Abstract
Background and Aims
Earlier studies have suggested that the tetraploid Primula egaliksensis (2n = 40) originated from hybridization between the diploids P. mistassinica (2n = 18) and P. nutans (2n = 22), which were hypothesized to be the maternal and paternal parent, respectively. The present paper is aimed at verifying the hybrid nature of P. egaliksensis using cytogenetic tools, and to investigate the extent to which the parental genomes have undergone genomic reorganization.
Methods
Genomic in situ hybridization (GISH) and fluorescent in situ hybridization (FISH) with ribosomal DNA (rDNA) probes, together with sequencing of the internal transcribed spacer (ITS) region of the rDNA, were used to identify the origin of P. egaliksensis and to explore its genomic organization, particularly at rDNA loci.
Key Results
GISH showed that P. egaliksensis inherited all chromosomes from P. mistassinica and P. nutans and did not reveal major intergenomic rearrangements between the parental genomes (e.g. interchromosomal translocations). However, karyological comparisons and FISH experiments suggested small-scale rearrangements, particularly at rDNA sites. Primula egaliksensis lacked the ITS-bearing heterochromatic knobs characteristic of the maternal parent P. mistassinica and maintained only the rDNA loci of P. nutans. These results corroborated sequence data indicating that most ITS sequences of P. egaliksensis were of the paternal repeat type.
Conclusions
The lack of major rearrangements may be a consequence of the considerable genetic divergence between the putative parents, while the rapid elimination of the ITS repeats from the maternal progenitor may be explained by the subterminal location of ITS loci or a potential role of nucleolar dominance in chromosome stabilization. These small-scale rearrangements may be indicative of genome diploidization, but further investigations are needed to confirm this assumption.
Key words: Diploidization, FISH, genome evolution, GISH, hybridization, ITS, polyploidy, Primula egaliksensis, 45S rDNA
INTRODUCTION
Polyploidization has played a major role in plant speciation (Soltis and Soltis, 1993; Bretagnolle et al., 1998; Otto and Whitton, 2000; Levin, 2002; Soltis et al., 2003). At least 70 % of all angiosperms are thought to have polyploid origins (reviewed by Soltis, 2005), including species of small genome size and chromosome number such as Arabidopsis thaliana (Henry et al., 2006). Two types of polyploids are commonly recognized according to the degree of homology among co-existing genomes. Autopolyploids, which arise within a single species, contain more than two sets of homologous chromosomes in their nuclear genome, while allopolyploids contain more than two sets of homeologous chromosomes that diverged from each other prior to interspecific hybridization (Ramsey and Schemske, 1998).
Polyploids generally undergo rapid genome restructuring following their formation (Wendel, 2000; Adams and Wendel, 2005; Chen and Ni, 2006) as demonstrated, for instance, by the studies on synthetic and natural allopolyploids of Nicotiana (Skalická et al., 2003, 2005; Kovarík et al., 2004; Lim et al., 2004a), wheat (Levy and Feldman, 2004) and Arabidopsis (Pontes et al., 2004). Genomic changes range from intergenomic chromosome translocations to interlocus recombinations (e.g. unequal crossing-over and gene conversion), and can be accompanied by epigenetic modifications (e.g. cytosine methylation) at specific loci leading to altered gene expression patterns (Wendel, 2000; Liu and Wendel, 2003; Wang et al., 2004; Chen and Ni, 2006). Altogether these changes may constitute a response to the cellular stress imposed by the novel hybrid chromosome composition and/or doubling of the gene dosage (i.e. genomic shock; McClintock, 1984; Madlung and Comai, 2004) and result in the gradual diploidization of polyploid taxa (Leitch and Bennett, 2004; Ma and Gustafson, 2005). Yet there are also examples of polyploids that have undergone only few changes in overall genome structure since their formation, e.g. in Gossypium (Liu et al., 2001) and Spartina (Baumel et al., 2002; Ainouche et al., 2004).
The varied response of plant genomes to polyploidization is reflected in the nature and arrangement of ribosomal DNA (rDNA) repeats of polyploids. Ribosomal genes occur in hundreds or thousands of copies that are tandemly repeated over one or more loci (Long and Dawid, 1980). Some hybrids show a strict conservation in number, but not necessarily in size, of both parental rDNA loci (e.g. in Nicotiana; Matyásek et al., 2003; Kovarík et al., 2004), while others display gain (e.g. in Gossypium; Hanson et al., 1996) or loss (e.g. in Arabidopsis; Pontes et al., 2004) of rDNA loci. At the sequence level, similar contrasting scenarios of rDNA loci evolution were observed as sequences from putative allopolyploids were found to be either additive for differing parental nucleotides (e.g. Campbell et al., 1997), or homogenized towards one parental repeat type (e.g. Wendel et al., 1995; Kovarík et al., 2004), due to processes of intra- and interlocus recombination (referred to as concerted evolution; Elder and Turner, 1995) or loss of rDNA loci.
The genus Primula (totalling approx. 430 species; Richards, 2002) constitutes an ideal model system to investigate the effects of hybridization and polyploidization on plant evolution. On the one hand, primulas have been hybridized for gardening purposes since the 16th century and several sections are known for their high levels of interspecific hybridization in the wild (e.g. sects. Auricula and Primula; Richards, 2002). On the other hand, the ploidy level varies from diploid to 14-ploid, with some sections being entirely polyploid (e.g. sects. Auricula and Parryi; Richards, 2002). Interestingly, morphological, caryological, distributional and phylogenetic data suggest that most polyploids belonging to sect. Aleuritia are of allopolyploid origin (Bruun, 1932; Vogelmann, 1956; Hultgård, 1990, 1993; Kelso, 1991, 1992; A. Guggisberg, G. Mansion and E. Conti, unpubl. res.).
