Abstract
Background and Aims
Crassula hunua and C. ruamahanga have been taxonomically controversial. Here their distinctiveness is assessed so that their taxonomic and conservation status can be clarified.
Methods
Populations of these two species were analysed using morphological, chromosomal and DNA sequence data.
Key Results
It proved impossible to differentiate between these two species using 12 key morphological characters. Populations were found to be chromosomally variable with 11 different chromosome numbers ranging from 2n = 42 to 2n = 100. Meiotic behaviour and levels of pollen stainability were both variable. Phylogenetic analyses showed that differences exist in both nuclear and plastid DNA sequences between individual plants, sometimes from the same population.
Conclusions
The results suggest that these plants are a species complex that has evolved through interspecific hybridization and polyploidy. Their high levels of chromosomal and DNA sequence variation present a problem for their conservation.
Key words: Chromosome variation; Crassula; Crassula hunua; Crassula ruamahanga; Crassulaceae, conservation; phylogenetics; taxonomy; New Zealand flora
INTRODUCTION
Remarkably little attention has been paid to the widespread chromosome polymorphism that is often found when extensive studies are made of plant populations (Parker and Wilby, 1989; Vaughan et al., 1997; Murray and Young, 2001) in the context of plant conservation (Allendorf and Luikart, 2006). For example, recent books (Avise and Hamrick, 1996; Henry, 2006) make no mention of the possible significance of chromosome variation in plant conservation. Intra-specific chromosome variation can range from structural variants, such as inversions and translocations, which occur as polymorphisms in plant populations, through to variation in the number of whole or partial genomes that are present (polyploidy). Translocation polymorphisms, for example, have been found in populations in a variety of genera in different plant families [Campanulaceae (James, 1965); Onagraceae (Bloom, 1974); Poaceae (Sieber and Murray, 1981)] and can, at least in some cases, be shown to be adaptive mechanisms for conserving genetic variation (Cleland, 1972; James, 1982). Similarly, examples of euploid and, to a lesser extent, aneuploid variation have also been found in taxonomically diverse groups (Murray, 1976; Murray and Young, 2001); these polyploids often have different distribution patterns to the diploids and extend the range of the species (Levin, 2002). All of this chromosome variation has an impact on plant conservation, for ensuring that maximum genetic variation is retained in a species or that reintroduction programmes do not mix plants of different ploidy levels and thus contribute to reduced fertility via inter-ploidy hybrids (Young and Murray, 2000). This latter problem has long been recognized by animal conservation scientists who are often working with captive populations that, upon cytogenetic analysis, are found to be chromosomally differentiated (Benirschke and Kumamoto, 1991). There is also a problem of defining the conservation units when populations are found to be chromosomally variable.
Taxonomy also has a major role to play in conservation, in providing an accurate and authentic name for the plants of interest, providing a description so that the plant can be identified, and establishing species relationships and distinctiveness. Taxonomic uncertainty can lead to unnecessary conservation effort as segregates can be described as distinct species that on further study and inspection can be seen as part of a larger continuum (Pillon and Chase, 2007). In this paper, extensive chromosome variation is described in what were believed to be two threatened species of Crassula, that, as a result of these investigations, are combined as a single taxon.
The taxonomic history of Crassula hunua and C. ruamahanga
Crassula hunua A.P.Druce and C. ruamahanga A.P.Druce were first described by Kirk (1899) as Tillaea pusilla Kirk and T. acutifolia Kirk. He distinguished T. pusilla from the other New Zealand species by the ‘leaves spreading, obtuse peduncles thickened upwards’ and T. acutifolia by the ‘leaves acute, sepals exceeding the petals’. Kirk (1899) also distinguished plants from the Wairoa Falls, Hunua, near Auckland as T. pusilla var. brevia Kirk, stating that these differed from the type by the ‘peduncles usually shorter than the leaves, not thickened upwards, carpels more obtuse’.
Cheeseman (1906, 1925) accepted both species, and added to the descriptions and distinguishing characters provided by Kirk (1899). For T. pusilla s.l., he noted that plants were ‘intricately branched, matted. Leaves thin, obtuse or subacute, 1/15–1/12 inches. Petals rather longer than the calyx’, whereas T. acutifolia had ‘Leaves thin, acute or apiculate, 1/15–1/10 inches. Petals shorter than the calyx’. Cheeseman (1906, 1925) considered both species to be allied, keying them out closely together, and remarking that T. acutifolia ‘has precisely the habit of T. pusilla, but appears to differ in the narrower and more acute leaves, and in the calyx-lobes exceeding the petals’. He also added the caution for T. acutifolia that he had ‘seen no specimens except those in Mr Kirk's herbarium, which are few and incomplete’. Cheeseman (1906, 1925) did not mention Tillaea pusilla var. brevia.
Allan (1961) also accepted both species, but unlike Cheeseman (1906, 1925), in the key to Tillaea he artificially heightened the differences between them by comparing them with species from section Glomeratae Haw. and he placed T. pusilla in two locations. First, he aligned it with the very different T. debilis Colenso, because both possess acute to subacute sepals, and then distinguishing T. pusilla by the ‘petals acute to subacute; leaves linear to linear-lanceolate; carpels 2–4-seeded’. Secondly, he placed it close to T. helmsii Kirk and T. multicaulis Petrie on the basis of the ‘calyx seg[ment]s or sepals obtuse’ species from which it differs by the ‘fl[ower]s minute, inconspicuous; leaves thin’. Allan (1961) placed T. acutifolia with the distinctive T. sieberiana Schult. et Schult.f. because both had acute sepals, separating T. acutifolia by the ‘stems prostrate, ascending only at tips, greenish leaves thin’. Regarding T. pusilla var. brevia, Allan (1961) repeated Kirk's protologue and typified the name but he did not venture anything further about its taxonomic status.
This remained the state of knowledge for New Zealand Tillaea until Toelken (1981), revised the Australian species of Tillaea and transferred them to Crassula. Soon after, Druce and Given (1984) followed suit for the New Zealand species and transferred T. pusilla and T. acutifolia to Crassula L., as C. pusilla (Kirk) A.P.Druce & D.R.Given and C. acutifolia (Kirk) A.P.Druce & D.R.Given. However, these combinations were predated by C. acutifolia Lam (1786), and C. pusilla Schönland (1913), and were therefore illegitimate. Consequently, Druce and Sykes (in Connor and Edgar, 1987) effected the full and legitimate transfer of both taxa to Crassula by adopting the nomina nova Crassula hunua A.P.Druce (≡ Tillaea pusilla Kirk) and C. ruamahanga A.P.Druce (≡ Tillaea acutifolia Kirk). Neither Druce and Given (1984) nor Druce and Sykes (in Connor and Edgar, 1987) offered any further advance on the distinction of either species, beyond cautioning that the ‘indigenous New Zealand species are imperfectly known and described … [and] are the subject of study by A. P. Druce’.
Subsequently, the New Zealand indigenous and naturalized Crassula were given a full treatment by Sykes (in Webb et al., 1988). There C. hunua and C. ruamahanga were retained as distinct species, being distinguished from each other by differences in leaf and petal shape, length and width, and these more detailed and comparative descriptions included better information about the distribution of the species. Since Sykes's treatment, Moar (1993) provided a detailed description of the pollen of C. hunua, noting that ‘similar grains occur in C. kirkii (Allan) A.P.Druce et Given, C. multicaulis (Petrie) A.P.Druce et Given, and C. ruamahanga A.P.Druce’, and Webb and Simpson (2001) described and illustrated seed morphology of C. hunua and C. ruamahanga. Murray and de Lange (1999) and de Lange et al. (2004a) reported that they share the same chromosome number, 2n = 42, but this was based on single accessions of each species.
Conservation status of C. hunua and C. ruamahanga
Since the early 1980s there has been the recognition that neither C. hunua nor C. ruamahanga were common, an observation, which when coupled with the limited number of herbarium specimens available for either species, prompted their first conservation listing of ‘rare’ (Given, 1990). Subsequent revisions of the New Zealand Vascular Threatened Plant List have placed C. hunua as ‘Vulnerable’ (Cameron et al., 1993), ‘Endangered’ (Cameron et al., 1995; de Lange et al., 1999), and ‘Acutely Threatened/Nationally Critical’ (de Lange et al., 2004b), whereas C. ruamahanga has been regarded as ‘rare’ (Cameron et al., 1993, 1995), ‘Naturally Uncommon/Sparse’ (de Lange et al., 1999) and ‘At Risk/Sparse’ (de Lange et al., 2004b). These listings have stimulated an interest in the ecology, abundance and distribution of both species and, for the highly threatened C. hunua, the need to develop sensible management protocols has been a high priority (Dopson et al., 1999). As a result of this interest many new locations for both species have been discovered, improving information on their distribution, ecology, the nature of the threats they face and their conservation status (de Lange et al., 1998; de Lange, 2000). However, these collections have also heightened problems in the circumscription and delimitation of these two species (Sykes, 2005). In particular, a number of important diagnostic characters, such as sepal and leaf size and shape, showed a tendency to grade between species (de Lange, 2000). Most recently, Sykes (2005), in a revised key to the indigenous and naturalized New Zealand species of Crassula, keyed out these species together. He commented that ‘Crassula ruamahanga and C. hunua virtually intergrade and cannot be separated properly if the variation over the whole of their range is taken into consideration … with several [collections] from the Chatham Islands occupying an intermediate position … it is therefore impossible to adhere to the present taxonomy’.
With this level of taxonomic uncertainty surrounding C. hunua and C. ruamahanga, and the high conservation status of C. hunua, further investigations of morphological, chromosomal and nuclear and plastid DNA sequence variation have been undertaken to re-evaluate the taxonomic and conservation status of C. hunua and C. ruamahanga.
MATERIALS AND METHODS
Plant material
All herbarium specimens in AK and CHR that had been determined as C. hunua and C. ruamahanga, together with type material from AK, WELT and WELTU, were examined, a total of 47 specimens (a full list of the herbarium voucher specimens that were used for the morphological analysis is available online as Supplementary Information). The live material used for chromosomal and DNA analysis is listed in Table 1. For consistency, in the Results and Discussion sections the names C. hunua and C. ruamahanga, as applied to the original collections, both herbarium and live, are used, and the names used by earlier authors are treated as synonyms.
Table 1.
