Abstract
Background
Supernumerary B chromosomes (Bs) are a major source of intraspecific variation in nuclear DNA amounts in numerous species of plants. They favour large genomes, and create polymorphisms for DNA variation in natural populations. By studying Bs we can gain useful knowledge about the organization, function and evolution of genomes. There are also significant biological questions concerning the origin and structural organization of Bs, and the way in which these selfish elements can establish themselves by exploiting the replicative machinery of their host genome nucleus.
Scope
It is a sine qua non that Bs originate from the A chromosomes, in a variety of ways. We can study their modes of drive and ask how it is that chromosomes which apparently lack genes can have control over their own drive process which leads to their survival in natural populations. Molecular cytogenetic studies are opening up new avenues of investigation. Population equilibria for B frequencies are determined by a balance between accumulation and harmful effects. Bs are also subject to meiotic loss due to polysomy and to elimination at meiosis as univalents. These balancing forces can be seen in the context of host/parasite interaction, based on a dissection of the genetic elements in both As and Bs (in maize) which interact to bring about a stable equilibrium, at least for a snapshot in time.
Conclusions
Aside from their intrinsic enigmatic properties, B chromosomes make useful experimental tools to study genome organization. Thus far they have not been exploited for their applications, other than through the use of A-B translocations used for gene mapping in maize; but there are opportunities to use them to modulate the frequency and distribution of recombination, to diploidize allopolyploids, to study centromeres and to be developed as plant artificial chromosomes; given that they can be structurally modified and their inheritance stabilized.
Key words: B chromosomes, DNA polymorphisms, host/parasite interaction, mitotic/meiotic drive, applications, genome organization/evolution, centromeres
INTRODUCTION
Supernumerary B chromosomes (Bs) were first discovered by E. B. Wilson in the leaf-footed plant bug insect Metapodius (now called Acanthocephal) a century ago (Wilson, 1907a,b), and many of their distinctive features were described at that time. In the plant world they first came to notice in rye and in maize in the 1920s. In rye (Secale cereale, 2n = 2x = 14 +Bs) their supernumerary nature was clearly defined by Gotoh (1924), who named them k-chromosomes, to distinguish them from the l-chromosomes of the basic A chromosome complement. Shortly afterwards they were reported in maize (Zea mays, 2n = 2x = 20 + Bs) by Kuwada (1925) and Longley (1927). Longley named them ‘supernumerary chromosomes’, and then Randolph (1928) later classified them as ‘B chromosomes’. Bs are now known in well over one and a half thousand species of plants, and their properties have been well documented (Jones and Rees, 1982; Jones, 1995; Puertas, 2002; Jones and Houben, 2003; Camacho, 2004; Burt and Trivers, 2006).
The main diagnostic features of Bs are that they are absent from some individuals of a population, and therefore supernumerary, and they fail to pair with the A chromosomes (As) at meiosis. In some cases they have mechanisms of meiotic (Lilium callosum) or mitotic drive (rye and maize), or no known mechanism of accumulation (Centaurea scabiosa), and in other cases they are consistently univalent yet still survive in populations. One of the big enigmas of Bs is the near impossibility to show any selective advantage that can explain their widespread natural polymorphisms. Selfish drive, where it occurs, can help to explain population polymorphisms, but where it does not we are left with an enigma. Bs have manifold effects upon all measurable aspects of the nuclear phenotype as well as on the whole plant and, in general, it is a truism that in higher numbers they are deleterious, especially to fertility.
In the present context, we are especially interested in the way in which they contribute to intraspecific variation in nuclear DNA amounts, as well as to the qualities of the additional DNA which they contribute. We need to ask again do these Bs carry any genes, are they transcriptionally silent or are they just gene-empty and a drag on the metabolic activity of the nucleus.
The present paper is dedicated to a century of B chromosomes, in the context of intraspecific variation in nuclear DNA in plants. The thought line leads us to consider their significance as a component of the genome, and to present some thoughts on what lessons we can learn from them more generally about certain issues in plant cytogenetics. In dealing with these questions we will, of necessity, focus our discussion on the three species where most of the molecular biology of Bs is currently taking place, namely maize, rye, and Brachycome dichromosomatica. What we know about these three species will give us clues about what we might expect to find in many other species as well, in the fullness of time. The vast majority have been left behind in a time domain of classical studies in population cytogenetics, with a sign saying revisit later with new ideas and new investigative tools.
POLYMORPHISMS FOR NUCLEAR DNA VALUES
Bs are widespread in natural populations of species in which they occur. In rye, for instance, they can be found in every region where the species grows in the wild or under semi-wild conditions (Jones and Puertas, 1993), and likewise in maize (Longley, 1938; McClintock et al., 1981) and in many other cases; the exclusions being from cultivars, and from inbreeders where they are totally absent.
The distribution of Bs among different groups of angiosperms is not random: there is considerable heterogeneity between different groups, with hot spots of occurrence; and their presence is correlated with genome size. Based on a survey of 979 species with Bs there is a large disparity between their presence in monocots (8·0 %) and eudicots (3·0 %). Within the monocots the heterogeneity is striking: there are Bs in 27·2 % of species in the Commelinales, in contrast with the sister order Zingiberales with only 4·3 %. There is also much more variability in B-frequency among monocot orders than among eudicots, and they are rare or absent among non-monocot basal angiosperms (Nymphaeaceae, Magnoliales, Laurales). Heterogeneity also extends to families: the two families with the largest number of +B species are the highly speciose Poaceae and Asteraceae; and there are hot spots of occurrence in Liliales and Commelinales (Levin et al., 2005). There is virtually no difference in frequency between diploid and polyploids (Jones and Rees, 1982; Palestis et al., 2004; Trivers et al., 2004), but there is trend suggesting that Bs have a higher frequency in families with a large genome size (Trivers et al., 2004). It is not clear why large genomes should favour the presence of Bs over their absence, although we can speculate that larger amounts of noncoding DNA may create a more conducive, or more tolerant, environment for the origin of Bs, which themselves share overall similarity with As, as in rye, except for the B-specific terminal region (Timmis et al., 1975; Tsujimoto and Niwa, 1992; Wilkes et al., 1995; Houben et al., 1996). There are no reports of Bs in arabidopsis, but they have been found in rice (Cheng et al., 2000).