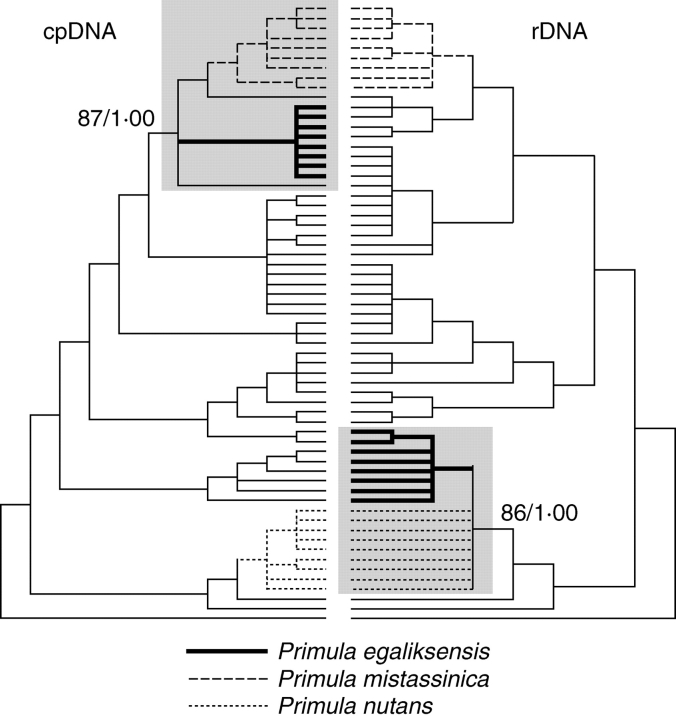
The tetraploid Primula egaliksensis (2n = 40) is widely distributed across North America and is taxonomically ascribed to sect. Armerina owing to its entire petiolate leaves, narrow elongated capsules and the absence of farina (Kelso, 1991; Richards, 2002). Karyological and morphological evidence suggest, however, that it may be an intersectional allopolyploid involving one species (out of 27) of sect. Aleuritia and one species (out of 14) of sect. Armerina (Kelso, 1991, 1992). This hypothesis relies primarily on the additivity of chromosome numbers, the intermediacy of gland types, pollen sizes, colpi numbers and exine reticulation patterns (Kelso, 1991, 1992). Within sect. Aleuritia, four diploid species occur in North America (P. alcalina, P. anvilensis, P. mistassinica and P. specuicola), but chloroplast DNA- (cpDNA) based phylogenies suggest that P. egaliksensis is more closely related to P. mistassinica (2n = 18) than to any other species of Primula (Fig. 1; Mast et al., 2001, 2006; Guggisberg et al., 2006; A. Guggisberg, G. Mansion and E. Conti, unpubl. res.). Within sect. Armerina, P. nutans (2n = 22) is the only diploid that occurs in North America and phylogenetic evidence based on rDNA sequences indicates a common origin for P. nutans and P. egaliksensis (Fig. 1; Guggisberg et al., 2007). Hence P. mistassinica and P. nutans are the most likely progenitors of P. egaliksensis, the former probably acting as the maternal parent. Distributional data further suggest that P. egaliksensis originated in North America, because it is rarely found outside of this area and the current ranges of its putative parents only overlap in north-western Canada. Finally, the diversity of cpDNA haplotypes recovered by sequencing of multiple accessions of P. egaliksensis advocates for a recurrent origin of this taxon (Guggisberg et al., 2006).
Fig. 1.
Phylogenetic placement of Primula egaliksensis in relation to its putative parents, i.e. P. mistassinica and P. nutans, in the cpDNA- and rDNA-based phylogenies inferred by Bayesian analysis. Clades of interest are highlighted in grey and followed by bootstrap support values/ posterior probabilities. Redrawn from A. Guggisberg, G. Mansion and E. Conti, unpubl. res.
The numerous studies on synthetic and natural polyploids have shown that polyploid genomes are dynamic in their response to polyploidization, underlining the need for additional case studies looking at genomic, genetic and epigenetic changes following chromosome doubling. Cytogenetic investigations are powerful in verifying the origin of polyploid taxa and may provide preliminary insights into the extent of genomic rearrangements. The present paper aims to (1) confirm the hypothesized allopolyploid origin of P. egaliksensis, (2) investigate the genomic organization of this tetraploid by genomic in situ hybridization (GISH), and (3) understand the evolutionary history of the 45S rDNA loci – consisting of the 18S, 5·8S and 26S ribosomal genes and the internal transcribed spacers (ITS) – using fluorescent in situ hybridization (FISH) and ITS sequencing.
MATERIALS AND METHODS
Plant material
The plant material used in this study is listed in Table 1. Different accessions were used for DNA extractions and chromosome spreads because none of the sampled populations contained both the leaf and seedling tissues necessary for the two sets of analyses.
Table 1.
Plant material of Primula used in this study. Voucher specimens were deposited at the herbarium of Z, Zürich, Switzerland
| Taxon | Tissue type | Provenance | Herbarium voucher |
|---|---|---|---|
| P. egaliksensis | Leaf | Alaska/USA | Guggisberg & Mansion 100703-1b1 |
| Leaf | Newfoundland/Canada | Guggisberg & Mansion 240604-6b2 | |
| Root tip | Alaska/USA | Guggisberg & Mansion 060703-1 | |
| Root tip | Alaska/USA | Guggisberg & Mansion 150703-1 | |
| P. mistassinica | Leaf | Wisconsin/USA | Anderson 210503-3 |
| Root tip | Alberta/Canada | Eveleigh 300603-3 | |
| P. nutans | Leaf | Alaska/USA | Guggisberg & Mansion 240603-2 |
| Root tip | Yukon Territory/Canada | Guggisberg & Mansion 250603-2 | |
| Root tip | Yukon Territory/Canada | Bennett, Line & Hett-Seccombe 030604-1 |
DNA extraction, PCR amplification, cloning and sequencing
Total genomic DNA (gDNA) was extracted using the DNeasy Plant Mini Kit (Qiagen, Switzerland). The two ITS and the 5·8S ribosomal gene of the rDNA cistron (hereafter called the ITS region, covering 638 bp) were amplified with ITS.LEU and ITS4 primers (Baum et al., 1998) using 2 mm MgCl2, 200 µm dNTPs and 0·2 µm of each primer in a standard PCR buffer using an annealing temperature of 52 °C (with a ramp speed of 1 °C s–1). PCR products destined for sequencing were purified (GFX PCR DNA and Gel Band Purification Kit, Biosciences Amersham, Switzerland) and cloned into pCR®II-TOPO® (TOPO TA Cloning® kit, Invitrogen, Switzerland) before sequencing (ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit, ABI Prism 3100 automated sequencer, Applied Biosystems, USA). Because preferential amplification of one sequence variant may lead to its over-representation in the final reaction mixture (cf. PCR drift and PCR selection; Wagner et al., 1994), three PCR reactions of each sample were pooled for use in ligations (Mason-Gamer, 2004).
