Abstract
Background and Aims
The aims of this study were to set up proliferation conditions for hairy roots of Coffea arabica regenerated after transformation by Agrobacterium rhizogenes strain A4-RS, and to carry out the morphological and molecular characterization of hairy root clones maintained over the long term.
Methods
Auxin supply, light conditions and sucrose concentration were modified with the aim of establishing efficient root proliferation conditions. The morphological variability among 62 established hairy root clones was phenotyped by scanning the roots and analysing the images using ‘whinRHIZO’ software procedures. PCR analysis of integration in transformed root cells of rol and aux oncogenes from the T-DNA of the Ri plasmid was used to study the molecular variability among clones.
Key Results
Auxin supply was necessary to obtain and stimulate growth and branching, and IBA applied at 0·5 µm was the most efficient auxin. Significant differences were shown among the 62 clones for total root length and for the percentage of fine roots. These variables were stable across subcultures and could hence be used for efficient characterization of hairy root clones. The majority of hairy root clones (86 %) exhibited non-significant phenotype differences with non-transformed roots. Eight clones were significantly different from the non-transformed controls in that they possessed a low proportion of fine roots. Two other hairy root clones grew significantly faster than the other clones. The PCR analysis revealed a low variability in the integration of rol and aux oncogenes in transformed root cells. The TR-DNA was never integrated as aux1 and aux2 genes were not found, although rolB and rolC genes from the TL-DNA were always present.
Conclusions
The discovery of low morphological variability among coffee hairy roots together with the identification of morphological variables allowing easy identification of phenotypically altered clones represent two important results. They make hairy roots a possible, and efficient, tool for functional-genomic studies of coffee root genes.
Key words: Agrobacterium rhizogenes, aux genes, Coffea arabica, genetic transformation, hairy roots, rol genes, root morphology
INTRODUCTION
Axenic cultures of transformed roots regenerated after Agrobacterium rhizogenes-mediated genetic transformation have been widely developed for various horticultural crops (Mugnier, 1987; Tepfer, 1990). Agrobacterium rhizogenes is a pathogenic soil bacterium that induces ‘hairy root’ disease on dicotyledonous plants, characterized by root proliferation at the infection site (Gaudin et al., 1994; Meyer et al., 2000). The term ‘hairy root’ is commonly used for A. rhizogenes-transformed roots and refers to the particular phenotype of those roots, often characterized by a highly branching root pattern and plagiotropic development attributed to increased endogenous auxin content (Nilsson and Olsson, 1997). Hairy roots have the capacity to grow even when removed from the plant and offer the interesting property of the easy regeneration of whole plants, avoiding callus formation and thus circumventing problems of somaclonal variation in a range of plant species (Tepfer, 1990). The Agrobacterium genes involved in rhizogenesis through the modification of plant cell growth and developmental regulation are commonly called rol and aux genes. These genes are located in the TR-DNA and TL-DNA regions, respectively, of the Ri (root-inducing) plasmid of A. rhizogenes agropine strains. Some of the genes are involved in auxin biosynthesis and/or auxin sensitivity, which cause differences in hairy root growth and morphology when compared to non-transformed roots (Meyer et al., 2000; Christey, 2001).
Noteworthy phenotype and growth variations have often been observed among hairy root clones derived from independent transformation events. In most cases, the variability that generally affects branching intensity, root diameter and growth rate has been described visually. Phenotype variations have been attributed to differences in the integration of genes from the TL-DNA and TR-DNA regions of A. rhizogenes in the host genome (Ambros et al., 1986; Mano et al., 1986, 1989; Jouanin et al., 1987). In Catharanthus, for example, Batra et al. (2004) demonstrated that an absence of aux genes did not affect either hairy root morphology or growth, whereas an absence of rolA&B genes induced callusing and slow-growing morphology. Since each hairy root is a cellular clone resulting from a single transformation event (Constantino et al., 1984), hairy root clone cultures and individuals regenerated from such roots retain the phenotype properties conferred by the specific T-DNA transferred to the plant cell (Mano et al., 1986, 1989; Jouanin et al., 1987; Meyer et al., 2000). The synergistic activity when all rol-gene products are simultaneously expressed is important in the induction of hairy roots and enhances development of the root rol-phenotype (Spena et al., 1987; Schmulling et al., 1988; Spanò et al., 1988). Phenotypic alterations such as short internodes, brittle and wrinkled leaves and stunted growth have been reported in A. rhizogenes-transformed coffee plantlets regenerated from somatic embryos of C. canephora (Spiral et al., 1993; Kumar et al., 2006), and from transformed roots of C. arabica (Sugiyama et al., 1995). However, no information has been provided about the phenotype of transformed roots. In other plant species, such morphological alterations have been reported for the root system and have been commonly described in terms of root thickness, excessive branching and amount of biomass production (Handa, 1991; Nguyen et al., 1997; Nin et al., 1997; Chaudhuri et al., 2005). This intra- or inter-clone variability among hairy root clones or the degree of phenotype alteration when compared to non-transformed roots could be major constraints for the utilization of hairy roots in functional-analysis studies. The high morphological variability results in a decrease in the accuracy of bio-assays and related analysis, and consequently requires the need to work with many lines and replicates that must be transferred regularly to fresh medium plates (Plovie et al., 2003).
Hairy roots have been used extensively to produce secondary metabolites for commercial use (see review by Hamill and Lidgett, 1997), in root nodule research (Diouf et al., 1995; Akasaka et al., 1998; Boisson-Dernier et al., 2001), to study genes specifically involved in plant morphology and development (Mikami et al., 1999) and as a system to validate and study genes of resistance to root-specific pathogens, such as nematodes. For example, hairy roots have been used successfully to study Mi, Hs1pr°–1 and Gpa2 gene function in tomato, sugar beet and potato, respectively (Kifle et al., 1999; Hwang et al., 2000; van der Vossen et al., 2000). One of the main constraints with hairy roots for functional-analysis studies is their frequent morphological variability. For example, this was shown to be responsible for large variations in nematode multiplication rates and hence complicated the interpretation of results, thus making it necessary subsequently to work with many clones (Plovie et al., 2003).
In coffee, protocols for A. rhizogenes-mediated transformation and plant regeneration were first described by Spiral et al. (1993), then by Alpizar et al. (2006) and Kumar et al. (2006). Although an efficient protocol for routine regeneration of hairy roots has been recently established for coffee (Alpizar et al., 2006), the transformed roots were unable to proliferate on semi-solid medium and died after two or three subcultures. Similar observations were made by Kumar et al. (2006). To our knowledge, suitable conditions for the effective maintenance of hairy root axenic cultures of coffee have yet to be described. The availability of long-term hairy root cultures in coffee would be very useful to study the root gene function through functional genomics. The purpose of this study therefore was: (1) to establish the culture conditions for efficient proliferation of hairy roots; (2) to phenotype numerous clones and determine if morphological variability observed was stable over time; and (3) to assess the links between the morphological variability and the integration pattern of rol and aux genes in transformed roots.
MATERIALS AND METHODS
Regeneration of hairy roots
Hairy roots of Coffea arabica L. ‘Caturra’ were regenerated by inoculating germinated zygotic embryos with the Agrobacterium rhizogenes A4RS strain according to procedures previously described by Alpizar et al. (2006). The A4RS strain, an agropine mannopine-type strain (Jouanin et al., 1986), was derived from the wild strain A4 modified for resistance to rifampycin and spectinomycin antibiotics. Briefly, zygotic embryos were infected by wounding the hypocotyl with a contaminated scalpel previously soaked in an Agrobacterium culture for 48 h. Co-cultivation with Agrobacterium was carried out by placing the inoculated embryos on a MS medium (Murashige and Skoog, 1962) supplemented with sucrose (30 g L–1) and solidified by adding 2·8 g L–1 of phytagel. Cultures were placed in 50-mm diameter Petri dishes in the dark for 12 d at 20 °C. Co-cultivated embryos were decontaminated by immersion in MS liquid medium with cefotaxime (500 µg mL–1) for 2 h and washed twice. They were then subcultured every 4 weeks onto MS germination medium containing decreasing cefotaxime concentrations (500, 200, 100 µg mL–1). Hairy roots appeared at the wound site after 8–10 weeks.