Notwithstanding the wealth of data we now have about the distribution of Bs, and at the same time our lack of understanding of these significant issues in plant systematics, the fact remains that where B do occur they impact strongly upon nuclear DNA values and all of the consequences that follow from that for individuals and for natural populations. The genome of rye has a 1C nuclear DNA value for its A chromosome set of 8·28 pg = 8114 Mbp, and a single B has a value of 800 Mbp, which is four times the genome size of arabidopsis. When we consider that natural populations of rye carry mostly 2 and 4 Bs we begin to appreciate the extent and the range of polymorphism for nuclear DNA due to these supernumerary elements (Jones, 1976). The mean B frequency of +B plants in a number of populations of rye ranges from 6·6 % to 54·0 % (Jones and Rees, 1982) and the additional DNA in a rye plant with 4Bs is a massive 3200 Mbp. The B in maize accounts for approx. 4 % of the total chromosome volume and there are correlations between the number of Bs and the size of heterochromatic knobs, which can mask the contribution the Bs to make total genome size (Rosato et al., 1998). Genome size databases do not normally indicate the existence of B chromosomes.
In most species, Bs are found in low numbers (0–4,5) in natural populations. Examples of high numbers are Silene maritima (0–15), Brachycome lineariloba (0–22), and Allium schoenoprasum. As many as 34 Bs have been recorded in Zea mays, in experimental material (for references, see Jones and Rees, 1982). As far as the size of Bs is concerned there is no known species in which the Bs exceed the size of the largest of the A chromosomes, and only a few cases where they are equal to and indistinguishable from the As at mitosis as, for example, in Clarkia elegans (Lewis, 1951), Sorghum nitidum (Raman and Krishnaswami, 1960) and Rumex thysiflorus (Zuk, 1969). At the other extreme, examples of small micro Bs are Hypochoeris maculata (Parker, 1976) and Campanula rotundifolia (Böcher, 1960).
HOW TO BE SUPERNUMERARY
What Bs tell us about intraspecific variation in nuclear DNA amount is that, in some species at least, part of the genome may ‘escape’ and establish its own existence with its own rules of heredity and evolution. In this sense to be supernumerary has advantages, albeit at the expense to the host genome from which it arose. To share the nucleus, together with its mechanisms of division, and then to have mainly negative effects upon nuclear physiology and the phenotype, requires the means of survival against the gradient of these negative effects; since we cannot as yet advance any convincing arguments that Bs have any selective advantages, except under certain severely drastic experimental conditions (Rees and Hutchinson, 1973; Holmes and Bougourd, 1991). In addition there has to be, by definition, an absolute barrier to recombination between A and B chromosomes. Neither can it be demonstrated that they contribute any genes [leaving aside RNA genes in some species, e.g. Crepis capillaris (Leach et al., 2005)], other than those elements needed for their own selfish survival in rye and in maize, and if they did there could be a greater conflict than that which already exists. Notwithstanding this comment there are reports of some newly discovered putative B-located genes in mammals, namely CCT6B, FHIT and a hypothetical XP transcript in the yellow-necked mouse Apodemus flavicollis (Tanic et al., 2005), and the C-KIT oncogene in the red fox, Vulpes vulpes (Graphodatsky et al., 2005).
What does it take to be supernumerary and to survive, and what conflicts does this generate?
HOW TO ‘ESCAPE’ THE GENOME
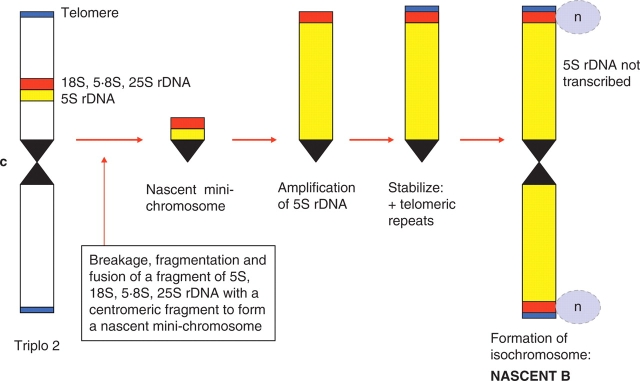
Nobody really knows where B chromosomes come from, other than to say that they originate from the As in some way, although there are some plausible ideas and models (Jamilena et al., 1994; Dhar et al., 2002; Berdnikov et al., 2003; Jones and Houben, 2003). The most convincing case is that for the fully documented origin of a nascent B in Plantago lagopus (2n = 2x = 12) (Dhar et al., 2002). The story began with finding a spontaneous trisomic of chromosome 2 in 1984, and then tracking it through several generations. The extra chromosome went through a number of rapid structural changes, including the formation of a ring chromosome, and finally stabilized as a heterochromatic isochromosome with features of a B. It showed preferential transmission, absence of any phenotypic effects, had a functional centromere and did not pair with any chromosomes of the standard complement. The sequence of events leading to the origin of this apparent B were unravelled by monitoring its life history over several generation and then charcaterizing it by FISH with several probes. It was ‘born’ by the massive amplification of 5S rDNA, as a component of a nascent mini-chromosome which included a centromere (Fig. 1). Telomeres were added de novo, which is known to happen, but it is not known how the nascent B undergoes preferential transmission.
Fig. 1.
Diagrammatic representation of the possible mode of origin of an apparent B chromosome from triplo 2 in Plantago lagopus, based on Dhar et al. (2002). c, Centromere.
Bs could also escape as small centric fragments following unequal translocation and a reduction in chromosome number to give a new species, as we speculate may have happened during the evolution of Crepis fuliginosa (2n = 2x = 8 + Bs) from C. neglecta (2n = 2x = 6) (Jones and Rees, 1982), and as proposed following an aneuploid reduction process in Haplopappus gracilis (2n = 2x = 4 + Bs) (Jackson, 1960).
Genomic rearrangements following interspecific hybridization offer another opportunity for supernumeraries to arise, e.g. the derivatives Coix gigantea (2n = 2x = 20) hybridizing naturally with C. aquatica (2n = 2x = 10). Coix gigantea has four pairs of small chromosomes, about the same size as those of C. aquatica. In the derivatives of hybrids, one or two of these small chromosomes appeared as alien extras in the genome of C. aquatica, giving plants with 2n = 11, and various other hybrid combinations. In plants where the additional small chromosome did not pair with the As of the C. aquatica genome it showed meiotic behaviour typical of a single univalent B as found in many +B species, and lacked obvious phenotypic effects. This single B could also undergo centromere misdivision to give smaller heterochromatic fragment chromosomes. In the absence of cytological observations in these hybrid derivatives the various forms of these additional chromosomes found in population samples of C. aquatica would almost certainly be taken to be B chromosomes (Sapre and Deshpande, 1987).