Sequencher 4·2 (Gene Codes Corp., USA) was used to check the quality of the electropherograms and compile the contiguous sequences for each PCR product. The starting and ending points of each sequence were determined by comparison with the partial rDNA sequence of Rhododendron kanehirai (GenBank AF172290) and deposited in GenBank (accession no. DQ993783-DQ993802, EU095369- EU095392). Cloned ITS sequences of P. egaliksensis were finally aligned with 18 sequences of P. mistassinica (GenBank DQ993741-DQ993749) and P. nutans (GenBank DQ993763-DQ993771) using Se-Al v2·0a9 (available at http://tree.bio.ed.ac.uk/) in order to determine the direction of concerted evolution at sites distinguishing the putative parents. The alignment was searched for variable positions using PAUP* 4·0·10b (Swofford, 1999).
Chromosome preparation and in situ hybridization
Seeds were imbibed in water, stratified for 6 weeks at 4 °C, and germinated on Murashige and Skoog basal medium in a constantly illuminated growth chamber at 20 °C. Two-week-old seedlings were treated with auxin (2·5 µm naphthalene acetic acid in medium) for 1 week to induce the formation of root meristems. Mitotic cells were stopped at the metaphase stage upon exposure to colchicine (0·05 % in liquid medium) for 2 h. Root tip samples of approx. 1 cm length were excised and fixed in ice-cold ethanol : acetic acid (3 : 1, v/v) and kept at –20 °C until use. Spread nuclei were prepared essentially as described by Lysak et al. (2006), but with 50 % acetic acid.
The preparations were stained with 1 mg mL–1 4,6-diamino-2-phenylindole (DAPI) in Vectashield antifade (Vector Laboratories, Canada) and screened for the quality of metaphase, prophase and interphase nuclei under epifluorescence microscopy (Axioplan, Zeiss, Germany and DM6000, Leica, Germany). The best preparations were rinsed in PBS (10 mm sodium phosphate, pH 7·0, 143 mm NaCl), dehydrated in ethanol series (70 %, 90 %, 100 %, 2 min each), and air dried before FISH or GISH analyses.
For FISH and GISH experiments, slide and probe preparation, hybridization and detection were carried out as described by Lysak et al. (2006) with the following minor modifications: the RNase treatment was done for 30 min instead of 60 min; chromosomes were treated with 25 mg mL–1 pepsin in 0·01 m HCl for 2 min at 37 °C before hybridization; the probes were pre-denatured for 15 min at 75 °C and cooled on ice for 5 min before hybridization onto the chromosomes; post-hybridization washes were done at 45 °C for 10 min each, followed by rinses at room temperature; for GISH, the chromosomes were denatured in 70 % formamide in 2× SSC for 2 min at 80 °C, fixed in 70 % ethanol at –20 °C for 5 min and dehydrated in 90 % and 100 % ethanol at room temperature. The hybridization mixture consisted of 50–100 ng of labelled probes, 50 % deionised formamide, 10 % dextran sulphate, 2× SSC, and 50 µg sheared salmon sperm DNA. Pre-denaturation of the genomic probe mix was found to be essential to eliminate cross-hybridization at heterochromatic regions, possibly due to quick re-annealing of conserved repeat sequences in the probe mixture before target chromosomes were reached.
The 45S rDNA probe was prepared from the plasmid pTa71 (a gift from P. Fransz, University of Amsterdam) containing the 18S–25S ribosomal genes isolated from Triticum aestivum (Barker et al., 1988), and labelled with digoxigenin (Nick Translation Mix, Roche, Switzerland). The ITS probes of P. egaliksensis (ITS-PE), P. mistassinica (ITS-PM) and P. nutans (ITS-PN) were prepared from PCR products as described above, and labelled with biotin (Nick Translation Mix, Roche, Switzerland). ITS probes from P. mistassinica (ITS-PM) and P. nutans (ITS-PN) were used on their respective genomes, but also on nuclei of P. egaliksensis. For GISH, gDNA of P. mistassinica (gDNA-PM) and P. nutans (gDNA-PN) was extracted as described above, and sheared to approx. 1000 bp by sonication for 10 s (UW2200, Bandelin Electronic, Germany) before labelling with either digoxigenin- or biotin-labelled nucleotides (Nick Translation Mix, Roche, Switzerland). Digoxigenin-labelled probes were detected using a Mouse-anti-Digoxigenin (Roche, Switzerland) and a Goat-anti-Mouse antibody coupled to Alexa 488 (Molecular Probes, Invitrogen, Switzerland) as primary and secondary antibody, respectively (Lysak et al., 2006). Biotin-labelled probes were detected with Avidin and Goat-anti-Avidin coupled to Texas Red as described by Lysak et al. (2006). All images were processed using Adobe Photoshop® (Adobe Systems, Switzerland) and treated for colour contrast and brightness.
