Abstract
Background and Aims
Much of our understanding of the genetic control of meiosis has come from recent studies of model organisms, which have given us valuable insights into processes such as recombination and the synapsis of chromosomes. The challenge now is to determine to what extent these models are representative of other groups of organisms, and to what extent generalisations can be made as to how meiosis works. Through a comparative proteomic approach with Arabidopsis thaliana, this study describes the spatial and temporal expression of key structural and recombinogenic proteins of cereal rye (Secale cereale).
Methods
Antibodies to two synaptonemal complex-associated proteins (Asy1 and Zyp1) and two recombination-related proteins (Spo11 and Rad51) of A. thaliana were bound to meiocytes throughout meiotic prophase of rye, and visualized using conventional fluorescence microscopy and confocal laser scanning microscopy. Western analysis was performed on proteins extracted from pooled prophase I anthers, as a prelude to more advanced proteomic investigations.
Key Results
The four antibodies of A. thaliana reliably detected their epitopes in rye. The expression profile of Rad51 is consistent with its role in recombination. Asy1 protein is shown for the first time to cap the ends of bivalents. Western analysis reveals structural variants of the transverse filament protein Zyp1.
Conclusions
Asy1 cores are assembled by elongation of early foci. The persistence of foci of Spo11 to late prophase does not fit the current model of molecular recombination. The putative structural variants of Zyp1 may indicate modification of the protein as bivalents are assembled.
Key words: Rye, Secale cereale, meiosis, proteins, immunocytology, western analysis
INTRODUCTION
Our understanding of the genetic control of meiosis has come largely from the study of model organisms, most notably Saccharomyces cerevisiae. With unprecedented access to genes and proteins resulting from the completion of genome sequencing projects, together with the adoption of comparative genomic strategies, meiosis of the model plant Arabidopsis thaliana has now succumbed to closer scrutiny (Zickler and Kleckner, 1999; Anderson and Stack, 2002). Many proteins controlling key events in meiosis have been identified and described in this organism, such as Asy1, Zyp1, Spo11, Rad51, Dmc1, Mlh1, Mlh3, Msh4, and Swi1 (Doutriaux et al., 1998; Jean et al., 1999; Grelon et al., 2001; Armstrong et al., 2002; Mercier et al., 2003; Higgins et al., 2004, 2005; Jackson et al., 2006). Many of these proteins, particularly those involved in recombination, are highly conserved. So the challenge now is to determine to what extent meiotic events in other plant groups can be explained via knowledge transfer from A. thaliana, and to demonstrate functional equivalence or otherwise of orthologous proteins in a variety of plant species.
This paper focuses upon our examination of the meiotic proteome of cereal rye (Secale cereale). Rye is a member of the Poaceae, which includes many major crop species, such as wheat, barley, maize, rice and the forage grasses. The molecular genetics of meiosis of members of this group is rather intractable, but comparative genomic and proteomic approaches in our study of rye have given us important insights into how meiosis works in this plant. Mikhailova et al. (2006) demonstrated that antibodies raised against two proteins (Asy1 and Zyp1) associated with the synaptonemal complex (SC) of A. thaliana reliably identified the orthologous proteins in rye. Furthermore, the antibody to Asy1 successfully bound its epitope in maize and barley as well, confirming the utility of the proteomic resources of A. thaliana in deciphering meiosis in Poaceae (Hamant et al., 2006). Interestingly, the expression and organization of the Asy1 protein is different in rye and maize compared with A. thaliana. In the latter, Asy1 protein is loaded onto the entire lengths of chromosome axes during leptotene (Armstrong et al., 2002). However, in rye the linear tracts of Asy1 are discontinuous at leptotene, and continue to load onto chromosome axes well into zygotene after the chromosomes have begun synapsis (Mikhailova et al., 2006). By contrast, the Asy1 protein of maize (Hamant et al., 2006) and orthologous Pair2 protein of rice (Nonomura et al., 2004, 2006) load at leptotene in a similar manner to A. thaliana, but are removed from chromosome axes as synapsis proceeds (Hamant et al., 2006).