Development of culture conditions for hairy root maintenance
Root fragments of approx. 40 mm in length were excised from hairy roots derived from independent transformation events and were cultured on a medium containing full-strength MS salts and the following vitamins: 10 mg L–1 L-cystein, 10 mg L–1 thiamine-HCl, 1 mg L–1 pyridoxine-HCl, 2 mg L–1 glycin, 1 mg L–1 nicotinic acid. The medium was supplemented with 30 g L–1 sucrose and solidified by 2·8 g L–1 of phytagel. Hairy root clones were established by transferring 40-mm root tips every 4 weeks onto fresh medium in 9-cm diameter Petri dishes. Cultures were kept at 26 °C in the dark and at 55–60 % relative humidity.
These culture conditions were modified with the aim of establishing efficient conditions for hairy root proliferation, as follows
Auxin addition. Different auxins and concentrations were tested: IBA (indole 3-butyric acid) and NAA (napthelene acetic acid) were added to the proliferation medium prior to autoclaving, whereas IAA (indole 3-acetic acid) was filter-sterilized. Auxins were added to the medium at concentrations of 0, 0·125, 0·25, 0·5 and 5 µm.
Light conditions. Three different light conditions were compared: darkness, 20 µmol m–2 s–1 (intermediate light) and 50 µmol m–2 s–1 (full light) with a 16-h photoperiod.
Sucrose concentration. The proliferation medium including 0·25 µm IBA was supplemented with 0·5, 1, 2, 3, 4, 6 and 8 % (w/v) sucrose.
For all experiments, the same five root clones (X1, X6, X23, X25, X34) were used for each culture condition and were cultured in separate Petri dishes, used as replicates. Each hairy root clone was maintained for two subcultures in the tested culture conditions before assessment of growth parameters. The growth parameters determined were root branching, defined as the number of lateral roots per cm of initial mother root (number of lateral roots cm–1) and growth rate, defined as the average growth of lateral roots over 4 weeks of culture (mm d–1). The growth rate and branching were measured at the end of the second subculture. Ten randomly chosen root branches from five different Petri dishes (replicates) were evaluated for each culture condition. Vitrification was also evaluated in two experiments using a scale of 0 to 5, where zero corresponded to non-vitrified roots and 5 to totally vitrified roots. Vitrification was characterized by translucent, thick roots with frequent callusing.
Analysis of hairy root morphological variability
Morphological variability was analysed using 62 hairy root clones established for 6 months under the optimum culture conditions described in this work (see Results). Surprisingly, the optimized conditions enabled the proliferation of non-transformed roots for the first time. Three non-transformed root clones were therefore established from independent plants and used as non-transgenic controls.
For the assessment of root morphological characteristics, root images of 40-mm long branched root fragments from both hairy root clones and non-transformed root clones (controls) were acquired during two 3-week subcultures. The roots from three different Petri dishes, corresponding to three replicates, were evaluated for each root clone. The images were acquired using a scanner (HP ScanJet, 6000C/T) and were analysed using the software procedures of WhinRHIZO V5·0 (Instrument Regent Inc., Quebec, Canada). For each root clone, the variables measured were: number of lateral roots per cm of mother root (to evaluate root branching), the total root length (cm) at the end of each 3-week subculture, and the percentage of fine roots with a diameter less than 0·5 mm.
Statistical analysis
Data from the evaluation of culture-condition assays were analysed by an ANOVA and mean values were compared using Duncan's test at P = 0·001 for the light, auxin and sucrose concentration experiments. Data from the analysis of morphological variability between axenic root clones showed a normal distribution, with the exception of growth rate for which values were log(x + 1)-transformed to standardize variances prior to analysis. Data collected from the second subculture were analysed by an ANOVA, with the clone factor considered as a classification criterion, followed by a Duncan multiple-comparison at P < 0·05. All of the measurements taken after the second subculture were then compared with the corresponding measurements from the first subculture. The comparisons were performed by correlations, and by a discriminant canonical analysis in order to determine whether distribution and correlations between the variables observed would be affected by subcultures. The redundancy between the two sets of measurements enabled an evaluation of phenotype stability over time. All the statistical analyses were performed using Statistica software (2004, Statsoft, France).
Histology
Five representative root samples (cut at 3 cm from the root tip) were selected on different clones from each of the following root phenotypes: non-transformed roots, transformed roots with a non-altered phenotype, transformed roots exhibiting the two altered phenotypes, namely fast-growing and slow-growing with low percentage of branched fine roots. The samples were fixed for 24 h in a solution containing 1 % glutaraldehyde, 2 % paraformaldehyde and 1 % caffeine in a 0·2 mm phosphate buffer at pH 7·2. For histological studies, the samples were dehydrated in a graded series of ethanol, embedded in a 7100 resin (LKB) and cut into 3-mm longitudinal sections. The sections were double-stained with PAS (periodic acid–Schiff)-NBB (naphthol blue black). PAS specifically stains polysaccharides red (walls and starch) and NBB stains soluble and insoluble proteins blue (Fischer, 1968).
Staining with IV Sudan (Jensen, 1962) was carried out as follows: sections were immersed for 15 min in IV Sudan, saturated in 70 % methanol, then quickly washed three times with 50 % ethanol for observation. This reagent reveals suberin, which appears as orange/brown in colour. Sections of fresh roots were examined for their natural fluorescence under UV conditions (excitation filter: WL 270–380 nm; stop filter: WL 410–580). Blue auto-fluorescence indicated the presence of phenol compounds, including lignin.
Polymerase chain reaction (PCR)
DNA from transformed roots was extracted using the Dneasy® Plant Mini Kit No. 69104 (Qiagen®). To verify that the transformed root clones studied were not contaminated by the agrobacteria, PCR was carried out using primers designed to amplify a fragment of the virD1 gene, which is located outside the T-DNA region of the Ri plasmid. The primers 5′-ATGTCGCAAGGACGTAAGCCGA-3′ and 5′-GGAGTCTTTCAGCATGGAGCAA-3′ were used for amplification of a 450-bp sequence from the virD1 gene (Hamill et al., 1991). All the root clones analysed for the presence of the rol and aux genes were previously proved to be free of bacteria. The presence of rolA, rolB, rolC, rolD, aux1 and aux2 genes was determined by PCR analysis using the primers shown in Table 1. The PCR mixture consisted of 5 ng of plant DNA, 2·5 µL of 10X Taq buffer, 1·5 µL of 25 mm MgCl2, 1·0 µL of 5 mm dNTP, 1·25 units of Taq DNA polymerase and 1 µL from each 10 pmol primer, in a final volume of 25 µL. Three independent PCR analyses were performed for each hairy root clone with a PTC-100TM thermocycler (MJ Research Inc., San Francisco, CA, USA). For DNA amplification, samples were heated to 94 °C for 5 min, followed by 29 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min and then 56 °C for 10 min. The amplified products were separated by electrophoresis on 1·5 % agarose gels stained with 0·5 mg L–1 ethidium bromide in 0·5X TAE (Tris-acetate/EDTA electrophoresis buffer) and visualized by fluorescence under UV light.
Table 1.