RESULTS
GISH demonstrates the hybrid origin of P. egaliksensis
The investigation of root tip prophase and metaphase cells of P. mistassinica, P. nutans and P. egaliksensis confirmed previous reports of chromosome numbers. A diploid chromosome number was counted in both P. mistassinica (2n = 18; Fig. 2A) and P. nutans (2n = 22; Fig. 2B), and a tetraploid chromosome number was counted in P. egaliksensis (2n = 40; Fig. 2C). In addition, most chromosomes of P. mistassinica were characterized by heterochromatic knobs (i.e. highly condensed chromatin) corresponding to brightly stained chromosomal regions (cf. arrowhead and inset in Fig. 2A) that were distinct from pericentromeric regions. Noticeably, these genomic structures were absent in both P. nutans and P. egaliksensis.
Fig. 2.
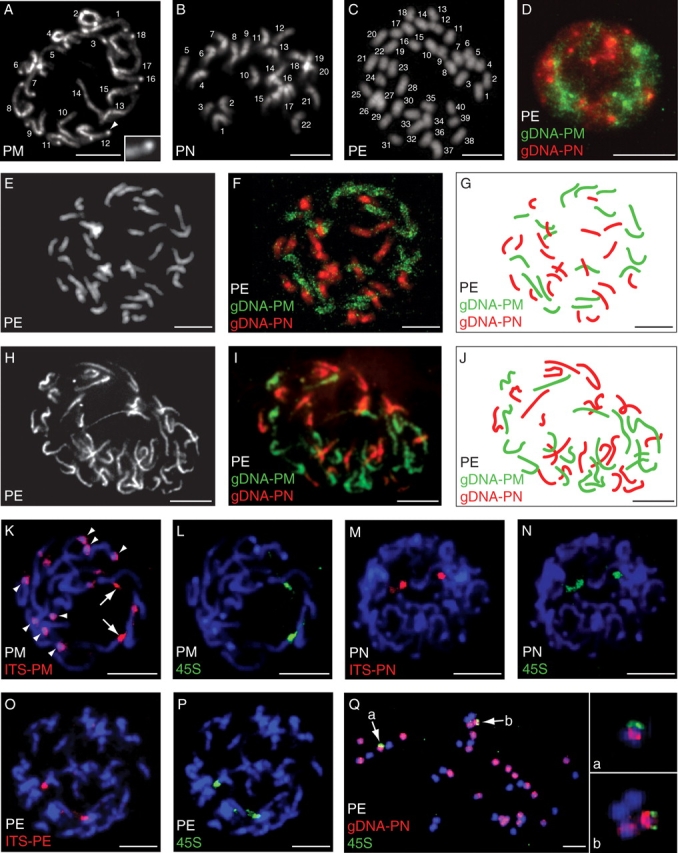
(A–C) DAPI-stained root tip prophases/metaphases of (A) Primula mistassinica showing 2n = 18 chromosomes, (B) P. nutans showing 2n = 22 chromosomes, and (C) P. egaliksensis showing 2n = 40 chromosomes; the arrowhead in (A) points to the heterochromatic knob shown in the inset. (D) GISH to interphase nuclei of P. egaliksensis using gDNA from P. mistassinica (green) and P. nutans (red). (E–J) Root tip prophases/metaphases of P. egaliksensis stained with DAPI (E, H), and resulting GISH signal (F, G, I, J) following in situ hybridization of gDNA from P. mistassinica (green) and P. nutans (red). (K–P) FISH localization of ITS (red) and 45S rDNA (green) loci on root tip prophases of (K, L) P. mistassinica, (M, N) P. nutans, and (O, P) P. egaliksensis counterstained with DAPI; in (K), arrows point to cross-hybridization of ITS and 45S rDNA probes, and arrowheads highlight heterochromatic knobs. (Q) Root-tip metaphase of P. egaliksensis counterstained with DAPI after GISH with gDNA of P. nutans (red) and FISH with 45S rDNA probes (green); arrows (a, b) point to the two 45S rDNA-bearing chromosomes shown in the insets. Scale bar = 10 µm. PE, P. egaliksensis; PM, P. mistassinica; PN, P. nutans.
In order to assess whether P. egaliksensis inherited the genome complements from both P. mistassinica and P. nutans, GISH was carried out. Control experiments using labelled gDNA of P. nutans to hybridize P. mistassinica nuclei, and vice versa, showed very little or no cross-hybridization with our experimental conditions (see Materials and Methods; data not shown). Hence, the genomes of the putative parents were differentially labelled and hybridized to P. egaliksensis spread nuclei of root tips. The results unambiguously demonstrate the hybrid origin of P. egaliksensis, for interphase nuclei as well as metaphase chromosomes of P. egaliksensis clearly revealed the presence of both P. nutans (red) and P. mistassinica (green) genomes (Fig. 2D–J).
Parental chromosomes of P. egaliksensis remain globally preserved
GISH on P. egaliksensis metaphase plates enabled the counting of 18 chromosomes hybridized with gDNA of P. mistassinica and 22 chromosomes hybridized with gDNA of P. nutans, as expected if all parental chromosomes were inherited by the allopolyploid (Fig. 2E–J). As evidenced by the relative homogenous hybridization pattern on each chromosome, no chromosomes of P. egaliksensis presented hybridization signals from both parents, suggesting that no major intergenomic rearrangements occurred between the parental genomes (e.g. interchromosomal translocations).
