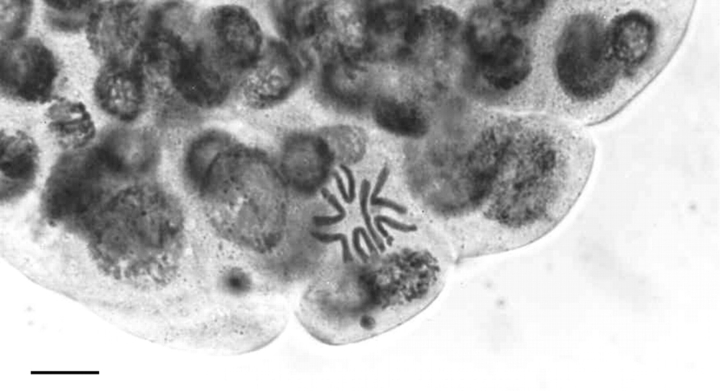
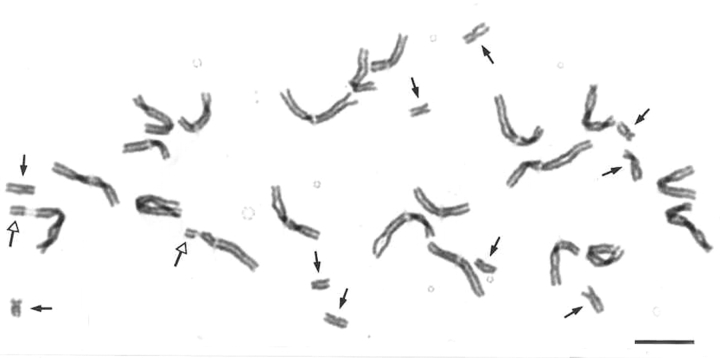
Perhaps Mike's most significant contribution to plant breeding is his work on wide hybridization and uniparental chromosome elimination. In plant breeding the ability to produce doubled haploids (dihaploids) considerably speeds up plant breeding as it produces homozygous lines. In the early 1970s it was shown that dihaploid barley could be generated by crossing barley with its wild relative Hordeum bulbosum as this led to the elimination of the entire H. bulbosum genome. Some of Mike's early work involved studying the timing and rate of chromosome elimination in such hybrids (Fig. 4; Bennett et al., 1976). Doubling the chromosome number produced homozygous dihaploids in a single generation, much more quickly than by back-crossing for at least eight generations as in conventional plant breeding. The first commercial barley cultivar made using this approach was called ‘Mingo’ (Ho and Jones, 1980). Although it was hoped that a similar method would be applicable to wheat breeding, studies revealed that this was not to be the case mainly due to dominant alleles of one or two crossability genes (Kr1 and Kr2) present in most breeding lines and commercial varieties of winter wheat (Snape et al., 1979). However, inspired by a preliminary report suggesting that embryos could be produced by pollinating wheat with maize (Zenkteler and Nitzshe, 1984), Mike and Laurie (also at the PBI) went on to show that it was possible to overcome crossability problems by crossing wheat with maize to generate wheat × maize hybrids (Fig. 5; Laurie and Bennett, 1988; see also Laurie and Bennett, 1986; 1989). Within three cell cycles all maize chromosomes were eliminated, resulting in a haploid wheat genome that could then be doubled to generate dihaploid wheat. This method is still used in plant breeding research (Forster et al., 2007). Mike went on to analyse chromosome elimination in a number of other cereal hybrids (reviewed in Bennett, 1995b) and to examine disposition of parental genomes in such hybrids.
Fig. 4.
Haploid metaphase nucleus showing the seven chromosomes of Hordeum vulgare ‘Vada’ following elimination of the H. bulbosum genome in the hybrid embryo, 72 h post-pollination (from Bennett et al., 1976). Scale bar = 10 µm.
Fig. 5.
Metaphase in a hybrid zygote 24 h after pollination of wheat (Triticum aestivum ‘Chinese Spring’, 2n = 42) with maize (Zea mays ‘Seneca 60’, 2n = 20). Solid arrows point to the ten maize chromosomes. The remaining larger 21 chromosomes come from wheat (open arrows indicate the wheat satellites; from Laurie and Bennett, 1988). Scale bar = 10 µm.
