Abstract
The central actions of leptin are essential for homeostatic control of adipose tissue mass, glucose metabolism, and many autonomic and neuroendocrine systems. In the brain, leptin acts on numerous different cell types via the long-form leptin receptor (LepRb) to elicit its effects. The precise identification of leptin’s cellular targets is fundamental to understanding the mechanism of its pleiotropic central actions. We have systematically characterized LepRb distribution in the mouse brain using in situ hybridization in wildtype mice as well as by EYFP immunoreactivity in a novel LepRb-IRES-Cre EYFP reporter mouse line showing high levels of LepRb mRNA/EYFP coexpression. We found substantial LepRb mRNA and EYFP expression in hypothalamic and extrahypothalamic sites described before, including the dorsomedial nucleus of the hypothalamus, ventral premammillary nucleus, ventral tegmental area, parabrachial nucleus, and the dorsal vagal complex. Expression in insular cortex, lateral septal nucleus, medial preoptic area, rostral linear nucleus, and in the Edinger-Westphal nucleus was also observed and had been previously unreported. The LepRb-IRES-Cre reporter line was used to chemically characterize a population of leptin receptor-expressing neurons in the midbrain. Tyrosine hydroxylase and Cre reporter were found to be coexpressed in the ventral tegmental area and in other midbrain dopaminergic neurons. Lastly, the LepRb-IRES-Cre reporter line was used to map the extent of peripheral leptin sensing by central nervous system (CNS) LepRb neurons. Thus, we provide data supporting the use of the LepRb-IRES-Cre line for the assessment of the anatomic and functional characteristics of neurons expressing leptin receptor. J. Comp. Neurol. 514:518–532, 2009.
Indexing terms: green fluorescent protein, Cre recombinase, tyrosine hydroxylase, in situ hybridization histochemistry, CNS distribution
Leptin (Zhang et al., 1994) is an adipose tissue hormone that acts centrally to regulate multiple physiologic systems including those governing energy homeostasis, neuroendocrine axes, and reproduction (Ahima et al., 2000; Harris, 2000; Zigman and Elmquist, 2003; Bjorbaek and Kahn, 2004; Badman and Flier, 2005). The importance of the central component of leptin signaling in energy homeostasis is now established (Schwartz et al., 2000; Spiegelman and Flier, 2001; Niswender and Schwartz, 2003; Benoit et al., 2004). Exogenous leptin administered intracerebroventricularly reduces food intake and body weight at doses found to be subthreshold in the periphery (Campfield et al., 1995; Seeley et al., 1996). Additionally, selective elimination of central leptin receptors recapitulates many of the phenotypes of the universal db/db knockout (Cohen et al., 2001). Reciprocally, abnormalities of the db/db mouse are markedly improved by transgenic replacement of central LepRbs. Consistent with this, long-form leptin receptor (LepRb) mRNA, the splice variant of the leptin receptor that is required for leptin’s biologic effects, has been localized in numerous brain regions.
The hypothalamus, in particular the arcuate nucleus (ARH), shows dense LepRb expression and the ablation of the LepRb in specific ARH cell groups (Balthasar et al., 2004; van de Wall et al., 2008) established the functional importance of leptin signaling at the ARH. This conclusion is further supported by the results obtained using selective reactivation of ARH LepRbs (Morton et al., 2003,Morton et al., 2005; Coppari et al., 2005) However, the inability of these manipulations to fully recapitulate or rescue the db/db obesity phenotype indicates that other brain regions besides the ARH are required for mediating the full spectrum of leptin’s effects. Thus, the identification and characterization of all the leptin-responsive brain regions, not just those in the hypothalamus, will be required to define the neural basis of leptin action (Robertson et al., 2008). The delineation of the aggregate sites of leptin action would facilitate the targeted genetic ablation of leptin receptor expression, establishing the minimal set of neurons required for mediating leptin’s biologic effects.
While several studies have attempted to define the sites of LepRb in rodent brain (Huang et al., 1996; Mercer et al., 1996a, 1998) the relatively low abundance of LepRb mRNA and the absence of selective leptin receptor antibodies have hindered a comprehensive description of LepRb distribution in mouse brain. In an effort to overcome these limitations and to provide a means for directly identifying LepRb-expressing cell populations, we have developed a novel mouse model that expresses Cre recombinase from the LepR locus, specifically in LepRb-expressing cells. Crossing the LepRb-IRES-Cre mouse (DeFalco et al., 2001) with the Cre reporter mouse lines R26R-EYFP and R26R-LacZ allows us to label LepRb-expressing neurons, obviating the need for performing in situ hybridization and avoiding the problems inherent in detecting the low level of LepRb mRNA expression.