ITS sequences of P. egaliksensis are mainly homogenized towards the P. nutans repeat type
Sequencing of 44 ITS clones from two accessions of P. egaliksensis revealed 22 different ITS sequences. These clones varied at 21 of the 27 positions distinguishing the putative progenitors of P. egaliksensis, i.e. P. mistassinica and P. nutans (Table 2). Thirty-six clones (82 %; numbers 1–13, 15, 17–18, 20–21 of P. egaliksensis 1 and numbers 1–15, 17–19 of P. egaliksensis 2) were completely homogenized towards the ITS repeat type found in P. nutans. Of the remaining eight clones, six (numbers 14, 16, 19, 22 of P. egaliksensis 1 and 16, 20 of P. egaliksensis 2) were mainly, but not completely, homogenized towards the ITS repeat type found in P. nutans, while two (21–22 of P. egaliksensis 2) were mainly homogenized towards the ITS repeat type found in P. mistassinica. Hence only 5 % of the clones presented a nucleotide composition characteristic of P. mistassinica ITS sequences. Finally, two clones (numbers 22 and 20 of P. egaliksensis 1 and 2, respectively) showed a deletion of 34 bp in the ITS1 (e.g. positions 21 and 44 of the ITS alignment; Table 2). From this analysis, we can conclude that most, but not all, ITS sequences of P. egaliksensis are homogenized towards the paternal repeat type (P. nutans), suggesting that P. egaliksensis has progressively lost the maternally inherited ITS sequences. The incomplete homogenization may indicate that this is still a dynamic process.
Table 2.
Clonal variation within Primula egaliksensis at the 27 nucleotide positions of the ITS region distinguishing P. mistassinica and P. nutans. Site numbering follows ITS alignment. Sites identical to the first sequence are represented by dots and gaps are represented by dashes. Site 378 belonging to the 5·8S gene and separating the ITS1 from ITS2 is highlighted in bold.
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | ||||||||||
| 1 | 2 | 4 | 5 | 5 | 6 | 8 | 9 | 9 | 0 | 0 | 0 | 1 | 2 | 3 | 4 | 9 | 0 | 3 | 7 | 4 | 9 | 5 | 7 | 9 | 9 | 1 | |
| Taxon | 1 | 1 | 4 | 2 | 7 | 8 | 5 | 0 | 7 | 5 | 6 | 9 | 9 | 6 | 0 | 2 | 3 | 7 | 2 | 8 | 1 | 8 | 4 | 4 | 5 | 8 | 7 |
| P. nutans | C | C | T | T | G | A | A | T | C | T | G | T | T | T | C | T | A | G | T | C | A | C | C | C | T | G | C |
| P. egaliksensis 1 c1-13/15/17-18/20-21 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · |
| c14 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | T | · | · | C | A | · |
| c16 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · |
| c19 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · |
| c22 | · | – | – | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | T | · | · | · | · | · |
| P. egaliksensis 2 c1-15/17-19 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · |
| c16 | · | · | · | · | · | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · |
| c20 | · | – | – | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | T | · | · | · | · | · |
| c21 | T | T | C | G | · | C | G | C | T | C | · | C | C | C | T | A | G | C | · | · | · | · | · | · | · | · | · |
| c22 | T | T | C | G | · | C | G | C | T | C | · | C | C | C | T | A | G | C | G | · | G | T | · | · | C | A | · |
| P. mistassinica | T | T | C | G | T | C | G | C | T | C | A | C | C | C | T | A | G | C | G | T | G | T | T | T | C | A | T |
rDNA loci of P. egaliksensis have undergone drastic restructuring
Because our sequencing results suggested that the rDNA of P. egaliksensis have undergone concerted evolution, perhaps involving intra- and interlocus recombination, we investigated the number and position of rDNA loci using FISH. Hybridization with the ITS-PM probe revealed at least 14 signals in P. mistassinica (Fig. 2K) and most of them co-localized with the heterochromatic knobs (indicated by arrowheads in Fig. 2K). However, only two ITS signals (indicated by arrows in Fig. 2K) belonged to the unique pair of 45S rDNA loci detected by FISH (Fig. 2L). Conversely, in P. nutans, the ITS-PN probe co-localized with the two 45S rDNA loci (Fig. 2M, N). A similar situation was found in the hybrid P. egaliksensis, where the ITS-PE probe co-localized with the two 45S rDNA loci (Fig. 2O, P). Likewise, the parental ITS probes (ITS-PN and ITS-PM) co-localized on two loci of P. egaliksensis chromosomes (data not shown). The latter experiment indicates that the parental ITS probes are not different enough (max. 5 % divergence) to be distinguished with our experimental conditions (approx. 80 % washing stringency) and suggest that P. egaliksensis lost the numerous ITS-bearing heterochromatic knobs observed in P. mistassinica (see Discussion). Clearly, P. egaliksensis has lost one pair of rDNA loci from one parent. To determine the origin of the retained loci, we combined GISH and 45S-FISH and found that the rDNA probe hybridized with chromosomes from P. nutans (Fig. 2Q).
DISCUSSION
Previous studies have suggested that P. egaliksensis is an allotetraploid originating from hybridization of the diploids P. nutans and P. mistassinica (Kelso, 1991, 1992; A. Guggisberg, G. Mansion and E. Conti, unpubl. res.). The present paper unambiguously confirms this hypothesis and provides insights into the evolution of the parental genomes.
Genomic origin and organization of P. egaliksensis
The chromosome number of P. egaliksensis was previously found to be the sum of the chromosomes found in P. mistassinica and P. nutans (Kelso, 1991), but the origin of the chromosomes has never been investigated. Using GISH, we can now state that P. egaliksensis possesses 40 chromosomes corresponding to the entire genome complements of P. mistassinica and P. nutans, thereby confirming its allotetraploid nature.