Primers used for PCR analysis of rol and aux genes (Ri plasmid) in 55 Coffea arabica hairy root clones established after A. rhizogenes-mediated transformation
| Genes | Primers | PCR product size (bp) | Reference |
|---|---|---|---|
| rolA | 5′-ACGGTGAGTGTGGTTGTAGG-3′ | 403 | Slightom et al. (1986) |
| 5′-GCCACGTGCGTATTAATCCC-3′ | |||
| rolB | 5′-GCTCTTGCAGTGCTAGATTT-3′ | 383 | Furner et al. (1986) |
| 5′-GAAGGTGCAAGCTACCTCTC-3′ | |||
| rolC | 5′-CTCCTGACATCAAACTCGTC-3′ | 586 | Furner et al. (1986) |
| 5′-TGCTTCGAGTTATGGGTACA-3′ | |||
| rolD | 5′-TGCCTTGAGGTCATTCATCAAGGCC-3′ | 741 | Slightom et al. (1986) |
| 5′-ATGGACTGAAGGAGCACTCATTGGC-3′ | |||
| aux1 | 5′-CTCCGATTCCTTTCCAACCG-3′ | 791 | Camilleri and Jouanin (1991) |
| 5′-CGCACGTTATCCTCATACCC-3′ | |||
| aux2 | 5′-CTGTCAACGGAGGCTGTTGGG-3′ | 722 | Camilleri and Jouanin (1991) |
| 5′-ACCCTAGTCTCATCCCAGGG-3′ |
RESULTS
Effect of auxin type and concentration
The effects on the growth of C. arabica hairy roots of adding different auxins (IBA, IAA and NAA) to the culture medium were compared (Table 2). Little or no branching and growth were observed in the absence of exogenous auxin; low concentrations of the three auxins enabled hairy roots to branch and grow. With IBA and NAA, the optimum concentrations were 0·50 µm and 0·25 µm for branching and growth, respectively. The 5 µm concentration inhibited branching and growth. In contrast, with IAA the best branching pattern was obtained for 5 µm, although the growth rate was very low at that concentration. The incorporation of 0·5 µm IBA most efficiently stimulated the growth rate and branching of coffee hairy roots. Root vitrification was enhanced by NAA and IAA, even at low concentrations (0·125 µm), while with IBA significant vitrification symptoms only occurred using higher concentrations (0·5 and 5·0 µm). Using 0·25 µm IBA was the best compromise between root growth and vitrification status, as it led to high growth intensity and branching rates along with the lowest vitrification rate. That auxin treatment was adopted in all the following experiments.
Table 2.
Effect of different exogenous auxins (IAA, IBA and NAA) on hairy roots of C. arabica. Each value represents the average ± s.e. of five replicates
| Auxin type | Concentration (μm) | Branching (number of lateral roots cm–1) | Growth rate (mm d–1) | Vitrification index (0–5)* |
|---|---|---|---|---|
| No auxin (control) | 0 | 2·3 ± 1·0b | 0·05 ± 0·06d | 0·0c |
| IAA | 0·125 | 8·8 ± 4·2ab | 0·48 ± 0·11abc | 3·0 ± 0·5a |
| 0·25 | 5·8 ± 2·0b | 0·50 ± 0·29abc | 3·2 ± 0·6a | |
| 0·5 | 9·2 ± 3·8ab | 0·21 ± 0·11bcd | 3·6 ± 0·8ab | |
| 5 | 14·8 ± 2·1ab | 0·14 ± 0·07cd | 4·2 ± 0·5a | |
| IBA | 0·125 | 5·0 ± 1·8b | 0·44 ± 0·15abcd | 0·2 ± 0·3c |
| 0·25 | 11·3 ± 3·1ab | 0·60 ± 0·1ab | 1·2 ± 0·6bc | |
| 0·5 | 18·5 ± 5·0a | 0·68 ± 0·08a | 3·2 ± 0·6ab | |
| 5 | 0·6 ± 0·1c | 0·0d | 5·0 ± 0·0a | |
| NAA | 0·125 | 4·8 ± 1·2b | 0·28 ± 0·11abcd | 2·4 ± 0·9abc |
| 0·25 | 19·0 ± 6·4a | 0·55 ± 0·19abc | 4·0 ± 0·4a | |
| 0·5 | 11·4 ± 5·8ab | 0·26 ± 0·15abcd | 4·6 ± 0·6a | |
| 5 | 0·0c | 0·0d | 5·0 ± 0·0a |
* Index using a scale where 0 = none, and 5 = totally vitrified.
Values with different letters are significantly different at P < 0·001 according to Duncan's test.
Effect of light intensity
Light conditions had significant and marked effects on the branching, growth rate and vitrification of C. arabica hairy roots (Table 3). The branching and growth rate of hairy roots in full light (50 µmol m–2 s–1) were the weakest out of the three different culture conditions that were studied. Significant enhancement of root growth was observed with an intermediate light intensity (20 µmol m–2 s–1); darkness led to efficient branching but intermediate light gave the highest root growth rate. None of the light conditions caused marked vitrification symptoms in coffee hairy roots.
Table 3.
Effect of light intensity on root branching, growth rate of lateral roots and root vitrification of C. arabica hairy roots grown in a MS medium containing 0·25 µm IBA. Each value represents the average ± s.e. of five replicates
| Light intensity (μmol m–2 s–1) | Branching (number of lateral roots cm–1) | Growth rate (mm d–1) | Vitrification index (0–5)* |
|---|---|---|---|
| 0 | 11·3 ± 3·2a | 0·6 ± 0·09b | 1·2 ± 0·6a |
| 20 | 10·4 ± 2·7a | 0·76 ± 0·08a | 0·0 ± 0·0b |
| 50 | 0·4 ± 0·4b | 0·01 ± 0·01c | 0·8 ± 0·3a |
* Index using a scale where 0 = none, and 5 = totally vitrified.
Means within a column followed by the same letter were not significantly different at P < 0·001 according to Duncan's multiple test.
Effect of sucrose
Sucrose concentration moderately affected the growth of coffee hairy roots. Optimum sucrose concentrations for branching were found over a wide range, between 1 and 4 % (data not shown). The highest growth rates were observed at low sucrose concentrations (1 and 2 %) and higher concentrations caused a progressive reduction in root growth. It was thus decided to incorporate 2 % sucrose in the proliferation medium for the following experiments.
Morphological variability among hairy root clones
Morphological differences could be observed visually between hairy roots (Fig. 1). The ANOVA of morphological variability among 62 hairy root clones and three non-transformed root clones of C. arabica showed significant differences between the clones for total root length (P < 0·001) and for the percentage of fine roots (P < 0·001). For these two variables, it was observed that the average behaviour of a clone at the end of the second growth period (42 d) was similar to that of the same clone after 21 d (Fig. 2). A correlation analysis between principal components at the end of the first subculture and the second subculture showed that canonical R was very high for the variables percentage of fine roots and total root length (R = 0·93 and 0·80, respectively). The redundancy between the two data sets (i.e. the share of variance explained by canonicals) amounted to 89%. Consequently, we assumed that the phenotypes were stable over time for these two parameters. The branching pattern, expressed as the number of lateral roots per cm of mother root, did not display significant differences between hairy root clones. The average behaviour of a clone after the second subculture period was not well correlated with that of the same clone after the first subculture (Fig. 2). Consequently, the branching pattern was not used to characterize particular altered phenotypes in the following analyses.
Fig. 1.
