Abstract
Background
The evolution and biology of rDNA have interested biologists for many years, in part, because of two intriguing processes: (1) nucleolar dominance and (2) sequence homogenization. We review patterns of evolution in rDNA in the angiosperm genus Nicotiana to determine consequences of allopolyploidy on these processes.
Scope
Allopolyploid species of Nicotiana are ideal for studying rDNA evolution because phylogenetic reconstruction of DNA sequences has revealed patterns of species divergence and their parents. From these studies we also know that polyploids formed over widely different timeframes (thousands to millions of years), enabling comparative and temporal studies of rDNA structure, activity and chromosomal distribution. In addition studies on synthetic polyploids enable the consequences of de novo polyploidy on rDNA activity to be determined.
Conclusions
We propose that rDNA epigenetic expression patterns established even in F1 hybrids have a material influence on the likely patterns of divergence of rDNA. It is the active rDNA units that are vulnerable to homogenization, which probably acts to reduce mutational load across the active array. Those rDNA units that are epigenetically silenced may be less vulnerable to sequence homogenization. Selection cannot act on these silenced genes, and they are likely to accumulate mutations and eventually be eliminated from the genome. It is likely that whole silenced arrays will be deleted in polyploids of 1 million years of age and older.
Key words: Diploidization, epigenetics, nucleolar dominance, polyploidy, rDNA, ribosomal DNA, sequence homogenization
INTRODUCTION
In most eukaryotes 5S and 18–5·8–26S nuclear ribosomal DNA (rDNA) units occur in tandem arrays at one or several loci. Each large rDNA unit contains the 18S, 5·8S and 26S rDNA subunits, the internal transcribed spacers (ITS) sequences and the intergenic spacer (IGS). The genes are highly conserved, whereas ITS divergence is sufficient to resolve species relationships within most genera (e.g. Nicotiana; Chase et al., 2003). The IGS, which contains the transcription-start site and genetic and epigenetic features that influence the regulation of the downstream genes, diverges more rapidly than ITS. Substantial differences in structure may even occur within a species (Kovarik et al., 2005).
Of particular interest to evolutionary biologists is the pattern of divergence of the whole rDNA array, which is often influenced by sequence homogenization that functions to replace existing genic units with variants of that unit over time, a process known as concerted evolution (Dover et al., 1982; Eickbush and Eickbush, 2007). Concerted evolution itself may be important to maintain sufficient numbers of active rDNA units, and Ohta (1989) suggested from computer models that homogenization acts to reduce mutational load and is favoured by selection. Patterns of ribosomal RNA (rRNA) gene expression are strongly influenced by epigenetic events such as cytosine methylation and histone acetylation, silencing potentially initiated by small interfering RNAs (Preuss and Pikaard, 2007). In hybrid organisms and allopolyploid species (interspecific hybrids with chromosome number duplication) silencing of entire loci is common, often leading to one parental rDNA array being expressed in preference to another, a phenomenon known as nucleolar dominance (Cermeno et al., 1985; Preuss and Pikaard, 2007).
Ribosomal DNA and the nuclear domain of its activity, the nucleolus, have long intrigued biologists; Alberts et al. (2002) stated in a 1898 review that there were already 700 references. Despite a huge volume of ongoing research (web-of-science search on 2 July, 2007, for ‘rDNA or ‘ribosomal DNA’ or ‘nucleolus’ revealed 25 437 hits), there are still critical and fundamental unanswered questions. These include: (a) why do many eukaryotes have more rRNA genes than required for ribosome biosynthesis; (b) why are large numbers of rDNA units often, but not always, epigenetically silenced, frequently leading to nucleolar dominance; (c) why are rDNA evolution and divergence often, but not always, associated with sequence homogenization, leading to concerted evolution; and (d) are any or all of these fundamental processes or phenomena interrelated?
This paper aims to review progress made in addressing these questions using polyploid species of the genus Nicotiana as a model system. It is not a general review of rDNA, nucleolar dominance or concerted evolution, which have been well reviewed elsewhere (e.g. Coen et al., 1982; Shaw and Jordan, 1995; Pikaard, 1999, 2000; Wendel, 2000; Raska et al., 2006a, b; Volkov et al., 2007). Instead, it reviews patterns of evolution in rDNA in related Nicotiana polyploids of different ages, from synthetic allopolyploids that mimic natural species to natural polyploids that formed 4·5 million years ago. Together these data enable us to propose a hypothesis that explains the relationship between nucleolar dominance, concerted evolution and rDNA locus number. We will discuss the implications of predictions of this hypothesis for other polyploid systems.
NATURAL ALLOPOLYPLOIDS OF NICOTIANA AND THEIR GENETIC HISTORY
As currently circumscribed, the genus Nicotiana includes 76 species distributed in America, Australia and Africa (Knapp et al., 2004). Phylogenetic reconstructions of plastid and nuclear DNA data have established that the genus Nicotiana is monophyletic and inform understanding of species relationships, patterns of divergence and likely parents of allopolyploid species (Chase et al., 2003; Clarkson et al., 2004; Knapp et al., 2004). The base chromosome number of the genus is n = 12, but more than 30 species (including the entire Australian section Suaveolentes) are allopolyploids, although some species in section Suaveolentes show secondary chromosome number reductions (chromosome numbers range from 32 to 48) (Goodspeed, 1954; Knapp et al., 2004). Most of the presumed relationships between diploids and polyploids have been confirmed using molecular cytogenetics and mapping of repetitive DNA probes (Kenton et al., 1993; Lim et al., 2000b, 2004, 2005, 2006a). Table 1 shows the closest living diploid relatives of the progenitors of Nicotiana allopolyploid species and sections (see also Fig. 1 in Leitch et al., 2008).
Table 1.
Likely ages and diploid progenitors of extant Nicotiana polyploids
| Allopolyploid | Maternal parent* | Paternal parent† | Maximum likely age (years before present) |
|---|---|---|---|
| N. tabacum | N. sylvestris | N. tomentosiformis | <200 000 |
| N. rustica | N. paniculata | N. undulata | <200 000 |
| N. arentsii | N. undulata | N. wigandioides | <200 000 |
| Nicotiana section Polydicliae | N. obtusifolia | N. section Petunioides/N. attenuata‡ | ∼1000 000 |
| Nicotiana section Repandae | N. sylvestris | N. obtusifolia | ∼4500 000 |
| Nicotiana section Suaveolentes | N. sylvestris | Unknown | >10 000 000 |
* Closest living relative of the maternal parent, based on analysis of plastid and nuclear sequence (Clarkson et al., 2004).