The hybridization experiments did not detect major intergenomic rearrangements between the parental genomes (e.g. interchromosomal translocations) in P. egaliksensis, suggesting that the chromosomes have retained their parental integrity. Yet karyological comparisons advocate for intragenomic rearrangements, because the tetraploid lacks the heterochromatic knobs characteristic of its maternal progenitor, P. mistassinica. Heterochromatic knobs are heavily stained heterochromatic features that contain a low density of expressed genes interspersed with numerous retrotransposons and tandemly repeated elements (reviewed by Bennetzen, 2000). The observation that all P. egaliksensis chromosomes inherited from P. mistassinica do not present these knobs may suggest active mechanisms of chromosome rearrangement near heterochromatic knobs, as shown in maize (Buckler et al., 1999). Alternatively, this result may argue for P. mistassinica being polymorphic for heterochromatic knobs, as it is found in maize (Brown, 1949), indicating that the original parent of P. egaliksensis may come from a different population of P. mistassinica (with no knobs) than the one sampled here (with knobs).
The most compelling evidence of genome restructuring in the allotetraploid P. egaliksensis comes from the analysis of rDNA loci. While the sequencing of ITS clones highlighted a vast homogenization towards the paternal parent P. nutans, FISH and GISH analyses demonstrated that the 45S rDNA loci from the maternal parent P. mistassinica were lost, in conjunction with the ITS-bearing heterochromatic knobs. The subterminal location of ITS loci in P. mistassinica and P. nutans may explain the rapid elimination of ITS copies from the maternal parent in the allopolyploid P. egaliksensis. Indeed, studies on cotton, tobacco and peonies indicated that loci located near the telomeres are more prone to recombination (leading to possible homogenization) than sites situated near the centromeres (Wendel et al., 1995; Cronn et al., 1996; Hanson et al., 1996; Zhang and Sang, 1999; Fulnecek et al., 2002). Alternatively, the loss of rDNA genes may be related to nucleolar dominance, a common epigenetic phenomenon in interspecific hybrids whereby only rDNA genes inherited from one parent are transcribed (Reeder, 1985; Pikaard, 2000). Recent investigations on Brassica × Orychophragmus hybrids have suggested that nucleolar dominance may play a role in chromosome stabilization by inducing genome-specific rearrangements (Li and Ge, 2007). However, remnant nucleotidic signatures of P. mistassinica in P. egaliksensis ITS clones imply that the loss of maternal ITS sequences occurred later than in the first hybrid generation, since recombination apparently occurred between P. nutans and P. mistassinica ITS repeats.
Primula egaliksensis vs. other allopolyploid models
According to the nucleo-cytoplasmic interaction (NCI) hypothesis proposed by Gill (1991), newly formed polyploids must undergo rapid structural chromosomal changes to lift the ‘sterility resulting from the adverse interaction between the male nuclear genome and both the nuclear and cytoplasmic genomes of the female’. Molecular and cytogenetic investigations on synthetic allotetraploid lines of tobacco support this hypothesis, because first-generation polyploids are the genomic sum of their parents, but interchromosomal translocations can be detected after three generations, along with changes in number and composition of rDNA loci (Skalická et al., 2003, 2005; Lim et al., 2006, 2007).
Primula egaliksensis is an intersectional hybrid (Kelso, 1991, 1992; Richards, 2002), and thus the lack of major intergenomic rearrangements between the parental genomes may be a consequence of the considerable genetic divergence between the putative parents. In synthetic allopolyploids, frequencies of intergenomic recombination were shown to be positively correlated with degrees of divergence between the putative progenitors (Song et al., 1995), but studies on hybrids of distantly related mouse strains also indicated that substantial chromosomal divergence suppresses recombination (Shao et al., 2001). Hence, there might be a positive correlation between the genetic divergence of the parents and the frequency of interchromosomal translocations in the hybrids until this genetic divergence reaches a level where homeologous recombination becomes extremely rare or even impossible.
The NCI hypothesis further predicts that the paternal genome should evolve more rapidly than the maternal one because it functions within an alien maternal genomic environment (Gill, 1991). This assumption has been supported by recent studies on synthetic tobacco (Skalická et al., 2003, 2005), but comparable assays on natural Nicotiana allopolyploids showed that genetic changes may also be targeted at the maternal genome donor (Lim et al., 2000; Kovarík et al., 2004; Clarkson et al., 2005). The present data on the natural allotetraploid P. egaliksensis argue for genome restructuring primarily affecting the genome of the maternal progenitor (P. mistassinica). Yet paternally targeted rearrangements at the gene level cannot be excluded since our investigations were restricted to whole-chromosome structure and rDNA loci.
Primula egaliksensis is supposed to have originated during the Pleistocene glaciations (i.e. between 1·8 million to 10 000 years ago) as a result of repeated contact between its putative parents following glacial advancement and retreat (Kelso, 1991, 1992; Richards, 2002; Guggisberg et al., 2006; A. Guggisberg, G. Mansion and E. Conti, unpubl. res.). The efficiency of in situ hybridization of the parental genomes onto P. egaliksensis chromosomes suggests a relatively high degree of conservation of the progenitor sequences in the hybrid and corroborates the hypothesis of a ‘geologically young’ hybrid with a maximum age of approx. 1 million years. Indeed, recent studies on natural allopolyploids of Nicotiana showed that the effectiveness of GISH is considerably reduced after 1 million years of genome evolution and fails after 5 million years of divergence (Clarkson et al., 2005; Lim et al., 2007).
The concomitant loss of ITS-bearing heterochromatic knobs and 45S rDNA sites of P. mistassinica in P. egaliksensis may be indicative of ongoing genome diploidization (Leitch and Bennett, 2004; Ma and Gustafson, 2005), attesting to the dynamic nature of polyploid taxa (Soltis and Soltis, 1993; Wendel, 2000; Soltis et al., 2003). Future investigations on the P. egaliksensis species complex will be aimed at (1) dating the origin of the allopolyploid; (2) assessing the copy number of 45S rDNA repeats in order to ascertain that the rDNA loci of P. egaliksensis were lost and not fused; and (3) identifying the ITS repeats borne on heterochromatic knobs of P. mistassinica, as they may not be linked to functional 45S rDNA loci (Maggini et al., 1991; Stupar et al., 2002; Lim et al., 2004b).