Zyp1 of A. thaliana is a transverse filament protein that loads between axial elements as chromosomes synapse (Higgins et al., 2005), mirroring the construction of the central element of the SC with the orthologous Zip1 protein of S. cerevisiae (Sym et al., 1993; Dong and Roeder, 2000). However, in rye long linear tracts of Zyp1 protein are pre-fabricated during leptotene, and are assembled into the mature tripartite structure during zygotene (Mikhailova et al., 2006). These observations of Asy1 and Zyp1 in rye contrast with the traditional models of SC assembly, and may reflect adaptations to the inordinately large genome size of this species. Members of Poaceae typically have significantly larger genomes than A. thaliana, and contain relatively large amounts of repetitive DNA (Devos et al., 1993; Devos and Gale, 2000; Moore, 2000; Alkhimova et al., 2004; Schulman et al., 2004; Varshney et al., 2004; Mikhailova et al., 2006). Obese genomes may impose constraints upon meiosis that are not encountered by organisms with small genomes. Clearly, although the repertoire of proteins involved in meiosis may be similar in this group of cereals and grasses, their expression, function and interaction may be subtly different.
Orthologues of key recombinogenic proteins have also been identified in the cereals, such as Dmc1 of rice (Ding et al., 2001; Kathiresan et al., 2001) and Rad51 of maize (Franklin et al., 1999; Pawlowski et al., 2003). This paper profiles the expression of Spo11 and Rad51 of rye, using antibodies raised against their orthologues of A. thaliana. It also highlights the latest data from both the immunolocalization of Asy1 and Zyp1, and a 2D western analysis of the Zyp1 protein as a pilot study for more advanced proteomics.
MATERIALS AND METHODS
Plant material
Spring variety ‘Shkolnaya’ and inbred populations of winter rye (Secale cereale L.; 2n = 2x = 14), segregating for mutations sy9 and sy10 were used in this study. The origins and growth conditions of these lines are detailed in Mikhailova et al. (2001) and Mikhailova et al. (2006).
Immunocytology
Immunocytology was performed according to Mikhailova et al. (2006), with the following modifications: sera containing anti-Spo11 and anti-Rad51 antibodies were raised in rabbit and diluted 1 : 150 with blocking buffer. The optimal concentration of each primary antibody was determined empirically. All appropriate controls using corresponding pre-immune sera at lower dilutions resulted in no non-specific binding to cells. The immunocytology of 3D nuclei is as described in Franklin et al. (1999) with the following modifications: two anthers from each rye floret were fixed separately and one anther from the same floret was used for determining the stage. Unlike in maize, 1× Buffer A was substituted by fixative at the first stage of preparing the anthers for polyacrylamide embedding. Incubations with the primary antibody were performed as described in Mikhailova et al. (2006). Fluorescence microscopy and confocal laser scanning microscopy were performed as described in Mikhailova et al. (2006). Cells were also imaged using a Leica TCS SP5 confocal laser scanning microscope.
Protein extraction from meiocytes
Staged prophase I anthers were collected from spring rye and frozen in liquid nitrogen and ground to a fine powder. 200 mL of buffer G (7 m urea, 2 m thiourea, 2 % CHAPS, 2 % ASB14, 100 mm DTT, 0·5 % ampholytes pH range 3–10, 40 mm Tris HCL, 0·002 % bromophenol blue) was added to the powdered anthers, along with a complete mini protease inhibitor tablet (Roche), and placed in a sonicating water bath for 40 s. The sample was ultracentrifuged at 100 000 g for 30 min at 4 °C. The supernatant containing semi-soluble and soluble proteins was collected and stored at –20 °C. An equal volume of ice-cold 20 % (w/v) trichloroacetic acid (TCA) in acetone was added to the sample and left for 1 h at –20 °C to precipitate the proteins in the sample. The sample was centrifuged at 9500 g for 15 min at 4 °C, returned to –20°C for a further 10 min and centrifuged as before for 1 min. The supernatant was discarded and washed in an excess of ice-cold 100 % acetone; the sample was mixed and placed in an ultrasonic water bath for 30 s to dissociate the pellet. The sample was left for 20 min at –20 °C and centrifuged as above. A further two acetone washes were performed to remove all traces of the TCA and the remaining pellet of protein was kept at –20 °C until the final traces of acetone had evaporated. The pellet was re-suspended in 50 mL of buffer G and placed in a sonicating water bath until the protein pellet had dissociated. The sample was stored at –20 °C. Protein concentration was determined by the Bradford assay (Bradford, 1976).