† Closest living relative of the paternal parent, based on analysis of plastid and nuclear sequence (Clarkson et al., 2004).
‡ Chase et al. (2003) analysed plastid sequence divergence and reported that one of the progenitors was most likely the progenitor of Nicotiana section Petunioides.
Fig. 1.
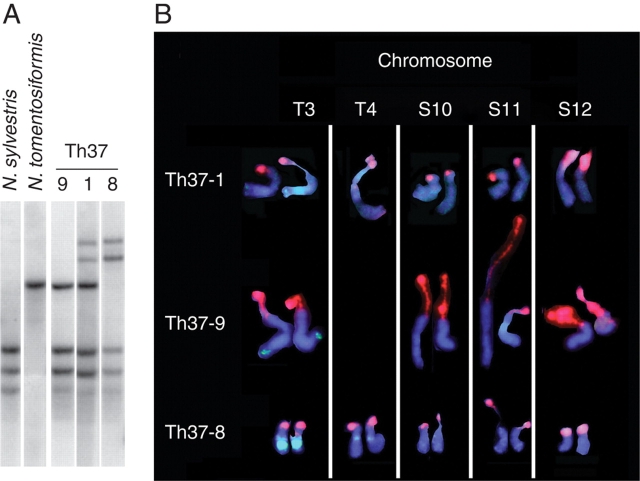
(A) Southern hybridization showing rDNA polymorphisms in three plants (numbers 1, 8 and 9) of the S4 generation of Burk's (1973) synthetic N. tabacum Th37 line and of the diploid species N. sylvestris and N. tomentosiformis. Genomic DNA digested with BstNI and probed with 26S rDNA (from Skalicka et al., 2003). (B) FISH showing the number and distribution of 35S rDNA loci (biotin-labelled, Cy3 detected, red fluorescence) to chromosomes [counterstained blue (DAPI) for DNA] in the same Th37 plants (from Skalicka et al., 2003).
To determine the influence of time on the divergence of rDNA and distribution of loci carrying rDNA (nucleolus organizer regions), it is necessary to estimate the time elapsed since the allopolyploids were formed. To do this, Clarkson et al. (2004) dated the nodes of a phylogenetic tree that included all diploid taxa, allotetraploid N. tabacum and four allotetraploid species of Nicotiana section Repandae. By combining data sets, they were able to generate trees with good bootstrap support. They determined likely maximum ages of all nodes on the tree (cf. Richardson et al., 2001) using calibrations based on ages of oceanic volcanic islands on which there are endemic species of Nicotiana. Section Repandae includes four species, two of which (N. nesophila and N. stocktonii) occur only on the Revillagigedos Islands in the Pacific Ocean (San Benedicto, Socorro and Clarión) far off the coast of Mexico. The oldest of these islands is Clarión, dated at approx. 1·2 million years. Assuming that N. nesophila and N. stocktonii diverged from mainland taxa subsequent to formation of this island, then the maximum age of the node that separates these species from N. repanda on the mainland is approx. 1·2 million years (cf. Clarkson et al., 2004). Likewise, if it is assumed that N. cordifolia formed on the island of Masafuera (Juan Fernández Islands off the coast of Chile in the Pacific Ocean), then dating enables predictions of the maximum likely age of the node that separates N. cordifolia from its sister taxon on the South American mainland, N. solanifolia, at 2·4 million years old (Stuessy et al., 1984). Using these as calibration points, it is possible to estimate the maximum age of allopolyploid groups (cf. Table 1; for more details on the dating procedure, see Clarkson et al., 2005).
Estimates of ages for the allopolyploids enable us to examine trends in evolution of rDNA over time. First we explore the consequence of de novo polyploidy on rDNA structure, organization and activity by reviewing what is known of synthetic hybrids and allopolyploids, the latter made by interspecific hybridization and chromosome doubling. We compare these data with natural polyploids divided into two categories based on age: young natural allopolyploids (<200 000 years old) and ancient allopolyploids (1–5 million years old).
rDNA AND SYNTHETIC HYBRIDS AND ALLOPOLYPLOIDS
Synthetic hybrids and allopolyploids enable us to follow genetic changes occurring in response to de novo allopolyploidy. Any deviation from ‘genetic additivity’ can be considered to be induced by allopolyploidy. In some cases, de novo allopolyploidy is thought to induce a ‘genomic shock’ responsible for the activation of transposons (McClintock, 1984), retrotransposons (Kashkush et al., 2003; Melayah et al., 2004; Petit et al., 2007), genomic translocations and insertions/deletions (Gill, 1991), and epigenetic reprogramming (Kashkush et al., 2002; Levy and Feldman, 2004; Ozkan et al., 2001). In synthetic allopolyploids of N. tabacum (Th37 line, constructed by Burk, 1973), we observed losses of repetitive sequences (including tandem and dispersed repeats and retroelements) predominantly from the paternal genome in early generations (Lim et al., 2004a; Skalicka et al., 2005; Petit et al., 2007). Targeted losses of restriction sites [studied via restriction fragment length polymorphism (RFLP) changes], predominantly in the paternal genome, were previously observed in synthetic allopolyploids of Brassica (Song et al., 1995). However, not all synthetic allopolyploids show such changes, and there was no change in the profile of amplified fragment length polymorphisms (AFLP) in synthetic allopolyploids of Gossypium (Liu et al., 2001).
The first reports of genetic change occurring to rDNA in association with hybrids and allopolyploids in Nicotiana were in N. tabacum × Atropa belladonna hybrids, in which novel rDNA units were observed (Borisjuk et al., 1988). Similarly in an established (1904) horticultural hybrid N. × sanderae (N. alata × N. forgetiana; Goodspeed, 1954), new IGS polymorphisms were observed (Lim et al., 2006a). In the synthetic tobacco line Th37, only three out of 20 plants analysed showed an additive pattern of rDNA RFLPs observed in the parents. In 75 % of plants, the IGS from N. tomentosiformis, accounting for approx. 2000 rDNA units, had been entirely replaced by a novel hybrid-specific rDNA cluster of comparable abundance (Skalicka et al., 2003). All novel rDNA variants were of N. tomentosiformis origin but 1–3 kb longer (Fig. 1A), primarily due to amplifications of the SR II and SR VI repetitive subregions of the IGS (subregions first identified by Volkov et al., 1999). Such structural changes in rDNA units have also been encountered in intergeneric somatic hybrids of N. tabacum × A. belladona (Borisjuk et al., 1988) and interspecific somatic hybrids of Medicago (Fabaceae) (Cluster et al., 1996).