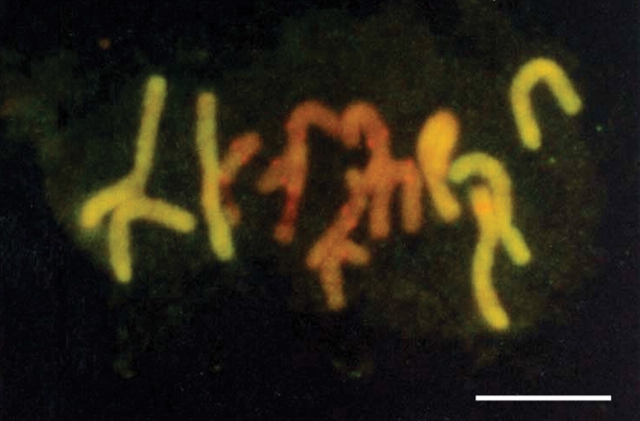
Understanding nuclear organization was a central theme in Mike's research from the late 1970s to the 1990s, including investigations of the somatic association of homologous chromosomes, genome separation and the arrangement of heterologous chromosomes (e.g. Heslop-Harrison and Bennett, 1983a, b; 1990). Arising from this work was the proposal of the ‘Bennett model’ as a means to predict the mean spatial order of chromosomes in a simple haploid genome (Fig. 6; Bennett, 1982; see also Bennett, 1981, 1983). This model was used to predict the arrangement of chromosomes in various cereals (Bennett, 1982; Heslop-Harrison and Bennett, 1983a, b), leading Mike to suggest that there were fundamental mechanisms controlling nuclear architecture and chromosome order (Bennett, 1982, 1984a). Given that there appeared to be common principles determining the spatial order of chromosomes, Mike proposed that the term ‘natural karyotype’ should be used to define such order (Bennett, 1984a). In 1986 Mike, with Heslop-Harrison, won a major grant from the BP Venture Research Unit (a branch of BP with a philosophy for ‘blue skies’ research). This enabled them to extend research on genome organization and in particular to develop and apply fluorescent in situ hybridization to plant chromosomes (e.g. Fig. 7; Leitch et al., 1991; see also Schwarzacher et al., 1989; Heslop-Harrison and Bennett, 1990; Leitch et al., 1990).
Fig. 6.
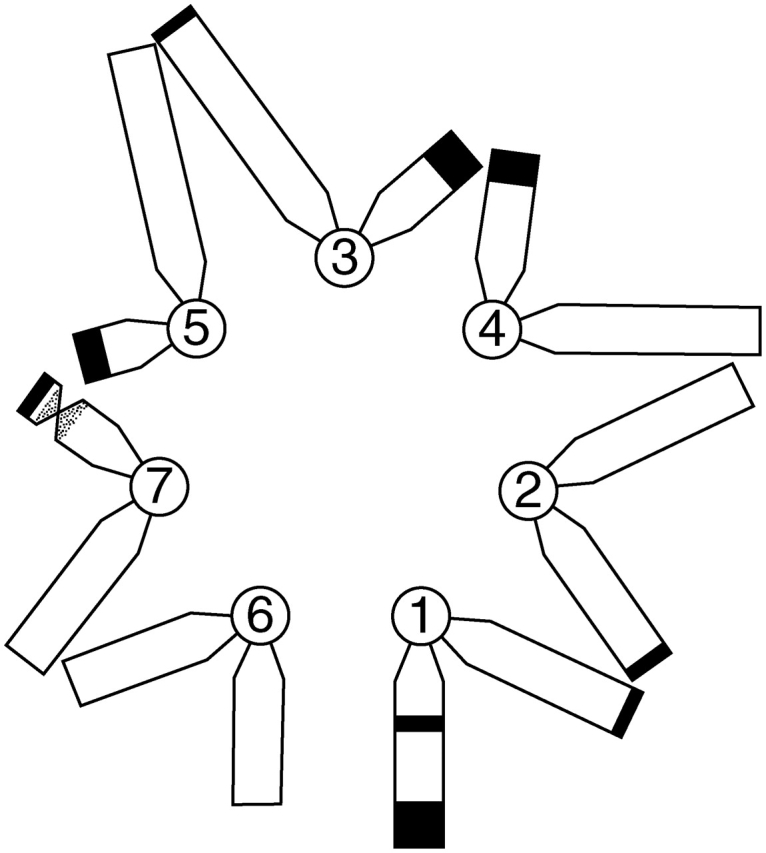
The natural karyotype of Secale africanum (2n = 14) ordered according to the Bennett model (from Bennett, 1982).
Fig. 7.
Metaphase chromosome spread of the hybrid barley × wild rye (Secale africanum) following genomic in situ hybridization (GISH) showing seven barley chromosomes (orange fluorescence) surrounded by seven wild rye chromosomes (yellow fluorescence; from Leitch et al., 1991). Scale bar = 10 µm.
More recently, identification of extensive synteny and colinearity in several grass genomes and their alignment into a circularized ‘ancestral grass genome’ (Moore et al., 1995) led Mike to question to what extent such circularized genomes related to the natural karyotype predicted by the Bennett model (Bennett, 1996). Addressing such a question was, however, only possible for maize as accurate cytological data were not available for the other grasses studied by Moore et al. (1995). The data from Moore et al. supported previous cytogenetic studies that maize is a tetraploid comprising two sets of five chromosomes each. By applying the rules of the Bennett model, Mike showed that he was able to predict the same order of chromosomes for the larger of the two maize genomes (comprising maize chromosomes 1, 4, 2, 3 and 6) as that shown in the circularized ancestral maize genome of Moore et al. (1995). With the smaller maize genome (comprising chromosomes 5, 7, 10, 8 and 9) there was just one difference between the order predicted and that given by Moore et al. (1995). Further work is clearly needed to extend these studies to the other cereals studied by Moore et al. as this will allow their natural karyotypes to be predicted and compared with the ancestral grass genome. Such wider comparisons may then establish to what extent the natural karyotype, conserved syntenic blocks and ancestral genome order are related.