Protein separation on 2D gels
Two-dimensional gel electrophoresis was carried out according to the principal methods outlined by Gorg et al. (1988). 7-cm non-linear pH 2–9 immobilized pH gradient (IPG) strips (BioRad) were rehydrated overnight at 20 °C with 125 mL of sample diluted with buffer G to 250 mg mL−1. The strips were loaded into a Protean IEF system (BioRad) and focused under the following conditions: 100 V for 1 h, linear increase from 100 V to 4000 V over 2 h, holding step at 4000 V until 10 000 Vh was reached. The IPG strips were washed with equilibration buffer (6 m urea, 30 % glycerol, 2 % SDS, 0·002 % bromophenol blue in 1·5 m Tris–HCl pH 8·8) containing 1 % DTT for 15 min at 20 °C on a rocker, followed by a second identical wash, although the DTT was replaced with iodoacetamide (25 mg mL−1). The second dimension was separated on a 12·5 % SDS–PAGE resolving gel overlaid with a 5 % stacking gel using a Protean Mini II kit (BioRad) according to manufacturer's instructions. Resulting gels were stained with Coomassie blue staining solution (0·25 % Coomassie blue, 50 % methanol, 10 % acetic acid) for 1 h with agitation. De-staining solution (40 % methanol, 10 % acetic acid) was applied and left until the desired level of staining was obtained.
Western analysis
Western analysis was based upon principles outlined by Towbin et al. (1979). Gels were blotted onto Hybond-P membrane (Amersham Biosciences) according to the manufacturer's instructions. Membranes were incubated with blocking buffer [1× TBS-Tween 20 (TBST), 1 % non-fat milk powder] with gentle agitation for 4 h. Primary antibodies to Zyp1 N and Zyp1C were diluted separately to 1 : 250 in blocking buffer and incubated with the membranes for 2 h. The membranes were washed 3 × 5 min in 1× TBST and incubated with a secondary antibody conjugated with alkaline phosphatase diluted to 1 : 20 000 in blocking buffer. After 3 × 5 min washes in 1× TBST the membranes were incubated with a 1 : 1 dilution of BCIP/NBT (Roche):double distilled water. The reaction was stopped with several washes in sterile distilled water when the required level of staining was achieved. Gels and membranes were scanned using a GS-800 calibrated densitometer (BioRad).
RESULTS
Immunolocalization of key meiotic proteins in prophase I
The temporal and spatial expression of two structural proteins (Asy1 and Zyp1) and two recombinogenic proteins (Spo11 and Rad51) were tracked through meiotic prophase of rye using immunolocalization. At early leptotene, numerous foci of Asy1 protein are detectable in meiocytes (Fig. 1A). As leptotene proceeds, densely packed linear tracts of Asy1 protein assemble, together with numerous foci of Spo11 protein (Fig. 1B). Some of the latter are paired, whilst others are co-localized with Asy1 cores or are unattached in the nucleoplasm. Organized but less numerous Rad51 foci also appear at this stage, although their numbers may be underestimated due to the dense, diffuse labelling of the nucleoplasm (Fig. 1C).
Fig. 1.
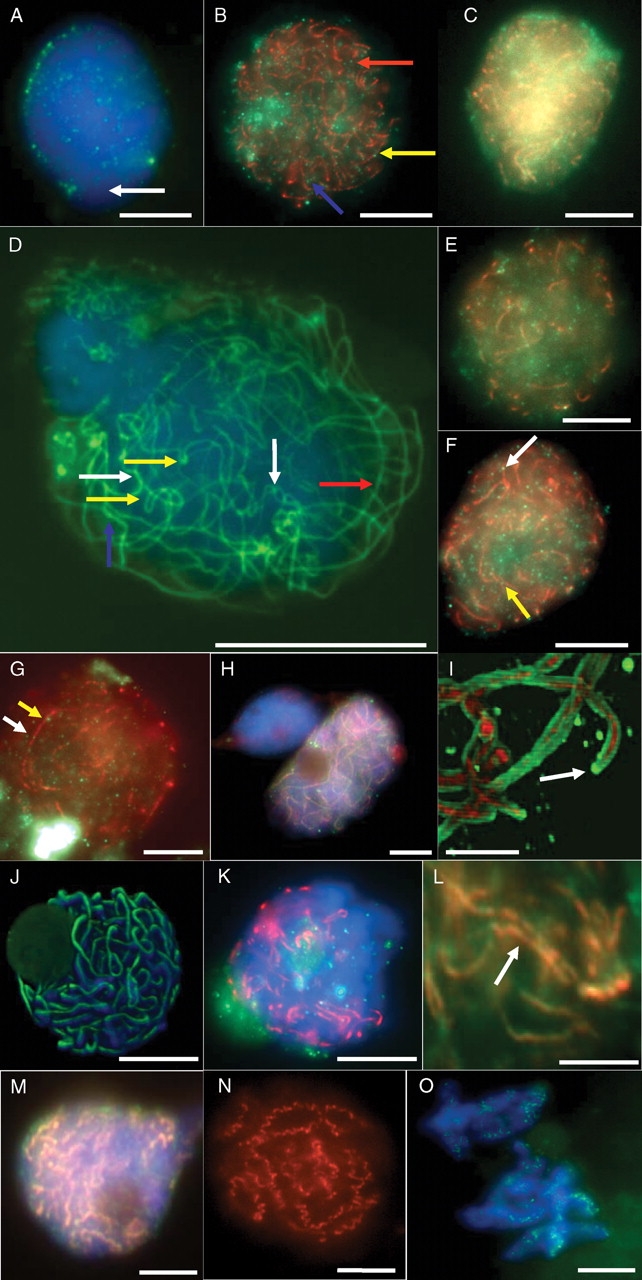