The chromosome number, pollen stainability, origin, herbarium voucher number and GenBank accession numbers of the Crassula plants used in this study
| n | 2n | Pollen (%) | Origin | Voucher | GenBank numbers |
||
|---|---|---|---|---|---|---|---|
| ITS | cpDNA | ||||||
| C. hunua | |||||||
| II + I | 78 | 60 | North I., North Auckland, Clevedon Bridge, Wairoa River | AK 294547 | EF436533 | – | |
| – | 42 | – | North I., South Auckland, Hunua, Wairoa Falls | AK 288129* | – | – | |
| II + I | 98 | 96 | North I., South Auckland, Hunua, Wairoa Falls, | AK 298407 | EF436528 | – | |
| 47II | (94) | 97 | #1 North I., South Auckland, Hunua, Wairoa Falls, | AK 298402 | – | – | |
| 49II | (98) | 93 | #2 North I., South Auckland, Hunua, Wairoa Falls, | AK 294737 | EF436529 | EF436513 | |
| 47II | (94) | 96 | #3 North I., South Auckland, Hunua, Wairoa Falls, | AK 298404 | – | – | |
| 47II | (94) | 95 | #4 North I., South Auckland, Hunua, Wairoa Falls, | AK 298405 | – | – | |
| II + I | (94) | 98 | #5 North I., South Auckland, Hunua, Wairoa Falls, | AK 298406 | – | – | |
| 49II | (98) | 97 | #6 North I., South Auckland, Hunua, Wairoa Falls, | AK 294739 | – | – | |
| 49II | (98) | 91 | #7 North I., South Auckland, Hunua, Wairoa Falls, | AK 294738 | – | – | |
| 47II | 94 | 2 | North I., South Auckland, Auckland City, Mt Albert Fowlds Park (naturalized in bowling green turf) | AK 294551 | EF436537 | EF436510 | |
| II + I | 70 | 58 | Chatham I., Te Whanga Lagoon | AK 291565 | EF436541 | EF436520 | |
| 47II | (94) | 88 | Chatham I., Lake Huro, Mangape Stream | AK 298410 | EF436532 | – | |
| C. ruamahanga | |||||||
| 47II | (94) | 99 | North I., South Auckland, Kawhia Harbour, Rakaunui Scenic Reserve | AK289902 | EF436535 | EF436517 | |
| – | 42 | – | North I., Wellington, Lake Wiritoa, Scoutlands | AK234448 | – | – | |
| 45II | 90 | 88 | North I., Wellington, Lake Wiritoa, Scoutlands | AK 294740 | EF436536 | EF436523 | |
| 32II | (64) | 85 | North I., Hawke's Bay, Mangarouhi Stream | AK286171 | AY787411 | EF436506 | |
| 34II | (68) | 83 | North I., Wellington, Carter's Bush | AK 289899 | EF436542 | EF436514 | |
| 48II | (96) | 93 | North I., Wellington, Wainuioru River, Admiral Farm Station | AK 289901 | EF436543 | EF436516 | |
| II + I | (70) | 34 | North I., Wellington, Lake Wairarapa Domain | AK 289134 | EF436534 | EF436512 | |
| – | 42 | – | North I., Wellington, Lake Wairarapa, near Hinaburn | AK 294741 | – | – | |
| 50II | (100) | 98 | South I., Westland, Barrytown Flats, Maher's Swamp | AK 289900 | EF436545 | EF436515 | |
| II + I | (70) | 2 | South I., Westland, Lake Kaniere | AK 294552 | EF436525 | EF436524 | |
| II + I | c.84 | 85 | South I., Southland, Invercargill, Andersons Park | AK 287202 | EF436538 | EF436511 | |
| II + I | (78) | 14 | Stewart I., Rakeahua Valley Mouth | AK 290559 | EF436526 | EF436518 | |
| II + I | (78) | 29 | Stewart I., Rakeahua Valley | AK 290558 | EF436527 | EF436519 | |
| C. colligata ssp. colligata | |||||||
| – | – | – | Three Kings Is., West I. | AK 298030 | EF436549 | – | |
| C. helmsii | |||||||
| II + I | (42) | – | Australia, Victoria, unspecified location | AK 289904 | EF436531 | EF436522 | |
| – | 14 | – | South I., West Coast, Westport, Cape Foulwind | AK 256182* | AY787405 | EF436504 | |
| C. kirkii | |||||||
| – | 84 | – | North I., Wellington, Wainuiomata River Mouth | AK 284526* | – | – | |
| II + I | 78 | – | North I., Cape Turakirae Scientific Reserve | AK 286752 | EF436539 | EF436505 | |
| C. manaia | |||||||
| – | – | – | North I., Taranaki, Stent Road, near Warea | AK 288422 | EF436548 | – | |
| C. mataikona | |||||||
| – | – | – | North I., Wellington, Ohairu Bay | AK 285422 | – | – | |
| – | – | – | North I., Taranaki, Stent Road, near Warea | AK 288421 | – | – | |
| C. moschata | |||||||
| – | – | – | North I., Wellington, Moa Point (cultivated) | AK 288299 | EF436546 | – | |
| C. multicaulis | |||||||
| 28II | 56 | – | South Island, Otago, Rock & Pillar Range, above Taieri River | AK 290642 | EF436544 | EF436521 | |
| C. peduncularis | |||||||
| – | – | – | North I., Tararua, Cape Turakirae Scientific Reserve | AK 286751 | AY787409 | EF436508 | |
| C. perfoliata | |||||||
| – | – | – | North I., North Auckland, Sunnynook | AK 285781 | AY787411 | – | |
| C. sieberiana | |||||||
| – | – | – | North I., Hawkes Bay, Napier, Ahuriri Lagoon | AK 296072 | – | EF436507 | |
| – | – | – | North I., South Auckland, Cornwallis, Puponga Point | AK 285560 | EF436547 | – | |
| C. sinclairii | |||||||
| 15II | 30 | – | South I., Taramoa, Oreti Beach, Long White Lagoon | AK 297928 | |||
| Lake Wairarapa | AK288396 | EF436540 | EF436509 | ||||
Values in the 2n column in brackets have been calculated from the n values to aid comparisons.
II, bivalents; I, univalents; –, measurements not made.
* Chromosome numbers taken from Murray and de Lange (1999) and de Lange et al. (2002).
Morphological analysis
Five vegetative and seven floral characters used previously to distinguish the two species were scored from the herbarium specimens (Table 2). (These non-succulent Crassula spp. preserve well as herbarium specimens). Ten measurements were made for each character from each herbarium specimen and the mean, standard deviation and range calculated. The vegetative characters scored were length of the third internode from the growing point, lamina length, lamina width, leaf apex angle relative to the midrib and apiculus. The floral characters were peduncle length, peduncle width, flower diameter, calyx lobe length, calyx lobe apex angle relative to the midrib, petal length and petal apex angle relative to the midrib.
Table 2.
Characters used by various authors to distinguish C. hunua and C. ruamahanga
| Present study |
|||||||
|---|---|---|---|---|---|---|---|
| Character | Taxon | Kirk (1899) | Cheeseman (1925) | Allan (1961) | Webb et al. (1988) | Mean ± s.d. | Range |
| Internode (mm) | C. hunua | Usually not >5 | 2·5 ± 0·9 | 1·5–3·6 | |||
| C. ruamahanga | Up to 10 | 4·2 ± 2·1 | 1·9–9·8 | ||||
| Leaf length* (mm unless indicated) | C. hunua | 1/16–1/14 inch | 1/15–1/10 inch | 1–2 | 0·7–2·8 | 1·7 ± 0·4 | 0·8–2·1 |
| C. ruamahanga | 1/16 inch | 1/15–1/12 inch | 1·5–2·5 | 1·3–5·0 | 2·0 ± 0·6 | 1·1–2·9 | |
| Leaf width (mm) | C. hunua | 0·3–0·6 | 0·5 ± 0·1 | 0·3–0·7 | |||
| C. ruamahanga | 0·4–1·0 | 0·5 ± 0·1 | 0·3–0·8 | ||||
| Leaf tip angle (degrees) | C. hunua | Obtuse | Obtuse or subacute; acute | Obtuse to subacute | Acute | 31·3 ± 17·3 | 8·4–58·4 |
| C. ruamahanga | Acute or apiculate | Acute or apiculate | Acute to apiculate (sometimes long-apiculate) | Sharply acute | 19·3 ± 7·5 | 5·6–31·5 | |
| Apiculus (degrees) | C. hunua | – | 0·04 ± 0·11 | 0·0–0·1 | |||
| C. ruamahanga | Shortly acuminate or apiculate | 0·08 ± 0·09 | 0·0–0·3 | ||||
| Peduncle length (mm) | C. hunua | Usually longer than leaves | Longer or shorter than leaves | Short | > Calyx, 0·5–1·3 | 0·9 ± 0·4 | 0·2–1·9 |
| C. ruamahanga | Sessile or on peduncles shorter than leaves | Shorter than leaves | Sessile or subsessile | > Calyx, 0·5–1·0 | 1·0 ± 0·6 | 0·1–5·4 | |
| Peduncle width (mm) | C. hunua | Thickened | 0·3 ± 0·1 | 0·1–0·7 | |||
| C. ruamahanga | 0·2 ± 0·1 | 0·1–0·6 | |||||
| Flower diameter (mm unless indicated) | C. hunua | Minute | 1/15 inch | ±1·75 | 2·2–3·0 | 1·6 ± 0·3 | 1·0–2·8 |
| C. ruamahanga | Minute | 1/20–1/15 inch | ±1·5 | 1·8–2·5 | 1·6 ± 0·4 | 0·9–2·7 | |
| Calyx length (mm) | C. hunua | < Petals | 0·5–0·7 | 0·7 ± 0·2 | 0·3–1·1 | ||
| C. ruamahanga | > Petals | 0·8–1·0 | 1·1 ± 0·3 | 0·5–1·7 | |||
| Calyx lobe apex angle (degrees) | C. hunua | – | Acute | Obtuse | Obtuse or apiculate | 35·7 ± 12·3 | 20·0–60·0 |
| C. ruamahanga | Acuminate | Acuminate | Acuminate | Acute | 22·2 ± 9·6 | 5·0–50·0 | |
| Petal length (mm) | C. hunua | Longer than sepals | > Calyx | > Calyx lobes | 1·0–1·4 | 1·0 ± 0·2 | 0·6–1·5 |
| C. ruamahanga | Shorter than sepals | < Calyx | < Calyx lobes | 1·0–1·3 | 1·2 ± 0·3 | 0·5–1·7 | |
| Petal apex angle (degrees) | C. hunua | - | Acute or subacute | Acute to subacute | Subacute | 31·0 ± 12·3 | 10·0–60·0 |
| C. ruamahanga | acute | - | Acute to subacute | Acute or sharply acute | 28·6 ± 18·9 | 8·0–64·0 | |
Character mean, standard deviation and range are presented from the present study (vegetative character sample sizes: C. ruamahanga n = 30; C. hunua n = 17; floral character sample sizes: C. ruamahanga n = 17; C. hunua n = 8).
* Leaf length differs in Cheeseman (1906, 1925) between the keys and descriptions for T. acutifolia and T. pusilla. The measurements presented here are those given by Cheeseman (1906, 1925) in the description.
Principal component analysis (PCA) for the vegetative characters of C. hunua and C. ruamahanga was undertaken. A correlation matrix scaled to have unit variance was used for the PCA because the variables were in two different units of measurement. The two-sample Wilcoxon rank-sum test was undertaken on each of the 12 characters to test the ability of the measurement data to discriminate between the specimens assigned to C. hunua and C. ruamahanga at 95 % and 99 % confidence levels [the Wilcoxon rank-test was selected because exploratory data analysis indicated the data did not have normal (Gaussian) distributions]. Scatter plots were generated to examine some characters. The summary statistics, scatter plots, PCA and Wilcoxon rank-test were undertaken using S-Plus (Statistical Sciences, 1998).
Chromosome and pollen analyses
Mitotic chromosomes were observed in root tips that were pre-treated with a saturated solution of paradichlorobenzene for approx. 18 h at 4 °C and fixed in ethanol:acetic acid (3:1, v/v). Initially the roots were hydrolysed in 1 m HCl at 60 °C for 15 min, stained by the Feulgen reaction then squashed on a slide in FLP orcein (Jackson, 1973) and observed under the microscope. Despite their thread-like nature, Crassula root tips required a longer than normal hydrolysis time and the chromosomes were still difficult to observe. Therefore a second method was used that involved washing the fixed root tips twice with 0·01 m citrate buffer (pH 4·8) and then digesting with a1 % (w/v) pectolyase (Sigma P3026) + 4 % cellulase (Onozuka R10) enzyme mix in citrate buffer on a slide in a humid chamber for 25 min at 38 °C. The enzyme was washed off the root tips with citrate buffer, followed by a wash with 45 % acetic acid, and then the root tips were gently macerated, heated and squashed in FLP orcein. For meiotic analysis, immature flower buds were fixed in ethanol:chloroform:acetic acid (6:3:1, v/v), and the anthers dissected from the flowers and squashed in FLP orcein. Images were captured with an Axiovision CCD camera and processed using Photoshop. Pollen stainability was measured as the number of grains in which the cytoplasm was well stained with Cotton Blue in a sample of 100 grains. The diameter of pollen grains (25 per plant) stained as above was measured using an eyepiece graticule and a ×40 objective.