Understanding the mechanism by which homologous chromosomes recognize each other and pair at meiosis has long intrigued biologists, including Mike, especially in polyploids in which the potential for pairing between homoeologous chromosomes can be high. Given the economic importance of wheat, the control of pairing here has received particular attention. This species contains 2n = 42 chromosomes comprising three ancestral genomes (each 2n = 14) known as the A, B and D genomes. Thus there are seven sets of six related chromosomes. The major locus controlling the correct pairing between only homologous chromosomes is the Ph1 locus, a single dominant locus located on chromosome 5B that was discovered by Riley and Chapman (1958). Since its discovery, its mode of action has received intense scrutiny. Indeed, Mike was originally employed by Riley at the PBI to investigate the problem of pairing in wheat and to understand the Ph genes. As part of this work, Mike became interested in premeiotic development and carried out detailed studies of the timing and ultrastructural developmental changes that occurred during this stage in several species including wheat, Lilium longiflorum and Trillium erectum (Bennett et al., 1973; Bennett, 1976). For example, he noted that the duration of successive premeiotic cell cycles increased in wheat from 25 to 55 h in the three cycles preceding meiosis (compared with 12·5 h in root-tip meristem cells; Bennett et al., 1973). Such mitotic cell divisions occur asynchronously in wheat, L. longiflorum and T. erectum, and yet meiosis proceeds synchronously. Thus one important aspect of premeiotic interphase involves synchronization via a developmental hold that accumulates meiocytes at the G1 stage. This is followed by synchronous DNA synthesis (S phase), which is again longer than in root-tip cells (e.g. 12–15 h compared with 3·8 h in root tips of Triticum aestivum; Bennett, 1976). Studies have also shown that activities during premeiotic interphase may be critical for correct chromosome alignment, and through electron microscopy Mike and colleagues revealed the presence of fibrillar material that was only present in the premeiotic interphase (and early stages of meiosis) of pollen mother cells and not in somatic cells surrounding the pollen mother cells of 19 members of Poaceae examined (Fig. 8; Bennett et al., 1979; see also Bennett et al., 1974). He went on to suggest that the fibrillar material may play a role in establishing or maintaining spatial co-orientation of chromosomes in the premeiotic interphase as a prerequisite for normal meiotic pairing, perhaps through movement of telomeres at the nuclear membrane formed after premeiotic mitosis (Bennett et al., 1979).
Fig. 8.
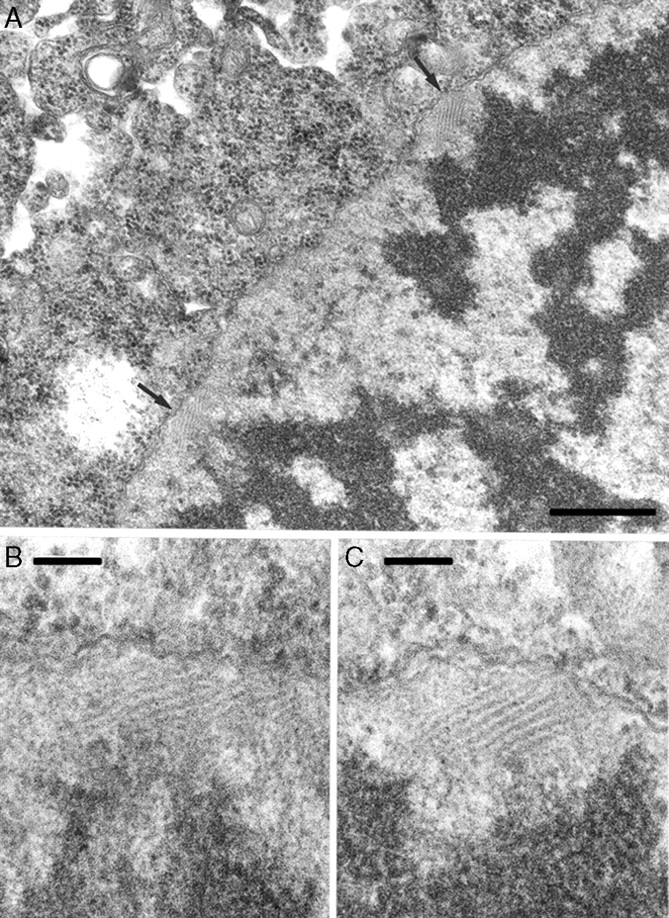
Electron micrographs of pollen mother cells of Triticum aesticum ‘Chinese Spring’ at premeiotic interphase showing (A) two bundles of fibrillar material (FM; arrowed) apparently linking chromatin to nuclear membrane; (B, C) FM from (A) at higher magnification showing bundles composed of microfibres (image taken from Bennett et al., 1979). Scale bars: (A) = 0·5 µm; (B, C) = 100 nm.