(A) Early leptotene in spring rye, showing Asy1 foci (green), and an area in the nucleus with relatively few foci (white arrow). The nucleus is counter-stained with DAPI (blue). (B) Mid-leptotene in spring rye showing Asy1 (red), Spo11 (green), a pair of Spo11 foci (red arrow), a Spo11 focus co-localized with an Asy1 core (yellow arrow), and a chain of three Spo11 foci (blue arrow). (C) Mid-leptotene in spring rye showing Asy1 (red) and Rad51 (green). (D) Zygotene in spring rye showing Asy1 (green), fold-back loops or aligned telomeres (yellow arrows), distant (red arrow) and close (blue arrow) alignment of Asy1 cores, and synapsed/paired regions (white arrows). The nucleus is counterstained with DAPI (blue). (E) Mid-zygotene in spring rye showing Asy1 (red) and Rad51 (green). (F) Mid-zygotene in spring rye showing Asy1 (red), Spo11 (green), a Spo11 focus co-localized with an Asy1 core (white arrow) and a Spo11 focus lying between two aligned Asy1 cores (yellow arrow). (G) Pachytene in spring rye showing Asy1 (red), Spo11 (green), a Spo11 focus adjacent to (yellow arrow) and co-localized with (white arrow) an Asy1 core. (H) Pachytene in spring rye showing complete co-localization of Asy1 (green) and Zyp1C (red). (I) Detail from a projection of an optical stack through pachytene in spring rye showing Asy1 (green), Zyp1C (red) and a cap of Asy1 (arrow). (J) Projection of an optical stack through pachytene of mutant sy1 showing Asy1 (green) and DAPI counterstain (blue). (K) Diplotene in spring rye showing Asy1 (red) and Spo11 (green). (L) Detail of diplotene in spring rye showing co-alignment (arrow) of desynapsed cores of Asy1 (red) and Zyp1 N (green). (M) Diplotene in spring rye showing widespread co-alignment (yellow) of Asy1 (red) and Zyp1 N (green). (N) Mid-diplotene in Sy10 showing spiralling of Asy1 cores only (red). (O) Late diakinesis showing fragments of Asy1 (green). All bars represent 10 µm, except those of (I) and (L) which represent 5 µm.
At the onset of zygotene, long linear tracts of Asy1 are maintained but at lower density (Fig. 1D). Some of the cores are aligned at a distance, but more commonly they are closely associated. Numerous pairing forks are visible at this stage, reflecting the synapsis of homologues. Paired cores are surprisingly indistinguishable in structure from those that are unpaired. Fold-back loops are apparent at this stage, suggesting some degree of irregular non-homologous alignment, although these structures could also be interpreted as closely aligned telomeres of homologues. The number of Rad51 foci increases initially as the meiocytes enter zygotene, then decreases as synapsis proceeds (Fig. 1E). Conversely, the number of Spo11 foci reduces as zygotene proceeds, although there are still some present that co-localize with Asy1 or span the gap between closely aligned Asy1 cores (Fig. 1F).
The numbers of foci of Spo11 and Rad51 (not shown) decline as the meiocytes enter pachytene (Fig. 1G). Some Spo11 foci either co-localize or are closely aligned with Asy1 cores, whilst a substantial number are unattached in the nucleoplasm. By pachytene, Asy1 and Zyp1 are co-aligned (Fig. 1H) into the typical tripartite structure of the SC when viewed as high resolution projections of optical stacks (Fig. 1I). The latter shows for the first time distinctive caps of Asy1 protein at the ends of the bivalents. Meiotic nuclei undisturbed by squashing confirm both the continuity of Asy1 cores at this stage, and reveal the structural relationship between this protein and the chromatin of the chromosomes (Fig. 1J). At diplotene, the linear tracts of Asy1 are fragmented and there is some retention of Spo11 foci (Fig. 1K). Some Asy1 and Zyp1 cores remain co-aligned at this stage, despite ongoing desynapsis (Fig. 1L, M). By mid-diplotene, cores of Asy1 adopt a spiral conformation (Fig. 1 N), before fragmenting and disappearing at late diakinesis (Fig. 1O).