In this report we systematically characterize LepRb mRNA distribution in the mouse brain using a sensitive free-floating in situ hybridization technique. Subsequently, we proceeded to validate the eutopic expression of EYFP and LacZ in progeny of LepRb-IRES-Cre × ROSA26-EYFP/LacZ intercrosses. The LepRb-IRES-Cre LacZ mice were then used to map the central nervous system (CNS) neuronal populations sensitive to peripherally administered leptin, as demonstrated by coexpression of LacZ with phosphorylated STAT3 (pSTAT3), a marker of leptin receptor activation. Finally, using LepRb-IRES-Cre-LacZ mice, we characterized the expression of leptin receptor in dopaminergic neurons of the mid/forebrain, a recently described target of peripheral leptin. (Figlewicz et al., 2003; Fulton et al., 2006; Hommel et al., 2006).
MATERIALS AND METHODS
Production of the LepRb-Cre EYFP and LacZ reporter mice
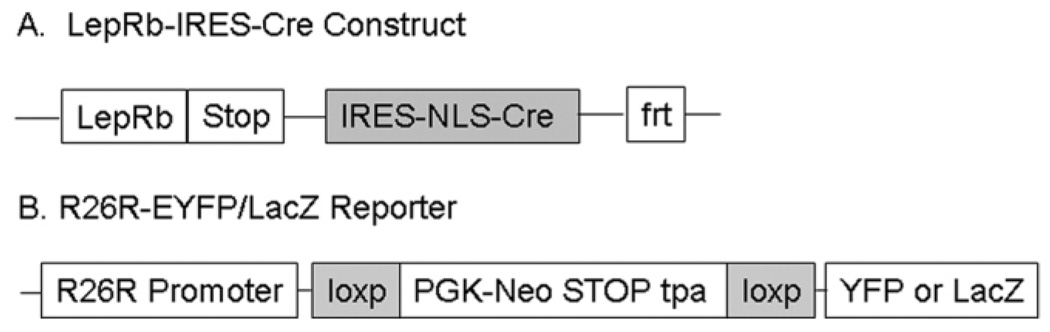
As described previously, LepRb-IRES-Cre mice were generated using homologous recombination to introduce an IRES-NLS-Cre cassette into the region immediately 3′ to the LepRb stop codon (Fig. 1A) (DeFalco et al., 2001). As a result of this placement, Cre recombinase is expressed selectively in cells that produce LepRb. In order to visualize the LepRb cells, we crossed the LepRb-IRES-Cre mice with reporter mice that express either EYFP or LacZ in a Cre-dependent manner (Srinivas et al., 2001). The reporter mouse was designed to contain a loxP-flanked transcriptional stop sequence preceding the EYFP/LacZ gene inserted into the ROSA26 locus (Fig. 1B). In the absence of Cre recombinase, a premature termination of the gene transcript results in no EYFP or LacZ. In the Cre recombinase-producing cells, the transcriptional termination sequence is excised allowing EYFP/LacZ production from the ROSA26 locus in LepRb-expressing cells. Importantly, there was no detectable difference in expression between the EYFP and LacZ reporter lines when crossed to LepRb-Cre (data not shown).
Figure 1.
Generation of mice expressing EYFP/LacZ selectively in LepRb-expressing cells. A: Schematic illustration of the LepRb-IRES-Cre construct. An IRES-NLS-Cre cassette was inserted just 3′ to the LepRb stop codon in the 3′ untranslated region of the LepR gene allowing Cre recombinase to be expressed from the IRES element only in LepRb-containing cells. B: The design for the R26R-EYFP/LacZ Cre recombinase reporter mouse. A floxed transcriptional termination sequence (tp-A) followed by the EYFP or lacZ gene was targeted to the ROSA26 locus. In the absence of Cre recombinase, the tp-A prevents EYFP or lacZ from being expressed. After crossing these mice with a Cre recombinase-expressing mouse strain, a Cre recombination event excises the tp-A sequence, thereby allowing EYFP or LacZ to be expressed under the constitutively active ROSA26 locus selectively in the Cre-producing cell types.