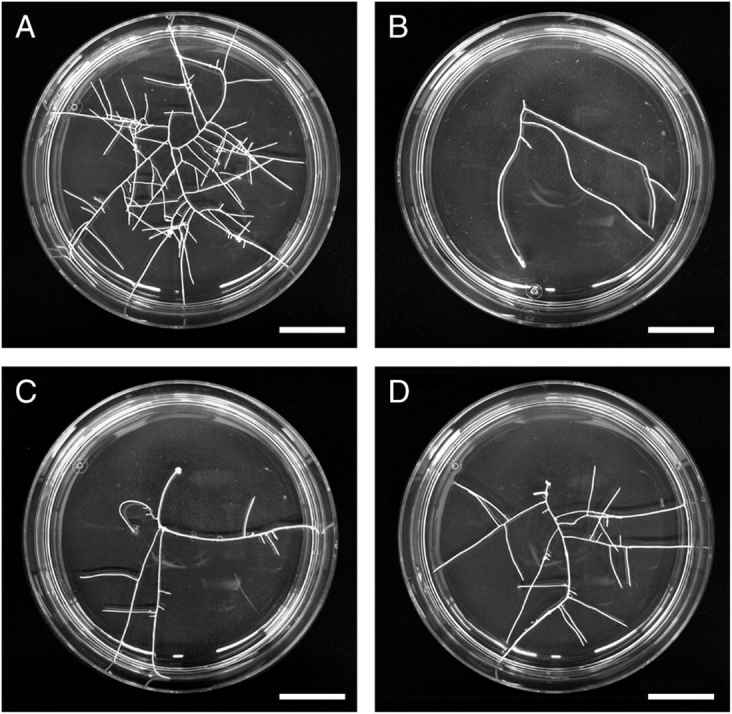
Different phenotypes of coffee hairy root clones in comparison with non-transformed roots (control). (A) X16 hairy root clone showing a high growth rate and a large percentage of branched fine roots; (B) non-transformed root clone (C1, control); (C) X43 hairy root clone with low growth rate and small percentage of branched fine roots; (D) X22 hairy root clone exhibiting a normal phenotype with a large percentage of branched fine roots and an intermediate growth rate. Scale bar = 10 mm.
Fig. 2.
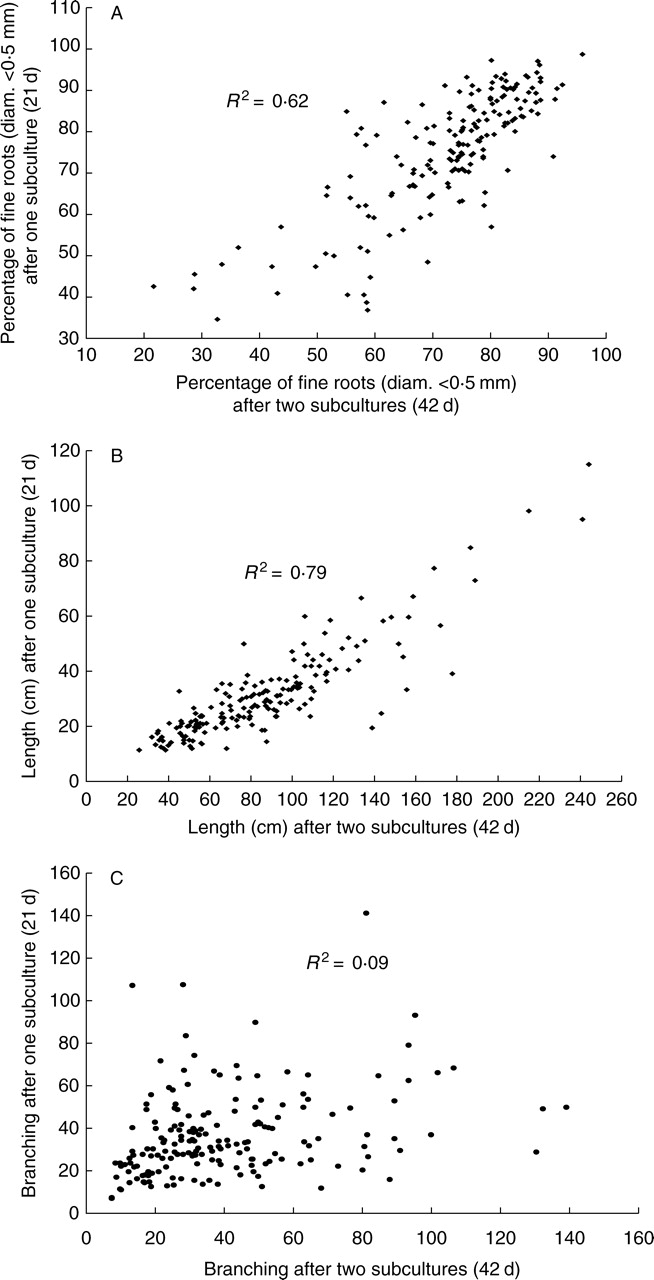
Correlations between growth variables characterizing the development of 62 coffee hairy root clones. The three variables were assessed at the end of the first subculture (21 d) and at the end of the following subculture (42 d). A coefficient of determination R2 was calculated and is indicated on the graphs.
The Duncan test following the ANOVA of the percentage of fine roots data revealed that 52 hairy root clones (amounting to 86 %) were significantly similar to the non-transformed controls for the proportion of fine roots. On the other hand, eight hairy root clones showed less than 75 % of fine roots and were significantly different from the non-transformed control roots. For the total root length variable, it was found that only two hairy root clones were significantly different from the other clones.
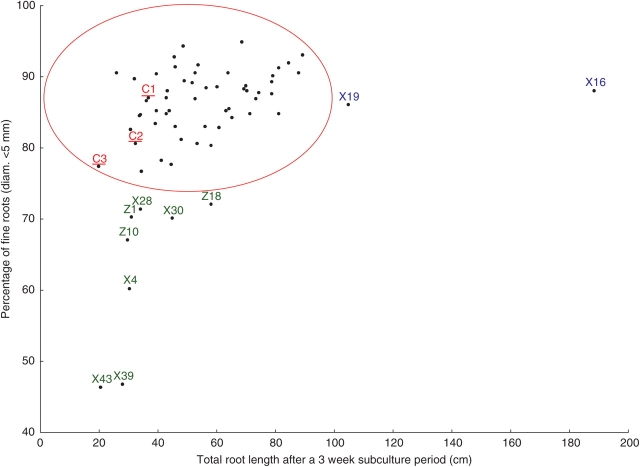
The data array in Fig. 3 shows that the majority of root clones formed a cluster with an average total root length of between 20–90 cm and a percentage of fine roots between 75–95%. Clones characterized by a high percentage of fine roots (including those of the cluster) showed higher total root lengths than clones with thicker roots. As expected, the two root clones (X19, X16) that were found to be significantly different from the non-transformed control roots for total root length and eight others (X4, X28, X30, X39, X43, Z1, Z10, Z18) different for percentage of fine roots were positioned distantly from the cluster of root clones showing a normal phenotype (Fig. 3), and could logically be considered as phenotypically altered.
Fig. 3.
Relation between total root length (cm) after a 3-week subculture period and the percentage of fine roots for 62 hairy root clones and three non-transformed clones (controls) of Coffea arabica. Data from the control clones are shown in red and underlined (C1, C2 and C3). Hairy root clones significantly different from the controls for the percentage of fine roots (i.e. possessing thick roots) are shown in green and those differing for the total root length are shown in blue (i.e. fast growing). The hairy root clones within the ellipse were not significantly different from the controls in terms of their morphology.
Histological observations of hairy root clones cultured in the same conditions revealed anatomical differences for the clones exhibiting the altered phenotype characterized by slow growth and a low percentage of fine roots (Fig. 4). The frequent thick roots from this altered phenotype systematically showed (1) a central cylinder with a greater diameter and (2) a greater number of cell layers in the cortical parenchyma (7 or 8) as compared with all other types of transformed or non-transformed coffee roots (4 or 5). UV observations did not reveal any differences in autofluorescence between the different root phenotypes. Similarly, staining with IV Sudan did not display differences regarding the presence of suberin (data not shown).
Fig. 4.