Western analysis of the Zyp1 protein
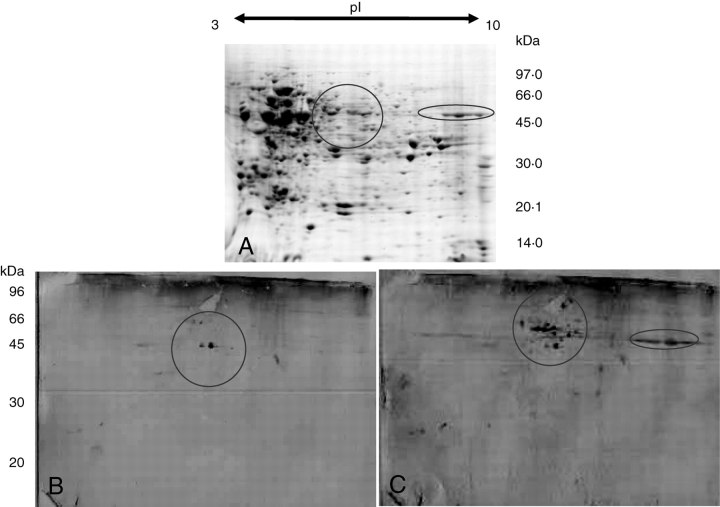
Proteins isolated from pooled, whole anthers at prophase I of spring rye were separated on a 7-cm 2D gel (Fig. 2A). Subsequent staining revealed 486 spots spread over the complete pI and molecular size ranges. All unstained proteins separated by 2D PAGE were transferred to two permeable PVDF membranes and probed separately with antibodies raised against the N and C termini of the Zyp1 protein of A. thaliana. The Zyp1C antibody identified two distinctive and well-defined spots next to each other at an approximate pI of 7 and an approximate molecular weight of 50 kDa (Fig. 2B). The Zyp1 N antibody identified the same two spots as the Zyp1C antibody, and an additional 12 spots in a similar pI range of 6–8 (Fig. 2C). All of the additional spots have a higher molecular weight than the two spots detected by the Zyp1C antibody, ranging in molecular weight from 60–100 kDa. The Zyp1 N also detected a smear of protein in another region of the blot at an approximate pI of 9 and at an approximate molecular weight of 40 kDa. None of the spots detected by either of the antibodies are easy to locate on the Coomassie-stained gel, although the area detected by the Zyp1 N antibody at the high pI could be localized on the stained gel (Fig. 2A).
Fig. 2.
(A) 2D gel of total proteins isolated from pooled prophase I anthers. Circles delimit the location of Zyp1 on the gel. (B) Western blot of a gel equivalent to (A) probed with Zyp1C antibody. (C) A similar blot probed with Zyp1 N antibody.
DISCUSSION
Analysis of structural proteins
Scrutiny of the temporal and spatial expression of Asy1 and Zyp1 of rye has revealed features in common with meiosis of other plant species, together with some notable differences peculiar to rye. Asy1 protein of A. thaliana (Armstrong et al., 2002) and its orthologue Pair2 of rice (Nonomura et al., 2004, 2006) appears at G2 of pre-meiotic interphase. Although it was not possible to time its appearance as precisely in rye, the abundance of foci at early leptotene clearly points to a similar initial expression profile. The paired nature of some Asy1 foci in rye is hitherto unreported in A. thaliana and rice, and its significance, if any, is unknown. The Asy1 protein is also excluded from the telomeric cluster at leptotene in A. thaliana (Armstrong et al., 2002), but its significance has not been elaborated in either species. The transition from Asy1 foci at early leptotene to elongated cores of protein at late leptotene could be effected by one of two processes. Armstrong et al. (2002) have suggested that in A. thaliana the foci could coalesce to form cores, or alternatively the foci could themselves elongate by the accretion of additional protein. The former mechanism is unlikely in rye, since the number of foci detected is insufficient to assemble the long linear tracts of the protein seen at later stages.