We selected three Th37 synthetic tobacco plants with different 35S rDNA RFLP profiles for cytogenetic analysis. (1) Plant Th37·9 had RFLP bands that were additive for those observed in the parents (Fig. 1A) and had an additive number of 35S rDNA loci (i.e. three loci from the maternal parent N. sylvestris on chromosomes S10, S11 and S12, and one from the paternal parent N. tomentosiformis on chromosome T3; Fig. 1B). (2) Plant Th37·1 had a hybrid-specific cluster of rDNA units (Fig. 1A) and a new rDNA locus on one of the two homologues of chromosome T4 of N. tomentosiformis-origin (Fig. 1B). (3) Plant Th37·8 had lost the paternal N. tomentosiformis-derived rDNA units, but the rDNA sites on both T3 homologues were still present; the units at these sites had been replaced by the new hybrid-specific cluster of rDNA units (Fig. 1A). In addition both homologues of chromosome T4 now carried a new site (Lim et al., 2000a).
Analysis of Th37 synthetic N. tabacum (Skalicka et al., 2003) revealed that: (a) in most plants there was a rapid amplification of 35S rDNA units (in four generations) involving many thousands of genes that were either new or in sub-threshold copy numbers for detection in N. tomentosiformis; (b) amplification of 35S rDNA units first occurred at a new chromosomal locus (on chromosome T4; similar amplification of units at a new locus has also been observed in synthetic allopolyploids of Arabidopsis suecica; Pontes et al., 2004); and (c) new 35S rDNA units in most plants had been transferred to chromosome T3. The mechanism for this last observation could have been (a) sequence homogenization, an ill-defined process involving recombination machinery and perhaps unequal recombination between homologous or non-homologous sequences, saltatory replication and rDNA unit amplification or (b) reciprocal translocations between the rDNA loci on chromosomes T3 and T4, followed by random segregation of chromosomes at meiosis that could lead to the loss of all chromosome T3-derived rDNA units in subsequent generations.
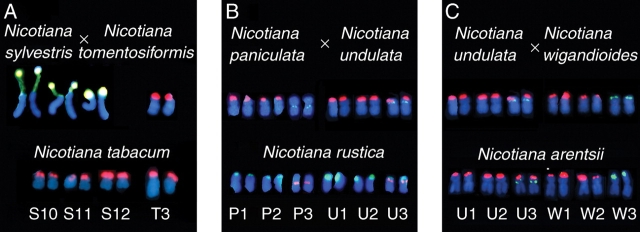
F1 N. sylvestris × N. tomentosiformis hybrids (Lim et al., 2006b) showed unaltered additivity even after duplication of chromosomes, suggesting that interspecific hybridization and chromosome duplication do not always lead to genetic changes. Similar observations were reported by Dadejova et al. (2007) in an analysis of two F1 hybrids, N. paniculata × N. undulata (synthetic N. rustica) and N. undulata × N. wigandioides (synthetic N. arentsii). Perhaps one or more meiotic cycles are needed to promote rapid and extensive reorganization of 35S rDNA units, copy numbers and/or chromosomal distributions.
rDNA AND YOUNG NATURAL ALLOPOLYPLOIDS (<200 000 YEARS OLD)
Three natural allopolyploids are thought to have formed within the last 200 000 years: N. tabacum, N. rustica and N. arentsii. The closest living descendents of the parents are shown in Table 1. Cytogenetic analyses using genomic in situ hybridization (GISH) revealed that each polyploid is 2n = 4x = 48, with 24 chromosomes from each diploid parent (Lim et al., 2004a, 2005). The chromosomes also carry satellite repeats observed in the parental species, and only N. tabacum exhibits intergenomic translocations, some of which are in all cultivars and some cultivar specific (Lim et al., 2004a, 2005). All species also have the sum of the rDNA loci expected from the numbers observed in the parents (Fig. 2). However, the allopolyploids have a decrease in rDNA copy number relative to expectation (based on measurements of rDNA copy numbers from one accession of each parental species) (Lim et al., 2000a; Dadejova et al., 2007). If there had been heteromorphy in the number of 35S rDNA loci in the early ancestors of these species, as is observed in synthetic N. tabacum (see section ‘rDNA in synthetic hybrids and allopolyploids’), it has been lost in modern populations (Kovarik et al., 2004). Likewise, the novel rDNA loci reported in synthetic A. suecica are not observed in natural A. suecica (Pontes et al., 2004). Thus, these synthetic allopolyploids display greater variation in rDNA locus numbers than their natural counterparts. Perhaps early population bottlenecks purged much of the genetic variation induced by de novo allopolyploidy.
Fig. 2.
FISH showing the number and chromosomal distribution of rDNA. (A) Nicotiana sylvestris showing 35S rDNA (yellow fluorescence, from Dadejova et al., 2007); N. tomentosiformis showing 35S rDNA (red fluorescence, from Dadejova et al., 2007); N. tabacum showing 35S rDNA (red fluorescence, new data, biotin labelled 35S rDNA probe, detected with Cy3 fluorescence using methods in Dadejova et al., 2007). (B) Nicotiana paniculata and N. undulata showing 35S (red fluorescence) and 5S (green fluorescence) rDNA, both from Dadejova et al. (2007); N. rustica showing 35S (green fluorescence) and 5S (red fluorescence) rDNA (from Lim et al., 2005). (C) Nicotiana undulata, N. wigandioides and N. arentsii showing 35S (red fluorescence) and 5S (green fluorescence) rDNA [from Dadejova et al. (2007) and Lim et al. (2005)].
RFLP analysis of the structure of the 35S rDNA units in N. tabacum revealed that the pattern of bands does not correspond to that found in extant N. tomentosiformis and N. sylvestris (Kovarik et al., 1997; Lim et al., 2000a), suggesting that N. tabacum has evolved its own distinct gene family. Sequence analysis revealed that the tobacco-specific units arose by reorganization of the parental N. tomentosiformis-inherited units followed by their subsequent amplification and homogenization between rDNA loci (Volkov et al., 1999). The sequence changes mainly involved amplification and reduction of subrepeats upstream and downstream of transcription start sites whereas point mutations were found to be rare.