Animals and Histology
The LepRb-Cre EYFP/LacZ adult male mice (20–30 g, 4–8 weeks old, n = 6) and C57BL/6 adult male mice (25–35 g, 6–8 weeks old; n = 4) were housed with ad libitum access to both food and water in a light- (12 hours on, 12 hours off) and temperature- (21.5–22.5°C) controlled environment. For the experiments investigating the CNS neurons responsive to peripherally administered leptin, animals were fasted overnight with access to water. The animals and procedures used for these studies were in accordance with the Rockefeller Animal Research Center guidelines as well as the guidelines and approval of the Harvard Medical School, Beth Israel Deaconess Medical Center, and UT Southwestern Institutional Animal Care and Use Committees. Mice were deeply anesthetized with an intraperitoneal injection of chloral hydrate (350 mg/kg) and perfused transcardially with DEPC-treated 0.9% saline followed by 10% neutral buffered formalin. Brains were removed, postfixed in 10% neutral buffered formalin overnight at 4°C, dehydrated in 20% sucrose made in DEPC-treated phosphate-buffered saline (PBS), pH 7.0 at 4°C, and cut coronally at 25 µM into five equal series on a freezing microtome. Tissue was stored at −20°C in antifreeze solution (Simmons et al., 1989) until processed.
Immunohistochemisty (IHC)
To identify nuclei expressing EYFP we performed IHC using a well-characterized polyclonal antiserum made in rabbit against the green fluorescent protein (GFP) purified directly from the jellyfish Aequorea victoria (Molecular Probes/ Invitrogen, Eugene, OR; Cat. no. A-6455, lot 71B1) and crossreacts with EYFP. The EYFP antiserum does not exhibit immunoreactivity in brains that are devoid of EYFP or GFP protein expression. IHC detection of LacZ was accomplished using polyclonal antisera made in rabbit (Cortex Biochemicals, San Leandro, CA; Cat. no. CR7001RP2, lot 595554) and chicken (Abcam, Cambridge, MA; Cat. no. ab9361-250, lot 397981) against purified bacterial β-galactosidase. The β-galactosidase antisera do not exhibit immunoreactivity in brains that are devoid of β-galactosidase expression. IHC detection of tyrosine hydroxylase was performed using antiserum made in sheep (Chemicon, Temecula, CA; Cat. no. AB1542, lot 0607035104) against native rat tyrosine hydroxylase from pheochromocytoma. In prior studies, confirmation of antibody specificity involved the electrophoresis and immunoblotting of purified rat tyrosine hydroxylase and pheochromocytoma cell extracts with antibody AB1542, which subsequently demonstrated the presence of a single protein species of expected molecular weight for tyrosine hydroxylase in both cases (Haycock and Waymire, 1982). As well, in our experiments antibody specificity was confirmed through analysis of the reactivity pattern of AB1542 in the brainstem, which corresponded highly with the original histofluorescence studies of catecholaminergic brainstem neurons (Dahlstroem and Fuxe, 1964). Detection of pSTAT3 was accomplished using antiserum made in rabbit (Cell Signaling Technologies, Beverly, MA; Cat. no. 9131L, lot 9) against a synthetic phosphopeptide of sequence ADPGSAAPyLKTKFIC corresponding to the amino acid residues flanking Tyr705 (lowercase y) of mouse STAT3. Antibody specificity was tested using an enzyme-linked immunosorbent assay (ELISA), demonstrating specific binding of the antibody with a phosphorylated STAT3 peptide and the absence of binding when incubated with the nonphospho-STAT3 peptide. As well, preabsorption of pSTAT3 antiserum using a synthetic phospho-STAT3 peptide (which was identical to that of the immunogen) completely blocked antibody reactivity in pSTAT3-expressing tissue sections (Cell Signaling Technologies, pers. commun.). Lastly, in our own experiments, phosphorylation of STAT3 in the CNS was only observed upon leptin administration, as fasted animals exhibited a near total absence of STAT3 phosphorylation in saline-injected controls (data not shown). The IHC for GFP was performed on tissue from three mice using an IHC protocol as previously reported (Elmquist and Saper, 1996; Elias et al., 1998; Liu et al., 2003). Sections were rinsed 10 times for 6 minutes each in PBS, pH 7.4 and then for 30 minutes in 0.3% hydrogen peroxide in PBS to quench endogenous peroxidase activity. Following a series of PBS washes, tissue was incubated for 1 hour in 3% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA) with 0.25% Triton X-100 in PBS (PBT) with 0.02% sodium azide. Sections were incubated overnight at room temperature in GFP antisera diluted to 1:20,000 in PBT with 0.02% sodium azide. After washing in PBS, tissue was incubated in biotinylated donkey antirabbit (Jackson ImmunoResearch Laboratories) diluted to 1:1,000 in 3% donkey serum in PBT for 1 hour at room temperature. Tissue was then rinsed in PBS and incubated in ABC (Vectastain Elite ABC kit; Vector Labs, Burlingame, CA) diluted 1:500 in PBS for 1 hour. Sections were washed in PBS then reacted in 0.04% DAB (Sigma, St. Louis, MO) and 0.01% hydrogen peroxide dissolved in PBS. Sections were mounted on gelatin-coated slides, air-dried, dehydrated in increasing concentrations of ethanol, cleared in xylenes, and coverslipped with Permaslip (Alban Scientific, St. Louis, MO). An adjacent series of sections was thionin-stained to identify GFP-positive nuclear groups. As control, we performed an IHC staining using the same antisera in wildtype tissue.