Because of the continuity of Asy1 cores at zygotene in rye, it is highly likely that this protein in rye is assembled in close proximity to the axial elements, as has been shown in A. thaliana (Armstrong et al., 2002) and rice (Nonomura et al., 2004, 2006). Therefore, the protein faithfully represents the axes of meiotic prophase chromosomes and can be used with confidence to track the behaviour and assembly of chromosomes. It is highly likely that alignment of Asy1 cores during zygotene represents axial associations (Zickler and Kleckner, 1999) or pre-meiotic pairing (Zickler, 2006). The putative fold-back loops of Asy1 cores at zygotene may simply represent ectopic mis-pairing of the many repeat regions within the genome of rye. Alternatively, they may mark the aligned ends of homologous chromosomes. The latter is perhaps more likely given that bivalents are capped by Asy1 protein (Fig. 1I). This observation also supports the assertion that the Asy1 protein is not a component of the lateral elements, since SCs terminate without a loop. It is our intention to investigate the structure of the ends of bivalents more thoroughly, by combining immunolocalization of Asy1 with in situ hybridization of telomeric arrays.
Simultaneous immunolocalization of Asy1 and Zyp1 proteins in rye has challenged the assumption as to how meiotic bivalents are assembled. In contrast to observations in other organisms, long linear tracts of the two proteins are formed before synapsis. Furthermore, when SCs dismantle at diplotene, and in the largely asynaptic mutant sy10 (Mikhailova et al., 2006), Asy1 and Zyp1 cores are in close alignment. The inference is that SCs of rye are not formed and disassembled by simply laying down and removing Zyp1 protein between synapsed homologous chromosomes. Rather, the SC is formed by unpaired linear cores of pre-formed Zyp1 that are complexed with axial elements and interact to form the central element of the SC. Indeed, the Zyp1 protein of rye could justifiably be referred to as an axial/lateral element protein that forms the central element.
Analysis of recombinogenic proteins
The detection of Spo11 foci in nuclei that contain continuous cores of Asy1 protein is consistent with the role of the former in catalysing double-strand breaks to initiate recombination (Giroux et al., 1989; Keeney, 2001; Lichten, 2001; Mikhailova et al., 2006). The only other report of immunolocalization of Spo11 is in mouse, in which it was suggested that foci of the protein at leptotene identify sites of early recombination nodules (Romanienko and Camerini-Otero, 2000). Expression of Spo11 in A. thaliana may be much earlier than in rye (F. C. H. Franklin, Birmingham University, UK, pers. comm.).
Although we have not quantified relative numbers of foci, the indication is that Spo11 protein appears before Rad51. Numerous Rad51 foci were reported at leptotene in lily (Anderson et al., 1997) and early zygotene in maize after the bouquet had formed (Franklin et al., 1999). As the bouquet forms earlier in rye at the onset of leptotene (Mikhailova et al., 2001), it is possible that the appearance of Rad51 foci is coincident with the formation of the bouquet. The highest number of Spo11 foci in rye was detected at early zygotene, after which their numbers declined progressively. This has been attributed to the loss of structures containing Spo11 through genetic interference (Anderson and Stack, 2005). In contrast, Rad51 foci increase from early zygotene, as has been seen in lily (Anderson et al., 1997) and maize (Franklin et al., 1999). Pairs of Spo11 and pairs of Rad51 foci were often observed in rye. Franklin et al. (1999) reported pairs of the latter in maize, and offered two possible explanations. Firstly, they represent the sites of Rad51 connected by single-stranded DNA between two chromosomes. They suggest that the intervening space between the two foci may once have been occupied by a recombination nodule. However, this does not explain the function of single foci observed in rye. Secondly, they suggest that paired foci could represent the sites of double Holliday junctions.
A few Spo11 foci remain associated with axes at pachytene, as seen in mouse (Romanienko and Camerini-Otero, 2000) and Sordaria (Tesse et al., 2003). These are likely to mark the sites of late recombination nodules, although their numbers and the expected numbers of chiasmata were not correlated. Many unattached foci persist to this stage. These may represent redundant conglomerates of unloaded protein, or could be fulfilling an additional role beyond inducing double-strand breaks. Spo11 foci are seen as late as diplotene in rye, and in mouse (Romanienko and Camerini-Otero, 2000). The persistence of Spo11 to this stage is inconsistent with models of double-strand break processing, which advocate the prompt and early removal of Spo11 by Mre11 (Keeney and Neale, 2006).
Western analysis
Western analysis with Zyp1C and Zyp1 N antibodies detects the same two spots with an approximate pI of 7 and an approximate molecular weight of 50 KDa. This verifies that the antibodies are recognizing most probably the same proteins, with the same molecular weight but with two different forms. This observation is similar to that of Lammers et al. (1995), who showed phosphorylation differences between the orthologous SCP3 protein of rat during meiosis. Two forms of the Zyp1 protein in rye are consistent with the identification of two Zyp1 proteins in A. thaliana (Higgins et al., 2005). However, as in C. elegans (MacQueen et al., 2002; Colaiacovo et al., 2003), the proteins identified in rye are about half the expected molecular weight of transverse filament proteins in other organisms, such as mammals, yeast, Drosophila, and A. thaliana (Meuwissen et al., 1992; Sym et al., 1993; Page and Hawley, 2001; Higgins et al., 2005). Whilst there is little sequence homology between these other proteins, secondary structure is more conserved and the molecular weights are about the same. The reason for this discrepancy in size of the Zyp1 protein of rye is not known, but may be the result of significant divergence in structure of the protein, perhaps reflecting its alternative configuration in the central element of the SC.