In Brachycome dichromosomatica the Bs are a conglomerate of mainly tandem repeat sequences derived from different A chromosome sites, and could not therefore have originated by a single excision of an A fragment (Houben et al. 2001). We propose instead that B-founder sequences were ‘released’ from a polymorphic A chromosome region, and were then stabilized by the addition of other sequences such as extrachromosomal DNA (eccDNA) and sequences necessary for their function as chromosomes (e.g. telomeric and centromeric sequences). Indeed, it should be noted that Bs contain similar types of coding and non-coding repeats as found in eccDNA of various organisms (Cohen et al., 2003) and eccDNA with similarity to tandem repeat sequences shared by A and B chromosomes has recently been identified (S. Cohen, A. Houben and D. Segal, unpubl. res.).
The composition of the nascent B would effectively prevent meiotic pairing with any of the As, and licence it to begin its own evolutionary pathway. Supernumerary A chromosome segments in B. dichromosomatica could also serve as potential regions to ‘donate’ founder sequences (Houben et al., 2001), but where the centromere would come from remains an open question, although their rare de novo formation is possible (Nasuda et al., 2005). In such a dilemma, we fall back on some epigenetic event to induce its activity. The Bs in maize (Page et al., 2001; Cheng and Lin, 2003) and rye (Wilkes et al., 1995) also share many sequences with the As, and a mode of origin similar to that proposed for the micro-B of B. dichromosomatica is the best guess (Jones and Houben, 2003).
Bs thus tell us a story about how a supernumerary chromosome can arise and create new and autonomous elements as components of the genome: (a) remodel a trisomic, starting with a small centric fragment, add other repetitive sequences to give stability and isolation from recombination, and be genetically silent – which seems to be the case in all plants studied to date; (b) arise as a small centric fragment following an unequal translocation and reduction in chromosome number; (c) arise as a by-product of interspecific hybridization; or (d) excise as a small fragment, and then recruit sequences, including a centromere to enable passage through the cell cycle, and a telomere to stabilize and protect the ends of the new fragile B.
No doubt there are many such events going continuously, resulting from errors in meiosis, hybridization, genome restructuring and other unknown processes, the products of which are aborted and never mature as B chromosomes. In any event the origin of a B is a rare event, because in species that we know well they appear to have a monophyletic origin, based on their sequence similarity across a range of cytodemes, as in Brachycome dichromosomatica (Houben et al., 1999) and their virtually invariant cytological form over a broad range of geographic regions (rye; Jones and Puertas, 1993). Rare or not, herein lies the potential for one major source of the origin of intra-specific DNA variation.
HOW TO SURVIVE
To be born as a by-product of genome reorganization is one thing, to survive the event and secure a future in the nucleus, with all the physiological disturbance this is likely to cause the host nucleus, is another story. We have already touched on the first priority, which is ‘divorce’ from the rest of the genome by meiotic isolation, i.e. lack of pairing and no adultery by recombination with the A chromosomes – of mutual necessity. Homology distinctiveness can be due to several features of a nascent B: (a) sequences may be common with the As, but not necessarily in register; (b) size difference; (c) epigenetics differences in the state of chromatin, and differences in replication timing, as in Brachycome dichromosomatica (Houben et al., 1997; Marschner et al., 2007); (d) B-specific sequences, as in rye (Langdon et al., 2000); (e) nuclear disposition, whereby small Bs are often isolated at the periphery of the nucleus in prophase of meiosis.
Given that a newly formed B may well be present as a ‘single’, without a pairing partner, how can it survive meiosis without simply being excluded from the nucleus at the end of anaphase I or II? It seems that it can do it by dividing at anaphase I (AI) and then suffering some loss at anaphase II (AII). Where there is a pairing partner, or polysomy with several Bs, including structural variants, the pairing is not as regular as for the disomic As, but nonetheless Bs will transmit into the gametophytes. In many cases Bs do not pair at all, and have the capacity to pass though meiosis as univalents, as in Centaurea scabiosa, and still be maintained in populations, by means unknown (for details on meiosis see Jones and Rees, 1982; Jones, 1995). Loss at meiosis, as well as various detrimental effects on the phenotype, can be overcome to achieve a population equilibrium by various mechanisms of mitotic and meiotic drive (Jones, 1991a, 1995).
In Lilium callosum, B transmission through the pollen is normal/Mendelian, and there is drive at female meiosis based on spindle asymmetry. Single Bs lie outside the metaphase plate and with a preference for the micropylar end of the spindle rather than the chalazal end; they pass to the micropylar pole of the egg mother cells in about 80 % of meioses, and then into the egg (Kayano, 1957). In this case, we may be looking an opportunistic positional effect without any obvious active process being involved. Preferential meiotic segregation also occurs for the Bs in Phleum nodosum (Fröst, 1969) and Plantago serraria (Fröst, 1959) in a similar way. The situation in P. serraria is interesting in that the Bs move to the poles at anaphase I (AI) ahead of the paired As, implying a difference in centromere/spindle microtubule interaction between As and the non-chiasmate Bs in the same cell.
The more common drive process for Bs in plants, especially in the Gramineae, is that based on directed nondisjunction in the gametophyte phase of the life cycle. In rye it takes place both at first pollen mitosis and at first egg cell mitosis (rye is unique in this respect), and in maize at the second pollen mitosis, and in other situations in various other species (Jones and Rees, 1982). We will use rye (pollen) and maize as our models, since these two cases offer the best information on this significant biological question concerning centromeres, spindle microtubules, and genetic control processes that cause direct nondisjunction to take place in specific developmental stages.