DNA extraction for sequencing
Sequences of the nrDNA ITS regions and intervening 5·8 S RNA coding region were generated for 20 representatives of C. ruamahanga or C. hunua. ITS sequences were also generated for the other New Zealand members of Crassula section Helophytum: two individuals of C. helmsii (one from Australia and one from New Zealand) and a single individual each of C. kirkii and C. multicaulis. New Zealand members of Crassula section Glomeratae (two individuals of C. sieberiana and single individuals of C. colligata, C. mataikona and C. manaia) were also sequenced and used as outgroups for ITS. As a further outgroup, ITS sequences were also generated from a cultivated individual of C. perfoliata (subgenus Crassula; section Crassula), the type species of the genus.
Genomic DNA was extracted using QIAGEN Dneasy® Plant Kits (QIAGEN GmbH, D40724 Hilden, Germany) and samples of fresh, young leaves of cultivated accessions of the species (Table 1). The ITS region was amplified from genomic DNA using ITS4 and ITS5 primers (White et al., 1990) and the plastid trnL intron and trnL-F spacer sequences using primers c and f (Taberlet et al., 1991). Each 50 μL reaction contained 20 mm Tris–HCl (pH 8·4), 50 mm KCl, 500 nmol of each primer, 2 mm MgCl2, 200 µmol of each dNTP, 1 µL of genomic DNA (40–100 ng) and 1 unit of Platinum Taq (Invitrogen, Life Technologies, Gaithersberg, MD, USA). The same reaction conditions were used for both regions with an initial denaturation of 2 min at 94 °C, then 30 cycles of 94 °C for 1 min, 60 °C for 30 s and 72 °C for 1 min. Amplifications were then followed by a final extension of 72 °C for5–7 min. The PCR products were purified using a High Pure PCR Purifying Kit (Roche Molecular Biochemicals, Mannheim, Germany) and the products were sequenced at the Centre for Gene Technology, University of Auckland. Sequencing reactions were performed using the same primers as the PCR amplifications and the 3·1 ABI Prism™ BIG DYE Terminator Sequencing Kit. Sequencing products were then run on an ABI Prism™ 3100 DNA Sequencer.
Forward and reverse sequences were aligned and edited using the Contig Express® module of Vector NTI Advance (Informax™, Invitrogen™ Life Science Software, Frederick, MD, USA) and the edited DNA sequences were compared using ClustalX (Thompson et al., 1997).
Parsimony analysis of ITS sequences was undertaken using PAUP*4.0b10 (Swofford, 2003). Branch and bound searches were employed, guaranteeing that all shortest trees were found. DELTRAN character optimization was used. Bootstrap values were generated from 1000 replicate branch and bound searches. The Neighbor Net algorithm as implemented in Splitstree 4·0 (Huson and Bryant, 2006) was used to analyse ITS sequences with the inclusion of mixed sequences with multiple base signals coded using ambiguity codes. In calculating genetic distances between pairs of sequences, SplitsTree 4·0 treats a difference between a single base signal and a mixed signal coded using ambiguity codes as half of a change from one base to another. Plastid trnL intron and trnL-F spacer sequences were analysed using median network analysis as implemented in Spectronet 1·1 (Huber et al., 2002).
RESULTS
Morphology
For the vegetative characters, ten measurements were obtained from each of the 47 available herbarium specimens, but only 25 of these specimens could be scored for floral characters. In addition, many specimens had only a small number of flowers so all ten measurements of all characters were obtained for only five of the 25 specimens. Therefore, for the vegetative data, a PCA and summary statistics were undertaken, but for the incompletely sampled floral data only summary statistics were used.
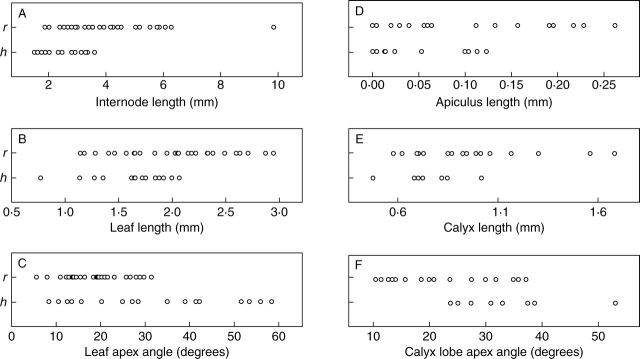
Only three of the five vegetative characters showed statistically significant mean differences (internode length, lamina length, leaf apex angle), but in each case the range of values either overlapped or one was a subset of the other (Table 2 and Fig. 1). Similarly only two of the seven floral characters had significantly different mean values, but again they showed either an overlapping range (calyx lobe length) or the range of one was a subset of the other (calyx lobe angle; Table 2). The means for the other characters were not significantly different (Table 2).
Fig. 1.
Comparative measurements of (A) internode length; (B) leaf length; (C) leaf apex angle; (D) apiculus length; (E) calyx length; (F) calyx lobe apex angle in the selected specimens of Crassula hunua (h) and C. ruamahanga (r).
As mean internode length and leaf length are the only two vegetative characters that were significantly different a scatter plot was used to assess their combined effect; all C. hunua samples grouped on the lower left of the graph whereas the C. ruamahanga samples were scattered across the graph (Fig. 2A). Similarly, as mean calyx length and calyx lobe apex angle are the only two floral characters that are significantly different they were also combined in a scatter plot (Fig. 2B). Again the samples of C. hunua grouped on the lower half of the graph and the C. ruamahanga ones were scattered across the graph. Thus, although mean internode length, leaf length and calyx length and calyx lobe apex angle are significantly different for C. hunua and C. ruamahanga, each character shows considerable overlap and they do not allow the delimitation of discrete groups corresponding to C. hunua and C. ruamahanga.
Fig. 2.
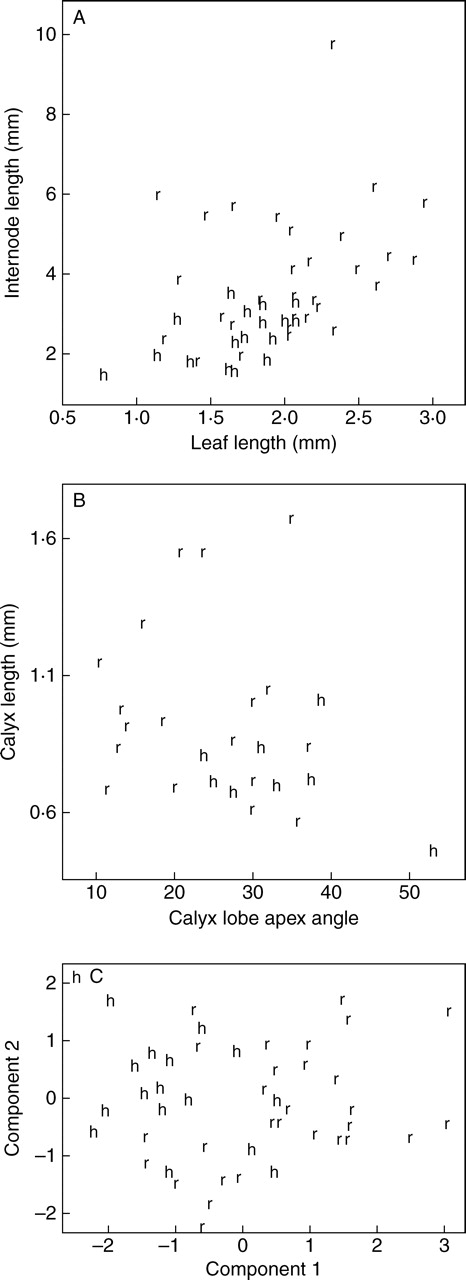
Scatter plots of leaf length against internode length (A), calyx lobe apex angle against calyx length (B) and principle component analysis (C) of the morphological variation in Crassula hunua (h) and C. ruamahanga (r).
The principal component analysis of the five vegetative characters did not distinguish discrete groups that correspond to C. hunua or C. ruamahanga (Fig. 2C). The first and second co-ordinates explained 36 % and 20 % of the total variation, respectively. Two additional PCA analyses were also undertaken to examine patterns of variation, but the results of these analyses are not presented. For the first one, to assess whether there was a geographic component to the morphological variation, the specimens were coded for latitude by dividing New Zealand into 13 bands of equal degrees of latitude from north to south. For the second, specimens from the restricted geographic area around Wairoa Falls (ten specimens collected and determined as C. hunua) and Wairarapa (seven specimens determined as C. ruamahanga) were coded separately from all other specimens. In neither analysis was there any geographic pattern, and specimens from different latitudes and geographic locations were intermixed. Examples of plants from the geographical range of the species are shown in Fig. 3.
Fig. 3.
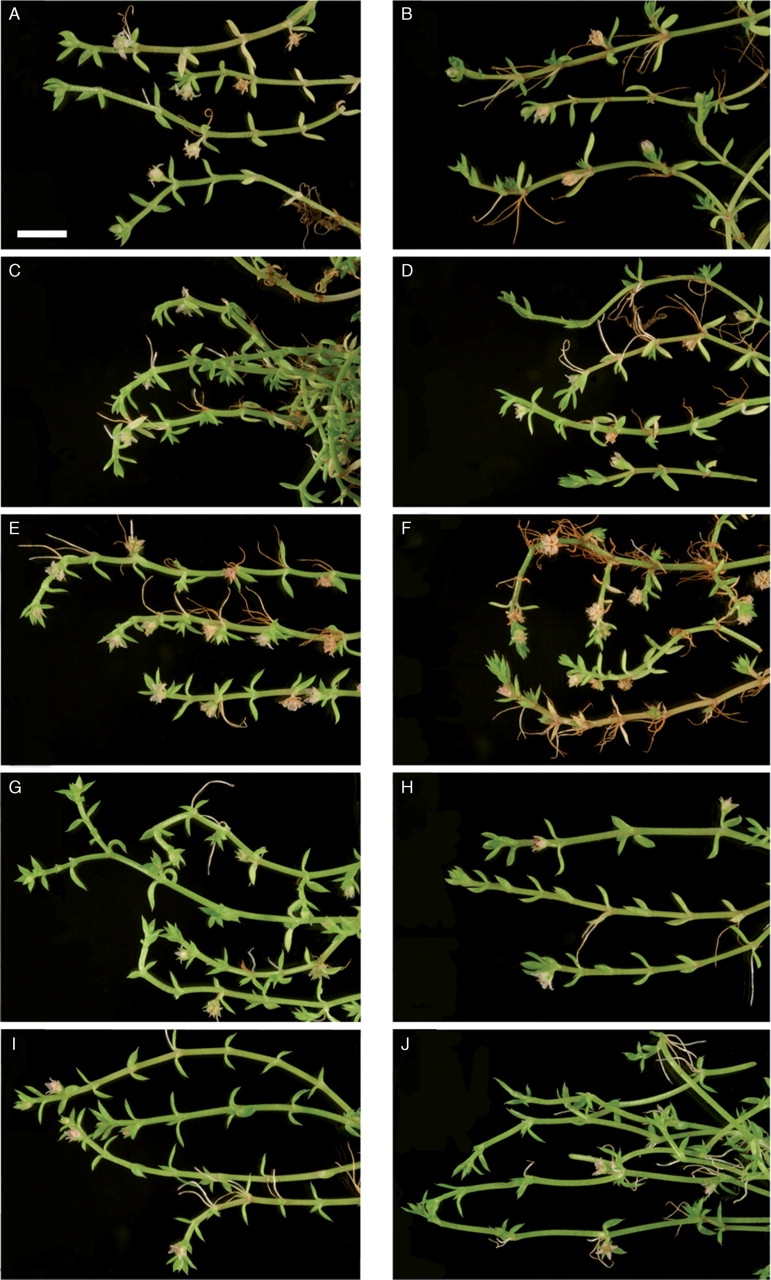
Examples of cultivated accessions of Crassula hunua and C. ruamahanga (plants cultivated under uniform conditions for 2 years) arranged geographically north to south: (A) C. hunua, Clevedon Bridge; (B) C. hunua, Fowlds Park; (C) C. hunua, Wairoa Falls (type locality for C. hunua); (D) C. ruamahanga, Kawhia Harbour, Rakaunui Scenic Reserve; (E) C. ruamahanga, Lake Wiritoa; (F) C. ruamahanga, Carter's Bush Scenic Reserve; (G) C. ruamahanga, Maher's Swamp; (H) C. ruamahanga, Lake Kaniere; (I) C. ruamahanga, Anderson's Park, Invercargill; (J) C. ruamahanga, Rakeahua Valley.