Fluorescent IHC for LacZ, phospho-STAT3, and tyrosine hydroxylase was performed on 3–4 mice, as described above. Sections were rinsed 10 times for 6 minutes each in PBS, pH 7.4. Tissue was then incubated for 1 hour in 3% normal donkey serum (Jackson ImmunoResearch Laboratories) with 0.25% Triton PBT with 0.02% sodium azide. Sections were incubated overnight at room temperature in antisera (anti-lacZ 1:2,000, anti-TH 1:2,000, anti-pSTAT3 1:2,000) in PBT with 0.02% sodium azide. After washing in PBS, tissue was incubated in fluorescently labeled Alexa 594 or Alexa 488 donkey secondary antisera (Jackson ImmunoResearch Laboratories) diluted to 1:200 in 3% donkey serum in PBT for 1 hour at room temperature. Tissue was then rinsed in PBS and mounted on gelatin-coated slides. Dried slides were washed in ddH20 and coverslipped using Vectashield (Vector Labs).
In situ hybridization histochemistry (ISHH)
To determine the distribution of LepRb expression in the mouse brain, we performed single-label free-floating ISHH on wildtype mice brain tissue (n = 4). The mouse LepRb-specific riboprobe (a generous gift from Dr. Christian Bjørbæk) was generated using reverse-transcriptase polymerase chain reaction (RT-PCR) of mouse whole brain RNA (Ambion, Austin, TX) and the RT-PCR kit (Advantage; Stratagene, La Jolla, CA). The following primers were used to amplify a 400-base pair fragment corresponding to the intracellular domain of the long-form isoform of the leptin receptor: primer 1: 5′-AAAGAGCTCCTTCTCTGGGTCTCAGAGCAC-3′ and primer 2: 5′-AAAAAGCTTCTCACCAGTCAAAAGCACACCAC-3′ where the single-underlined sequence represent restriction enzyme recognition sites (primer 1: SacI site and primer 2: HinDIII site) and the double-underlined sequences are complementary to the mouse LepRb. The resulting PCR products were digested with SacI and HinDIII restriction enzymes and cloned into the pGEM-11Zf (+) plasmid (Promega, Madison, WI). Inserts were confirmed by DNA sequencing using T7 and Sp6 primers, Sequenase (Amersham Life Sciences, Arlington Heights, IL), and the 35Sequetide system (New England Nuclear, Boston, MA). Plasmids were linearized with HinDIII or SacI for generation of sense and antisense probes, respectively. These linearized templates were transcribed in vitro into cRNA using either T7 or Sp6 polymerase (Ambion).