Further insights into the control of chromosome pairing have recently come to light through the use of molecular tools to dissect and characterize the chromosomal region containing the Ph1 locus. Griffiths et al. (2006) used deletion line mapping to localize the Ph1 locus to a 2·5 Mb region on chromosome 5B of wheat and found a segment of subtelomeric heterochromatin inserted into a cluster of Cdc2 (Cdk-like) -related genes. Further characterization of the deletion lines using deletion mutants, together with expression profiling of genes in the region of the Ph1 locus forms the focus of the paper by Al-Kaff et al. (2008) in this Special Issue, leading to the suggestion that the Ph1 locus may be defined to a Cdk-like gene cluster related to Cdk2 in humans. Such genes in humans are involved in meiosis (Marston and Amon, 2004; Cohen et al., 2006), perhaps in licensing origins of replication and chromatin remodelling, essential for the onset of meiosis (Alexandrow and Hamlin, 2005).
Most of our understanding of meiosis at the molecular level has come from the study of model organisms, and in plants much of this knowledge originates from Arabidopsis thaliana, given the wealth of genetic resources available (Mezard et al., 2007; Wijeratne and Ma, 2007). However, to what extent information from A. thaliana is representative of other plants remains to be determined. To address this issue, Phillips et al. (2008) use a comparative proteomic approach to study meiosis in rye by analysing the organization of two synaptonemal complex-associated proteins and two recombination-related proteins throughout meiotic prophase using antibodies isolated from A. thaliana. The results demonstrate that resources available for A. thaliana can be used to study meiosis in cereals, with elements in common and striking differences being highlighted. Such studies, although still in their infancy, clearly emphasize the importance of applying such approaches to enable unifying features of meiosis to be identified and distinguished from species-specific events.
In 1987 Mike moved to Kew to become the Keeper of the Jodrell Laboratory. Surrounded by researchers whose focus was systematics, evolution and conservation rather than plant breeding, he expanded his research further. Techniques such as karyotype analysis and genomic in situ hybridization (GISH), which had been used to analyse material of plant breeding interest, were now applied to study organization, evolution and diversity of genomes of wild species (Parokonny et al., 1992; Bailey et al., 1993; Kenton et al., 1993; Bennett, 1995a; Takahashi et al., 1999). Nevertheless, in reality this only represented a natural extension of his understanding of the need for genomic data for biosystematics (e.g. see Bennett, 1984a).
In his role as Keeper, Mike was instrumental in embedding the novel fields of molecular systematics and conservation genetics in the work programme of the laboratory and of Kew as a whole. Since its establishment in 1992, the Molecular Systematics Section of the Jodrell has become an internationally renowned centre for phylogenetic and related studies, leading to the publication of a new classification of the angiosperms as a result of a major international collaboration (APG, 1998; APG II, 2003). The papers by Kovarik et al. (2008) and Leitch et al. (2008) in this Special Issue illustrate the application of phylogenetic analyses to questions of ribosomal DNA evolution and genome size in Nicotiana allopolyploids, respectively, and de Lange et al. (2008) use a combination of phylogenetic, cytogenetic and morphological studies as the basis for conservation recommendations in Crassula species from New Zealand.
Through his supervision of Tony Cox's PhD (Cox, 1995), Mike was directly involved in one of the first conservation genetics studies at Kew, focusing on the lady's slipper orchid (Cypripedium calceolus). Pulling together the themes of conservation genetics and genome-size measurement, one product of the Conservations Genetics Group in the Jodrell (now part of the Genetics Section) has been the demonstration that AFLP (amplified fragment length polymorphisms; Vos et al., 1995) are not readily applicable to wild species with large genomes, including C. calceolus (Fay et al., 2005). Following the work of Garner (2002), who found a correlation between increasing genome size and decreasing amplification success with nuclear microsatellites in animals, Barbará et al. (2007), based in the Jodrell, found a significant negative effect of genome size on cross-species amplification of nuclear microsatellites in a wide range of eukaryotes, including plants.
In his analyses of conservation status and genome sizes from the Plant DNA C-values database, Vinogradov (2003) found a ‘spectacular “dose-dependent” relationship’, with threatened species having larger genomes on average than their less-threatened relatives. Thus it appears that large genomes can be a double disadvantage: species with large genomes are likely to be rare/threatened and, at the same time, assessing genetic diversity in these species is problematic.