ACKNOWLEDGEMENTS
For help with the collection of tissue samples, we thank Bruce Bennett, Guilhem Mansion, Pamela Eveleigh, Wendy Maovlic and all colleagues listed in Table 1. We also acknowledge John Richards for his advice on seed germination, Guilhem Mansion and two anonymous reviewers for comments on earlier versions of the manuscript, and Prof. Ingo Schubert for help in the interpretation of heterochromatic knobs. This work was supported by the University of Zürich and by the EPIGENOME Network of Excellence of the European Union to UG. Finally, AG would like to thank the Georges und Antoine Claraz-Schenkung and the Swiss Academy of Sciences for funding the field work.
LITERATURE CITED
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ainouche AK, Baumel A, Salmon A. Spartina anglica C. E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biological Journal of the Linnean Society. 2004;82:475–484. [Google Scholar]
- Barker RF, Harberd NP, Jarvis MG, Flavell RB. Structure and evolution of the intergenic region in a ribosomal DNA repeat unit of wheat. Journal of Molecular Biology. 1988;201:1–17. doi: 10.1016/0022-2836(88)90434-2. [DOI] [PubMed] [Google Scholar]
- Baum DA, Small RL, Wendel JF. Biogeography and floral evolution of baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Systematic Biology. 1998;47:181–207. doi: 10.1080/106351598260879. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche M, Kalendar R, Schulman AH. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C. E. Hubbard (Poaceae) Molecular Biology and Evolution. 2002;19:1218–1227. doi: 10.1093/oxfordjournals.molbev.a004182. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. The many hues of plant heterochromatin. Genome Biology. 2000;1 doi: 10.1186/gb-2000-1-1-reviews107. reviews107·1–107·4. doi: 10·1186/gb-2000-1-1-reviews107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretagnolle F, Felber F, Calame FG, Küpfer P. La polyploïdie chez les plantes. Botanica Helvetica. 1998;108:5–37. [Google Scholar]
- Brown WL. Numbers and distribution of chromosome knobs in United States maize. Genetics. 1949;34:524–536. doi: 10.1093/genetics/34.5.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun HG. Cytological studies in Primula, with special reference to the relation between the karyology and taxonomy of the genus. Symbolae Botanicae Upsalienses. 1932;1:1–239. [Google Scholar]
- Buckler ES, IV, Phelps-Durr TL, Keith Buckler CS, Dawe RK, Doebley JF, Holtsford TP. Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics. 1999;153:415–426. doi: 10.1093/genetics/153.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CS, Wojciechowski MF, Baldwin BG, Alice LA, Donoghue MJ. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae) Molecular Biology and Evolution. 1997;14:81–90. doi: 10.1093/oxfordjournals.molbev.a025705. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Ni Z. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays. 2006;28:240–252. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, Lim KY, Kovařík A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytologist. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Cronn RC, Zhao X, Paterson AH, Wendel JF. Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA diploid and allopolyploid cottons. Journal of Molecular Evolution. 1996;42:685–705. doi: 10.1007/BF02338802. [DOI] [PubMed] [Google Scholar]
- Elder JFJ, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Quarterly Review of Biology. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Fulnecek J, Lim KY, Leitch AR, Kovarík A, Matyasék R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity. 2002;88:19–25. doi: 10.1038/sj.hdy.6800001. [DOI] [PubMed] [Google Scholar]
- Gill BS. Nucleo-cytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. In: Sasakuma T, Kinoshita T, editors. Nuclear and organellar genomes of wheat species. Proceedings of Dr. H. Kihara memorial international symposium on cytoplasmic engineering in wheat. Yokohama, Japan: Kihara Memorial Foundation; 1991. pp. 48–53. [Google Scholar]
- Guggisberg A, Mansion G, Kelso S, Conti E. Evolution of biogeographic patterns, ploidy levels, and breeding systems in a diploid-polyploid species complex of Primula. New Phytologist. 2006;171:617–632. doi: 10.1111/j.1469-8137.2006.01722.x. [DOI] [PubMed] [Google Scholar]
- Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Stelly DM, Price HJ. Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma. 1996;105:55–61. doi: 10.1007/BF02510039. [DOI] [PubMed] [Google Scholar]
- Henry Y, Bedhomme M, Blanc G. History, protohistory and prehistory of the Arabidopsis thaliana chromosome complement. Trends in Plant Science. 2006;11:267–273. doi: 10.1016/j.tplants.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Hultgård U-M. Polyploidy and differentiation in N. European populations of Primula subgenus Aleuritia. Sommerfeltia. 1990;11:117–135. [Google Scholar]
- Hultgård U-M. Primula scandinavica and P. stricta – patterns of distribution, variation, reproductive strategies and migrations. Opera Botanica. 1993;121:35–43. [Google Scholar]
- Kelso S. Taxonomy of Primula sects. Aleuritia and Armerina in North America. Rhodora. 1991;93:67–99. [Google Scholar]
- Kelso S. The genus Primula as a model for evolution in the Alaskan flora. Arctic and Alpine Research. 1992;24:82–87. [Google Scholar]
- Kovarík A, Matyásek R, Lim KY, Skalická K, Koukalová B, Knapp S, Chase M, Leitch AR. Concerted evolution of 18-5·8-26S rDNA repeats in Nicotiana allotetraploids. Biological Journal of the Linnean Society. 2004;82:615–625. [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Levin DA. The Role of chromosomal change in plant evolution. New York, NY: Oxford University Press; 2002. [Google Scholar]
- Levy AA, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society. 2004;82:607–613. [Google Scholar]