Chromosome number, pollen size and stainability
Crassula chromosomes are small, and mitotic spreads were particularly difficult to obtain as the fine roots contained few dividing cells due to the small size of their meristems. In addition, the roots were particularly hard and needed a long hydrolysis time with the Feulgen method that then tended to degrade the chromosomes. The enzyme-based method yielded much better chromosome spreads (compare A and B in Fig. 4) though the small number of dividing cells in a root tip meristem remained a problem. Consequently, a switch was made to observe meiosis; these observations were considerably more successful, although there were again only a small number of cells at the appropriate stage in the small anthers, and only five to ten cells were analysed for each plant. Differentiating bivalents from univalents was difficult as the chromosomes are so small and numerous.
Fig. 4.
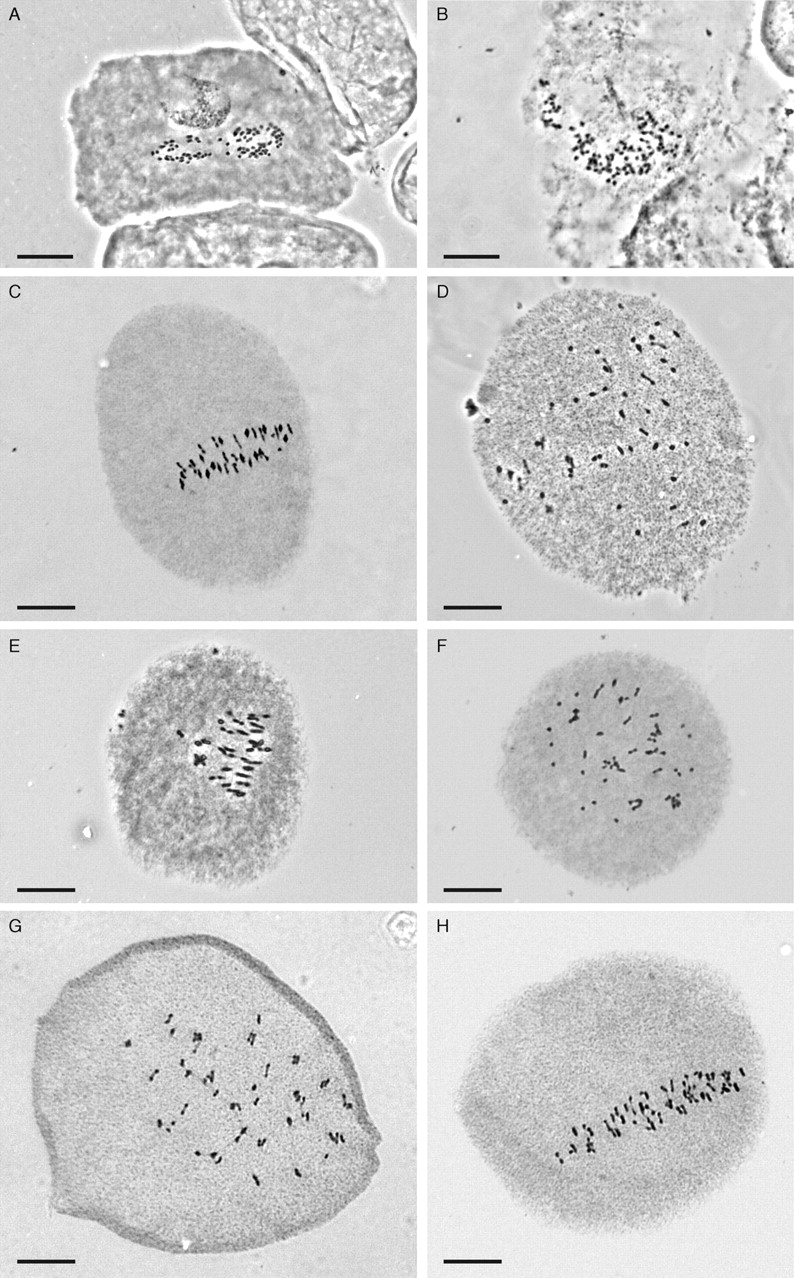
(A, B) Mitotic and (C–H) meiotic chromosomes of Crassula hunua and C. ruamahanga: (A) C. hunua, Clevedon Bridge 2n = 78; (B) C. hunua, Wairoa Falls, 2n = 98; (C) C. ruamahanga, Mangarouhi Stream, 2n = 64, all II; (D) C. hunua, Te Whanga Lagoon, 2n = 70, II + I; (E) C. ruamahanga, Carter's Bush, 2n = 68, all II; (F) C. ruamahanga, Stewart I., 2n = 78, all II; (G) C. hunua #2, Wairoa Falls, 2n = 98, all II; (H) C. hunua #5, Wairoa Falls, 2n = 94, II + I. Scale bars = 10 µm.
Plants of C. hunua and C. ruamahanga from 16 localities spanning much of the range of the species were examined and 11 different chromosome numbers were found (Fig. 4 and Table 1). These ranged from 2n = 42 in plants from Lake Wairarapa to 2n = 100 in a plant from Maher's Swamp in Westland. Plants collected from the same locality can have different chromosome numbers as seen in the largest sample of plants in the present study, from the type locality for C. hunua at Wairoa Falls, Hunua. In most plants encompassing a wide range of chromosome numbers, meiosis appeared to be regular and bivalents could be seen clearly aligned on the metaphase plate (Fig. 4C, E, G). In other plants, however, univalents were clearly identified as chromosomes that were not aligned on the metaphase plate and were smaller than the bivalents (Fig. 4D, F, H). Only bivalents, in the majority of cases ring bivalents with two chiasmata, and univalents were observed in the present sample of plants. No multivalents were found.
Chromosome numbers were also determined for three other species in section Helophytum (Eckl. & Zeyh.) Toelken for which they were previously unknown (Table 1). Each had a different chromosome number, C. sinclairii had 2n = 30, C. multicaulis had 2n = 56 and C. helmsii had 2n = 14 in a New Zealand plant and 2n = 42 in one from Australia. Univalents were observed at meiotic metaphase I in the Australian C. helmsii, but in the other species meiosis was regular.
The pollen stainability of the C. hunua and C. ruamahanga plants was also variable, ranging from 2 % to 98 %, and low stainability was not always correlated with the regularity of chromosome pairing at meiotic metaphase I. For example, the Bowling Green plant of C. hunua showed regular bivalent formation but only 2 % pollen stainability, whereas the plant of C. ruamahanga from Lake Wiritoa, which also showed regular bivalent formation, had 88 % pollen stainability. Pollen diameters were measured in the plants with relatively high stainability (>80 %) and these showed a clear correlation with chromosome number (r = 0·808).
DNA sequences
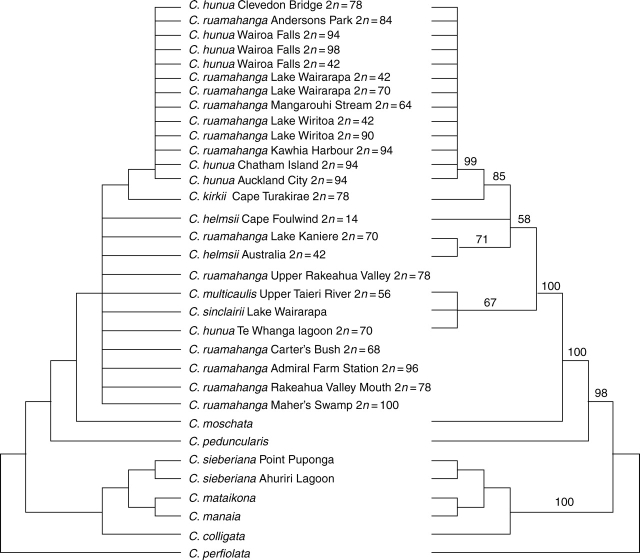
A heuristic search of the ITS sequence matrix recovered 25 664 trees of length 259, a strict consensus tree generated from these is shown on the left-hand side of Fig. 5. One of the shortest trees is also shown as a phylogram (Fig. 6). Many of the C. ruamahanga and C. hunua ITS sequences displayed multiple signals at several sites indicating the amplification of multiple sequence types. Where these multiple signals coincide with potentially parsimony informative sites in the sequence alignment these sequences were removed from the analysis and a branch and bound search conducted. This found three shortest trees of length 259 (CI = 0·853, RI = 0·953, excluding uninformative characters). The strict consensus of these three trees is shown on the right-hand side of Fig. 5. A further 12 sequences were identical. All but one of these was removed prior to parsimony analysis, but they were added back to the redrawn tree (Fig. 5). Also shown in Fig. 5 are the chromosome numbers for each individual, where these are known.
Fig. 5.
Strict consensus of the most parsimonious trees for the full ITS sequence set (left) and reduced set of sequences with those showing additive signal at potentially parsimony informative sites removed (right). Numbers above branches are bootstrap values greater than 50 % derived from 1000 replicates.
Fig. 6.