A summary of Mike's time at Kew would not be complete without mention of his reputation for being an effective manager of major scientific infrastructure development projects (two extensions to the Jodrell Laboratory and the Millennium Seed Bank at Wakehurst Place). These provide a physical legacy of his contribution to the work of the Royal Botanic Gardens, Kew, at both sites (Fig. 9).
Fig. 9.

Views of (A) the Millennium Seed Bank at Wakehurst Place (completed 2000) and (B) the Wolfson wing of the Jodrell Laboratory (completed 2006).
Mike Bennett retired in August 2006, and we hope that this Special Issue provides a snapshot of his scientific career, with links to many of his scientific publications. We wish Mike a long and happy retirement, although we suspect that he has not finished writing yet!
ACKNOWLEDGEMENTS
We would like to thank Professors Mark Chase and Andrew Leitch for their useful comments in preparing this manuscript. Comments from two referees also led to improvements. We also thank the Royal Botanic Gardens, Kew, Annals of Botany and the Linnean Society who co-sponsored the meeting ‘Plant Genome Horizons – Vistas & Visions’. Some of the figures are reproduced with kind permission from Springer Science and Business Media.
LITERATURE CITED
- Alexandrow MG, Hamlin JL. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. Journal of Cell Biology. 2005;168:875–886. doi: 10.1083/jcb.200409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kaff N, Knight E, Bertin I, Foote T, Hart N, Griffiths S, Moore G. Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling. Annals of Botany. 2008;101:863–872. doi: 10.1093/aob/mcm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APG. An ordinal classification for the families of flowering plants. Annals of the Missouri Botanical Garden. 1998;85:531–553. [Google Scholar]
- APG II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- Bailey JP, Bennett ST, Bennett MD, Stace CA. Genomic in situ hybridization identifies parental chromosomes in the wild grass hybrid × Festulpia hubbardii. Heredity. 1993;71:413–420. [Google Scholar]
- Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C. Cross-species transfer of nuclear microsatellite markers: potential, limitations, and alternatives. Molecular Ecology. 2007;16:3759–3767. doi: 10.1111/j.1365-294X.2007.03439.x. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Moles AT, Leitch IJ, Bennett MD, Dickie JB, Knight CA. Correlated evolution of genome size and seed mass. New Phytologist. 2007;173:422–437. doi: 10.1111/j.1469-8137.2006.01919.x. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Experimental control of chromosome structure and behaviour. University of Wales; 1968. PhD thesis. [Google Scholar]
- Bennett MD. Natural variation in nuclear characters of meristems in Vicia faba. Chromosoma. 1970;29:317–335. doi: 10.1007/BF00325946. [DOI] [PubMed] [Google Scholar]
- Bennett MD. The duration of meiosis. Proceedings of the Royal Society of London Series B. 1971;178:277–299. [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London Series B. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Meiotic, gametophytic and early endosperm development in Triticale. In: MacIntyre R, Campbell M, editors. Triticale. Ottawa, Canada: International Development Research Centre; 1974. pp. 137–148. [Google Scholar]
- Bennett MD. The cell in sporogenesis and spore development. In: Yeoman M, editor. Cell division in higher plants. New York & London: Academic Press; 1976. pp. 161–198. [Google Scholar]
- Bennett MD. Heterochromatin, aberrant endosperm nuclei and grain shriveling in wheat-rye genotypes. Heredity. 1977;a 39:411–419. [Google Scholar]
- Bennett MD. The time and duration of meiosis. Philosophical Transactions of the Royal Society of London Series B. 1977;b 277:201–277. doi: 10.1098/rstb.1977.0012. [DOI] [PubMed] [Google Scholar]
- Bennett MD. The spatial order of barley chromosomes. In: Asher MJC, Ellis RP, Hayter AM, Whitehouse RNH, editors. Barley genetics VI. Edinburgh: Edinburgh University Press; 1981. pp. 751–757. [Google Scholar]
- Bennett MD. Nucleotypic basis of the spatial ordering of chromosomes in eukaryotes and the implications of the order for genome evolution and phenotypic variation. In: Dover GA, Flavell RB, editors. Genome evolution. London: Academic Press; 1982. pp. 239–261. [Google Scholar]
- Bennett MD. The spatial distribution of chromosomes. In: Brandham PE, Bennett MD, editors. Kew chromosome conference II. London: George Allen & Unwin; 1983. pp. 71–79. [Google Scholar]
- Bennett MD. The genome, the natural karyotype and biosystematics. In: Grant WF, editor. Plant biosystematics. a. Canada: Academic Press; 1984. pp. 41–66. [Google Scholar]
- Bennett MD. Premeiotic events and meiotic chromosome pairing. In: Evans CW, Dickinson HG, editors. Controlling events in meiosis. b. Cambridge: Company of Biologists; 1984. pp. 87–121. [PubMed] [Google Scholar]