Directed nondisjunction of Bs in rye pollen
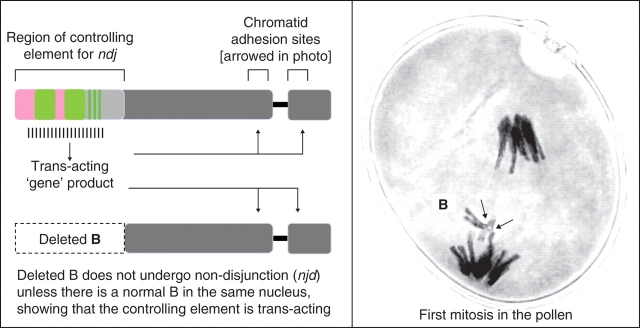
The phenomenon was first described cytologically by Hasegawa in 1934, for the standard B. It was later shown that a variant iso-chromosome for the long arm of the B has the same capacity for directed nondisjunction as the standard-B (shown in Fig. 2), but that a small iso-B for the short arm, which lacks the terminal region of the long arm, does not (Müntzing, 1945, 1946, 1948). In his 1946 paper Müntzing demonstrated that in the standard-B, as well as in the large and small isos, the centromeres divided normally at anaphase of the pollen mitosis, but that in the standard and large iso there are sticking sites on either side of the centromere which prevent normal anaphase separation of the chromatids and thereby cause the nondisjunction of these two types. Müntzing found the different behaviour of the small iso to be ‘inexplicable’; but following a detailed pachytene analysis of the deficiency-B, Müntzing and Lima-de-Faria (1952) realized that the sticking regions are actually present in both the deficiency-B and the small iso-B, but are not active, and that the reason for their failure to nondisjoin could be the absence of the terminal part of the long arm, which might supply some vital function to which the sticking sites are sensitive. Lima-de-Faria (1962) later devised an experiment in which he made crosses to place a deficiency-B and a standard-B together in the same plant, and then proved that the terminal half of the long arm of the standard-B interacted with the sticking regions of both chromosomes, causing the deficiency as well as the standard-B to nondisjoin. The sensitive sticking regions adjacent to the centromere are thus controlled by a trans-acting element/gene located in the distal half of the long arm of the standard-B (and long iso-B), as illustrated in Fig. 2.
Fig. 2.
Genetic organization and nondisjunction properties of the rye B chromosome. The B-specific E3900 sequences are shown in pink and the D1100 in green (based on Jones and Pašakinskienė, 2005).
The region which carries the element controlling nondisjunction is comprised of a concentration of B-specific sequences from two families, E3900 and D1100, assembled from a variety of repetitive elements, some which are also represented in the A genome (Sandery et al., 1990; Blunden et al., 1993; Houben et al., 1996; Langdon et al., 2000). No genes have been found in the region, which begs the question about what genetic process controls nondisjunction, since its occurrence happens with very high frequency (Matthews and Jones, 1983; Ortiz et al., 1996).
As far as we can speculate from cytological studies, the B centromeres act in the normal way at anaphase of the first pollen mitosis, and can be seen to be separated and pulling to opposite poles (Fig. 2). The B chromatids appear to be transiently held together at sensitive sticking sites (receptors) on either side of the centromere, and since the spindle is asymmetrical, the equator is closer to the pole which will passively (?) include the B chromatids in the generative nucleus. The question to be answered is how the B-specific region signals the receptors to remain conjoined just long enough to facilitate directed nondisjunction? This is a fundamental question in terms of genome evolution, since the mechanism had to arise de novo, and then be rapidly established in a highly conserved way to allow the rye Bs to survive following their origin in many diverse populations. Further more, the autonomous nature of drive in rye B was clearly demonstrated by Lindström (1965), who showed that this enigmatic chromosome behaved in just the same way in the pollen of wheat as it does in rye, although its pairing properties at meiosis in this alien environment are compromised. The same thing happens when the B of S. cereale is transferred to S. vavilovii (Puertas et al., 1985).
Mitotic drive in maize
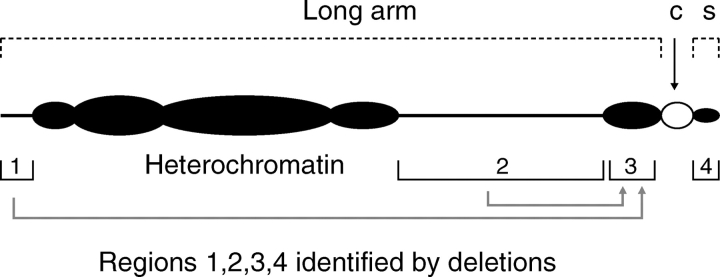
The Bs of maize undergo nondisjunction at the second pollen grain mitosis, with the B-containing sperm enjoying an approx. 70 % advantage in fertilizing the egg (Roman, 1947, 1948; Carlson, 1969, 1978, 1986; González-Sánchez et al., 2003). It is this selective fertilization which constitutes the drive, and which raises questions about how the egg selects the +B sperm? One possible reason to explain preferential fertilization is that the Bs are positioned differently in the sperm nuclei to the As, being located at the tip; this was established by FISH using the 157-bp B-specific ZmB satellite repeat sequence (Shi et al., 1966; Rusche et al., 1997), but it is not known how this positional effect confers the advantage. A–B translocations and deletion derivates have been used to dissect the B into various sized pieces, and to evaluate the contribution of different regions of its long arm, and of its centromere region, in control of the nondisjunction process. The sticking region for nondisjunction is the heterochromatic region 3 adjacent to the centromere (Fig. 3); and it is also known that only a small fraction of the B-specific ZmB repeat sequence, approx. 700 kb domain, is needed to interact with the centromeric histone H3 variant CenH3 to provide for the function of the B centromere (Jin et al., 2005). Recent work confirms the trans acting nature of region 1 in the long arm; since, when the B centromeric region is separated from the tip of the long arm, through a translocation event, the B–A chromosome containing the B centromere retains the ability to non-disjoin, but only if the reciprocal A–B chromosome containing the B long arm is present in the same cell (Carlson, 1978; Lamb et al., 2006). It is also noteworthy, in relation to the sensitive sticking region 3, that centromere function and nondisjunction are independent components of the B chromosome accumulation mechanism, since an epigenetically silenced B centromere (of a dicentric B–A translocation chromosome) still shows the property of nondisjunction (Han et al., 2007). The B of maize is thus providing us with an invaluable insight into aspects of the genome concerning the mechanism of nondisjunction, as well as centromere organization and function.
Fig. 3.
Diagram of the structure of the maize B in relation to nondisjunction. Regions 1 and 2 are trans acting, and are thought to signal the sensitive sticking sites of essential region 3. Deletion of regions 1 or 2 causes complete elimination of nondisjunction. Region 4 modifies the rate of nondisjunction, but its loss does not eliminate it. c, Centromere; s, short arm. (Based on Carlson, 1986.).
Host/parasite interaction
B chromosomes are the ultimate genome parasites, occupying the nucleus of their host and exploiting all of the nuclear machinery needed for their replication and transmission although, in some cases, they have a different replicative chronology to that of the A chromosomes (Marschner et al., 2007). They differ from transposons, and other forms of selfish DNA, in that they are autonomous elements, and can vary their transmission rate, their number within individual plants and their frequency in natural populations. They have the capacity to spread themselves and to optimize their survival strategy, but within certain constraints imposed upon them by their hosts.