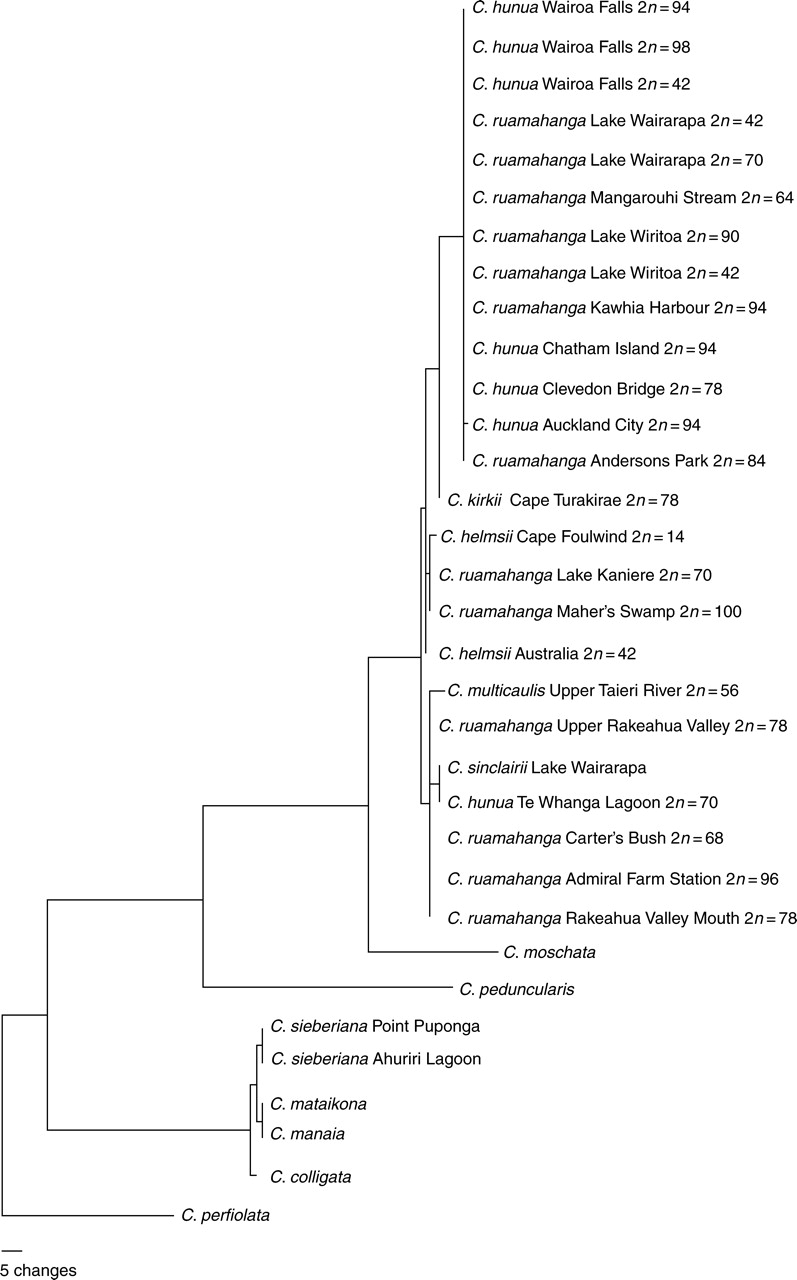
Phylogram for one selected most parsimonious tree for the full ITS sequence set.
In all trees the C. ruamahanga and C. hunua ITS sequences appear as part of a strongly supported clade that also includes the C. helmsii, C. multicaulis, C. sinclairii and C. kirkii sequences (for simplicity referred to as the C. hunua clade). Crassula moschata is the sister to this clade, and the sequences of C. perfoliata (section Crassula), C. peduncularis (section Helophytum) and those of the section Glomeratae samples are much more distantly related (Fig. 6).
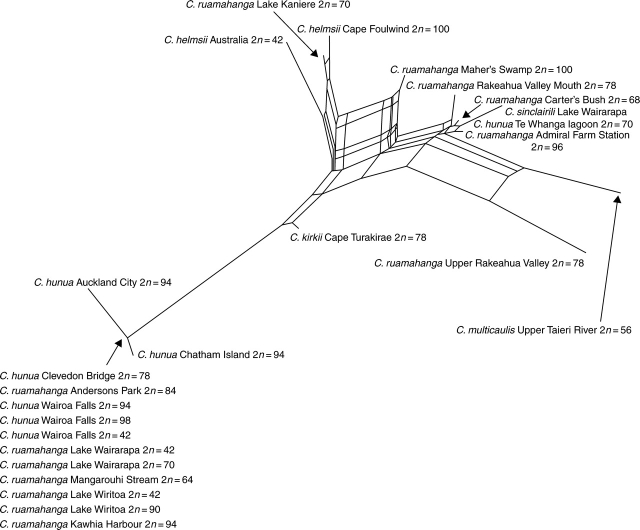
The sequence variation within the C. hunua clade is complicated, and the presence of multiple sequences within individuals makes these data unsuitable for analysis under most phylogenetic analysis programs. However, the program Splitstree 4 is capable of recovering genetic distance information from direct sequences by including ambiguity codes to represent mixed signals. Interpretation of the graphs produced by SplitsTree 4, as all representations of phylogenetic relationships, must be cautious, but the Neighbor Net graph shown (Fig. 7) provides a visual representation of the patterns of similarity in the sequences. ITS sequence variation is also shown in Table 3 (variable characters only).
Fig. 7.
Neighbor Net graph for ITS sequences of Crassula hunua clade (as defined in text).
Table 3.
Variable nucleotide positions in the ITS sequences of Crassula hunua and C. ruamahanga
| nrDNA ITS1 and ITS2 base pair position |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Specimen sequenced | 152 | 170 | 203 | 215 | 224 | 236 | 284 | 523 | 534 | 584 | 602 | 685 | 725 | 728 | 732 | 745 |
| Crassula ruamahanga | A | G | C/T | A | T | C | C | T | A | T | C | C/T | T | A | T | T |
| North Island, Southern Wairarapa, Lake Wairarapa, Lakeside Reserve (AK 284533) | ||||||||||||||||
| C. ruamahanga | A | G/T | C/T | A | T | C | C | T | A | C/T | C/T | C/T | T | A | T | T |
| South Island, Barrytown Flats, Mahers Swamp (AK 289900) | ||||||||||||||||
| C. ruamahanga | A | G | T | A | T | C | C | T | A | C | T | T | T | T | T | T |
| South Island, Invercargill, Anderson's Park (Thomson's Bush) (AK 287202) | ||||||||||||||||
| C. hunua | A | T | T | A | T | C | C | T | A | C | T | T | T | T | T | T |
| Chatham Island, Te Whaanga Lagoon (AK 291565) | ||||||||||||||||
| C. ruamahanga | A | T | T | A | T | C | C | T | A | C | C | T | T | T | T | T |
| Stewart Island, Rakeahua Valley(AK 290558) | ||||||||||||||||
| C. ruamahanga | A | G/T | T | A | T | C | C | T | A | C/T | C/T | T | T | T | T | T |
| North Island, Southern Wairarapa, Carter's Bush Scenic Reserve (AK 289899) | ||||||||||||||||
| C. ruamahanga | A | G/T | T | A | T | C | C | T | A | C/T | C/T | T | T | T | T | T |
| North Island, Eastern Wairarapa, Admiral Road Farm Station (AK 289901) | ||||||||||||||||
| C. ruamahanga | A | G/T | T | A | T | C | C | T | A | C/T | C/T | T | T | T | T | T |
| Stewart Island, Rakeahua Valley Mouth (AK 290559) | ||||||||||||||||
| C. ruamahanga | G | A | T | T | T | T | T | T | T | T | C | T | C | A | – | T |
| North Island, Northern Wairarapa. Mangarouhi Stream (AK 286171) | ||||||||||||||||
| C. hunua | G | A | T | T | T | T | T | – | T | T | C | T | C | A | – | T |
| North Island, Hunua, Wairoa Falls (AK 288129) | ||||||||||||||||
| C. hunua | G | A | T | C/T | C/T | T | T | T | T | T | C | T | C | A | – | C |
| North Island, Auckland City, Mt Albert, Rocky Nook Bowling Club (AK 294551) | ||||||||||||||||
| C. ruamahanga | G | A | T | T | T | T | T | T | T | T | C | T | C | A | – | T |
| North Island, Kawhia Harbour, Rakaunui Scenic Reserve (AK 289902) | ||||||||||||||||
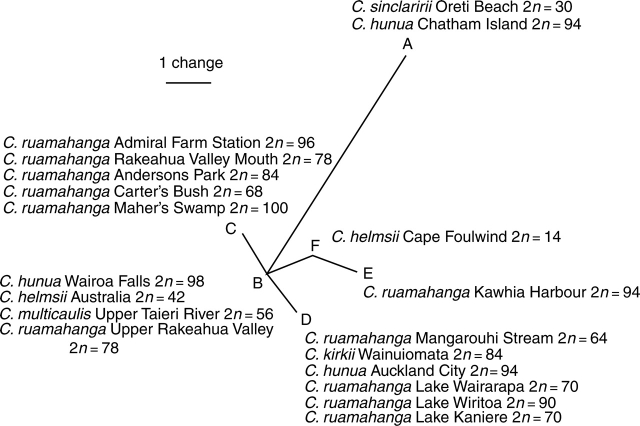
Plastid trnL intron and trnL-F spacer sequences were generated for a subset of 12 of the C. hunua and C. ruamahanga samples, the two C. helmsii provenances, one of the two C. kirkii samples and the C. multicaulis, C. peduncularis, C. sieberiana and C. sinclairii individuals. Further sequences were not generated because it was clear no simple pattern was emerging. Within the New Zealand members of the C. hunua clade, six distinct plastid haplotypes were identified. Relationships among these haplotypes were assessed by median network analysis using the Spectronet 1·1 program and are shown in the median network (Fig. 8). Haplotype A was found in C. sinclairii and C. hunua Te Whanga Lagoon. Haplotype B was found in C. hunua Wairoa Falls and C. multicaulis and C. helmsii Australia. Haplotypes C and D were found in various geographically diverse C. ruamahanga and C. hunua plants and haplotype D was also found in C. kirkii. Haplotype E was only found in C. ruamahanga from Rakaunui Inlet and haplotype F was only found in C. helmsii from Westport. In general there was little correspondence between plastid and ITS sequence groupings.
Fig. 8.
Median network of plastid trnL intron and trnL-F intergenic spacer sequences for the Crassula hunua clade as defined in the text. Haplotypes are labelled A–F.
DISCUSSION
Morphology
The present analyses of morphological characters that have traditionally been used to distinguish C. hunua and C. ruamahanga do not support the continued recognition of two species. Although the means of several characters were significantly different, the range of variation for the characters showed that there is either no difference between species or the variation of one species is essentially a subset of the variation of the other. Furthermore, as shown by the PCA of vegetative characters (Fig. 2C) and the scatter plots (Fig. 2A and B), combining characters does not support the recognition of discrete groups. These combined analyses are particularly informative, since they each involved characters for which means are statistically significantly different.
The failure to retrieve any discrete groups in the analyses of morphological characters presented here infers that the natural variation of the C. hunua/C. ruamahanga complex is mosaic with no obvious taxonomic or geographic pattern. Therefore, on the basis of their morphology we consider that it is not possible to distinguish two species.
Chromosome and pollen
Crassula hunua and C. ruamahanga present a unique example of chromosome diversity in the New Zealand endemic flora. There is a greater than 2-fold variation in chromosome number in the plants that were sampled with no obvious variation around a single basic number. de Lange et al. (2004a) suggested that the basic number for the genus was x = 7, as at that time all the New Zealand species that had been determined contained multiples of seven chromosomes. In a phylogenetic study based on matK sequence data, Mort et al. (2001) proposed that x = 8 is ancestral in Crassula and Crassulaceae but their study did not include species from section Tillaea. Regardless of whether the basic number is x = 7 or 8, there appears to be both euploid and aneuploid variation in the C. hunua/C. ruamahanga complex. This raises questions about the meiotic pairing behaviour of the plants, as no multiple chromosome configurations were observed. On the basis of their meiotic pairing behaviour, the plants fall into two groups, one in which there is regular bivalent formation, despite a range of chromosome numbers, and a second in which there is an appreciable frequency of univalent formation. High polyploids and aneuploids would be expected to show multivalent formation and the lack of multivalents is not a consequence of low chiasma frequencies; the majority of bivalents have two chiasmata, so autopolyploids would be expected to form multivalents (Jackson and Casey, 1982). Bivalent promoting mechanisms are also common in polyploid plants (Sears, 1976; Jackson, 1982; Jauhar and Joppa, 1996) and may be operational in C. hunua/ruamahanga, but clearly more work is needed to fully elucidate the nature of the chromosome variation that has been discovered. The formation of univalents at meiosis can also be indicative of hybridity as the genomes of the different parental species may fail to pair with each other. The phylogenetic analyses, discussed below, are suggestive of hybridization between species of section Helophytum, so a further possibility is that some of the plants show allopolyploid meiotic behaviour.