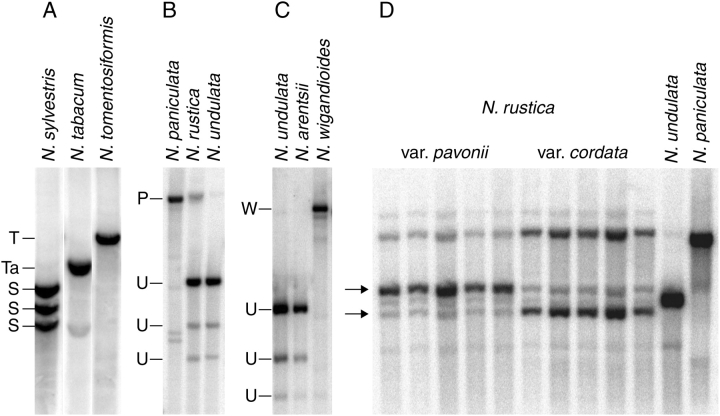
Similarly, N. rustica and N. arentsii have also lost parental units (Fig. 3), although retaining the expected number of rDNA loci. However, the extent of parental gene replacement varied significantly between species. The extent of rDNA sequence homogenization decreases in the order N. arentsii > N. tabacum > N. rustica. Nicotiana rustica still maintains a considerable number of unconverted N. paniculata-origin units. In both N. tabacum and N. rustica upstream IGS subrepeats were fully homogenized, whereas downstream subrepeats were only partly homogenized, enabling IGS to be resolved into several distinctive families (Matyasek et al., 2003; Kovarik et al., 2004). IGS subrepeats may have a role in homogenization, perhaps by promoting or facilitating recombination between units. In Nicotiana, shorter upstream SRII subrepeats (10–23 bp) could undergo recombination more rapidly than the longer SRVI subrepeats (approx. 135 bp), or perhaps they diverge more rapidly. In Drosophila melanogaster the IGS contains topoisomerase II sites that are thought to promote recombination and homogenization. The putative AT-rich topoisomerase II binding element has been cloned from N. tomentosiformis IGS and shown to promote amplification of a linked reporter transgene (Borisjuk et al., 2000).
Fig. 3.
Southern hybridization showing rDNA polymorphisms in polyploid species N. tabacum (A), N. rustica (B, D) and N. arentsii (C) with those from the diploids most closely related to the parents of the allopolyploids. Genomic DNA digested with (A) BstNI and probed with 26S rDNA (Lim et al., 2000a); (B) EcoRV and probed with 18S rDNA (Matyasek et al., 2003); (C) EcoRV and probed with 18S rDNA (Kovarik et al., 2004). (D) EcoRV and probed with 26S rDNA (new data using methods described in Dadejova et al., 2007). Ribosomal DNA units characteristic of a species are indicated: T = N. tomentosiformis, Ta = N. tabacum, S = N. sylvestris, P = N. paniculata, U = N. undulata, W = N. wigandioides. In (D) arrows indicate variety-specific bands corresponding to a major gene family derived from N. undulata. Note that the parental IGS types (U and P) are under-represented in a hybrid.
All N. tabacum cultivars show relatively uniform RFLP profiles, with >90 % of novel hybrid-specific units and <10 % of unconverted N. sylvestris-types of unit (Skalicka et al., 2003). However, a feral tobacco collected in Bolivia had a higher proportion of N. sylvestris-type unit (20 %), indicating that cultivation and breeding may accelerate the action of homogenization (Kovarik et al., 2004) or that cultivated strains were derived from a narrow range of genotypes compared with variation exhibited by plants in South America (truly wild plants of tobacco are unknown – they are always associated with human habitation); certainly we can expect inbreeding to promote a reduction in heterozygosity and fixation of alleles. Dadejova et al. (2007) examined seven varieties of cultivated N. rustica, which contained rDNA units designated as P- and U-units depending on the ancestry from the N. paniculata or N. undulata progenitor (Fig. 3B). All varieties displayed a reduced number of N. paniculata-origin units relative to prediction. However, the proportion of N. paniculata-origin rDNA varied (10–40 %) between, but not within, varieties (Fig. 3D). In addition, analysis of the IGS subregion downstream from the 26S gene showed significant length polymorphism between varieties, probably reflecting IGS rearrangements and further homogenization in the course of variety diversification. Such IGS polymorphisms in crop plants can be potentially exploited for genotyping purposes in breeding programmes.
It may be surprising that molecular reorganization of rDNA units and their amplification within and between rDNA loci are more rapid genetic events than changes in numbers of rDNA loci. Nicotiana is not exceptional; allopolyploids of Gossypium that are several million years old have also maintained the expected rDNA locus number, whereas the parental units at these loci have been overwritten with new, variant sequences (Wendel et al., 1995). However, over millions of years of allopolyploid divergence in Nicotiana, we observe a reduction in rDNA locus number. Thus in cotton, locus number may be a more conserved genetic trait than in Nicotiana (see below). Variation in locus number also seems to accompany evolution of many diploid (Dubcovsky and Dvorak, 1995; Lim et al., 2000b, 2007; Siroky et al., 2001; Dobigny et al., 2003) and even autopolyploid species (Weiss-Schneeweiss et al., 2007). No apparent reduction of locus number was observed in Brassica (Hasterok et al., 2006), recent Nicotiana (Kenton et al., 1993; Moscone et al., 1996; Lim et al., 2000a; Matyasek et al., 2003) and Tragopogon (Kovarik et al., 2005) allopolyploids. These studies indicated that locus number changes are not necessarily associated with the early evolution of allopolyploids.
The presence of activity at any particular rDNA locus, giving rise to nucleolar dominance, can be influenced genetically by the action of homogenization (termed here genetic rDNA dominance) or epigenetically by, for example, cytosine methylation and histone acetylation (termed here epigenetic rDNA dominance). Homogenization of rDNA arrays is probably an ongoing process in many eukaryotes and may function to maintain a high proportion of functional rDNA units (Ohta, 1989). It also leads to the divergence of the IGS, which is not under such strong selective constraints. Nevertheless, proteins that bind to the IGS, including those that function in the regulation of the rDNA unit, must presumably co-evolve with the changing structure of the IGS.
rDNA AND OLD ALLOPOLYPLOIDS (APPROX. 1–5 MILLION YEARS OLD)
Two allopolyploid sections in Nicotiana are considered here: (1) section Polydicliae, comprising N. clevelandii and N. quadrivalvis, estimated to have formed approx. 1 million years ago (from progenitors of diploid N. obtusifolia + N. attenuata); and (2) section Repandae, comprising N. nesophila, N. repanda, N. stocktonii and N. nudicaulis, estimated to have formed approx. 4·5 million years ago (from progenitors of diploid N. sylvestris + N. obtusifolia) (Clarkson et al., 2005; Lim et al., 2007). All these polyploids have the expected chromosome number (2n = 4x = 48). GISH to N. quadrivalvis is effective in that two parental genomes can be resolved using N. obtusifolia and N. attenuata total genomic DNAs as probes (Lim et al., 2007). However, there is considerable mixing of probe labels between genomes, indicating substantial mobility of DNA between chromosome sets after about 1 million years of divergence. Homogenization of retroelements between subgenomes is also revealed over similar time frames in analyses of Gossypium allopolyploids (Zhao et al., 1998). In contrast, GISH to N. nesophila failed, with little DNA labelling by probes of N. sylvestris and N. obtusifolia-origin chromosomes. These data led us to consider that the majority of the repetitive fraction of the genome has been ‘turned over’ and replaced by new or variant sequences over a time frame of approx. 5 million years (Lim et al., 2007).