The in situ hybridization procedure was a modification of that previously reported by our laboratory (Priestley et al., 1993; Elias, 1998; Yamamoto, 2003; Liu, 2003). Tissue was rinsed with DEPC-treated PBS, pH 7.0, for 1 hour before being pretreated with 1% sodium borohydride (Sigma) in DEPC-PBS for 15 minutes. After washing in DEPC-PBS, tissue was briefly rinsed in 0.1M TEA, pH 8.0, and incubated for 10 minutes in 0.25% acetic anhydride in 0.1M TEA. The tissue was then rinsed in DEPC-treated 2× SSC before hybridization. cRNA probes were diluted to 106 cpm/mL in 50% formamide, 10 mM Tris-HCl, pH 8.0 (Gibco-BRL, Bethesda, MD), 5 mg tRNA (Invitrogen, Carlsbad, CA), 10 mM dithiothreitol, 10% dextran sulfate, 0.3M NaCl, 1 mM EDTA, pH 8.0, and 1 × Denhardt’s solution (Sigma) and applied to the tissue. Sections were incubated overnight at 57°C. Tissue was rinsed four times in 4× sodium chloride/sodium citrate (SSC) before being incubated in 0.002% RNase A (Roche Applied Bioscience, Indianapolis, IN) diluted in 0.5M NaCl, 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA (RNase buffer) for 30 minutes at 37°C. After 30 minutes in RNase buffer, and two rinses at room temperature in 2× SSC, tissue was washed three times in 50% formamide in 0.2× SSC for 10 minutes at 50°C. Tissue was then washed 2× SSC at 50°C, 0.27× SSC at 55°C, and 0.2× SSC at 60°C for 1 hour at each wash. After rinsing twice in 2× SSC at room temperature, tissue was mounted on SuperFrost Plus slides (Fisher, Pittsburgh, PA), dehydrated in 3-minute incubations of increasing concentrations of ethanol (50%, 70%, 80%, 95%, and 100%), and delipidated for 5 minutes in chloroform. After washes in 100% and 95% ethanol, tissue was air-dried and slides were placed in X-ray film cassettes with BMR-2 film (Kodak, Rochester, NY) for 2 days. Slides were then dipped in NTB2 photographic emulsion (Kodak), dried, and stored in desiccant-containing, foil-wrapped slide boxes at 4°C for 4 weeks. Slides were developed with D-19 developer, counterstained with thionin, and dehydrated in an increasing concentration of ethanol, cleared in xylenes, and coverslipped with Permaslip. A control experiment where an antisense LepRb riboprobe was used on tissue pretreated with RNase A (200 µg/ml) demonstrated the specificity of the protocol.
Dual-label in situ hybridization histochemistry/ immunohistochemistry (ISHH/IHC)
We performed dual-label ISHH/IHC in LepRb-LacZ/EYFP mice to confirm the colocalization of LacZ/EYFP immunoreactivity and LepRb mRNA (n = 3) throughout the brain. The free-floating ISHH was performed as described above followed by IHC using the immunoperoxidase method as described in the IHC section. Brain tissue was then mounted on SuperFrost Plus slides, dehydrated, and delipidated before being placed in X-ray film cassettes with BMR-2 film for 2 days. Slides were treated as previously mentioned without the thionin counterstaining. LacZ/EYFP-positive cells were analyzed as expressing LepRb based on the following criteria: the number of silver grains over the immunoreactive cell body was 5 times above background hybridization levels and the silver grains above the background level conformed to the shape of the immunoreactive cell body.
Data analysis, estimates of cell counts, and photomicrographs
Sections were analyzed with a Zeiss Axioplan light microscope, a Zeiss Stemi 2000-C dissecting microscope, and Zeiss Apotome microscope. Cell counts were performed on every fifth section of each mouse brain then multiplied by 5 to obtain an estimate of the absolute cell number for each CNS location. Estimates of cell counts were performed using a 10× objective. The data were not corrected for double counting, nor was the stereological technique used since the cell size and section thickness did not vary between animals. It is also important to note that the double-label studies presented are inherently qualitative, thus our results provide data for relative comparisons of cell numbers between brain nuclei and are not accurate counts of absolute cell numbers. pSTAT and TH cell counts were performed on brains from 3–4 mice. Data are presented as average ± SEM. Adobe Photoshop 7.0 (San Jose, CA) was used to adjust only sharpness, brightness, and contrast as well as to combine select images into plates. Unless otherwise noted, nomenclature corresponds to the Paxinos and Franklin (2001) mouse brain atlas.
RESULTS
Distribution of GFP-immunoreactivity in LepRb-EYFP mice
We assayed brain tissue from progeny of a LepRb-IRESCre × ROSA26 (loxSTOPlox) EYFP mice for GFP-like immunoreactivity to determine whether GFP immunoreactive (GFP-IR) cells localized to the same brain regions as have been previously shown using ISH to map sites of LepRb RNA expression. Hereafter we will refer to the LepRb-IRES-Cre × ROSA26 (loxSTOPlox) EYFP mice as LepRb-EYFP mice. Reporter mice expressing bacterial β-galactosidase (LacZ) from the ROSA26 locus were also used, with the LepRb-IRES-Cre × ROSA26 (loxSTOPlox) LacZ progeny denoted as LepRb-LacZ. It is important to note that there was no observed difference between LacZ and EYFP reporter expression when crossed to the LepRb-IRES-Cre line. In the LepRb-IRES-Cre mouse line, an IRES-Cre cassette was knocked into the endogenous LepR locus just 3′ to the LepRb stop codon using gene targeting. In general, the pattern of GFP immunostaining was consistent with previous reports of the distribution of LepRb mRNA except as noted below (Table 1; Fig. 2).
Table 1.