Crassula helmsii, C. kirkii, C. hunua and C. ruamahanga all show intraspecific chromosome variation, a relatively rare phenomenon in the New Zealand native flora. Murray et al. (1989) calculated that approx. 3 % of species that had been examined chromosomally showed evidence of intraspecific chromosome variation, and a recalculation, using the comprehensive list produced by Dawson (2000), in which chromosome numbers are reported for >80 % of species, gives a similar percentage. However, this may be an underestimate as in many cases only single plants of a species have been examined and it is likely that more extensive sampling will reveal intraspecific variation in other taxa. Most of the variation that has been found relates to polyploid races and these have been well documented in several genera including Veronica [as Hebe] (Murray et al., 1989; Dawson and Beuzenberg, 2000), Pratia (Murray et al., 2004) and several genera of Poaceae (Murray et al., 2005). Aneuploid variation, as found in C. hunua/ruamahanga, is virtually unknown in the New Zealand angiosperm flora, but well-documented examples are reported from elsewhere and include several groups of Crassulaceae and other families (Lewis and Oliver, 1971; Uhl and Moran, 1999; Murray and Young, 2001).
DNA sequences
Two major groups of ITS sequences can be defined for the purpose of this discussion. One includes only C. ruamahanga and C. hunua sequences (group I). Only two of the sequences in group I displayed any additivity in their sequences, and each of these had only one mixed base each, both at positions invariant in the rest of the samples. Group II includes sequences of individuals identified as C. hunua and C. ruamahanga and the sequences of C. kirkii, C. helmsii, C. multicaulis and C. sinclairii. All of the plants identified as C. hunua or C. ruamahanga with group II ITS sequences displayed a considerable degree of mixed sequence with the exception of the C. ruamahanga sample from Te Whanga Lagoon, Chatham Island which displayed an ITS sequence identical to that displayed by C. sinclairii except for one mixed base in the C. sinclairii sequence (Table 3). The Te Whanga Lagoon C. ruamahanga and C. sinclairii samples also shared the distinctive A plastid haplotype not found in any other plants, providing corroboration of a close relationship between the plants. Some mixed sequences are attributable to combinations of other sequences represented in the graph (Fig. 7). For example, the C. ruamahanga Maher's Swamp sequence can be obtained by combining the sequences of the C. helmsii or C. ruamahanga Lake Kaniere samples with those of the C. ruamahanga Rakeahua Valley Mouth, Carter's Bush, Te Whanga Lagoon or Admiral Farm samples. Therefore, it is possible that at least some of the plants displaying multiple sequences are recent hybrids or have a hybrid ancestry. No mixing between group I and II sequences was detected.
Given the high ploidy of many of the plants sampled in this study, it is possible that direct sequencing of PCR products has not provided a true reflection of the full diversity of ITS sequences present in individuals. Those individuals that displayed a single sequence may also have multiple ITS sequence types not detected in this study as a result of imbalanced copy number between homoeologous nrDNA repeats (possibly as a result of concerted evolution favouring one homoeologue) or PCR bias. Failure of direct sequencing to recover all the ITS sequence types present in a genome has been documented (Rauscher et al., 2002), as have differences in copy number of different homoeologous nrDNA repeats (Rauscher et al., 2004). In general, where hybridization and especially allopolyploidy are important factors in the evolution of a group, ITS sequence evolution can be idiosyncratic (Álvarez and Wendel, 2003).
Together with the chromosome and pollen stainability data, these complex sequence patterns are most readily explained by hypothesizing a high frequency of hybridization between biological species and a mixture of allo- and possibly autopolyploidy along with the persistence of hybrid individuals in nature through vegetative reproduction. Despite the lack of sequences displaying an additive combination of group I and group II sequence types, one possibility is that the group I ITS sequences represent the C. ruamahanga/hunua ancestral sequence and the individuals identified as C. ruamahanga or C. hunua that display group II sequences have hybrid origins with some of their ancestors including one or more of the other Crassula species of section Helophytum. Another possibility is that two cryptic lineages are represented in C. ruamahanga and C. hunua, one of which (group II) has undergone more hybridization with other species of section Helophytum than the other, but both of which have given rise to a polyploid series. Other more complicated explanations are also plausible, and it seems likely that at least some of the individuals displaying group I ITS sequences also have hybrid origins, given that group I includes individuals that show univalent formation and/or poor pollen stainability. In particular, the possession of a plastid C haplotype, not otherwise sampled from plants with group I ITS sequences, may indicate a hybrid origin of the C. ruamahanga Anderson Park, Invercargill sample. Other processes, such as the independent sorting of ancestral polymorphisms at different genetic loci may also have contributed to the phylogenetic incongruence observed between nuclear and plastid markers in this study and the discrepancies between relationships indicated by analysis of either sequence data set with morphologically defined taxa [for general discussion of these see Avise (2000); and for examples see Smissen et al. (2004), Fehrer et al. (2007) and references therein].
Unravelling the details of this hypothetical reticulate evolution in the C. hunua clade will require extensive population level sampling to address issues such as the level of sexual reproduction in different populations, the fertility of different hybrid combinations and phylogenetic relationships. AFLP or other multilocus DNA fingerprinting techniques may be informative in addressing these questions, and may potentially provide an assessment of nuclear DNA phylogenetic relationships independent of ITS. Clearly in Crassula it is not logical to rely too heavily on sequences from a single region of the genome. Cloning of ITS PCR products to explore the diversity of sequences present in those specimens displaying mixed signals in direct sequences might allow the identification of a larger number of variants within individuals, and allow for more robust phylogenetic analysis of these. The use of sequence-specific primers would allow a further test of whether PCR-bias or copy number differences are resulting in a distorted view of ITS sequence diversity within individuals. While these experiments might give a better picture of the complexities of ITS sequence evolution in the group, given their cytogenetic diversity, it is doubtful that these data would be of direct relevance to the taxonomic and conservation issues of Crassula in New Zealand. Despite the sampling and technical limitations of the DNA sequence data presented here, it is clear that issues of population and breeding biology within these Crassula taxa need to be addressed before DNA sequence variation can be meaningfully incorporated into historical accounts of evolution within the C. ruamahanga clade. However, the DNA sequence data, particularly the incongruence between nuclear and plastid markers do suggest a complex phylogenetic history in accord with the cytological data presented here. The other four Crassula species, C. helmsii, C. kirkii, C. multicaulis and C. sinclairii, which also group in the C. ruamahanga clade, show clear morphological distinctions from C. ruamahanga and are therefore not included in the recircumscribed C. ruamahunga.
Taxonomy
As a result of the morphological, chromosomal and DNA sequence data presented here, and in the context of the preceding discussion, it is not possible to retain the two species C. hunua and C. ruamahanga. We consider these to be conspecific and therefore accept only one species in this complex and reduce the other to synonymy. However, C. ruamahanga as recircumscribed probably includes a number of ‘biological species’ and hybrids between them, but accepting a widespread and variable species is the only practical and realistic taxonomic option. As both C. hunua A.P.Druce and C. ruamahanga A.P.Druce were validly and effectively published simultaneously by the same author, under Article 11·5, Note 2, of the International Code of Botanical Nomenclature (McNeill et al., 2006) either name has equal priority, and so a choice can be effected by adopting one of these names, or its final epithet in the required combination, and simultaneously rejecting or relegating to synonymy the other. Accordingly, Crassula ruamahanga A.P.Druce was selected as the preferred name because the subacute, acute to sharply acute leaves normally associated with this name is the more usual vegetative state found in populations of this species throughout New Zealand. Plants with the alternative condition, obtuse leaf apices are virtually restricted to the Wairoa Falls, Hunua. It should be noted that this latter condition, while frequent at the Hunua Falls, has proved unstable over time in cultivation, with plants from that site reverting to the acute to sharply acute leaves state ‘typical’ of C. ruamahanga.
Crassula ruamahanga A.P.Druce, N. Z. J. Bot. 25, 128 (1987) emend. de Lange et Heenan ≡ Tillaea acutifolia Kirk, Stud. Fl. N. Z. 143 (1899) ≡ Crassula acutifolia (Kirk) A.P.Druce et Given, N. Z. J. Bot. 22, 583 (1985) non. C. acutifolia Lam., Encyc. II, 175 (1786)
Type collections. ‘NORTH Island: Hurunuiorangi (flowers not seen). SOUTH Island: Winton Forest, Southland, T. K[irk], Dec.’
Lectotype (vide Allan, Fl. New Zealand 1:199. 1961). Hurunuiorangi, May 17, 1877, T. Kirk. WELT SP050125a
=Crassula hunua A.P.Druce, N. Z. J. Bot. 25, 128 (1987)
≡C. pusilla (Kirk) A.P.Druce et D.R.Given, N. Z. J. Bot. 22, 583 (1985) non C. pusilla Schönland Rec. Albany Mus. II, 451 (1913)
≡Tillaea pusilla Kirk, Stud. Fl. N.Z. 143 (1899)
Type collections. ‘NORTH Island: banks of streams, &c. Kawakawa, Bay of Islands, T. K[irk]. Auckland Cheeseman! Dec.’
Lectotype (vide Allan 1961 : 198). ‘WELT SP050132!, Tillaea pusilla T. Kirk, Stud. Fl. N.Z., Auckland, Mr Cheeseman.’
Notes. It has not been possible to locate the Kawakawa River specimens that Kirk (1899) cited in his protologue, and that Cheeseman (1906, p. 142) evidently saw. Kirk (1899) states clearly that he used material gathered by Cheeseman from Auckland in December, yet the label details on WELT SP050132 suggests he was unsure if Cheeseman had collected that specimen. Cheeseman (1906, p. 142) makes it clear he did, but not from Auckland, which he does not mention at all, stating rather that he collected it from the Wairoa Falls, near Hunua. Whatever the exact details, it is ‘Auckland’ we must assume in a broad geographic sense and not the more specific Wairoa Falls, Hunua which must stand as the type locality for Tillaea pusilla. = Tillaea pusilla var. brevis Kirk, Stud. Fl. N.Z. 143 (1899) (‘brevia’)
Type collection. ‘NORTH Island: Wairoa Falls, Hunua, T. K[irk].’
Lectotype (vide Allan 1961). WELT SP050134!, Tillaea pusilla var. brevia T. Kirk, Stud. Fl. N.Z., Wairoa Falls, Hunua, T. Kirk, Dec 1868.
Notes. Allan (1961, p. 198) typified this taxon in the following manner ‘Type: specimens in W[ELT] labelled as var. brevia Wairoa Falls, Hunua, Dec 1868’. At WELT there is now only one gathering, WELT SP050134 (L. Perrie, WELT herbarium, Te Papa Tongarewa – The Museum of New Zealand, New Zealand, ‘pers. comm.’), and this must stand as the lectotype. We prefer to regard it as lectotype because we cannot be sure that there are no other specimens of this gathering in herbaria known to hold Kirk material, and Allan's wording suggests there might have been. WELT SP050135, though matching Kirk's protologue as to location, name and authors hand was collected in March 1867. Although Kirk (1899) does not provide a date for his type collections in his protologue, by Allan's full and direct reference to a specimen including collection date as lectotype, WELT SP050135 must now stand as a paralectotype.
Notes. Because C. ruamahanga now includes both C. hunua and C. ruamahanga and our circumscription differs substantially from that of the taxon originally described by Kirk (1899), and, in addition, characters either not seen or unavailable to Kirk have been included, formal recognition of this is required (see Article 47, Recommendation 47A.1. of the ICBN; McNeill et al., 2006).