An analysis of the number of 5S and 35S rDNA loci in species of sections Polydicliae (Fig. 4) and Repandae (Fig. 5) revealed a marked reduction in number from expectation. This reduction results in the total number of sites typical of Nicotiana diploids (i.e. two or three 35S rDNA sites and one 5S rDNA site). These allopolyploid species in sections Polydicliae and Repandae also show unique rDNA RFLPs that differ from diploid parents, indicating that each species has evolved unique rDNA family/ies. Nicotiana nesophila, N. repanda and N. stocktonii possess similar ITS sequences, suggesting their recent diversification from a common allopolyploid ancestor (Chase et al., 2003). However, each species has unique IGSs (Clarkson et al., 2005), indicating either that (a) homogenization of IGS occurs more rapidly than that of ITS, or (b) homogenization is equally rapid, but changes to ITS are selected against more strongly than those to the IGS.
Fig. 4.
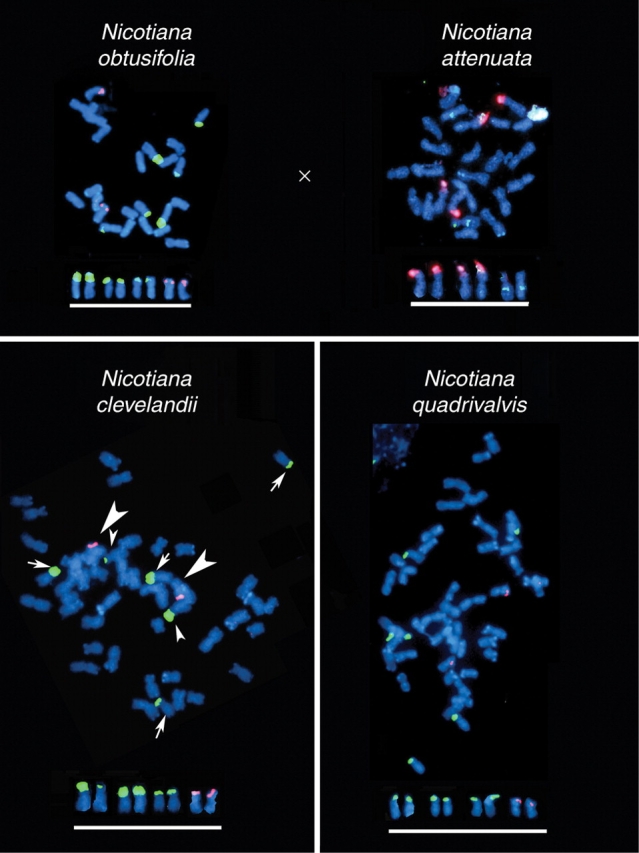
FISH showing the number and distribution of rDNA loci in diploid progenitors (N. obtusifolia and N. attenuata) and derived allopolyploids (N. clevelandii and N. quadrivalvis): N. obtusifolia, N. clevelandii and N. quadrivalvis showing 35S (yellow fluorescence) and 5S (red fluorescence) rDNA. N. attenuata showing 35S (red fluorescence) and 5S rDNA (yellow fluorescence). Data from N. obtusifolia were taken from Clarkson et al. (2005), the remaining data are new [digoxigenin-labelled probes were detected by FITC fluorescence (yellow); biotin-labelled probes with Cy3 fluorescence (red)]. Probes and methods are described in Clarkson et al. (2005). Scale bar = 10 µm (metaphases).
Fig. 5.
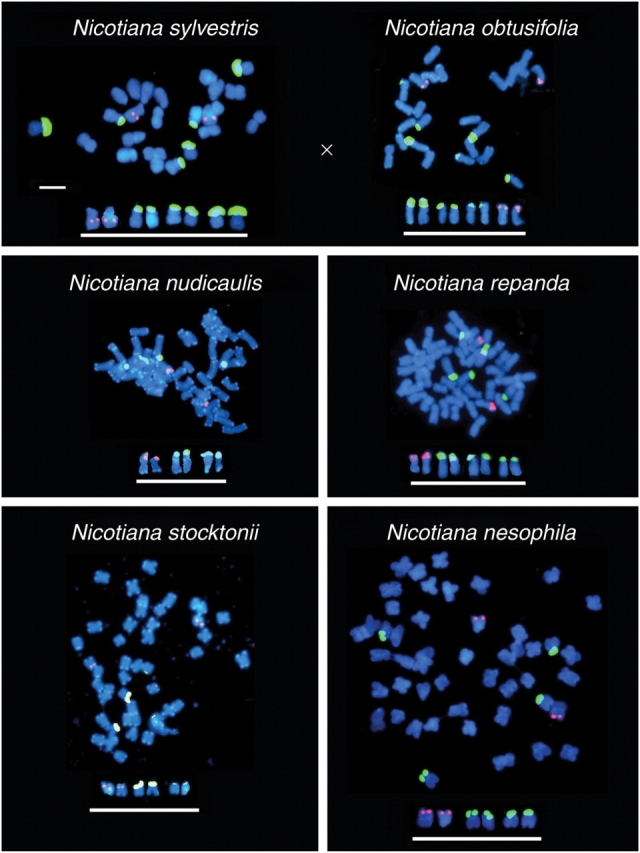
FISH showing the numbers and distributions of 35S (yellow fluorescence) and 5S (red fluorescence) rDNA loci. Data for the diploid progenitor species N. sylvestris and N. obtusifolia are taken from Clarkson et al. (2005) while those from the derived allopolyploid species (N. nudicaulis, N. repanda, N. stocktonii and N. nesophila) are new (using methods described in Clarkson et al., 2005). Scale bar = 10 µm (metaphases).