- Bennett MD. Variation in genomic form in plants and its ecological implications. New Phytologist. 1987;106:177–200. [Google Scholar]
- Bennett MD. The development and use of genomic in situ hybridization (GISH) as a new tool in plant biosystematics. In: Brandham PE, Bennett MD, editors. Kew chromosome conference IV. a. Kew: Royal Botanic Gardens, Kew; 1995. pp. 167–183. [Google Scholar]
- Bennett MD. Losing genomes in hybrid plants. In: Oono K, Takaiwa F, editors. Modification of gene expression and non-Mendelian inheritance. b. Tsukuba, Japan: NIAR; 1995. pp. 253–276. [Google Scholar]
- Bennett MD. The nucleotype, the natural karyotype and the ancestral genome. Symposia of the Society for Experimental Biology. 1996;50:45–52. [PubMed] [Google Scholar]
- Bennett MD. Perspectives on polyploidy in plants – ancient and neo. Biological Journal of the Linnean Society. 2004;82:411–423. [Google Scholar]
- Bennett MD, Kaltsikes PJ. The duration of meiosis in a diploid rye, a tetraploid wheat and the hexaploid triticale derived from them. Canadian Journal of Genetics and Cytology. 1973;15:671–679. [Google Scholar]
- Bennett MD, Rees H. Natural and induced changes in chromosome size and mass in meristems. Nature. 1967;215:93–94. doi: 10.1038/215093a0. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Rees H. Induced and developmental variation in chromosomes of meristematic cells. Chromosoma. 1969;27:226–244. [Google Scholar]
- Bennett MD, Smith JB. The effects of polyploidy on meiotic duration and pollen development in cereal anthers. Proceedings of the Royal Society of London B. 1972;181:81–107. [Google Scholar]
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London B. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Rao MK, Smith JB, Bayliss MW. Cell development in the anther, the ovule, and the young seed of Triticum aestivum L. var. Chinese Spring. Philosophical Transactions of the Royal Society of London B. 1973;266 [Google Scholar]
- Bennett MD, Stern H, Woodward M. Chromatin attachment to nuclear membrane of wheat pollen mother cells. Nature. 1974;252:395–396. doi: 10.1038/252395a0. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB, Barclay I. Early seed development in the Triticeae. Philosophical Transactions of the Royal Society of London B. 1975;272:199–227. [Google Scholar]
- Bennett MD, Finch RA, Barclay IR. The time, rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma. 1976;54:175–200. [Google Scholar]
- Bennett MD, Smith JB, Simpson S, Wells B. Intranuclear fibrillar material in cereal pollen mother cells. Chromosoma. 1979;71:289–332. [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. DNA amounts in two samples of angiosperm weeds. Annals of Botany. 1998;82:121–134. [Google Scholar]
- Bennett MD, Price HJ, Johnston JS. Anthocyanin inhibits propidium iodide DNA fluorescence in Euphorbia pulcherrima: implications for genome size variation and flow cytometry. Annals of Botany. 2008;101:777–790. doi: 10.1093/aob/mcm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ST, Kenton AY, Bennett MD. Genomic in situ hybridization reveals the allopolyploid nature of Milium montianum (Gramineae) Chromosoma. 1992;101:420–424. [Google Scholar]
- Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, et al. Uneven chromosome contraction and expansion in the maize genome. Genome Research. 2006;16:1241–1251. doi: 10.1101/gr.5338906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocrine Review. 2006;27:398–426. doi: 10.1210/er.2005-0017. [DOI] [PubMed] [Google Scholar]
- Cox AV. The utility of 5S rDNA in phylogenetic reconstructions. Development of the polymerase chain reaction in plant systematics. UK: University of Reading; 1995. Ph.D. thesis. [Google Scholar]
- Fay MF, Cowan RS, Leitch IJ. The effects of nuclear DNA content (C-value) on the quality and utility of AFLP fingerprints. Annals of Botany. 2005;95:237–246. doi: 10.1093/aob/mci017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch RA, Bennett MD. The duration of meiosis in diploid and autotetraploid barley. Canadian Journal of Genetics and Cytology. 1972;14:507–515. [Google Scholar]
- Forster BP, Heberle-Bors E, Kasha KJ, Touraev A. The resurgence of haploids in higher plants. Trends in Plant Science. 2007;12:368–375. doi: 10.1016/j.tplants.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Francis D, Kidd AD, Bennett MD. DNA replication in relation to DNA C values. In: Bryant JA, Francis D, editors. The cell division cycle in plants. Cambridge: Cambridge University Press; 1985. pp. 61–82. [Google Scholar]
- Francis D, Davies MS, Barlow PW. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Annals of Botany. 2008;101:747–757. doi: 10.1093/aob/mcn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner TWJ. Genome size and microsatellites: the effect of nuclear size on amplification potential. Genome. 2002;45:212–215. doi: 10.1139/g01-113. [DOI] [PubMed] [Google Scholar]