Description. Diminutive, perennial herb forming diffuse to extensive, compact to loose, yellow-green, green to dark green, moss-like mats; stems much branched, prostrate, rooting at nodes, scarcely ascending at tips: internodes 1·5–9·8 mm, terete, 0·3–0·6 mm diam. Leaves connate at base, 0·7–2·9 × 0·3–1 mm, 0·3–0·5 mm thick, linear-oblong, linear-lanceolate, lanceolate or elliptic-lanceolate, flattened to slightly concave above, convex beneath, apex sometimes obtuse, usually subacute, acute to sharply acute; shortly acuminate or with an apiculus up to 0·3 mm long, caducous. Flowers solitary in leaf axils, star-like, sweetly fragrant, 4-merous, peduncle 0·2–5·4 mm long, 0·7–0·7 mm wide, not or scarcely elongating at fruiting. Calyx lobes 0·5–1·7 × 0·3–0·5 mm, triangular to triangular-ovate, obtuse, acute, ± acuminate, usually with an apiculus up to 0·1 mm long, caducous. Corolla 0·9–3·2 mm diam.; petals 0·5–1·7 × 0·3–0·6 mm, narrowly elliptic-ovate, ovate to triangular-ovate, white or pink-flushed, subacute, acute to sharply acute, ≥ calyx. Stamens 4, filaments 0·9–1·2 mm, white, terete; anthers 0·4–0·6 mm, rose-pink; pollen (vide Moar, 1993) pale yellow, usually anisopolar, tricolporate, sometimes tetracolporate; ectoaperture broad, long, generally branching, and often fusing to form a polar cap at one, or both, poles; polar axis 13–14 µm, equatorial axis 12–15 µm. Scales 4, 0·3–0·5 mm long, narrowly to broadly cuneate. Follicles 4, 0·9–1·2 × 0·6–1 mm, ovoid to ellipsoid, turgid, recurved, smooth. Style minute, approx. 0·01 mm, recurved. Seeds (vide Webb and Simpson, 2001) 2–4 per follicle, 0·4 × 0·25 mm, narrowly elliptic, elliptic-oblong, to oblong, more or less terete to almost square in section, apex and base truncate, base sometimes with a slight projection. Testa dull, dark henna, dark purple-brown, orange-brown or dark red-brown, almost smooth or with an indistinct regulate-colliculate pattern. Fl. (Aug-)Jan(–Apr). Fr. (Sep–)Jan(–May). Chromosome number 2n = 42 to 2n = 100 (vide Murray and de Lange, 1999; de Lange et al., 2004a; this paper).
Representative specimens. NORTH ISLAND: NORTH AUCKLAND: Northern Wairoa River, T. F. Cheeseman s.n., n. d., AK 4554. SOUTH AUCKLAND: Wairoa Falls, T. Kirk 153, n.d., AK 11433; Falls of the southern Wairoa, T. F. Cheeseman s.n., Apr 1896, AK 4555; Wairoa River Gorge, R. O. Gardner 4344, 3 Oct 1984, AK 168426; Waihou (Thames) River, at Hikutaia Wharf, R. Mason 7304, 30 Nov 1959, CHR 113044; Rakaunui Scenic Reserve, Rakaukeke Inlet, Tawairoa Stream, P. J. de Lange 4312, 3 Apr 2000, AK 252131. WELLINGTON: Wanganui, Lake Wiritoa, Scoutlands, C. C. Ogle 3451, 31 Jan 1999, CHR 518730; Northern Wairarapa, Mangarouhi Stream, P. J. de Lange 5994, 14 Apr 2004, AK 286171; Wairarapa, Carter's Bush, L. B. Moore s.n., 29 Nov 1958, CHR 124456; Ruamahanga River, SE of Carterton, A. P. Druce s.n., Oct 1973, CHR 208995; Lake Wairarapa, Lakeside Reserve (NE end), P. Enright s.n., 16 Nov 2003, AK 284533; Wellington Harbour, Mokopuna Island, P. J. de Lange 1580 & G. M. Crowcroft, 4 Sept 1992, AK 212059. SOUTH ISLAND: NELSON: Westport Domain, Buller River, R. Mason s.n. & N. J. Moar 1739, 25 Jan 1953, CHR 81569. WESTLAND: Barrytown Flats, Maher Swamp, P. J. de Lange 1002, 1 Sept 1991, CHR 473589. CANTERBURY: Ashburton River, H. H. Allan s.n., n.d., CHR 11950; South Canterbury, near Pareora River, R. Mason s.n., 9 May 1945, CHR 51479. OTAGO: Otago, Mihiwaka Hill, P. N. Johnson s.n., 3 May 1981, CHR 363947; SOUTHLAND: Winton Forest, T. Kirk s.n., n.d., AK 4557; Invercargill, Andersons Park (Thomson's Bush), B. D. Rance s.n., 25 Jan 2004, AK 287202; Waitutu Forest, east of Lake Poteriteri, C. C. Ogle 1068, 21 Jan 1984, CHR 417095; Waitutu Forest, west of Waitutu Hut, above Waitutu River, B. D. Rance s.n., 5 Feb 2007, AK 298466; Lake Manapouri at mouth of Spey River, M. J. A. Simpson s.n., 16 Feb 1959, CHR 111815; Lake Manapouri, Supply Bay, B. D. Rance s.n., 17 Jan 2007, AK 298442. STEWART (RAKIURA) ISLAND: Mouth of the Rakeahua Valley (ex cultivated), P. J. de Lange 6492, 29 Apr 2005, AK 290559; Rakeahua Valley (ex cultivated), P. J. de Lange 6491, 29 Apr 2005, AK 290558. CHATHAM ISLANDS: Rekohu (Chatham Island), Lake Huro, Mangape Stream outlet, P. J. de Lange CH223 & G. M. Crowcroft, 29 Mar 1996, AK 229937; Pitt Island, G. Walls s.n., 16 May 1999, CHR 535256.
For a revised key to New Zealand Crassula Section Helophytum, see Table 4.
Table 4.
Revised key to New Zealand Crassula Section Helophytum
| 1. | Flowers often in small clusters in the leaf axils, sometimes solitary, inconspicuous; petals closely appressed to carpels or with spreading tips, usually < calyx lobes, sometimes equal to calyx lobes, or nearly so | Section Glomeratae |
| Flowers always solitary in the leaf axils, conspicuous; petals spreading and corolla star-like, usually significant < calyx lobes, rarely nearly equal | 2. Section Helophytum | |
| 2. | Leaves often >7 × 2 mm; flowers 4–7 mm diam.; plants terrestrial | C. moschata |
| Leaves <7 × 1·6 mm; flowers < 4 mm diam.; plants aquatic to semi-aquatic, if terrestrial then leaves always smaller | 3 | |
| 3. | Plants strict summer annuals. Pedicels elongating to 13 mm long at fruiting; seeds >10 per follicle | C. peduncularis |
| Plants perennial. Pedicels ≤7 mm long, scarcely elongating at fruiting; seeds ≤5 per follicle | 4 | |
| 4. | Leaves ± triangular-lanceolate, clustered and imbricate at stem nodes, acuminate, petals distinctly rounded | C. multicaulis |
| Leaves linear to elliptic, or if ± lanceolate then not triangular lanceolate, usually opposite, occasionally clustered at stem nodes; petals obtuse, subacute to acute | 5 | |
| 5. | Petals 0·9–1·5 ± calyx lobes | 6 |
| Petals 2–3 × calyx lobes | 7 | |
| 6. | Leaves (0·7–)2(–2·8) × (0·3–)0·6(–1) mm, not noticeably succulent; calyx lobes (0·5–)0·8(–1·6) mm long; petals equal to or 2 × width | C. ruamahanga |
| Leaves (2·5–)8(–10) × 0·7–1·6 mm, obviously thick and succulent; calyx lobes 1–1·5 mm long, petals with length 1·7–1·8 × width | C. helmsii | |
| 7. | Plants mat forming but not obviously moss-like; leaves obtuse, subacute or sometimes ± apiculate, 1·5–5·5 × 0·8–1·7 mm; seeds 0·5–0·7 mm long | C. kirkii |
| Plants forming fine moss-like mats; leaves ± acute, 0·5–2 × 0·2–0·5 mm; seeds 0·3–0·45 mm long | C. sinclairii |
Conservation implications
With the recircumscription of C. ruamahanga to include plants previously known as C. hunua, a reassessment of the conservation status of C. ruamahanga is warranted. Previously, C. hunua sensu A.P.Druce had been assessed using the New Zealand Threat Classification System (Molloy et al., 2002) as ‘Acutely Threatened/Nationally Critical’ (de Lange et al., 2004b), because the species was believed to be restricted to two sub-populations (Wairoa Falls and the adjacent Gorge, and the Chatham Islands), which collectively occupy an area of ≤1 ha.
With the reduction of C. hunua into the synonymy of C. ruamahanga, the recircumscribed C. ruamahanga is now more widespread than before with extant populations ranging from the Wairoa Falls, Hunua, in the North Island, through most of the South Island and Stewart and Chatham Islands. Within this range, the species has a naturally sparse distribution but at some localities (e.g. in the southern Wairarapa and Southland) it may be locally common. The range of habitats occupied is varied with the species being most commonly recorded from ephemeral, often muddy or silty pools within lowland alluvial forest and from lake margin turf communities. The species may also be an urban weed, and it has been found in bowling green turf and in damp shaded sites in Auckland, Wellington, Christchurch and Dunedin. Even with the combination of the two taxa some range contraction is known, as C. hunua sensu A.P.Druce has not been recorded north of the Wairoa Falls, Hunua since 1906, when Cheeseman last reported it from the Wairoa River near Dargaville. In contrast to its apparent extinction from the Northland Peninsula, there have been discoveries of new populations, often from quite unexpected locations in the northern Wairarapa, Wanganui and Waikato regions (de Lange et al., 1998; de Lange, 2000). These discoveries have increased the number of known populations and it is possible that C. ruamahanga could be rediscovered in Northland. Therefore, it is concluded that the recircumscribed species is most appropriately assessed using the New Zealand Threat Classification System as ‘At Risk/Sparse’, a conservation rating which most accurately reflects the current level of knowledge about the ecology, distribution, range of habitat preferences and the nature of threats it faces.
The chromosomal and DNA sequence variation that has been uncovered in these studies pose questions concerning the conservation of C. ruamahanga. In addition, they highlight the need for further critical study at the population level using a range of different molecular markers. This study has also shown that the range of molecular markers (plastid DNA and nrDNA) used for taxonomic research, when undertaken on multiple samples, spanning a large geographic range, are not necessarily reliable in this group. For all these reasons it is important to conserve these plants at the maximum number of sites if the full range of variation is to be maintained.
SUPPLEMENTARY INFORMATION
Supplementary information is available online at www.aob.oxfordjournals.org and lists herbarium specimens used for morphological analysis of Crassula hunua and C. ruamahanga.
ACKNOWLEDGEMENTS
We acknowledge the considerable collecting efforts, advice and comments received from the following former or current Department of Conservation Staff, M. Thorsen, G. Foster, R. Stanley, P. Knightbridge, B. Rance, A. Baird, B. Gibb and I. Keenan. C. Ogle of Wanganui and P. Enright, of Featherston, Wairarapa, helped procure live material from their respective regions. We also thank R. K. Brummitt (Royal Botanic Gardens, Kew), H. R. Toelken (AD, Adelaide, South Australia) and C. Ogle for comments and advice and L. Perrie and B. V. Sneddon in locating type material. The late F. Pitt (WELT) helped resolve typification issues by clarifying aspects of the acquisition and curation of the Kirk herbarium, now housed at WELT, Te Papa, Museum of New Zealand, Wellington. Funds for this study were provided by the New Zealand Department of Conservation, Conservation Management Units Investigation 3838 and the Foundation for Research, Science and Technology.