EPIGENETIC SILENCING AND SEQUENCE HOMOGENIZATION
Epigenetic silencing of rRNA genes, known as nucleolar dominance, has been known for >70 years (Navashin, 1934). The consequence of nucleolar dominance in many polyploids is that one parental gene family is transcriptionally silenced. Previous cytogenetic data in N. tabacum indicated that unconverted parental N. sylvestris-origin units were highly methylated, perhaps located at a locus on chromosome S12 (Fig. 2A) (Lim et al., 2000a) that does not show secondary constrictions at metaphase (a hallmark of genetic inactivity). More recently Dadejova et al. (2007) used RT-PCR to investigate expression of rRNA genes in N. rustica and N. tabacum and observed that units that were not replaced by gene conversion were transcriptionally silenced (i.e. epigenetic rDNA dominance). These authors also showed that there was strong uniparental rDNA silencing in N. paniculata × N. undulata F1 hybrids, whereas N. sylvestris × N. tomentosiformis and N. undulata × N. wigandioides F1 hybrids showed little or no rRNA gene silencing (i.e. co-dominance). Occurrence of both nucleolar dominance and locus co-dominance has been described elsewhere, e.g. in polyploids and hybrids involving wheat (Lacadena et al., 1984; Cermeno et al., 1985).
We propose that nucleolar dominance, established early in allopolyploid formation including F1 hybrids, plays a significant role in the evolution of rDNA. We suggest that epigenetic silencing of rDNA loci makes them less vulnerable to homogenization and more likely to be lost, perhaps thousands or millions of years later. The data from Nicotiana point to a link between epigenetic (nucleolar dominance) factors and genetic outcome (homogenization).
In N. tabacum and N. rustica, rDNA units that are unconverted and inherited little changed genetically from the diploid progenitor are epigenetically silenced. Those units that are active are newly formed hybrid-specific units and carry a clear signature of homogenization in their ancestry (cf. Kovarik et al., 2004).
Unconverted rDNA units in natural polyploids occur outside the nucleolus and contain heterochromatin (Lim et al 2000a; Matyasek et al., 2003).
There is an inverse correlation between the degree of epigenetic silencing of parental rDNA units in F1 hybrids and the proportion of parental units that have been homogenized in natural allopolyploids (Fig. 6).
Fig. 6.
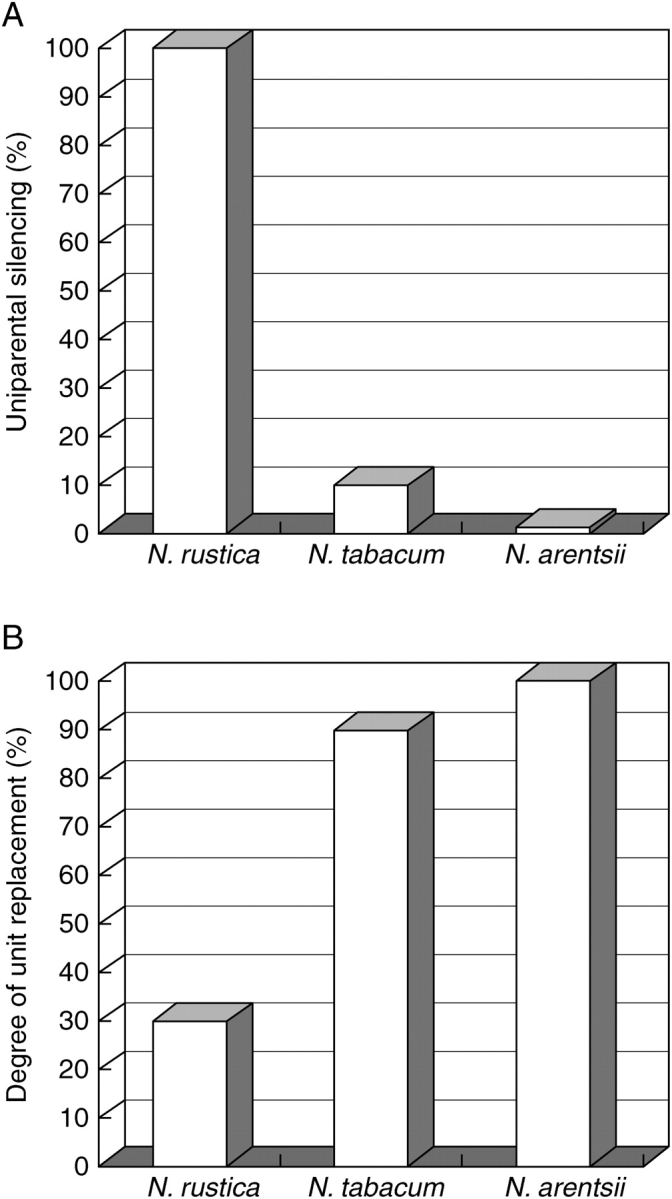
The relationship between epigenetic silencing in synthetic F1 hybrids and retention of parental rRNA genes in natural allopolyploids. Data and further details from Dadejova et al. (2007). (A) Uniparental silencing of rRNA genes in F1 hybrids made from the diploid species that most closely resemble the parents of the allopolyploids N. rustica, N. tabacum and N. arentsii. The percentage of silencing was calculated from the ratio of abundance of each parental rRNA transcript; zero indicates no silencing (codominance), 100 % indicates complete silencing of one parental homoeologue. (B) The proportion of rRNA genes that are replaced with alternative units by intergenomic homogenization (i.e. between parental loci) in natural polyploids. Zero indicates no intergenomic homogenization; 100 % indicates complete intergenomic homogenization. The replacement of parental units by intragenomic homogenization (within the locus) has not been considered. Note: a novel major repeat type based on rearrangements of the IGS has evolved in all allopolyploids (Fig. 3).
In early allopolyploids there may be over-expression of genes or imbalanced gene-product dosages arising from genome duplication. If this applies to rDNA, there are several ways that gene expression patterns can be tuned to meet cellular/organism physiology: (a) physical elimination of genes; (b) epigenetic inactivation of units (epigenetic rDNA dominance); (c) overwriting of one rDNA variant with another – perhaps by a version that will be silenced (genetic rDNA dominance); or (4) a combination of these. Active rRNA genes organized as decondensed chromatin may be most vulnerable to recombination events that can promote both homogenization and altered copy numbers (Kobayashi and Ganley, 2005; Ganley and Kobayashi, 2007). As a consequence, hybrid-specific units may evolve relatively rapidly, colonizing nucleolus organizer regions at all active loci, potentially from both subgenomes of an allopolyploid (depending on whether both are active). Epigenetic inactivation will increase levels of chromatin condensation and heterochromatin, which might act to protect rDNA from genetic recombination. Under this scenario, inactive, non-homogenized rDNA parental units may be transmitted over many generations even in high copy numbers. Recent findings indicate that rDNA may become dispersed in yeast mutants with abnormal methylation of histone proteins, production of siRNAs and factors involved in chromatin condensation (Peng and Karpen, 2007) showing linkage between genetic stability of repeats and epigenetic pathways. In the future it will be worth analysing epigenetic markers of chromatin (e.g. histone H3K9 methylation) that have survived unaltered during homogenization in allopolyploid species.