- Greilhuber J. Cytochemistry and C-values: the less-well-known world of nuclear DNA amounts. Annals of Botany. 2008;101:791–804. doi: 10.1093/aob/mcm250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Gustafson JP, Bennett MD. The effect of telomeric heterochromatin from Secale cereale on triticale (×Triticosecale). I. Canadian Journal of Genetics and Cytology. 1982;24:83–92. The influence of the loss of several blocks of telomeric heterochromatin on early endosperm development and kernel characteristics at maturity. [Google Scholar]
- Gustafson JP, Lukaszewski AJ, Bennett MD. Somatic deletion and redistribution of telomeric heterochromatin in the genus Secale and in Triticale. Chromosoma. 1983;88:293–298. [Google Scholar]
- Heslop-Harrison JS, Bennett MD. The positions of centromeres on the somatic metaphase plate of grasses. Journal of Cell Science. 1983;a 64 doi: 10.1242/jcs.64.1.163. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Bennett MD. The spatial order of chromosomes in root-tip metaphases of Aegilops umbellulata. Proceedings of the Royal Society of London B. 1983;b 218 [Google Scholar]
- Heslop-Harrison JS, Bennett MD. Nuclear architecture in plants. Trends in Genetics. 1990;6:401–405. doi: 10.1016/0168-9525(90)90300-u. [DOI] [PubMed] [Google Scholar]
- Ho KM, Jones GE. Mingo barley. Canadian Journal of Plant Sciences. 1980;60:279–280. [Google Scholar]
- Jasienski M, Bazzaz FA. Genome size and high CO2. Nature. 1995;376:559–560. [Google Scholar]
- Jones RN, Viegas W, Houben A. A century of B chromosomes in plants: so what? Annals of Botany. 2008;101:767–775. doi: 10.1093/aob/mcm167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R, Schulman AH. IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nature Protocols. 2006;1:2478–2484. doi: 10.1038/nprot.2006.377. [DOI] [PubMed] [Google Scholar]
- Kenton A, Parokonny AS, Gleba YY, Bennett MD. Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics. Molecular & General Genetics. 1993;240:159–169. doi: 10.1007/BF00277053. [DOI] [PubMed] [Google Scholar]
- Kidd AD, Francis D, Bennett MD. Replicon size, mean rate of DNA replication and the duration of the cell cycle and its component phases in eight monocotyledonous species of contrasting DNA C-values. Annals of Botany. 1987;59:603–609. [Google Scholar]
- King J, Thorogood D, Edwards KJ, Armstead IP, Roberts L, Skøt K, Hanley Z, King IP. Development of a genomic microsatellite library in perennial ryegrass (Lolium perenne) and its use in trait mapping. Annals of Botany. 2008;101:845–853. doi: 10.1093/aob/mcn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters. 2002;5:66–76. [Google Scholar]
- Knight CA, Beaulieu JM. Genome size scaling through phenotype space. Annals of Botany. 2008;101:759–766. doi: 10.1093/aob/mcm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik A, Dadejova M, Lim YK, Chase MW, Clarkson JJ, Knapp S, Leitch AR. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Annals of Botany. 2008;101:815–823. doi: 10.1093/aob/mcn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange PJ, Heenan PB, Keeling DJ, Murray BG, Smissen R, Sykes WR. Biosystematics and conservation: a case study with two enigmatic and uncommon species of Crassula from New Zealand. Annals of Botany. 2008;101:881–899. doi: 10.1093/aob/mcm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. Wheat × maize hybridization. Canadian Journal of Genetics and Cytology. 1986;28:313–316. [Google Scholar]
- Laurie DA, Bennett MD. Chromosome behaviour in wheat × maize, wheat × sorghum and barley × maize crosses. In: Brandham PE, editor. Kew chromosome conference III. London: HMSO; 1988. pp. 167–177. [Google Scholar]
- Laurie DA, Bennett MD. The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome. 1989;32:953–961. [Google Scholar]
- Le HT, Armstrong KC, Miki B. Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe. Plant Molecular Biology Reporter. 1989;7:150–158. [Google Scholar]
- Leitch AR, Mosgoller W, Schwarzacher T, Bennett MD, Heslop-Harrison JS. Genomic in situ hybridization to sectioned nuclei shows chromosome domains in grass hybrids. Journal of Cell Science. 1990;95:335–341. doi: 10.1242/jcs.95.3.335. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Schwarzacher T, Mosgoller W, Bennett MD, Heslop-Harrison JS. Parental genomes are separated throughout the cell cycle in a plant hybrid. Chromosoma. 1991;101:206–213. [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany. 1998;82:85–94. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta) Annals of Botany. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Annals of Botany. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA, Palestis BG, Jones RN, Trivers R. Phyletic hot spots for B chromosomes in angiosperms. Evolution. 2005;59:962–969. [PubMed] [Google Scholar]