Relative Densities of Long-form Leptin Receptor mRNA Expression in the Wildtype Mouse Brain and Localization of GFP-IR in the LepRb-EYFP Mouse Brain
| Cerebral cortex (layers) | mRNA (35S) | GFP-IR |
|---|---|---|
| Cingulate cortex (2) | ++ | + |
| Claustrum | +++ | ++ |
| Dorsal endopiriform nucleus | ++ | + |
| Insular, dysgranular (3, 4 and 6b) | + | + |
| Insular, granular (3, 4 and 6b) | + | + |
| Piriform | ++ | − |
| Motor, primary (3, and 6b) | ++ | − |
| Motor, secondary (3, 4 and 6b) | ++ | − |
| Somatosensory, primary (3, 4 and 6b) | ++ | + |
| Somatosensory, secondary (3, 4 and 6b) | ++ | + |
| Restrosplenial, agranular (3, and 6b) | + | − |
| Restrosplenial, granular (3, 4 and 6b) | + | − |
| Perirhinal | + | + |
| Ectorhinal | + | + |
| Lateral entorhinal | + | + |
| Auditory, secondary (3, 4 and 6b) | ++ | + |
| Temporal association (3, 4 and 6b) | + | − |
| Hippocampus and septum | ||
| Ammon’s horn, CA1 | ++ | − |
| Ammon’s horn, CA2 | + | − |
| Ammon’s horn, CA3 | ++ | + |
| Dentate gyrus | ++ | ++ |
| Lateral septal nucleus, intermediate | ++ | ++ |
| Lateral septal nucleus, ventral | + | − |
| Bed nucleus of stria terminalis | + | + |
| Caudate/Putamen | + | +/− |
| Thalamus | ||
| Paraventricular nucleus, anterior | + | + |
| Anteromedial nucleus | ++ | + |
| Central medial nucleus | + | − |
| Central lateral nucleus | + | − |
| Interoanteromedial nucleus | + | − |
| Paracentral nucleus | + | − |
| Subparafascicular nucleus | ++ | − |
| Reuniens nucleus | ++ | + |
| Submedial nucleus | + | − |
| Ventral posterolateral nucleus | + | − |
| Ventral posteromedial nucleus | + | − |
| Hypothalamus | ||
| Medial preoptic area | +++ | +++ |
| Median preoptic nucleus | ++ | ++ |
| Lateral preoptic area | + | + |
| Paraventricular nucleus | +/− | +/− |
| Supraoptic nucleus | + | − |
| Anterior hypothalamic area, posterior | + | + |
| Retrochiasmatic area | ++++ | ++++ |
| Arcuate nucleus | ++++ | +++ |
| Ventromedial nucleus, central | ++ | + |
| Ventromedial nucleus, dorsomedial | +++ | + |
| Lateral hypothalamic area | ++ | +++ |
| Dorsomedial nucleus, dorsal | +++ | +++ |
| Dorsomedial nucleus, ventral | +++ | +++ |
| Ventral premammillary nucleus | +++ | +++ |
| Posterior hypothalamic area | ++ | + |
| Supramammillary nucleus | + | − |
| Cerebellum | ++ | − |
| Midbrain, pons and medulla oblongata Precomissural nucleus | + | + |
| Ventral tegmental area | ++ | ++ |
| Ventrolateral tegmental area | + | + |
| Substantia nigra, compact part | +++ | + |
| Edinger Westphal nucleus | ++ | + |
| Rostral linear nucleus | ++ | ++ |
| Superior colliculus, deep gray | ++ | + |
| Superior colliculus, superficial gray | + | + |
| Deep mesencephalic nuclei | + | +/− |
| Periaqueductal gray | ++ | ++ |
| Dorsal raphe nucleus | +++ | +++ |
| Cuneiform nucleus | + | + |
| Lateral parabrachial nucleus, superior | +++ | ++ |
| Pontine Reticular nucleus, caudal part | + | − |
| Pontine Reticular nucleus, oral part | + | +/− |
| Supratrigeminal nucleus | + | − |
| Nucleus of the solitary tract, dorsolateral | +++ | − |
| Nucleus of the solitary tract, medial | +++ | +++ |
| Nucleus of the solitary tract, ventral | +++ | ++ |
| Nucleus of the solitary tract, ventrolateral | +++ | + |
| Nucleus of the solitary tract, intermediate | +++ | ++ |
| Dorsal motor nucleus of vagus | +++ | + |
| Intercalate nucleus of the medulla | + | + |
| Hypoglossal nucleus | +/− | − |
| Intermediate reticular nucleus | ++ | ++ |
| Medullary reticular nucleus, ventral | +/− | +/− |
Qualitative estimates of LepRb mRNA expression were based on hybridization signal strength and of GFP-IR were based on the number of labeled cells.