Twelve additional spots are delimited by the Zyp1 N antibody, all with a molecular weight greater than that of the two proteins identified by the Zyp1C antibody. Whilst this could be due to non-specific binding of the antibody, it could also have biological significance. The difference in size could indicate a modification of the C terminus of the Zyp1 protein some time during meiosis. This conclusion is corroborated by preliminary fluorescent immunolocalization with the two Zyp1 antibodies. The Zyp1 N antibody identifies the monomers of the protein on unsynapsed axial elements during leptotene, whereas the Zyp1C does not. Both antibodies recognize their epitopes during zygotene. This may indicate a modification of the C terminus as meiosis proceeds. All of the spots detected by the Zyp1 N antibody are of a higher molecular weight than the two proteins detected by the Zyp1C antibody. This may indicate that the proteins at leptotene contain a motif that is lost as the monomers of the Zyp1 protein synapse. The cleavage of the Zyp1 protein may herald the activation of the protein by an unknown complex of proteins responsible for the synapsis of the single Zyp1 cores. This initial form of the protein may have evolved to allow the loading of the Zyp1 monomers onto the unsynapsed cores during leptotene, ensuring that synapsis does not occur between non-homologous chromosomes.
Other spots detected by the Zyp1 N antibody may represent different forms of the Zyp1 protein. It is possible that some of the spots at the higher molecular weight represent Zyp1 proteins that have not been organized along chromosome axes. It is feasible that some modification of the Zyp1 protein occurs as the single Zyp1 monomers are incorporated into the axial elements during leptotene. The fact that the Zyp1C antibody fails to detect the 12 additional spots identified by the Zyp1 N antibody does not necessarily indicate that the C terminus of the protein is absent. The result may inform us that the antibody cannot recognize the unactivated C terminus, as the antibody was raised against the active form of the protein.
Future prospects
The experiments described above demonstrate the utility of using the resources amassed in A. thaliana for dissecting meiosis in a distantly related cereal. The work has highlighted common elements of structure and control, but has also revealed surprising differences in a process that is supposedly well understood. The study emphasizes the importance of investigating meiosis on a case-by-case basis, and to be careful in making generalizations. The need now is to use all technologies at our disposal to probe meiosis further, and to build up an inventory of meiotic genes and proteins. We may then be in a position to decipher the complex interactions of meiosis, and to understand its systems' biology.
ACKNOWLEDGEMENTS
We thank Chris Franklin, Sue Armstrong and Gareth Jones for the generous supply of antibodies, and the staff of the Core Facility ‘Chromas’ St. Petersburg University for expert help in laser scanning confocal microscopy. We acknowledge with gratitude financial support from the Leverhulme Trust, the Royal Society, the Russian Foundation for Basic Research (Grant No. 06–04–48419), and the Estonian Research Foundation (Grant No. ETF6904).