The B in rye has a powerful drive based on nondisjunction through both the male and female, and simulation studies (Matthews and Jones, 1982, 1983) predicted that this drive is strong enough to overcome their negative effects upon plant vigour and fertility, and that the main factor enabling variation in population equilibria for B-frequency is the level of pairing of Bs at meiosis, as bivalents or multivalents. When unpaired, as univalents, they tend to divide at AI and then to be eliminated at AII as micronuclei. Confirmation of the predicted variation in pairing levels was forthcoming when selection for high (H) and low (L) transmission genotypes was found be based on the level of B pairing at MI in lines of Korean rye (Jiménez et al., 1997). It was found that 2B plants of the L line form bivalents in only 20 % of MI cells, whereas in the H line there are more than 90 % of bivalents. Puertas et al. (1998, 2000) later proposed that what they called the ‘genes’ for transmission rate are actually the sites of chiasma formation, or the binding sites, in the Bs themselves. The corollary to this proposition is that the Bs in rye modulate there own transmission rates and, therefore, their population equilibrium frequencies and the nuclear DNA polymorphisms in the populations that carry them.
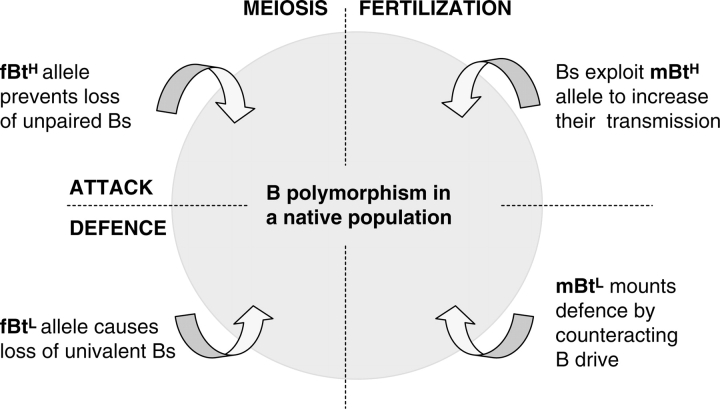
The behaviour of the Bs in maize is more complex than that in rye, in terms of host/parasite interactions, since the A genome of maize has some influence over the transmission rate of the Bs (as may well evolve in rye in the fullness of time). It has been known for some time that the nondisjunction of the maize Bs at the second mitosis of the pollen is a property of the Bs themselves (Lin, 1978), but preferential fertilization of the egg by the B-carrying sperm is the critical factor in modulating B-drive, and this is controlled by a gene(s) in the A chromosomes (González-Sánchez et al., 2003; Fig. 4). There is thus a conflict, and a co-evolution, between the A genome and its parasitic Bs, and as in rye this generates a polymorphism for nuclear DNA amounts in native maize populations.
Fig. 4.
Diagram showing a polymorphic system of attack and defence between A and B chromosomes in maize. MEIOSIS: fBtL, dominant allele for female low transmission, by promoting meiotic loss of unpaired Bs; fBtH, recessive allele for female high transmission, by preventing the loss of unpaired Bs at meiosis. FERTILIZATION: mBtH, allele for high transmission, by promoting preferential fertilization of the egg by +B sperm; mBtL, allele for low transmission by causing random fertilization by 0B and +B sperm. (Based on González-Sánchez et al., 2003.).
POTENTIAL APPLICATIONS
B chromosomes offer the potential for modifying and exploring the A genomes of their host species. They have utility in mapping the A genome, in maize, modulating recombination, exploring the structure of the centromere and the process on nondisjunction, as well as other aspects of genome evolution, as already discussed. They also have some applications in crop plants (Jones, 1991).
Genetic mapping in maize
Roman (1947) first used translocations to study the process of nondisjunction in the pollen of maize, and since that time A–B translocations have found great utility in gene mapping in the maize A chromosome complement (Beckett, 1991; Birchler, 1991).
Dissecting the maize centromere
Bs have also been widely used as an model system to investigate centromere organization. This is possible because when a B is univalent it often undergoes misdivision of its centromere at AI of meiosis, and produces misdivision products of varying sizes (Kaszás and Birchler, 1998). Such studies have shown that the centromere has a vast excess of sequences over that which is required for normal transmission. Jin et al. (2005) later undertook molecular and function dissection of the maize B and found that its functional boundary mapped to a relatively small CentC (satellite repeat) and CRM (centromere retrotransposons in maize)-rich region embedded with the megabases of the ZmBs repeat. Probes for CentC and CRM have also shown that these sequences from the A centromeres are present throughout the whole length of the B, as well as at its own centromeres, and these experiments have demonstrated that these DNA sequences on their own are not sufficient to mark the centromere; the CenH3 histone must also be present (Lamb et al., 2005).
Modulation of recombination in the A genome
Work in rye was instrumental in showing that Bs could alter the pattern of distribution of chiasmata in the A chromosomes (Jones and Rees, 1967), and Moss (1966) had early shown a greater variability among the progenies of plants with Bs than of those without. Ayonoadu and Rees (1968) later found the same effect in maize, and at about the same time Rhoades (1968) found genetic evidence for changes in recombination due the Bs, and much more evidence from maize followed later, as well as for other species (see Jones and Rees, 1982). Despite the potential significance of these basic studies we have yet to utilize this knowledge as a component in crop improvement.
Diploidization in allopolyploids
It was pure serendipity to discover that Bs could provide a diploidizing function in tetraploid hybrids between Lolium perenne × L. temulentum (2n = 4x = 28) (Evans and Macefield, 1972, 1973). The Bs were contributed to the diploid hybrid by L. perenne, and had a marked effect on reducing chiasma frequency in pollen mother cells. Following chromosome doubling, the majority of the chromosomes were in homologous pairs, as bivalents, whereas in the tetraploids without Bs there was both homologous and homoeologous pairing. A similar effect occurs in hybrids between wheat and other Triticeae (reviewed in Jenkins and Jones, 2004) but, as with Lolium, the difficulty in utilizing these pairing control effects is not being able to stabilize the inheritance of the B chromosomes.
Plant artificial chromosome
There is potential to modify the B chromosome of candidate species, such as maize and rye, to construct a stably inherited plant artificial chromosome to carry transgenes which would be outside the domain of the A chromosome genome, with all the advantages that this could entail. Birchler and colleagues have recently described how telomere-mediated chromosomal truncation could be used to down-size maize chromosomes, and to build in sites for foreign gene integration (Yu et al., 2006).