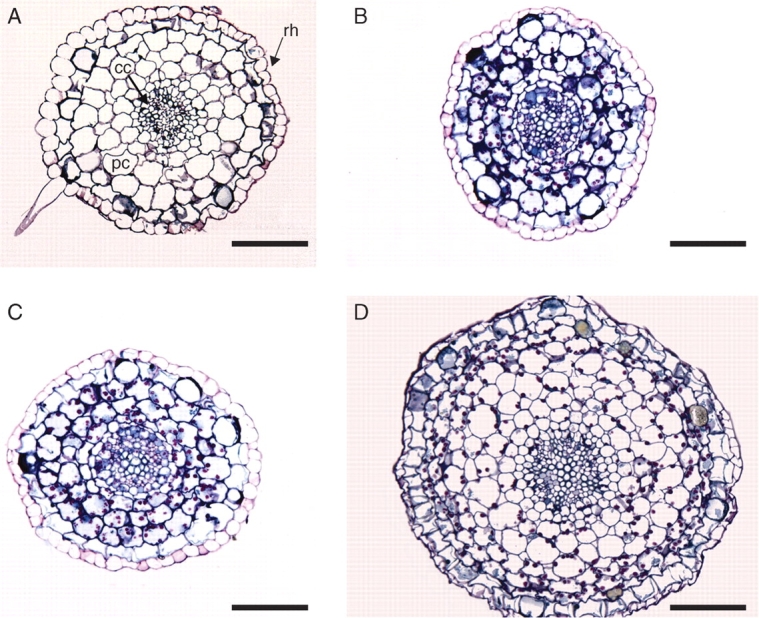
Histological cross-sections of normal, non-transformed roots and A. rhizogenes-transformed roots. (A) Normal root; (B) transformed root showing a non-altered phenotype (clone X13); (C) transformed root exhibiting the altered ‘fast growing’ phenotype (clone X19); and (D) transformed root with the altered phenotype characterized by slow growth and low percentage of fine roots (diameter <0·5 mm; clone X39). cc, central cylinder; rh, rhizoderm; pc, cortical parenchyma. Scale bar = 100 µm.
Molecular variability for the rol and aux genes in hairy root clones
No band was amplified from non-transgenic roots for rol and aux genes (Table 4). All the hairy root clones showed a similar pattern for the oncogenes and produced bands identical to the positive control for rol genes (Ri plasmid). The rolB and rolC genes were systematically integrated in the transformed coffee roots and the few differences concerning the presence of rolA and rolD genes could not be related to the morphological variability. For the clones showing altered root phenotypes (fast-growing or thick diameter), no correlations were found between the phenotype alterations and the absence/presence of rol and aux oncogenes from the T-DNA of the Ri plasmid established through PCR analysis. Interestingly, we noted that the TR-DNA from the Ri plasmid bearing the aux1 and aux2 genes was never integrated in coffee cells, although the TL-DNA bearing the rol genes is always transferred.
Table 4.
PCR analysis of aux and rol genes insertion in coffee hairy root clones. The presence of the different aux and rol genes belonging to the TR and TL T-DNA sub-fragments from the A. rhizogenes Ri plasmid was determined in 55 hairy root clones
| Genes from TL |
Genes from TR |
|||||||
|---|---|---|---|---|---|---|---|---|
| Hairy root clones | rolA | rolB | rolC | rolD | aux1 | aux2 | Phenotypic alteration | |
| A4RS strain (control) | + | + | + | + | + | + | ||
| Non-transformed roots | − | − | − | − | − | − | ||
| X1 | + | + | + | + | − | − | − | |
| X2 | + | + | + | + | − | − | − | |
| X3 | + | + | + | + | − | − | Thick roots | |
| X4 | + | + | + | + | − | − | − | |
| X6 | + | + | + | + | − | − | − | |
| X8 | + | + | + | + | − | − | − | |
| X9 | + | + | + | + | − | − | − | |
| X11 | + | + | + | + | − | − | − | |
| X12 | + | + | + | + | − | − | − | |
| X13 | + | + | + | + | − | − | − | |
| X14 | + | + | + | + | − | − | − | |
| X16 | + | + | + | − | − | − | Fast growing | |
| X17 | + | + | + | + | − | − | − | |
| X18 | + | + | + | + | − | − | − | |
| X19 | + | + | + | + | − | − | Fast growing | |
| X20 | + | + | + | + | − | − | − | |
| X22 | + | + | + | − | − | − | − | |
| X23 | + | + | + | + | − | − | − | |
| X24 | + | + | + | + | − | − | − | |
| X25 | + | + | + | + | − | − | − | |
| X26 | + | + | + | + | − | − | − | |
| X28 | + | + | + | + | − | − | Thick roots | |
| X29 | + | + | + | − | − | − | − | |
| X30 | + | + | + | + | − | − | Thick roots | |
| X32 | + | + | + | + | − | − | − | |
| X34 | + | + | + | + | − | − | − | |
| X36 | + | + | + | + | − | − | − | |
| X37 | + | + | + | − | − | − | − | |
| X38 | + | + | + | − | − | − | − | |
| X39 | + | + | + | − | − | − | Thick roots | |
| X40 | + | + | + | − | − | − | − | |
| X41 | + | + | + | + | − | − | − | |
| X43 | + | + | + | + | − | − | Thick roots | |
| X44 | + | + | + | + | − | − | − | |
| Y3 | − | + | + | + | − | − | − | |
| Y5 | + | + | + | + | − | − | − | |
| Y8 | + | + | + | + | − | − | − | |
| Y9 | + | + | + | + | − | − | − | |
| Y12 | + | + | + | + | − | − | − | |
| Y13 | + | + | + | + | − | − | − | |
| Z1 | + | + | + | + | − | − | Thick roots | |
| Z3 | + | + | + | + | − | − | − | |
| Z5 | − | + | + | − | − | − | − | |
| Z7 | + | + | + | + | − | − | − | |
| Z8 | − | + | + | − | − | − | − | |
| Z10 | − | + | + | + | − | − | Thick roots | |
| Z14 | + | + | + | + | − | − | − | |
| Z15 | + | + | + | + | − | − | − | |
| Z16 | + | + | + | + | − | − | − | |
| Z18 | − | + | + | + | − | − | Thick roots | |
| Z19 | − | + | + | + | − | − | − | |
| Z21 | + | + | + | + | − | − | − | |
| Z22 | − | + | + | − | − | − | − | |
| Z23 | + | + | + | − | − | − | − | |
| Z24 | + | + | + | + | − | − | − | |
DISCUSSION
The main cultivated varieties of C. arabica have been shown to be susceptible to sedentary endoparasitic root-knot nematodes (Meloidogyne spp.; Campos et al., 1990), which represent a major pest for coffee culture. Hairy roots have been proposed as an easy system for testing nematode-resistance in crop plants and were successfully used to study Mi and Hs1pro-1 gene function in tomato and sugar beet, respectively (Cai et al., 1997; Remeeus et al., 1998; Kifle et al., 1999; Hwang et al., 2000). The development of protocols for coffee hairy root regeneration by A. rhizogenes-mediated transformation and for subsequent proliferation has become a priority in order to rapidly validate different nematode-resistance genes, such as Mex-1 conferring resistance to Meloidogyne exigua (Noir et al., 2003). Although coffee hairy roots have been obtained by different workers (Alpizar et al., 2006; Kumar et al., 2006), the conditions for proliferation and maintenance remained to be established.
Negative effects of light exposure for hairy root multiplication
We have demonstrated that coffee hairy root cultures require an absence of light or low light intensity for successful proliferation. A review of the literature revealed contradictory information about the effect of photoperiod and intensity of light on hairy root development. There have been few reports of such a negative impact of light exposure and for many species hairy roots have been grown under light conditions. Handa (1991) reported the successful maintenance of hairy roots from 30 different plant species under full light conditions. C. Liu et al. (2002) found higher growth rates of Artemisia sp. hairy roots under continuous light conditions and suggested that long periods of illumination accelerated the uptake of nutrients from the culture medium and produced an accumulation of carbohydrates in root axenic cultures. Opposite results were found by Vanhala et al. (1998), who observed that darkness stimulated root biomass production in Hyoscyamus compared with a 12-h photoperiod. In the current study, we also observed that coffee hairy roots under full light intensity became green and thicker, but exhibited little or no branching.