highest density
high density
moderate density
low density
labeling inconsistently above background
no labeling
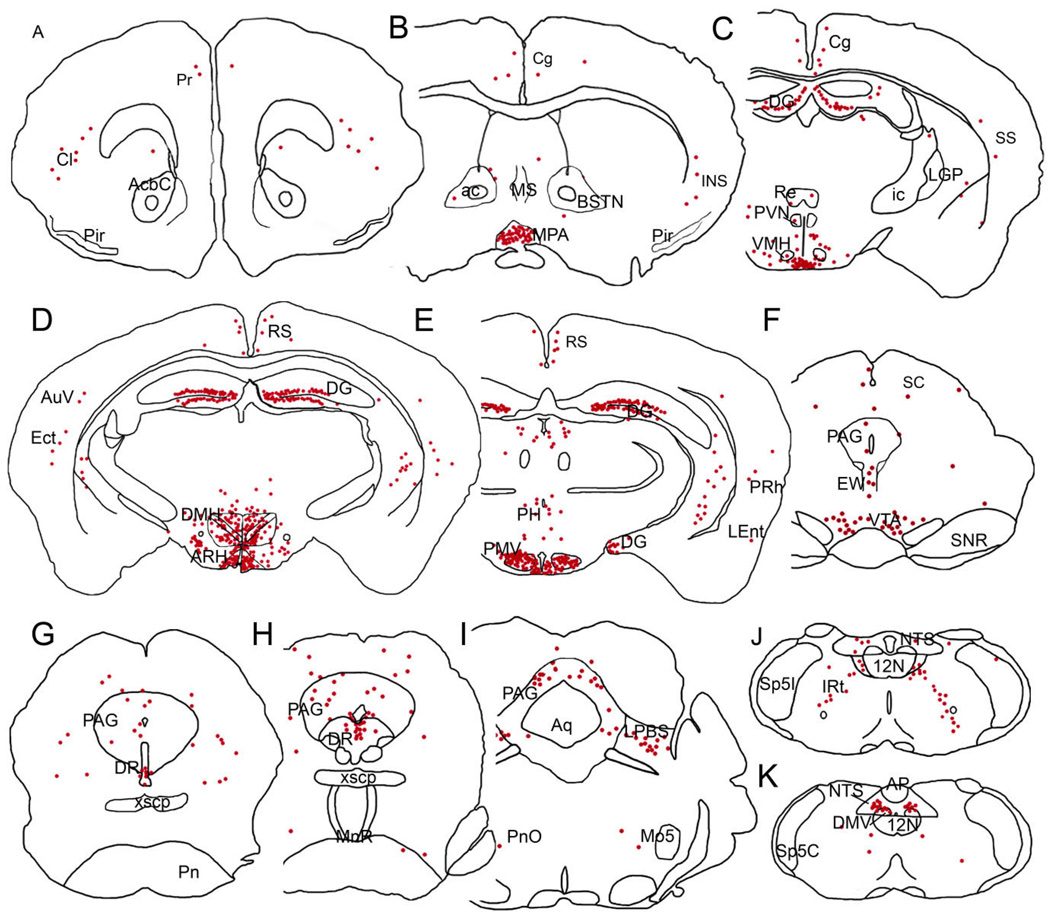
Figure 2.
A series of line drawings illustrating the GFP immunoreactivity distribution in the LepRb-EYFP mouse brain. Sections are arranged in a rostral-to-caudal manner (A–K). Each red dot represents ≈2 GFP-IR cells. 12N, hypoglossal nucleus; ac, anterior commissure; AcbC, accumbens nucleus, core; AP, area postrema; Aq, aqueduct; ARH, arcuate nucleus; AuV, secondary auditory nucleus; BSTN, bed nucleus of the stria terminalis; Cg, cingulated cortex; Cl, claustrum; DG, dentate gyrus; DMH, dorsomedial hypothalamic nucleus; DMV, dorsal motor nucleus of the vagus; DR, dorsal raphe; Ect, ectorhinal cortex; EW, Edinger-Westphal nucleus; ic, internal capsule; IRt, intermediate reticular nucleus; LEnt, lateral entorhinal cortex; LHA, lateral hypothalamic area; LGP, lateral globus pallidus; LPBS, superior lateral parabrachial subnucleus; MnR, median raphe nucleus; Mo5, motor trigeminal nucleus; MPA, medial preoptic area; MS, medial septal nucleus; NTS, nucleus of the solitary tract; PAG, periaqueductal gray; PH, posterior hypothalamic area; Pir, piriform cortex; PMV, ventral premammillary nucleus; Pn, pontine nuclei; PnO, pontine reticular nucleus, oral part; PVN, paraventricular hypothalamic nucleus; Re, reuniens thalamic nucleus; RS, retrosplenial cortex; SC, superior colliculus; SNR, substantia nigra, reticular part; Sp5C, spinal trigeminal nucleus, caudal part; Sp5I, spinal trigeminal nucleus, interpolar part; SS, somatosensorial cortex; VMH, ventromedial hypothalamic nucleus; VTA, ventral tegmental area; xscp, decussation of the superior cerebellar peduncle.