LITERATURE CITED
- Ayonoadu U, Rees H. The influence of B chromosomes on chiasma frequencies in Black Mexican sweet corn. Genetica. 1968;39:75–81. [Google Scholar]
- Beckett JB. Cytogenetic, genetic and plant breeding applications of B-A translocations in maize. In: Gupta PK, Tsuchiya T, editors. Chromosome engineering in plants: genetics, breeding, evolution. Amsterdam: Elsevier; 1991. pp. 493–529. [Google Scholar]
- Berdnikov VA, Gorel FL, Kosterin OE, Bogdanov VS. Tertiary trisomics in the garden pea as a model of B chromosome evolution in plants. Heredity. 2003;91:577–583. doi: 10.1038/sj.hdy.6800357. [DOI] [PubMed] [Google Scholar]
- Birchler JA. Chromosome manipulations in maize. In: Gupta PK, Tsuchiya T, editors. Chromosome engineering in plants: genetics, breeding, evolution. Amsterdam: Elsevier; 1991. pp. 531–559. [Google Scholar]
- Blunden R, Wilkes TJ, Forster JW, Jiménez MM Sandery MJ, Karp A, et al. Identification of the E3900 family, a 2nd family of rye chromosome B-specific repeated sequences. Genome. 1993;36:706–711. doi: 10.1139/g93-095. [DOI] [PubMed] [Google Scholar]
- Böcher TW. Experimental and cytogenetical studies on plant species. Biologiske Skrifter. 1960;11:1–69. [Google Scholar]
- Burt A, Trivers R. Genes in conflict: the biology of selfish genetic elements. Cambridge, MA: Harvard University Press; 2006. B chromosomes; pp. 325–380. [Google Scholar]
- Camacho JPM, editor. B chromosomes in the eukaryote genome. Cytogenetic and Genome Research. 2004;106:143–412. [Google Scholar]
- Carlson WR. Factors affecting preferential fertilisation in maize. Genetics. 1969;62:543–554. doi: 10.1093/genetics/62.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson WR. The B-chromosome of corn. Annual Review of Genetics. 1978;12:5–23. doi: 10.1146/annurev.ge.12.120178.000253. [DOI] [PubMed] [Google Scholar]
- Carlson WR. The B chromosome of maize. CRC Critical Reviews in Plant Sciences. 1986;3:201–226. [Google Scholar]
- Cheng YM, Lin BY. Cloning and characterization of maize B chromosome sequences derived from microdissection. Genetics. 2003;164:299–310. doi: 10.1093/genetics/164.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YM, Yu HX, Yan HH, Gu MH. B chromosome in a rice aneuploid variation. Theoretical and Applied Genetics. 2000;101:564–568. [Google Scholar]
- Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Research. 2003;13:1133–1145. doi: 10.1101/gr.907603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar MK, Friebe B, Koul AK, Gill BS. Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma. 2002;111:332–340. doi: 10.1007/s00412-002-0214-4. [DOI] [PubMed] [Google Scholar]
- Evans GM, Macefield AJ. Suppression of homoeologous pairing by B chromosomes in a Lolium species hybrid. Nature New Biology. 1972;236:110–111. doi: 10.1038/newbio236110a0. [DOI] [PubMed] [Google Scholar]
- Evans GM, Macefield AJ. The effect of B chromosomes on homoeologous pairing in species hybrids. I. Lolium temulentum × L. perenne. Chromosoma. 1973;41:63–73. [Google Scholar]
- Fröst S. The cytological behaviour and mode of transmission of accessory chromosomes in Plantago serraria. Hereditas. 1959;45:191–210. [Google Scholar]
- Fröst S. The inheritance of accessary chromosomes in plants, especially in Ranunculus acris and Phleum nodosum. Hereditas. 1969;61:317–326. [Google Scholar]
- González-Sánchez M, González-Sánchez E, Molina E, Chiavarino AM, Rosato M, Puertas MJ. One gene determines maize B chromosome accumulation by preferential fertilisation; another gene(s) determines their meiotic loss. Heredity. 2003;90:122–129. doi: 10.1038/sj.hdy.6800185. [DOI] [PubMed] [Google Scholar]
- Gotoh K. Über die Chromosomenzahl von Secale cereale L. Botanical Magazine Tokyo. 1924;38:135–152. [Google Scholar]
- Graphodatsky AS, Kukekova AV, Yudkin DV, Trifonov VA, Vorobieva NV, Beklemisheva VR, et al. The proto-oncogene C-KIT maps to canid B-chromosomes. Chromosome Research. 2005;13:113–122. doi: 10.1007/s10577-005-7474-9. [DOI] [PubMed] [Google Scholar]
- Han F, Lamb JC, Yu W, Gao Z, Birchler JA. Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. The Plant Cell. 2007;19:524–533. doi: 10.1105/tpc.106.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa N. A cytological study on 8-chromosome rye. Cytologia. 1934;6:68–77. [Google Scholar]
- Holmes DS, Bougourd SM. B chromosome selection in Allium schoenoprasum. II. Experimental populations. Heredity. 1991;67:117–122. [Google Scholar]
- Houben A, Kynast RG, Heim U, Hermann H, Jones RN, Forster JW. Molecular cytogenetic characterisation of the terminal heterochromatic segment of the B chromosome of rye (Secale cereale) Chromosoma. 1996;105:97–103. doi: 10.1007/BF02509519. [DOI] [PubMed] [Google Scholar]
- Houben A, Belyaev ND, Leach CR, Timmis JN. Differences of histone H4 acetylation and replication timing between A and B chromosomes of Brachycome dichromosomatica. Chromosome Research. 1997;5:233–237. doi: 10.1023/B:CHRO.0000032297.10876.86. [DOI] [PubMed] [Google Scholar]
- Houben A, Thompson N, Ahne R, Leach CR, Verlin D, Timmis JN. A monophyletic origin of the B chromosomes of Brachycome dichromosomatica (Asteraceae) Plant and Systematic Evolution. 1999;219:127–135. [Google Scholar]
- Houben A, Verlin D, Leach CR, Timmis JN. The genomic complexity of micro B chromosomes of Brachycome dichromosomatica. Chromosoma. 2001;110:451–459. doi: 10.1007/s00412-001-0173-1. [DOI] [PubMed] [Google Scholar]
- Jackson RC. Supernumerary chromosomes in Haplopappus gracilis. Evolution. 1960;14:135–136. doi: 10.1126/science.132.3436.1316. [DOI] [PubMed] [Google Scholar]
- Jamilena M, Ruiz Rejón C, Ruiz Rejón M. A molecular analysis of the origin of the Crepis capillaris B chromosome. Journal of Cell Science. 1994;107:703–708. doi: 10.1242/jcs.107.3.703. [DOI] [PubMed] [Google Scholar]