Requirement of auxin supply for hairy root proliferation
It has frequently been proposed that auxin – through an increased auxin content or a higher sensitivity to auxins – is the main factor controlling the vigorous growth and development of hairy roots in comparison with non-transformed roots. In several plant species, A. rhizogenes- transformed roots can proliferate after excision from the plant in media devoid of growth regulators. That autonomy is related to the integration and expression in transformed cells of aux and rol genes from the Ri plasmid that enable the production of endogenous auxin (Nilsson and Olsson, 1997). However, in some species including coffee, that situation does not occur and it becomes essential to add exogenous auxins to the culture media in order to obtain and/or stimulate hairy root development. We have shown that the type and concentration of exogenous auxin strongly influenced root development. Depending on the auxin concentration, positive or adverse effects on hairy roots growth have been described. Park and Facchini (2000) in Papaver and C. F. Liu et al. (2002) in Pueraria found that IAA and IBA at 1·0 µm gave the strongest stimulation of branching but also caused higher inhibition of root growth, whereas both auxins at 0·1 µm promoted better lateral-root formation and growth. Conversely, even if high exogenous auxin concentrations promoted hairy root branching by stimulating cell division in the pericycle, the process seemed to inhibit hairy root growth because an excessive accumulation of auxin in the apex meristematic cells could inhibit meristematic tip activity (Finlayson et al., 1996) or lead to alterations in signalling response from other endogenous hormones involved in hairy root emergence and growth, such as polyamine (Ben-Hayyim et al., 1996), ethylene (Lorbiecke and Sauter, 1999) or ABA (De Smet et al., 2003). In our study, it was found that the optimum concentration for root branching (0·25 µm IBA) also corresponded to an optimum for growth rate; moreover, that auxinic treatment, when applied on several subcultures, supported very satisfactory coffee hairy root proliferation over the long term.
Evidence of limited morphological variability among coffee hairy roots
The improvement of culture conditions for hairy root proliferation is commonly evaluated by assessing weight increase, mainly because most hairy root research focuses on increased root biomass in order to stimulate the production of secondary metabolites. In the case of a functional analysis of genes involved in plant root/pathogen interactions, hairy roots can be of great interest but it is imperative to dispose of transformed roots that are morphologically similar to non-transgenic roots. An analysis of morphological variability thus becomes very important before using hairy roots in gene validation approaches.
Using the optimal culture conditions for coffee hairy root proliferation described above has resulted for the first time not only in the establishment of long-term axenic cultures of numerous transformed roots (62 lines), but also of some non-transformed roots. The availability of a significant number of established hairy root clones was an opportunity to study the morphological variability conferred by the A. rhizogenes-mediated transformation and to define the morphological variables that might enable an efficient distinction to be made between phenotypes. Most of the literature on hairy roots highlights the degree of lateral branching, plagiotropism, presence of numerous hairs and the capacity of hairy roots to grow when isolated from the mother plant in hormone-free medium (Nilsson and Olsson, 1997). Our results revealed that coffee hairy roots did not display most of these particular properties and that the majority of hairy root clones exhibited limited phenotype differences when compared with normal non-transformed root clones; strong differences were restricted to some particular clones. Both coffee hairy roots and non-transformed roots did not express growth autonomy in the absence of auxin supply. Abundant lateral branching has often been mentioned as one of the most typical traits within the altered phenotype of hairy roots (Tepfer, 1984; Spanò et al., 1988; Guivarc'h et al., 1999). Coffee hairy roots showed a higher degree of lateral branching than non-transformed roots. However, surprisingly, a statistical analysis of images of coffee root clones revealed high intra-clonal variability for the branching variable, which led to that variable being discarded as an efficient differentiation between hairy root phenotypes. It is probable that the average number of laterals per cm of mother root was not sufficient to characterize the root branching pattern. In our study, with the exception of the two altered phenotypes, the majority of hairy root clones displayed a growth intensity (expressed by root length) that was not significantly different from that of non-transformed roots. Fast growth of hairy roots has often been reported in the literature; nevertheless, that difference remains slight when compared with other species (Bonhomme et al., 2000; Park and Facchini, 2000). However, C. F. Liu et al. (2002), using kudzu (Pueraria lobata), observed that lateral root elongation was similar for both types of roots. Moreover, coffee hairy roots belonging to a composite plant exhibited a normal positive geotropism when grown in soil (Alpizar et al., 2006).
The existence of morphological variability between transformed and non-transformed roots on the one hand, and between hairy root clones on the other, was revealed using two variables: the percentage of branched fine roots and the total root length. Both variables were confirmed to be stable over subcultures and to provide an immediate application – via routine measurements through image acquisition and analysis of hairy roots – to screen and discard aberrant phenotypes. Phenotypic and growth characteristics of most of the coffee hairy root clones (86 %) were similar. Histological observations revealed differences in tissue organization only for roots from the altered phenotype exhibiting slow growth and a low proportion of branched fine roots. It was demonstrated that the frequency of altered phenotypes is rather low in C. arabica. Moreover, using these two morphological variables, it is now possible to limit the heterogeneity of hairy roots with a view to using only those clones displaying the normal phenotype in functional-analysis studies of coffee root genes.
Evidence of a limited variability in the integration of rol and aux genes
Among the 55 coffee hairy root clones analysed by PCR for the presence of rol and aux genes, we found evidence of only weak variability in the integration of these oncogenes, which was limited to the rol genes. Indeed, the TL-DNA fragment of the Ri plasmid was proved to be systematically integrated in transformed coffee cells, whereas aux genes from the TR-DNA fragment were never found, suggesting that TR-DNA is never inserted. This result agrees with previous works on different species. As reported by Altamura (2004), it has been demonstrated in various experiments performed by different researchers that the aux genes do not play a main role in hairy root disease, whereas the rol genes are necessary and sufficient per se to induce hairy roots. The absence of rolA or rolD genes of the TL-DNA fragment in a few independent hairy root clones do not affect their developmental fate, suggesting that only rolB and rolC gene integration is indispensable for the induction of the adventitious transformed roots in coffee. The rolB gene has been the most studied in the literature because it is the only one able to induce root formation on all plants tested when transferred individually, or to totally suppress root formation if inactivated (Nilsson and Olsson, 1997). As consequence, the presence of 10 % of altered phenotypes in coffee hairy roots can not be related to either the presence or absence of oncogenes. Nevertheless, those altered phenotypes could be explained by particular expression levels of one or various rol genes within these clones, through differences in gene copy number, position effects or by an epigenetic control. The inability of coffee hairy roots to grow without an auxin supply could be related to the absence of aux genes, responsible for auxin biosynthesis (Camilleri and Jouanin, 1991). Several authors have proposed that the role of TR-DNA aux genes of agropine T-DNA is restricted to supplementing auxin when the endogenous auxin content is insufficient (Binns and Costantino, 1998; Altamura, 2004), even if it was also demonstrated that other mechanisms leading to a stimulation of auxin biosynthesis and/or sensitivity are under control of TL-DNA genes (Filippini et al., 1994).