LITERATURE CITED
- Allan HH. Flora of New Zealand. Vol. I. Wellington: Government Printer; 1961. [Google Scholar]
- Allendorf FW, Luikart G. Conservation and the genetics of populations. Malden, MA: Blackwell; 2006. [Google Scholar]
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press; 2000. [Google Scholar]
- Avise JC, Hamrick JL. Conservation genetics: case histories from nature. New York, NY: Chapman and Hall; 1996. [Google Scholar]
- Benirschke K, Kumamoto AT. Mammalian cytogenetics and conservation of species. Journal of Heredity. 1991;82:187–191. doi: 10.1093/oxfordjournals.jhered.a111063. [DOI] [PubMed] [Google Scholar]
- Bloom WL. Origin of reciprocal translocations and their effects in Clarkia speciosa. Chromosoma. 1974;49:61–76. [Google Scholar]
- Cameron EK, de Lange PJ, Given DR, Johnson PN, Ogle CC. New Zealand Botanical Society threatened and local plant lists (1993 revision) New Zealand Botanical Society Newsletter. 1993;32:14–28. [Google Scholar]
- Cameron EK, de Lange PJ, Given DR, Johnson PN, Ogle CC. New Zealand Botanical Society threatened and local plant lists (1995 revision) New Zealand Botanical Society Newsletter. 1995;39:15–28. [Google Scholar]
- Cheeseman TF. Manual of the New Zealand flora. Wellington: Government Printer; 1906. [Google Scholar]
- Cheeseman TF. Manual of the New Zealand Flora. 2nd edn. Wellington: Government Printer; 1925. [Google Scholar]
- Cleland RE. Oenothera cytogenetics and evolution. London: Academic Press; 1972. [Google Scholar]
- Connor HE, Edgar E. Name changes in the indigenous New Zealand Flora, 1960–1986 and Nomina Nova IV. New Zealand Journal of Botany. 1987;25:115–170. 1983–1986. [Google Scholar]
- Dawson MI. Index of chromosome numbers of indigenous New Zealand spermatophytes. New Zealand Journal of Botany. 2000;38:47–150. [Google Scholar]
- Dawson MI, Beuzenberg E. Contributions to a chromosome atlas of the New Zealand flora. 36. Miscellaneous families. New Zealand Journal of Botany. 2000;38:1–23. [Google Scholar]
- Dopson SR, de Lange PJ, Ogle CC, Rance BD, Courtney SP, Molloy J. Threatened Species Occasional Publication No. 13. Wellington: Department of Conservation; 1999. The conservation requirements of New Zealand's nationally threatened vascular plants. [Google Scholar]
- Druce AP, Given DR. New combinations in New Zealand Crassula (Crassulaceae) New Zealand Journal of Botany. 1984;22:583. [Google Scholar]
- Fehrer J, Gemeinholzer B, Chrtek J, Brautigam S. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae) Molecular Phylogenetics and Evolution. 2007;42:347–361. doi: 10.1016/j.ympev.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Given DR. Threatened and local plant lists – New Zealand botanical region, vascular plants. Christchurch: DSIR Land Resources; 1990. [Google Scholar]
- Henry RJ. Plant conservation genetics. New York, NY: Food Products Press; 2006. [Google Scholar]
- Huber KT, Langton M, Penny D, Moulton V, Hendy M. Spectronet: a package for computing spectra and median networks. Applied Bioinformatics. 2002;1:159–161. [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jackson RC. Chromosome evolution in Haplopappus gracilis: a centric transposition race. Evolution. 1973;27:243–256. doi: 10.1111/j.1558-5646.1973.tb00669.x. [DOI] [PubMed] [Google Scholar]
- Jackson RC. Polyploidy and diploidy: a new perspective on chromosome pairing and its evolutionary implications. American Journal of Botany. 1982;69:1512–1523. [Google Scholar]
- Jackson RC, Casey J. Cytogenetic analyses of autopolyploids: models and methods for triploids to octoploids. American Journal of Botany. 1982;69:487–501. [Google Scholar]
- James SH. Complex hybridity in Isotoma petraea. 1. The occurrence of interchange heterozygosity, autogamy and a balanced lethal system. Heredity. 1965;20:341–353. [Google Scholar]
- James SH. The relevance of genetic systems in Isotoma petraea to conservation practice. In: Groves RH, Ride WDL, editors. Species at risk: proceedings of a symposium on the biology of rare and endangered plants in Australia. Canberra: Australian Academy of Science; 1982. pp. 63–71. [Google Scholar]
- Jauhar PP, Joppa LR. Chromosome pairing as a tool in genome analysis: merits and limitations. In: Jauhar PP, editor. Methods of genome analysis in plants. Boca Raton, FL: CRC Press; 1996. pp. 9–37. [Google Scholar]
- Kirk T. The students' flora of New Zealand. Vol. 1899. Wellington: Government Printer; [Google Scholar]
- de Lange PJ. A new northern limit for Crassula ruamahanga. New Zealand Botanical Society Newsletter. 2000;60:20–21. [Google Scholar]
- de Lange PJ, Gardner RO. A taxonomic reappraisal of Coprosma obconica Kirk (Rubiaceae; Antherospermeae) New Zealand Journal of Botany. 2002;40:25–38. [Google Scholar]
- de Lange PJ, Ogle CC, Foster G, Champion PD. Managing Crassula ruamahanga at its type locality and a new northern limit for the species. New Zealand Botanical Society Newsletter. 1998;53:13–15. [Google Scholar]
- de Lange PJ, Heenan PB, Given DR, Norton DA, Ogle CC, Johnson PN, et al. Threatened and uncommon plants of New Zealand. New Zealand Journal of Botany. 1999;37:603–628. [Google Scholar]
- de Lange PJ, Murray BG, Datson PM. Contributions to a chromosome atlas of the New Zealand flora – 49 miscellaneous families. New Zealand Journal of Botany. 2004;a 42:873–904. [Google Scholar]
- de Lange PJ, Norton DA, Heenan PB, Courtney SP, Molloy BPJ, Ogle CC, et al. Threatened and uncommon plants of New Zealand. New Zealand Journal of Botany. 2004;b 42:45–76. [Google Scholar]
- Levin DA. The role of chromosome change in plant evolution. New York, NY: Oxford University Press; 2002. [Google Scholar]
- Lewis WH, Oliver RL. Meiotic chromosome variation in Claytonia virginiana. Journal of Heredity. 1971;62:379–380. [Google Scholar]
- McNeill J, Barrie FF, Burdet HM, Demoulin V, Hawksworth DL, Marhold K, et al. International Code of Botanical Nomenclature (Vienna Code) Regnum Vegetabile. 2006;146 [Google Scholar]
- Moar NT. Pollen grains of the New Zealand dicotyledonous plants. Lincoln: Manaaki Whenua Press; 1993. [Google Scholar]
- Molloy J, Bell B, Clout M, de Lange P, Gibbs G, Given D, et al. Classifying species according to threat of extinction – a system for New Zealand. Wellington: Department of Conservation; 2002. [Google Scholar]
- Mort ME, Soltis DE, Soltis PS, Francisco-Ortega J, Santos-Guerra A. Phylogenetic relationships and evolution of Crassulaceae inferred from matK sequence data. American Journal of Botany. 2001;88:76–91. [PubMed] [Google Scholar]
- Murray BG. The cytology of the genus Briza. II. Chiasma frequency, polyploidy and interchange heterozygosity. Chromosoma. 1976;57:81–93. [Google Scholar]
- Murray BG, de Lange PJ. Contributions to a chromosome atlas of the New Zealand flora. 35. Miscellaneous families. New Zealand Journal of Botany. 1999;37:511–522. [Google Scholar]
- Murray BG, Young AG. Annals of Botany. Vol. 87. Asteraceae: Gnaphalieae; 2001. Widespread chromosome variation in the endangered grassland forb Rutidosis leptorrhynchoides F. Muell; pp. 83–90. [Google Scholar]
- Murray BG, Braggins JE, Newman PD. Intraspecific polyploidy in Hebe diosmifolia (Cunn.) Cockayne et Allan (Scrophulariaceae) New Zealand Journal of Botany. 1989;27:587–589. [Google Scholar]
- Murray BG, Datson PM, Lai ELY, Sheath KM, Cameron EK. Polyploidy, hybridization and evolution in Pratia (Campanulaceae) New Zealand Journal of Botany. 2004;41:905–920. [Google Scholar]
- Murray BG, de Lange PJ, Ferguson AR. Nuclear DNA variation, chromosome numbers and polyploidy in the endemic and indigenous grass flora of New Zealand. Annals of Botany. 2005;96:1293–1305. doi: 10.1093/aob/mci281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Wilby AS. Extreme chromosomal heterogeneity in a small-island population of Rumex acetosa. Heredity. 1989;62:133–140. doi: 10.1038/hdy.1989.18. [DOI] [PubMed] [Google Scholar]
- Pillon Y, Chase MW. Taxonomic exaggeration and its effects on orchid conservation. Conservation Biology. 2007;21:263–265. doi: 10.1111/j.1523-1739.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- Rauscher JT, Doyle JJ, Brown AHD. Internal transcribed spacer repeat-specific primers and the analysis of hybridization in the Glycine tomentella (Leguminosae) polyploid complex. Molecular Ecology. 2002;11:2691–2702. doi: 10.1046/j.1365-294x.2002.01640.x. [DOI] [PubMed] [Google Scholar]
- Rauscher JT, Doyle JJ, Brown AHD. Multiple origins and nrDNA internal transcribed spacer homeologue evolution in the Glycine tomentella (Leguminosae) allopolyploid complex. Genetics. 2004;166:987–998. doi: 10.1534/genetics.166.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. Genetic control of chromosome pairing in wheat. Annual Review of Genetics. 1976;10:31–51. doi: 10.1146/annurev.ge.10.120176.000335. [DOI] [PubMed] [Google Scholar]
- Sieber VK, Murray BG. Structural and numerical chromosomal polymorphism in natural populations of Alopecurus L. (Poaceae) Plant Systematics and Evolution. 1981;139:121–136. [Google Scholar]
- Smisen RD, Breitwieser I, Ward JM. Phylogenetic implications of trans-specific chloroplast DNA sequence polymorphism in New Zealand Gnaphalieae (Asteraceae) Plant Systematics and Evolution. 2004;249:37–53. [Google Scholar]
- Statistical Sciences. S-Plus for Windows user's manual, Version 4·5. Release 2. Seattle, WA: Statistical Sciences; 1998. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- Sykes WR. Notes on Euphorbia and Crassula with a revised key to the latter wild in New Zealand. New Zealand Botanical Society Newsletter. 2005;79:8–16. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toelken HR. The species of Crassula L. in Australia. Journal of the Adelaide Botanic Garden. 1981;3:57–90. [Google Scholar]
- Uhl CH, Moran R. Chromosomes of Villadia and Altamiranoa (Crassulaceae) American Journal of Botany. 1999;86:387–397. [PubMed] [Google Scholar]
- Vaughan HE, Taylor S, Parker JS. The ten cytological races of the Scilla autumnalis species complex. Heredity. 1997;79:371–379. [Google Scholar]
- Webb CJ, Simpson MJA. Seeds of New Zealand gymnosperms and dicotyledons. Christchurch: Manuka Press; 2001. [Google Scholar]
- Webb CJ, Sykes WR, Garnock-Jones PJ. Flora of New Zealand. Vol. IV. Christchurch: DSIR Botany Division; 1988. pp. 571–583. [Google Scholar]
- White T, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. [Google Scholar]
- Young AG, Murray BG. Genetic bottlenecks and dysgenic gene flow into re-established populations of the grassland daisy Rutidosis leptorrhynchoides. Australian Journal of Botany. 2000;48:409–416. [Google Scholar]