- Jenkins G, Jones RN. B chromosomes in hybrids of temperate cereals and grasses. Cytogenetic and Genome Research. 2004;106:314–319. doi: 10.1159/000079305. [DOI] [PubMed] [Google Scholar]
- Jiménez MM, Romera F, González-Sánchez M, Puertas MJ. Genetic control of the rate of transmission of rye B chromosomes. III. Heredity. 1997;78:636–644. Male meiosis and gametogenesis. [Google Scholar]
- Jin W, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang J. Molecular and functional dissection of the maize B chromosome centromere. The Plant Cell. 2005;17:1412–1423. doi: 10.1105/tpc.104.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RN. Genome organisation in higher plants. In: Pearson PL, Lewis KR, editors. Chromosomes today. Vol. 5. Chichester: John Wiley, and Sons; 1976. pp. 117–130. [Google Scholar]
- Jones RN. Are B-chromosomes selfish? In. In: Cavalier-Smith T, editor. The evolution of genome size. Chichester: John Wiley and Sons; 1985. pp. 397–425. [Google Scholar]
- Jones RN. B-chromosome drive. American Naturalist. 1991;a 137:430–442. [Google Scholar]
- Jones RN. Cytogenetics of B chromosomes in crops. In: Gupta PK, Tsuchiya T, editors. Chromosome engineering in plants: genetics, breeding, evolution. b. Amsterdam: Elsevier; 1991. pp. 141–157. [Google Scholar]
- Jones RN. B chromosomes in plants. New Phytologist. 1995;131:411–434. doi: 10.1111/j.1469-8137.1995.tb03079.x. [DOI] [PubMed] [Google Scholar]
- Jones RN, Houben A. B chromosomes in plants: escapeees from the A chromosome genome? Trends in Plant Science. 2003;8:417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- Jones RN, Pašakinskienė I. Genome conflict in the Gramineae. New Phytologist. 2005;165:391–410. doi: 10.1111/j.1469-8137.2004.01225.x. [DOI] [PubMed] [Google Scholar]
- Jones RN, Puertas MJ. The B-chromosomes of rye (Secale cereale L.) In: Dhir KK, Sareen TS, editors. Frontiers in plant science research. Dehli: Bhagwati Enterprises; 1993. pp. 81–112. [Google Scholar]
- Jones RN, Rees H. Genotypic control of chromosome behaviour in rye. XI. The influence of B chromosomes on meiosis. Heredity. 1967;22:333–347. [Google Scholar]
- Jones RN, Rees H. B chromosomes. London: Academic Press; 1982. [Google Scholar]
- Kasás E, Birchler JA. Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics. 1998;150:1683–1692. doi: 10.1093/genetics/150.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayano H. Cytogenetic studies in Lilium callosum. III. Proceedings of the Japanese Academy; 1957. pp. 553–558. Preferential segregation of a supernumerary chromosome in EMCs. [Google Scholar]
- Kuwada Y. On the number of chromosomes in maize. Botanical Magazine Tokyo. 1925;39:227–234. [Google Scholar]
- Lamb JC, Kato A, Birchler JA. Sequences associated with A chromosome centromeres are present throughout the maize B chromosome. Chromosoma. 2005;113:337–349. doi: 10.1007/s00412-004-0319-z. [DOI] [PubMed] [Google Scholar]
- Lamb JC, Han F, Auger DL, Birchler JA. A trans-acting factor required for non-disjunction of the B chromosome is located distal to the TB-4Lb breakpoint on the B chromosome. Maize Genetics Cooperation Newsletter. 2006;80:51–54. [Google Scholar]
- Langdon T, Seago C, Jones RN, Ougham H, Thomas H, Forster JW, et al. De novo evolution of satellite DNA on the rye B chromosome. Genetics. 2000;154:869–884. doi: 10.1093/genetics/154.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach CR, Houben A, Field B, Pistrick K, Demidov D, Timmis JN. Molecular evidence for transcription of B chromosome ribosomal RNA genes in Crepis capillaris. Genetics. 2005;171:269–278. doi: 10.1534/genetics.105.043273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA, Palestis BG, Jones RN, Trivers R. Phyletic hot spots for B chromosomes in angiosperms. Evolution. 2005;59:962–969. [PubMed] [Google Scholar]
- Lewis H. The origin of supernumerary chromosomes in natural populations of Clarkia elegans. Evolution. 1951;5:142–157. [Google Scholar]
- Lima-de-Faria A. Genetic interaction in rye expressed at the chromosome phenotype. Genetics. 1962;47:1455–1462. doi: 10.1093/genetics/47.10.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BY. Regional control of nondisjunction of the B chromosome in maize. Genetics. 1978;90:613–627. doi: 10.1093/genetics/90.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström J. Transfer to wheat of accessory chromosomes from rye. Hereditas. 1965;54:149–155. [Google Scholar]
- Longley AE. Supernumerary chromosomes in Zea mays. Journal of Agricultural Research. 1927;35:769–784. [Google Scholar]
- Longley AE. Chromosomes of maize from North American Indians. Journal of Agricultural Research. 1938;56:177–195. [Google Scholar]
- McClintock B, Kato Y-TA, Blumenschein A. Chromosome constitution of races of maize. Chapingo, Mexico: Colegio De Postgraduados; 1981. [Google Scholar]
- Marschner S, Kumke K, Houben A. B chromosomes of B. dichromosomatica show a reduced level of euchromatic histone H3 methylation marks. Chromosome Research. 2007;15:215–222. doi: 10.1007/s10577-006-1114-x. [DOI] [PubMed] [Google Scholar]
- Matthews RB, Jones RN. Dynamics of the B chromosome polymorphism in rye. I. Simulated populations. Heredity. 1982;48:345–369. [Google Scholar]
- Matthews RB, Jones RN. Dynamics of the B chromosome polymorphism in rye. II. Estimates of parameters. Heredity. 1983;50:119–137. [Google Scholar]
- Moss JP. The adaptive significance of B chromosomes in rye. Chromosomes Today. 1966;1:15–23. [Google Scholar]
- Müntzing A. Cytological study of extra fragment chromosomes in rye. II. Transmission and multiplication of standard fragments and iso-fragments. Hereditas. 1945;31:457–477. [PubMed] [Google Scholar]
- Müntzing A. Cytological studies of extra fragment chromosomes in rye. III. The mechanism of non-disjunction at the pollen mitosis. Hereditas. 1946;32:97–119. [PubMed] [Google Scholar]