Homogenization is a powerful process that keeps the tandem arrays uniform, acting to remove non-functional units from the cluster (Ohta, 1989). We can anticipate that silenced rDNA units that are not homogenized would acquire changes leading to loss of function and could potentially be lost over evolutionary time. Certainly no alteration of inactive units would have any selective consequence. Thus, we can predict that the consequences of epigenetic silencing of rDNA loci are: (a) reduced or eliminated interlocus homogenization (and perhaps also intralocus homogenization); (b) divergence of units in the silenced loci, including substitutions that lead to loss of function, indels and loss of array uniformity; and (c) deletion of silenced genes (Fig. 7), perhaps through accumulation of indels, as for ancient retroelements (Ma et al., 2004), or, in those species in which rDNA arrays occupy a subterminal position, via deletions of chromosome termini. If this cascade of events does apply, then we might envisage that the lost rDNA loci in sections Repandae and Polydicliae were epigenetically silenced early in evolution of allopolyploid sections.
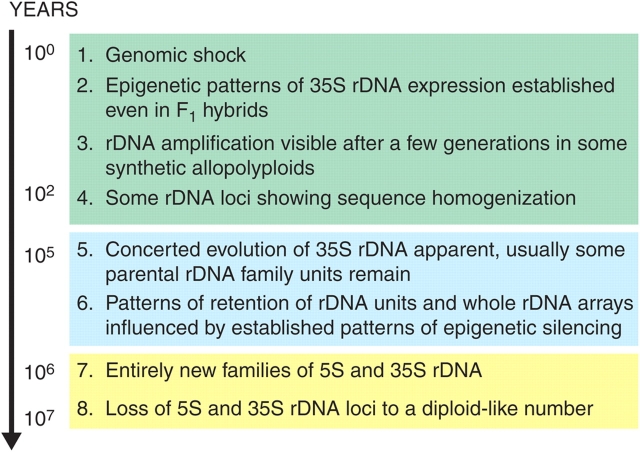
Fig. 7.
Summary of rDNA evolution after the formation of Nicotiana allopolyploids. The background colours separate allopolyploids according to categories established here: green background, de novo polyploids (from synthetic polyploids, few generations); blue background, young polyploids (<200 000 years old); yellow background, old polyploids (1–5 million years old).
ACKNOWLEDGEMENTS
We thank NERC, UK, the grant agency (521/07/0116) and Academy of Sciences (AVOZ50040507) of the Czech Republic for support.
LITERATURE CITED
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell. New York, NY: Garland Science; 2002. [Google Scholar]
- Borisjuk NV, Momot VP, Gleba YY. Novel class of rDNA repeat units in somatic hybrids between Nicotiana and Atropa. Theoretical and Applied Genetics. 1988;76:108–112. doi: 10.1007/BF00288839. [DOI] [PubMed] [Google Scholar]
- Borisjuk N, Borisjuk L, Komarnytsky S, Timeva S, Hemleben V, Gleba Y, et al. Tobacco ribosomal DNA spacer element stimulates amplification and expression of heterologous genes. Nature Biotechnology. 2000;18:1303–1306. doi: 10.1038/82430. [DOI] [PubMed] [Google Scholar]
- Burk LG. Partial self-fertility in a theoretical amphiploid progenitor of N. tabacum. Journal of Heredity. 1973;64:348–350. [Google Scholar]
- Cermeno MC, Orellana J, Santos JL, Lacadena JR. Nucleolar activity and competition (amphiplasty) in the genus Aegilops. Heredity. 1985;53:603–611. [Google Scholar]
- Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko Y, Joseph J, et al. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae) Annals of Botany. 2003;92:107–127. doi: 10.1093/aob/mcg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Molecular Phylogenetics and Evolution. 2004;33:75–90. doi: 10.1016/j.ympev.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytologist. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Cluster PD, Calderini O, Pupilli F, Crea F, Damiani F, Arcioni S. The fate of ribosomal genes in three interspecific somatic hybrids of Medicago sativa: three different outcomes including the rapid amplification of new spacer-length variants. Theoretical and Applied Genetics. 1996;93:801–808. doi: 10.1007/BF00224079. [DOI] [PubMed] [Google Scholar]
- Coen E, Strachan T, Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the Melanogaster species subgroup of Drosophila. Journal of Molecular Biology. 1982;158:17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Dadejova M, Lim KY, Souckova-Skalicka K, Matyasek R, Grandbastien MA, Leitch A, et al. Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids. New Phytologist. 2007;174:658–668. doi: 10.1111/j.1469-8137.2007.02034.x. [DOI] [PubMed] [Google Scholar]
- Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V. Evolution of rRNA gene clusters and telomeric repeats during explosive genome repatterning in Taterillus X (Rodentia, Gerbillinae) Cytogenetic and Genome Research. 2003;103:94–103. doi: 10.1159/000076296. [DOI] [PubMed] [Google Scholar]
- Dover GA, Strachan T, Coen ES. Molecular drive. Science. 1982;218:1069–1069. doi: 10.1126/science.7146895. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Ribosomal-RNA multigene loci — nomads of the Triticeae genomes. Genetics. 1995;140:1367–1377. doi: 10.1093/genetics/140.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley ARD, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Research. 2007;17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill BS. Nucleocytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. In: Sasakuma T, Kinoshita T, editors. Proceedings of the Kihara Memorial Symposium on Cytogenetic Engineering in Wheat: Yokohama, Japan: Kihara Memorial Foundation; 1991. pp. 48–53. [Google Scholar]
- Goodspeed TH. The genus Nicotiana. Waltham, MA: Chronica Botanica Company; 1954. [Google Scholar]
- Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, et al. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Annals of Botany. 2006;97:205–216. doi: 10.1093/aob/mcj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nature Genetics. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Kenton A, Parokonny AS, Gleba YY, Bennett MD. Characterization of the Nicotiana tabacum-L genome by molecular cytogenetics. Molecular and General Genetics. 1993;240:159–169. doi: 10.1007/BF00277053. [DOI] [PubMed] [Google Scholar]
- Knapp S, Chase MW, Clarkson JJ. Nomenclatural changes and a new sectional classification in Nicotiana (Solanaceae) Taxon. 2004;53:73–82. [Google Scholar]
- Kobayashi T, Ganley ARD. Recombination regulation by transcription-induced cohesion dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Koukalova B, Bezdek M, Opatrny Z. Hypermethylation of tobacco heterochromatic loci in response to osmotic stress. Theoretical and Applied Genetics. 1997;95:301–306. [Google Scholar]
- Kovarik A, Matyasek R, Lim KY, Skalicka K, Koukalova B, Knapp S, et al. Concerted evolution of 18–5·8–26S rDNA repeats in Nicotiana allotetraploids. Biological Journal of the Linnean Society. 2004;82:615–625. [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, Lim KY, Sherwood AM, Matyasek R, et al. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics. 2005;169:931–944. doi: 10.1534/genetics.104.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadena JR, Cermeño MC, Orellana J, Santos JL. Evidence for wheat–rye nucleolar competition (amphiplasty) in triticale by silver-staining procedure. Theoretical and Applied Genetics. 1984;67:207–213. doi: 10.1007/BF00317037. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson J, et al. The ups and downs of genome evolution in polyploid species of Nicotiana (Solanaceae) Annals of Botany. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AA, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society. 2004;82:607–613. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Bezdek M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;a 109:161–172. doi: 10.1007/s004120050424. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Lichtenstein CP, Leitch AR. Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma. 2000;b 109:245–258. doi: 10.1007/s004120000074. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biological Journal of the Linnean Society. 2004;a 82:599–606. [Google Scholar]
