Lost loci in Primula parentage probe
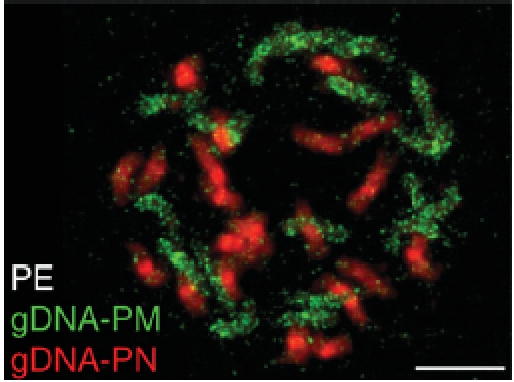
I have often heard it said that speciation is a slow process, taking place over many generations, even when populations are separated. There is certainly much truth in that statement but equally it ignores genetic events that create new species more or less instantly. One of those events is polyploidization. A glance at chromosome numbers in almost any angiosperm genus reveals that polyploidization has played a major role in angiosperm evolution. Indeed, Guggisberg et al. from Zurich, Switzerland (pp. 919–927) cite a review by Soltis (2005. Ancient and recent polyploidy in angiosperms. New Phytologist 166: 5–8), which states that at least 70 % of angiosperms have polyploid origins. Modern molecular and cytological techniques help investigate putative parental origins of polyploids as has been used here by the Zurich group for a North American Primula species, P. egaliksensis (2n = 40). This is believed to be an allopolyploid derived from a hybrid between the relatively widely diverged P. mistassinica (2n = 18) and P. nutans (2n = 22). Hybridization of either P. mistassinica or P. nutans DNA to chromosome spreads of P. egaliksensis revealed that both putative parental chromosome sets are present in P. egaliksensis, with no evidence for major chromosomal re-arrangements between the two genomes. However, focus on the genes encoding the major rRNAs (‘rDNA’) revealed some more local changes, in particular the loss in the hybrid of the maternal parent's 45S rDNA sequences. Further, heterochromatic knobs carrying the rDNA internal transcribed spacer (ITS), present in the maternal parent P. mistassinica, are absent in the hybrid, while sequencing of 44 clones of the ITS of the hybrid revealed that only 5 % were of P. mistassinica origin. The loss of the maternal rDNA sequences may have resulted from nucleolar dominance while the more general lack of chromosome re-arrangement and genome homogenization may be a result of the taxonomic distance between the two parents.
Roles of rols in root regulation
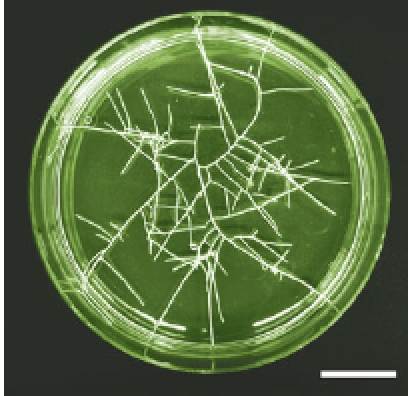
Like its close relative Agrobacterium tumefaciens, A.rhizogenes is a naturally occurring agent of plant genetic transformation; it is the causative agent of ‘hairy root’ disease in which roots proliferate at the infection site. Two groups of ‘oncogenes’ on the root-inducing plasmid, the aux genes and the rol genes, are important for this transformation. Interestingly, as pointed out by Alpizar et al.(Montpellier, France, pp. 929–940), the roots may be excised from the plant and grown readily in culture, and used for regeneration of whole plants and the study of root growth and physiology. In common with A. tumefaciens, A.rhizogenes shows differing abilities to infect different dicot plants. For both bacteria, coffee (Coffea arabica) is a difficult host (although methods for generating GM coffee have now been developed). The authors have previously achieved transformation of coffee with A. rhizogenes but the excised roots did not proliferate and soon died. In the new series of experiments, 62 different hairy root clones were grown under different conditions. All of the clones were shown by PCR to have the rolB and rolC oncogenes integrated into their genome but none of them had the bacterial aux genes. Added auxin was thus required for proliferation of all clones and was optimally supplied as 0·5 µm IBA. Sucrose was not essential but its inclusion at 2 % (w/v) was optimal for root growth with low (20 µmol m−2 s−1) light intensities or darkness favouring regeneration. Under optimal conditions, the 62 clones have been maintained and sub-cultured for over 3 years. Although there is some variation between clones in respect of root length and the proportion of fine roots, the majority of their phenotypes did not differ significantly from the roots of non-transformed plants. They therefore constitute a useful system for study of root characters such as resistance to nematodes.