LITERATURE CITED
- Alkhimova OG, Mazurok NA, Potapova TA, Zakian SM, Heslop-Harrison JS, Vershinin AV. Diverse patterns of the tandem repeats organization in rye chromosomes. Chromosoma. 2004;113:42–52. doi: 10.1007/s00412-004-0294-4. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Stack SM. Meiotic recombination in plants. Current Genomics. 2002;3:507–525. [Google Scholar]
- Anderson LK, Stack SM. Recombination nodules in plants. Cytogenetic and Genome Research. 2005;109:198–204. doi: 10.1159/000082400. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Offenberg HH, Verkuijlen WMHC, Heyting C. RecA-like proteins are components of early meiotic nodules in lily. Proceedings of the National Academy of Sciences USA. 1997;94:6868–6873. doi: 10.1073/pnas.94.13.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FCH. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. Journal of Cell Science. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve A. Synaptonemal assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Developmental Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. The Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, et al. Chromosomal rearrangements in the rye genome relative to that of wheat. Theoretical and Applied Genetics. 1993;85:673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Wang T, Chong K, Bai SN. Isolation and characterization of OsDMC1, the rice homologue of the yeast DMC1 gene essential for meiosis. Sexual Plant Reproduction. 2001;13:285–288. [Google Scholar]
- Dong H, Roeder GS. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. Journal of Cell Biology. 2000;148:417–426. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux M-P, Couteau F, Bergounioux C, White C. Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Molecular and General Genetics. 1998;257:283–291. doi: 10.1007/s004380050649. [DOI] [PubMed] [Google Scholar]
- Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, Cande WZ. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux CN, Dresser ME, Tiano HF. Genetic control of chromosome synapsis in yeast meiosis. Genome. 1989;31:88–94. doi: 10.1139/g89-017. [DOI] [PubMed] [Google Scholar]
- Gorg A, Postel W, Gunther S. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988;9:531–546. doi: 10.1002/elps.1150090913. [DOI] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. The EMBO Journal. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ. Genetics of meiotic prophase I in plants. Annual Review of Plant Biology. 2006;57:267–302. doi: 10.1146/annurev.arplant.57.032905.105255. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes and Development. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes and Development. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient. Arabidopsis. The EMBO Journal. 2006;25:1315–1323. doi: 10.1038/sj.emboj.7600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean M, Pelletier J, Hilpert M, Belzile F, Kunze R. Isolation and characterization of AtMLH1, a MutL homologue from Arabidopsis thaliana. Molecular and General Genetics. 1999;262:633–642. doi: 10.1007/s004380051126. [DOI] [PubMed] [Google Scholar]
- Kathiresan A, Khush GS, Bennett J. Two rice DMC1 genes are differentially expressed during meiosis and during haploid and diploid mitosis. Sexual Plant Reproduction. 2001;14:257–267. [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Current Topics in Developmental Biology. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochemical Society Transactions. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- Lammers JHM, van Aalderen M, Peters AHFM, van Pelt AAM, Gaemers IC, de Rooij DG, et al. A change in the phosphorylation pattern of the 30000–33000 M r synaptonemal complex proteins of the rat between early and mid-pachytene. Chromosoma. 1995;104:154–163. doi: 10.1007/BF00352179. [DOI] [PubMed] [Google Scholar]
- Lichten M. Meiotic recombination: breaking the genome to save it. Current Biology. 2001;11:R253–R256. doi: 10.1016/s0960-9822(01)00131-2. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes and Development. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, et al. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development. 2003;130:3309–3318. doi: 10.1242/dev.00550. [DOI] [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. The EMBO Journal. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailova EI, Sosnikhina SP, Kirillova GA, Tikholiz OA, Smirnov VG, Jones RN, Jenkins G. Nuclear dispositions of subtelomeric and percentromeric chromosomal domains during meiosis in asynaptic mutants of rye (Secale cereale L.) Journal of Cell Science. 2001;114:1875–1882. doi: 10.1242/jcs.114.10.1875. [DOI] [PubMed] [Google Scholar]
- Mikhailova EI, Phillips D, Sosnikhina SP, Lovtsyus AV, Jones RN, Jenkins G. Molecular assembly of meiotic proteins Asy1 and Zyp1 and pairing promiscuity in rye (Secale cereale L.) and its synaptic mutant sy10. Genetics. 2006;174:1247–1258. doi: 10.1534/genetics.106.064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. Cereal chromosome structure, evolution, and pairing. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:195–222. doi: 10.1146/annurev.arplant.51.1.195. [DOI] [PubMed] [Google Scholar]
- Nonomura K-I, Nakano M, Murata K, Miyoshi K, Eiguchi M, Miyao A, Hirochika H, Kurata N. An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Molecular and General Genomics. 2004;271:121–129. doi: 10.1007/s00438-003-0934-z. [DOI] [PubMed] [Google Scholar]
- Nonomura K-I, Nakano M, Eiguchi M, Suzuki T, Kurata N. PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. Journal of Cell Science. 2006;119:217–225. doi: 10.1242/jcs.02736. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes and Development. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Cande WZ. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell. 2003;15:1807–1816. doi: 10.1105/tpc.012898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Molecular Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Schulman AH, Gupta PK, Varshney RK. Organization of retrotransposons and microsatellites in cereal genomes. In: Gupta PK, Varshney RK, editors. Cereal genomics. The Netherlands: Kluwer Academic Publishers; 2004. pp. 83–118. [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proceedings of the National Academy of Sciences USA. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Korzun V, Börner A. Molecular maps in cereals: methodology and progress. In: Gupta PK, Varshney RK, editors. Cereal genomics. The Netherlands: Kluwer Academic Publishers; 2004. pp. 35–82. [Google Scholar]
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annual Review of Genetics. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]