Efficient culture conditions for long-term maintenance of A. rhizogenes-transformed roots were established in the present work for the first time. All the isolated root fragments cultured under these conditions succeeded in proliferating. Sixty-two hairy root clones have been successfully maintained over 3 years and were characterized at the morphological and molecular levels in order to evaluate the degree of intra- and interclonal variability. The phenotyping approach employed showed a low morphological variability between hairy root clones compared with other species and the existence of only two altered phenotypes, which represented 14 % of all clones. It was demonstrated that the values of two growth parameters were characteristic of each root clone and were stable over subcultures. They can thus be used to identify efficiently the clones exhibiting these particular phenotypes, with the aim of discarding them from subsequent functional-analysis studies and increasing the homogeneity of the plant material, and hence the accuracy of the analysis. Moreover, the analysis of rol and aux oncogene integration confirmed at the molecular level the existence of a low variability within A. rhizogenes-transformed roots of coffee. These results make hairy roots a viable and useful tool for coffee breeding and offer new research perspectives; hairy roots could be used for the production of plant secondary metabolites and for functional-genomic studies of coffee root genes.
ACKNOWLEDGEMENTS
Financial support for this study was provided by the European Union through a grant to E. Alpizar by the ‘Programme Alßan’ European Union Programme of High Level Scholarships for Latin America (ID: E03D16144CR). The authors gratefully acknowledge Marc Lartaud (Cirad-Bios) for its technical assistance.
LITERATURE CITED
- Akasaka Y, Mii M, Daimon H. Morphological alterations and root nodule formation in Agrobacterium rhizogenes-mediated transgenic hairy roots of peanut (Arachis hypogaea L.) Annals of Botany. 1998;81:355–362. [Google Scholar]
- Alpizar E, Dechamp E, Espeout S, Lecouls AC, Nicole M, Bertrand B, Lashermes P, Etienne H. Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell Reports. 2006;25:959–967. doi: 10.1007/s00299-006-0159-9. [DOI] [PubMed] [Google Scholar]
- Altamura MM. Agrobacterium rhizogenes rolB and rolD genes: regulation and involvement in plant development. Plant Cell Tissue and Organ Culture. 2004;77:89–101. [Google Scholar]
- Ambros PF, Matzke AJM, Matzke AM. Localization of Agrobacterium rhizogenes T-DNA in plant chromosomes by in situ hybridization. The EMBO Journal. 1986;5:2073–2077. doi: 10.1002/j.1460-2075.1986.tb04468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra J, Dutta A, Singh D, Kumar S, Sen J. Growth and terpenoid indole alkaloid production in Catharanthus roseus hairy root clones in relation to left- and right-termini-linked Ri T-DNA gene intregration. Plant Cell Reports. 2004;23:148–154. doi: 10.1007/s00299-004-0815-x. [DOI] [PubMed] [Google Scholar]
- Ben-Hayyim G, Martin-Tanguy J, Tepfer D. Changing root and shoot architecture with the rolA gene from Agrobacterium rhizogenes: interactions with gibberellic acid and polyamine metabolism. Physiologia Plantarum. 1996;96:237–243. [Google Scholar]
- Binns AN, Costantino P. The Agrobacterium oncogenes. In: Spink H, Kondorosi A, Hooykaas PJJ, editors. The rhizobiaceae. Dordrecht: Kluwer Academic Publishers; 1998. pp. 251–266. [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker GD. Agrobacterium rhizogenes transformed roots of M. truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Molecular Plant–Microbe Interactions. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- Bonhomme V, Laurain-Mattar D, Fliniaux MA. Effects of the rol C gene on hairy root: induction development and tropane alkaloid production by Atropa belladonna. Journal of Natural Products. 2000;63:1249–1252. doi: 10.1021/np990614l. [DOI] [PubMed] [Google Scholar]
- Cai D, Kleine M, Kifle S, Harloff HJ, Sandal NN, Marcker KA, et al. Positional cloning of a gene for nematode resistance in sugar beet. Science. 1997;275:832–834. doi: 10.1126/science.275.5301.832. [DOI] [PubMed] [Google Scholar]
- Camilleri C, Jouanin L. The TR-DNA region carrying the auxin synthesis genes of the Agrobacterium rhizogenes agropine-type plasmid pRiA4: nucleotide sequence analysis and introduction into tobacco plants. Molecular Plant–Microbe Interactions. 1991;4:155–162. doi: 10.1094/mpmi-4-155. [DOI] [PubMed] [Google Scholar]
- Campos VP, Sivapalan P, Gnanapragasam NC. Nematode parasites of coffee, cocoa and tea. In: Luc M, Sikora RA, Bridge J, editors. Plant-parasitic nematodes in subtropical and tropical agriculture. Wallingford, UK: CAB International; 1990. pp. 113–126. [Google Scholar]
- Chaudhuri KN, Ghosh B, Tepfer D, Jha S. Genetic transformation of Tylphora indica with Agrobacterium rhizogenes A4: growth and tylophorine productivity in different transformed root clones. Plant Cell Reports. 2005;24:25–35. doi: 10.1007/s00299-004-0904-x. [DOI] [PubMed] [Google Scholar]
- Christey MC. Use of Ri-mediated transformation for production of transgenic plants. In vitro Cellular and Developmental Biology - Plant. 2001;37:687–700. [Google Scholar]
- Constantino P, Spanò L, Pomponi M, Ancora G. T-DNA of Agrobacterium rhizogenes is transmitted to the progeny pf hairy root plants. Journal of Molecular and Applied Genetics. 1984;2:465–470. [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Foyer CH, Zhang H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. The Plant Journal. 2003;33:543–555. doi: 10.1046/j.1365-313x.2003.01652.x. [DOI] [PubMed] [Google Scholar]
- Diouf D, Gherbi H, Prin Y, Franche C, Duhoux E, Bogusz D. Hairy root nodulation of Casuarina glauca: a system for the study of symbiotic gene expression in actinorhizal tree. Molecular Plant–Microbe Interactions. 1995;8:532–537. doi: 10.1094/mpmi-8-0532. [DOI] [PubMed] [Google Scholar]
- Filippini F, Lo Schiavo F, Terzi M, Costantino P, Trovato M. The plant oncogene rolB alters binding of auxin to plant cell membranes. Plant Cell Physiology. 1994;35:767–771. [Google Scholar]
- Finlayson SA, Liu JH, Reid DM. Localization of ethylene biosynthesis in roots of sunflower seedlings. Physiologia Plantarum. 1996;96:36–42. [Google Scholar]
- Fisher DB. Protein staining of ribboned epon sections for light microscopy. Histochemie. 1968;16:92–96. doi: 10.1007/BF00306214. [DOI] [PubMed] [Google Scholar]
- Furner IJ, Huffman GA, Amasino RM, Garfinkel DJ, Gordon MP, Nester EW. An Agrobacterium transformation in the evolution of the genus Nicotiana. Nature. 1986;319:422–427. [Google Scholar]
- Gaudin V, Vrain T, Jouanin L. Bacterial genes modifying hormonal balances in plants. Plant Physiology and Biochemistry. 1994;32:11–29. [Google Scholar]
- Guivarc'h A, Boccara M, Prouteau M, Chriqui D. Instability of phenotype and gene expression in long-term culture of carrot hairy root clones. Plant Cell Reports. 1999;19:43–50. doi: 10.1007/s002990050708. [DOI] [PubMed] [Google Scholar]
- Handa T. Establishment of hairy root lines by inoculation with Agrobacterium rhizogenes. Bulletin of RIAR, Ishikawa Agriculture College. 1991;2:13–18. [Google Scholar]