- Lim KY, Skalicka K, Koukalova B, Volkov RA, Matyasek R, Hemleben V, et al. Dynamic changes in the distribution of a satellite homologous to intergenic 26–18S rDNA spacer in the evolution of Nicotiana. Genetics. 2004;b 166:1935–1946. doi: 10.1534/genetics.166.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Fulnecek J, Leitch AR. Molecular cytogenetics and tandem repeat sequence evolution in the allopolyploid Nicotiana rustica compared with diploid progenitors N. paniculata and N. undulata. Cytogenetic and Genome Research. 2005;109:298–309. doi: 10.1159/000082413. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Chase MW, Knapp S, McCarthy E, et al. Comparative genomics and repetitive sequence divergence in the species of diploid Nicotiana section Alatae. The Plant Journal. 2006;a 48:907–919. doi: 10.1111/j.1365-313X.2006.02930.x. [DOI] [PubMed] [Google Scholar]
- Lim KY, Souckova-Skalicka K, Sarasan V, Clarkson JJ, Chase MW, Kovarik A, et al. A genetic appraisal of a newly synthetic Nicotiana tabacum (Solanaceae) and the Kostoff synthetic tobacco. American Journal of Botany. 2006;b 93:875–883. doi: 10.3732/ajb.93.6.875. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Chase MW, Clarkson JJ, Grandbastien MA, et al. The sequence of events leading to near complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Ma JX, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Research. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyasek R, Lim KY, Kovarik A, Leitch AR. Ribosomal DNA evolution and gene conversion in Nicotiana rustica. Heredity. 2003;91:268–275. doi: 10.1038/sj.hdy.6800333. [DOI] [PubMed] [Google Scholar]
- Melayah D, Lim KY, Bonnivard E, Chalhoub B, De Borne FD, Mhiri C, et al. Distribution of the Tnt1 retrotransposon family in the amphidiploid tobacco (Nicotiana tabacum) and its wild Nicotiana relatives. Biological Journal of the Linnean Society. 2004;82:639–649. [Google Scholar]
- Moscone EA, Matzke MA, Matzke AJ. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma. 1996;105:231–236. [PubMed] [Google Scholar]
- Navashin M. Chromosomal alterations caused by hybridization and their bearing upon general genetic problems. Cytologia. 1934;5:169–203. [Google Scholar]
- Ohta T. The mutational load of a multigene family with uniform members. Genetical Research. 1989;53:141–145. doi: 10.1017/s0016672300028020. [DOI] [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. The Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nature Cell Biology. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M, Lim KY, Julio E, Poncet C, De Borne FD, Kovarik A, et al. Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (N. tabacum) Molecular Genetics and Genomics. 2007;278:1–15. doi: 10.1007/s00438-007-0226-0. [DOI] [PubMed] [Google Scholar]
- Pikaard CS. Nucleolar dominance and silencing of transcription. Trends in Plant Science. 1999;4:478–483. doi: 10.1016/s1360-1385(99)01501-0. [DOI] [PubMed] [Google Scholar]
- Pikaard CS. The epigenetics of nucleolar dominance. Trends in Genetics. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, et al. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proceedings of the National Academy of Sciences of the USA; 2004. pp. 18240–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S, Pikaard CS. rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochemica et Biophysica Acta – Gene Structure and Expression. 2007;1769:383–392. doi: 10.1016/j.bbaexp.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. International Review of Cytology. 2006;a 255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Current Opinion in Cell Biology. 2006;b 18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Richardson JE, Weitz FM, Fay MF, Cronk QCB, Linder HP, Reeves G, et al. Rapid and recent origin of species richness in the Cape flora of South Africa. Nature. 2001;412:181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annual Review of Cell and Developmental Biology. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Siroky J, Lysak MA, Dolezel J, Kejnovsky E, Vyskot B. Heterogeneity of rDNA distribution and genome size in Silene spp. Chromosome Research. 2001;9:387–393. doi: 10.1023/a:1016783501674. [DOI] [PubMed] [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Koukalova B, Leitch AR, Kovarik A. Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. American Journal of Botany. 2003;90:988–996. doi: 10.3732/ajb.90.7.988. [DOI] [PubMed] [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Matzke MA, Leitch AR, Kovařík A. Preferential elimination of repeated DNA sequences from the paternal, N. tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytologist. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Song KM, Lu P, Tang KL, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proceedings of the National Academy of Sciences of the USA; 1995. pp. 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuessy TF, Foland KA, Sutter JF, Silva M. Botanical and geological significance of potassium-argon dates from the Juan Fernandez Islands. Science. 1984;225:49–51. doi: 10.1126/science.225.4657.49. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Molecular Biology and Evolution. 1999;16:311–320. doi: 10.1093/oxfordjournals.molbev.a026112. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Komarova NY, Hemleben V. Ribosomal RNA in plant hybrids: inheritance, rearrangement, expression. Systematics and Biodiversity. 2007;5:1–16. [Google Scholar]
- Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, Mabuchi T, Park J-M, Jang C-G, et al. Chromosomal stasis in diploids contrasts with restructuring in auto- and allopolyploid taxa of Hepatica (Ranunculaceae) New Phytologist. 2007;174:669–682. doi: 10.1111/j.1469-8137.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium); Proceedings of the National Academy of Sciences of the USA; 1995. pp. 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XP, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, et al. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research. 1998;8:479–492. doi: 10.1101/gr.8.5.479. [DOI] [PubMed] [Google Scholar]