- Li Z-Y, Ge X-H. Unique chromosome behavior and genetic control in Brassica × Orychophragmus wide hybrids: a review. Plant Cell Reports. 2007;26:701–710. doi: 10.1007/s00299-006-0290-7. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarík A, Matyásek R, Bezdek M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;109:161–172. doi: 10.1007/s004120050424. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyásek R, Kovarík A, Leitch AR. Genome evolution in allotetraploid Nicotiana. Biological Journal of the Linnean Society. 2004;a 82:599–606. [Google Scholar]
- Lim KY, Skalická K, Koukalová B, Volkov RA, Matyásek R, Hemleben V, Leitch AR, Kovarík A. Dynamic changes in the distribution of a satellite homologous to intergenic 26-18S rDNA spacer in the evolution of Nicotiana. Genetics. 2004;b 166:1935–1946. doi: 10.1534/genetics.166.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Soucková-Skalická K, Sarasan V, Clarkson JJ, Chase MW, Kovarík A, Leitch AR. A genetic appraisal of a new synthetic Nicotiana tabacum (Solanaceae) and the Kostoff synthetic tobacco. American Journal of Botany. 2006;93:875–883. doi: 10.3732/ajb.93.6.875. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarík A, Matyásek R, Chase M, Clarkson JJ, Grandbastien MA, Leitch AR. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Liu B, Wendel JF. Epigenetic phenomena and the evolution of plant allopolyploids. Molecular Phylogenetics and Evolution. 2003;29:365–379. doi: 10.1016/s1055-7903(03)00213-6. [DOI] [PubMed] [Google Scholar]
- Liu B, Brubaker CL, Mergeai G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. [PubMed] [Google Scholar]
- Long EO, Dawid IB. Repeated genes in eukaryotes. Annual Review of Biochemistry. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Lysak M, Fransz P, Schubert I. Cytogenetic analyses of Arabidopsis. In: Salinas J, Sanchez-Serrano JJ, editors. Arabidopsis protocols. Methods in molecular biology. 2nd edn. Vol. 323. Totowa, NJ: Humana Press; 2006. pp. 173–186. [DOI] [PubMed] [Google Scholar]
- Ma X-F, Gustafson JP. Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenetics and Genome Research. 2005;109:236–249. doi: 10.1159/000082406. [DOI] [PubMed] [Google Scholar]
- Madlung A, Comai L. The effect of stress on genome regulation and structure. Annals of Botany. 2004;94:481–495. doi: 10.1093/aob/mch172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggini F, Cremonini R, Zolfino C, Tucci GF, D'Ovidio R, Delre V, et al. Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma. 1991;100:229–234. doi: 10.1007/BF00344156. [DOI] [PubMed] [Google Scholar]
- Mason-Gamer R. Reticulate evolution, introgression, and intertribal gene capture in an allohexaploid grass. Systematic Biology. 2004;53:25–37. doi: 10.1080/10635150490424402. [DOI] [PubMed] [Google Scholar]
- Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DMS, Conti E. Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. International Journal of Plant Sciences. 2001;162:1381–1400. [Google Scholar]
- Mast AR, Kelso S, Conti E. Are any primroses (Primula) primitively monomorphic? New Phytologist. 2006;171:605–616. doi: 10.1111/j.1469-8137.2006.01700.x. [DOI] [PubMed] [Google Scholar]
- Matyásek R, Lim KY, Kovarík A, Leitch AR. Ribosomal DNA evolution and gene conversion in Nicotiana rustica. Heredity. 2003;91:268–275. doi: 10.1038/sj.hdy.6800333. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Ecology and Systematics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Pikaard CS. The epigenetics of nucleolar dominance. Trends in Genetics. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, Viegas W, Pikaard CS. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome; Proceedings of the National Academy of Science, USA; 2004. pp. 18240–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Reeder RH. Mechanisms of nucleolar dominance in animals and plants. Journal of Cell Biology. 1985;101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. Primula. London: B. T. Batsford Ltd; 2002. [Google Scholar]
- Shao C, Stambrook PJ, Tischfield JA. Mitotic recombination is suppressed by chromosomal divergence in hybrids of distantly related mouse strains. Nature Genetics. 2001;28:169–172. doi: 10.1038/88897. [DOI] [PubMed] [Google Scholar]
- Skalická K, Lim KY, Matyášek R, Koukalová B, Leitch AR, Kovařík A. Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. American Journal of Botany. 2003;90:988–996. doi: 10.3732/ajb.90.7.988. [DOI] [PubMed] [Google Scholar]
- Skalická K, Lim KY, Matzke M, Leitch AR, Kovarík A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytologist. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Molecular data and dynamic nature of polyploidy. Critical Reviews in Plant Sciences. 1993;12:243–273. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since Plant speciation. New Phytologist. 2003;161:173–191. [Google Scholar]
- Soltis PS. Ancient and recent polyploidy in angiosperms. New Phytologist. 2005;166:5–8. doi: 10.1111/j.1469-8137.2005.01379.x. [DOI] [PubMed] [Google Scholar]
- Song K, Lu P, Tang K, Osborn TC. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution; Proceedings of the National Academy of Science, USA; 1995. pp. 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Song J, Tek AL, Cheng Z, Dong F, Jiang J. Highly condensed potato pericentromeric heterochromatin contains rDNA-related tandem repeats. Genetics. 2002;162:1435–1444. doi: 10.1093/genetics/162.3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 1999. version 4. [Google Scholar]
- Vogelmann HW. MI, USA: University of Michigan, Ann Arbor; 1956. A biosystematic study of Primula mistassinica Michx. PhD thesis. [Google Scholar]
- Wagner A, Blackstone N, Artwright P, Dick M, Misof B, Snow P, et al. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Systematic Biology. 1994;43:250–261. [Google Scholar]
- Wang J, Tian L, Madlung A, Lee H-S, Chen M, Lee JJ, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proceedings of the National Academy of Science, USA. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Sang T. Physical mapping of ribosomal RNA genes in peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: implications for phylogeny and concerted evolution. American Journal of Botany. 1999;86:735–740. [PubMed] [Google Scholar]