- Hamill JD, Lidgett AJ. Hairy root cultures – opportunities and key protocols for studies in metabolic engineering. In: Doran PM, editor. Hairy roots: culture and applications. Amsterdam: Harwood Academic Publishers; 1997. pp. 1–30. [Google Scholar]
- Hamill JD, Rounsley S, Spencer A, Todd G, Rhodes MJ. The use of the polymerase chain reaction in plant transformation studies. Plant Cell Reports. 1991;10:221–224. doi: 10.1007/BF00232562. [DOI] [PubMed] [Google Scholar]
- Hwang CF, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM. Evidence for a role of the N terminus and Leucine-Rich Repeat region of the Mi gene product in regulation of localized cell death. Plant Cell. 2000;12:1319–1329. doi: 10.1105/tpc.12.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen WA. Botanical histochemistry: principles and practices. San Francisco: WH Freeman and Company; 1962. [Google Scholar]
- Jouanin L, Tourneur J, Casse-Delbart F. Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid. 1986;16:124–134. doi: 10.1016/0147-619x(86)90071-5. [DOI] [PubMed] [Google Scholar]
- Jouanin L, Guerche D, Pamboukdjian N, Tourneur C, Casse-Delbart F, Tourneur J. Structure of T-DNA in plants regenerated from roots transformed by Agrobacterium rhizogenes strain A4. Molecular and General Genetics. 1987;206:387–392. [Google Scholar]
- Kifle S, Shao M, Jung C, Cai D. An improved transformation protocol for studying gene expression in hairy roots of sugar beet (Beta vulgaris L.) Plant Cell Reports. 1999;18:514–519. [Google Scholar]
- Kumar V, Satyanarayana KV, Itty S, Indu EP, Giridhar P, Chandrashekar A, Ravishankar GA. Stable transformation and direct regeneration in Coffea canephora P ex. Fr. by Agrobacterium rhizogenes mediated transformation without hairy-root phenotype. Plant Cell Reports. 2006;25:214–222. doi: 10.1007/s00299-005-0045-x. [DOI] [PubMed] [Google Scholar]
- Liu C, Guo C, Wang Y, Ouyang F. Effect of light irradiation on hairy root growth and artemisinin biosynthesis of Artemisia annua L. Process Biochemistry. 2002;38:581–585. [Google Scholar]
- Liu CF, Zhu J, Liu Z, Li L, Pan RC, Jin LH. Exogenous auxin effects on growth and phenotype of normal and hairy roots of Pueraria lobata (Wild) Ohwi. Plant Growth Regulation. 2002;38 [Google Scholar]
- Lorbiecke R, Sauter M. Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiology. 1999;119:21–29. doi: 10.1104/pp.119.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y, Nabeshima S, Matsui C, Ohkawa H. Production of tropane alkaloids by hairy root cultures of Scopolia japonica. Agricultural Biological Chemistry. 1986;50:2715–2722. [Google Scholar]
- Mano Y, Ohkawa H, Yamada Y. Production of tropane alkaloids by hairy root cultures of Duboisia leichhaardtii by Agrobacterium rhizogenes. Plant Science. 1989;59:191–201. [Google Scholar]
- Meyer A, Tempé J, Constantino P. Hairy root: a molecular overview. Functional analysis of Agrobacterium rhizogenes T-DNA genes. In: Stacey G, Keen NT, editors. Plant–microbe interactions. Vol. 5. Saint Paul, MN: APS Press; 2000. pp. 93–139. [Google Scholar]
- Mikami Y, Horiike G, Kuroyanagi M, Noguchi H, Shimizu M, Kobayashi Niwa Y. Gene for a protein capable of enhancing lateral root formation. FEBS Letters. 1999;451:45–50. doi: 10.1016/s0014-5793(99)00489-5. [DOI] [PubMed] [Google Scholar]
- Mugnier J. Establishment of new hairy root lines by inoculation with Agrobacterium rhizogenes. Plant Cell Reports. 1987;7:9–12. doi: 10.1007/BF00272966. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nguyen C, Ourgaud F, Forlot P, Guckert A. Establishment of hairy root cultures of Psoralea species. Plant Cell Reports. 1997;11:424–427. doi: 10.1007/BF00234375. [DOI] [PubMed] [Google Scholar]
- Nilsson O, Olsson O. Getting to the root: the role of the Agrobacterium rhizogenes rol genes in formation of hairy root. Physiologia Plantarum. 1997;100:403–473. [Google Scholar]
- Nin S, Bennici A, Roselli G, Mariotti D, Schiff S, Magherini R. Agrobacterium-mediated transformation of Artemisia absinthium L. (wormwood) and production of secondary metabolites. Plant Cell Reports. 1997;16:725–730. doi: 10.1007/s002990050310. [DOI] [PubMed] [Google Scholar]
- Noir S, Anthony F, Bertrand B, Combes MC, Lashermes P. Identification of a major gene (Mex-1) from Coffea canephora conferring resistance to Meloidogyne exigua in coffee. Plant Pathology. 2003;52:97–103. [Google Scholar]
- Park SU, Facchini PJ. Agrobacterium rhizogenes-mediated transformation of opium poppy, Papaver somniferum L., and California poppy, Eschscholzia californica Cham., root cultures. Journal Experimental Botany. 2000;51:1005–1016. doi: 10.1093/jexbot/51.347.1005. [DOI] [PubMed] [Google Scholar]
- Plovie E, De Buck S, Goeleven E, Tanghe M, Vercauteren I, Gheysen G. Hairy roots to test for transgenic nematode resistance: think twice. Nematology. 2003;5:831–841. [Google Scholar]
- Remeeus PM, van Bezooijen J, Wijbrandi J, van Bezooijen J. In vitro testing is a reliable way to screen the temperature sensitivity of resistant tomatoes against Meloidogyne incognita. Proceedings of 5th international symposium on crop protection; Faculteit Landbouwkundige en Toegepaste Biologische Wetenschappen, Universiteit Gent, Belgium: Universiteit Gent; 1998. pp. 635–640. 2b. [Google Scholar]
- Schmulling T, Schell J, Spena A. Single genes from Agrobacterium rhizogenes influence plant development. The EMBO Journal. 1988;7:2621–2629. doi: 10.1002/j.1460-2075.1988.tb03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom JL, Durand-Tardif M, Jouanin L, Tepfer D. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid: identification of open-reading frames. Journal of Biological Chemistry. 1986;261:108–121. [PubMed] [Google Scholar]
- Spanò L, Mariotti D, Cardarelli M, Branca C, Constantino P. Morphogenesis and auxin sensitivity of transgenic tobacco with different complements of Ri T-DNA. Plant Physiology. 1988;87:479–483. doi: 10.1104/pp.87.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena A, Schmulling T, Koncz C, Schell J. Independent and synergistic activities of the rolA, B, and C loci in stimulating abnormal growth in plants. EMBO Journal. 1987;6:3891–3899. doi: 10.1002/j.1460-2075.1987.tb02729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiral J, Thierry C, Paillard M, Pétiard V. Obtention de plantules de Coffea canephora Pierre (Robusta) transformées par Agrobacterium rhizogenes. Comptes Rendus de l'Académie des Sciences – Paris. 1993;316:1–6. [Google Scholar]
- Sugiyama M, Matsuoka C, Takagi T. Transformation of coffee with Agrobacterium rhizogenes. Proceedings of the 16th International Conference on Coffee Science; 9–14 April; Kyoto, Japan. Lausanne, Switzerland: ASIC,: 1995. pp. 853–859. [Google Scholar]
- Tepfer D. Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell. 1984;37:959–967. doi: 10.1016/0092-8674(84)90430-6. [DOI] [PubMed] [Google Scholar]
- Tepfer D. Genetic transformation using Agrobacterium rhizogenes. Physiologia Plantarum. 1990;79:140–146. [Google Scholar]
- van der Vossen EAG, Rouppe van der Voort JNAM, Kanyuka K, Bendahmane A, Sandbrink H, Baulcombe CD, et al. Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant Journal. 2000;23:567–576. doi: 10.1046/j.1365-313x.2000.00814.x. [DOI] [PubMed] [Google Scholar]
- Vanhala L, Eeva M, Lapinjoki S, Hiltunen R, Oksman-Caldenty KM. Effect of growth regulators on transformed root cultures of Hyoscyamus muticus. Plant Physiology. 1998;153:475–481. [Google Scholar]