- Ma X-F, Gustafson JP. Allopolyploidization-accommodated genomic sequence changes in triticale. Annals of Botany. 2008;101:825–832. doi: 10.1093/aob/mcm331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nature Reviews Molecular Cell Biology. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, et al. Sequence composition and genome organization of maize. Proceedings of the National Academy of Sciences (USA) 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezard C, Vignard J, Drouaud J, Mercier R. The road to crossovers: plants have their say. Trends in Genetics. 2007;23:91–99. doi: 10.1016/j.tig.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD. Cereal genome evolution – grasses, line up and form a circle. Current Biology. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- O'Meara BC, Ané C, Sanderson MJ, Wainwright PC. Testing for different rates of continuous trait evolution using likelihood. Evolution. 2006;60:922–933. [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palestis BG, Trivers R, Burt A, Jones RN. The distribution of B chromosomes across species. Cytogenetic and Genome Research. 2004;106:151–158. doi: 10.1159/000079281. [DOI] [PubMed] [Google Scholar]
- Parokonny AS, Kenton AY, Meredith L, Owens SJ, Bennett MD. Genomic divergence of allopatric sibling species studied by molecular cytogenetics of their F1 hybrids. Plant Journal. 1992;2:695–704. [Google Scholar]
- Pennington PD, Costa LM, Gutierrez-Marcos JF, Greenland AJ, Dickinson HG. When genomes collide: aberrant seed development following maize interploidy crosses. Annals of Botany. 2008;101:833–843. doi: 10.1093/aob/mcn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D, Mikhailova EI, Timofejeva L, Mitchell JL, Osina O, Sosnikhina SP, Jones RN, Jenkins G. Dissecting meiosis of rye using translational proteomics. Annals of Botany. 2008;101:873–880. doi: 10.1093/aob/mcm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce WP. The effect of phophorus on chromosome and nuclear volume in a violet species. Bulletin of the Torrey Botanical Club. 1937;64:345–355. [Google Scholar]
- Riley R, Chapman V. Genetic control of cytologically diploid behaviour of hexaploid wheat. Nature. 1958;182:713–715. [Google Scholar]
- Ross-Ibarra J. Genome size and recombination in angiosperms: a second look. Journal of Evolutionary Biology. 2007;20:800–806. doi: 10.1111/j.1420-9101.2006.01275.x. [DOI] [PubMed] [Google Scholar]
- Saeidi H, Rahiminejad MR, Heslop-Harrison JS. Retroelement insertional polymorphisms, diversity and phylogeography within diploid, D-genome Aegilops tauschii (Triticeae, Poaceae) sub-taxa in Iran. Annals of Botany. 2008;101:855–861. doi: 10.1093/aob/mcn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman AH. Molecular markers to assess genetic diversity. Euphytica. 2007;158:313–321. [Google Scholar]
- Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. In situ localization of parental genomes in a wide hybrid. Annals of Botany. 1989;64:315–324. [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytologist. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Snape JW, Chapman V, Moss J, Blanchard CE, Miller TE. The crossabilities of wheat varieties with Hordeum bulbosum. Heredity. 1979;42:291–298. [Google Scholar]
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ. Evolution of genome size in the angiosperms. American Journal of Botany. 2003;90:1596–1603. doi: 10.3732/ajb.90.11.1596. [DOI] [PubMed] [Google Scholar]
- Song KM, Lu P, Tang KL, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proceedings of the National Academy of Sciences (USA) 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi C, Marshall JA, Bennett MD, Leitch IJ. Genomic relationships between maize and its wild relatives. Genome. 1999;42:1201–1207. doi: 10.1139/gen-42-6-1201. [DOI] [PubMed] [Google Scholar]
- Trivers R, Burt A, Palestis BG. B chromosomes and genome size in flowering plants. Genome. 2004;47:1–8. doi: 10.1139/g03-088. [DOI] [PubMed] [Google Scholar]
- Vaughan DA, Balazs E, Heslop-Harrison JS. From crop domestication to super-domestication. Annals of Botany. 2007;100:893–901. doi: 10.1093/aob/mcm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Rijans M, Van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Selfish DNA is maladaptive: evidence from the plant Red List. Trends in Genetics. 2003;19:609–614. doi: 10.1016/j.tig.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Wang XY, Shi XL, Hao BL, Ge S, Luo JC. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytologist. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- Wijeratne AJ, Ma H. Genetic analyses of meiotic recombination in Arabidopsis. Journal of Integrative Plant Biology. 2007;49:1199–1207. [Google Scholar]
- Zenkteler M, Nitzshe W. Wide hybridization experiments in cereals. Theoretical and Applied Genetics. 1984;68:311–315. doi: 10.1007/BF00267883. [DOI] [PubMed] [Google Scholar]
- Zhao XP, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, Wendel JF, Paterson AH. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research. 1998;8:479–492. doi: 10.1101/gr.8.5.479. [DOI] [PubMed] [Google Scholar]