ALS well in mutant medic

‘Test the trait, not the breeding method’ has almost become a mantra in my own contributions to the UK's debate on GM crops. For me, this point is well illustrated in the paper by Oldach et al., Urrbrae, South Australia (pp. 997–1005). The authors describe the use of sulfonylurea (SU) herbicides with cereal crops. Although these herbicides are regarded as safe, nevertheless their rate of degradation in alkaline soils is slow; slow enough to affect the annual medics (Medicago spp.) grown in rotation with the cereals. An SU-tolerant cultivar of M. littoralis (‘Angel’) has been developed by mutagenesis-based breeding and the main aim of the authors has been to identify the molecular basis of this mutation. Analysis of segregation ratios of the F2 populations from crosses between ‘Angel’ and intolerant M. trunculata indicated a single dominant gene. Based on the mode of action of SU herbicides, this was likely to be a mutant form of ALS, the gene that encodes acetolactate synthase, an enzyme involved in synthesis of branched-chain amino acids. The sequence of the Arabidopsis thaliana ALS gene was used to interrogate the Medicago database, revealing two homologues located respectively on chromosomes 2 and 3. Linkage analysis using known markers then revealed that the herbicide-tolerance trait was associated with a region of the chromosome 3 containing the ALS locus. Sequencing of the wild-type and mutant ALS genes revealed a single amino-acid change from proline to leucine. These data facilitated the development of a diagnostic marker for SU tolerance while RT–PCR showed that ‘Angel’ does indeed express the mutant ALS. The way is thus open for use of the mutant allele in Medicago breeding programmes. However, a similar mutant ALS from Arabidopsis has been used to transform Nicotiana and Brassica napus by GM techniques. To return to my opening comment: which is more important here, the genetic trait or the breeding method?
Pump up the volume
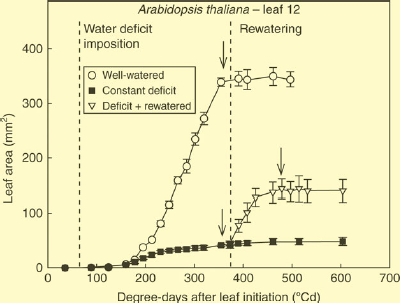
Scientific progress involves both expanding the boundaries of what we know and, from time to time, revising our ideas about what we thought we already knew. The paper by Lechner et al. from Balcarce, Argentina and Montpellier, France (pp. 1007–1015) provides a clear example of the second category. They grew Arabidopsis thaliana and Helianthus annuus plants under water deficit and under well-watered conditions. In both species, water deficit markedly reduced leaf growth rate, although the period of leaf expansion was similar in the two treatments. Final leaf area in the droughted plants was therefore much smaller than that of control plants. When droughted plants were re-watered, leaves still in the expansion phase showed a much increased expansion rate. However, the really surprising result was that leaves of droughted plants that had stopped expanding (and were assumed therefore to have reached their ‘final’ size) started expanding again. This was especially dramatic in A. thaliana, with some leaves increasing their area by up to 186 %. Increases in H. annuus were smaller (up to 27 %) but nevertheless significant. The increases in area did not involve cell division but were solely caused by cell expansion. Analysis of the response in relation to leaf age showed that the longer the gap between the cessation of expansion in droughted conditions and its re-initiation induced by re-watering, the smaller was the response, eventually declining to zero in the oldest leaves. This indicates that there is a ‘developmental window’ during which it is possible for leaf cells to resume expansion growth. Under the conditions used in these experiments, the window was 4 days in H. annuus and 11 days in A. thaliana. This window is likely to represent a period in which the biophysical/biochemical changes in the cell wall are making the wall more rigid but until full rigidity is achieved, the wall is able to respond to increased turgor pressure.