- Müntzing A. Cytological studies of extra fragment chromosomes in rye. V. A new fragment type arisen by deletion. Hereditas. 1948;34:435–442. [Google Scholar]
- Müntzing A, Lima-de-Faria A. Pachytene analysis of a deficient accessory chromosome in rye. Hereditas. 1952;38:1–10. [Google Scholar]
- Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proceedings of the National Academy of Sciences of the USA; 2005. pp. 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M, Puertas MJ, Jimeniz M, Romera F, Jones RN. B chromosomes in inbred lines of rye (Secale cereale L.). 2. Effects on metaphase I and first pollen mitosis. Genetica. 1996;97:65–72. [Google Scholar]
- Page BT, Wanous MK, Birchler JA. Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics. 2001;159:291–302. doi: 10.1093/genetics/159.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palestis BG, Trivers R, Burt A, Jones RN. The distribution of B chromosomes across species. Cytogenetic and Genome Research. 2004;106:151–158. doi: 10.1159/000079281. [DOI] [PubMed] [Google Scholar]
- Parker JS. The B chromosome system of Hypochoeris maculata. 1. B-distribution, meiotic behaviour and inheritance. Chromosoma. 1976;59:167–177. [Google Scholar]
- Puertas MJ. Nature and evolution of B chromosomes in plants: a non-coding but information-rich part of plant genomes. Cytogenetic and Genome Research. 2002;96:198–205. doi: 10.1159/000063047. [DOI] [PubMed] [Google Scholar]
- Puertas MJ, Romera F, Delapena A. Comparison of B chromosome effects in Secale cereale and Secale vavilovii. Heredity. 1985;55:229–234. [Google Scholar]
- Puertas MJ, González-Sánchez M, Manzanero S, Romera F, Jiménez MM. Genetic control of the rate of transmission of rye B chromosomes. IV. Localization of the genes controlling B transmission rate. Heredity. 1998;80:209–213. [Google Scholar]
- Puertas MJ, Jiménez G, Manzanero S, Chiavarino AM, Rosato M, Naranjo CA, Poggio L. Genetic control of B chromosome transmission in maize and rye. Chromosomes Today. 2000;13:79–92. [Google Scholar]
- Raman VS, Krishnaswami D. Accessory chromosomes in Sorghum nitidum Pers. Journal of the Indian Botanical Society. 1960;39:278–280. [Google Scholar]
- Randolph LF. Types of supernumerary chromosomes in maize. Anatomical Record. 1928;41:102. [Google Scholar]
- Rees H, Hutchinson J. Nuclear DNA variation due to B chromosomes. Cold Spring Harbor Symposium in Quantitative Biology. 1973;38:175–182. doi: 10.1101/sqb.1974.038.01.021. [DOI] [PubMed] [Google Scholar]
- Rhoades MM. Studies on the cytological basis of crossing over. In: Peacock WJ, Brock RD, editors. Replication and recombination of genetic material. Canberra: Australian Academy of Sciences; 1968. pp. 229–241. [Google Scholar]
- Roman H. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics. 1947;32:391–409. doi: 10.1093/genetics/32.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H. Directed fertilisation in maize. Proceedings of the National Acadamey of Sciences of the USA. 1948;34:450–454. doi: 10.1073/pnas.34.2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato M, Chiavarino AM, Naranjo CA, Hernandez JC, Poggio L. Genome size and numerical polymorphism for the B chromosome in races of maize (Zea mays ssp. mays. Poaceae) American Journal of Botany. 1998;85:168–174. [PubMed] [Google Scholar]
- Rusche ML, Mogensen Hl, Shi L, Keim P, Rougier M, Chaboud A, Dumas C. B chromosome behaviour in maize pollen as determined by a molecular probe. Genetics. 1997;147:1915–1921. doi: 10.1093/genetics/147.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandery MJ, Forster JW, Blunden R., Jones RN. Identification of a family of repeated sequences on the rye B-chromosome. Genome. 1990;33:908–913. doi: 10.1139/g93-095. [DOI] [PubMed] [Google Scholar]
- Sapre AB, Deshpande DS. Origin of B chromosomes in Coix L. through spontaneous interspecific hybridisation. Journal of Heredity. 1987;78:191–196. [Google Scholar]
- Shi L, Zhu T, Mogensen L, Keim P. Sperm identification in maize by fluorescence in situ hybridisation. The Plant Cell. 1996;8:815–821. doi: 10.1105/tpc.8.5.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanic N, Vujosevic M., Dedovic-Tanic N., Dimitrijevic B. Differential gene expression in yellow-necked mice Apodemus flavicollis (Rodentia, Mammalia) with and without B chromosomes. Chromosoma. 2005;113:418–427. doi: 10.1007/s00412-004-0327-z. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ingle J, Sinclair J, Jones RN. Genomic quality of rye B chromosomes. Journal of Experimental Botany. 1975;26:367–378. [Google Scholar]
- Tsujimoto H, Niwa K. DNA structure of the B-chromosome of rye revealed by in situ hybridization using repetitive sequences. Japanese Journal of Genetics. 1992;67:233–241. [Google Scholar]
- Trivers R, Burt A, Palestis BG. B chromosomes and genome size in flowering plants. Genome. 2004;47:1–8. doi: 10.1139/g03-088. [DOI] [PubMed] [Google Scholar]
- Wilkes TM, Francki MG, Langridge P, Karp A, Jones RN, Forster JW. Analysis of rye B-chromosome structure using fluorescence in situ hybridization (FISH) Chromosome Research. 1995;3:466–472. doi: 10.1007/BF00713960. [DOI] [PubMed] [Google Scholar]
- Wilson EB. The supernumerary chromosomes of Hemiptera. Science. 1907;a 26:870–871. [Google Scholar]
- Wilson EB. Note on the chromosome groups of Metapodius and Banasa. Biological Bulletin. 1907;b XII:303–313. [Google Scholar]
- Yu W, Lamb JC, Han F, Birchler JA. Telomere-mediated chromosomal truncation in maize. Proceedings of the National Academy of Sciences of the USA. 2006;103:17331–17336. doi: 10.1073/pnas.0605750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk J. The additional heterochromatic chromosome and its influence on sex chromosome pairing in Rumex. Heredity. 1969;24:69–74. [Google Scholar]