Hypothalamus
Consistent with previous reports of LepRb expression, EYFP staining in the ARH was very dense throughout both the rostral-caudal and medial lateral axes (Fig. 2C–E). Notably, the paraventricular nucleus (PVN) was inconsistently labeled with only a few GFP-IR cells and staining in the ventrolateral hypothalamus (VMH) was relatively sparse, localized mostly to the central division. We also found large numbers of GFP-IR cells in the lateral hypothalamic area (LHA), in the dorsal and ventral parts of the dorsomedial nucleus (DMH), and in the ventral premammillary nucleus (Fig. 2E, Fig. 6C,E,F). Very few GFP cells were also found in the compact region of the DMH (Fig. 2D, Fig. 6C). In addition, we found a robust GFP staining in the medial preoptic area (MPA, Fig. 2B, Fig. 6G) and median preoptic nucleus (MnPO). The anterior hypothalamic area, in particular the posterior part, showed a small population of GFP-IR cells (Fig. 2C). In the posterior aspects of the hypothalamus, moderate staining was found in the posterior hypothalamic area (Fig. 2E).
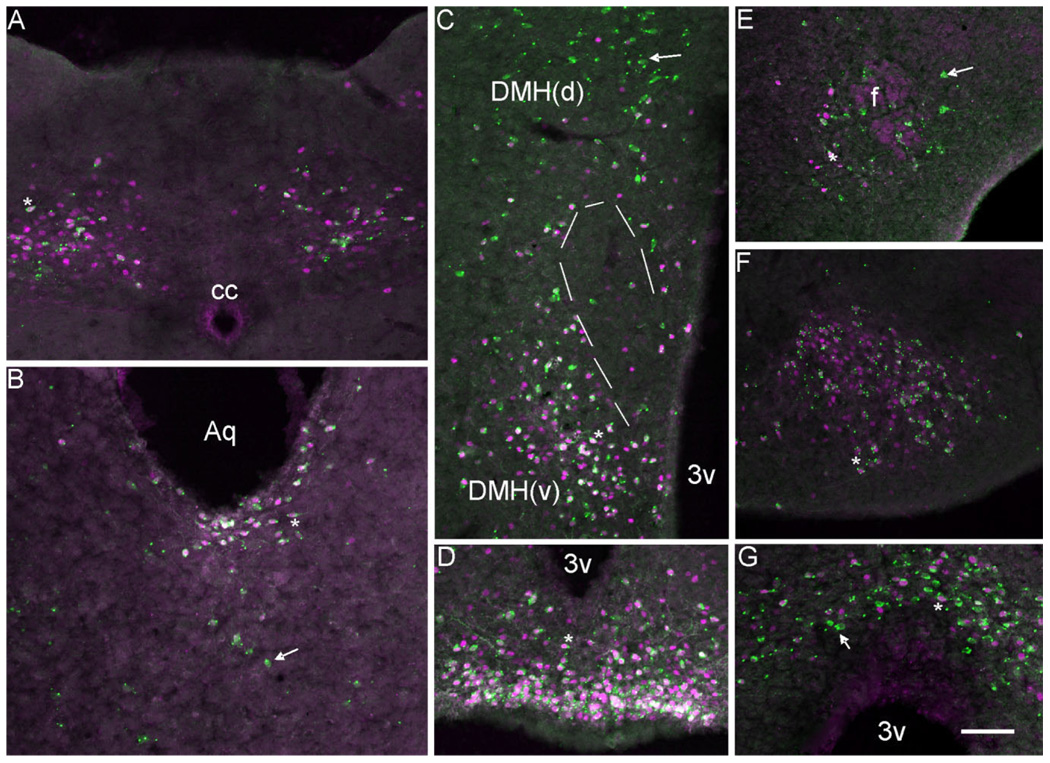
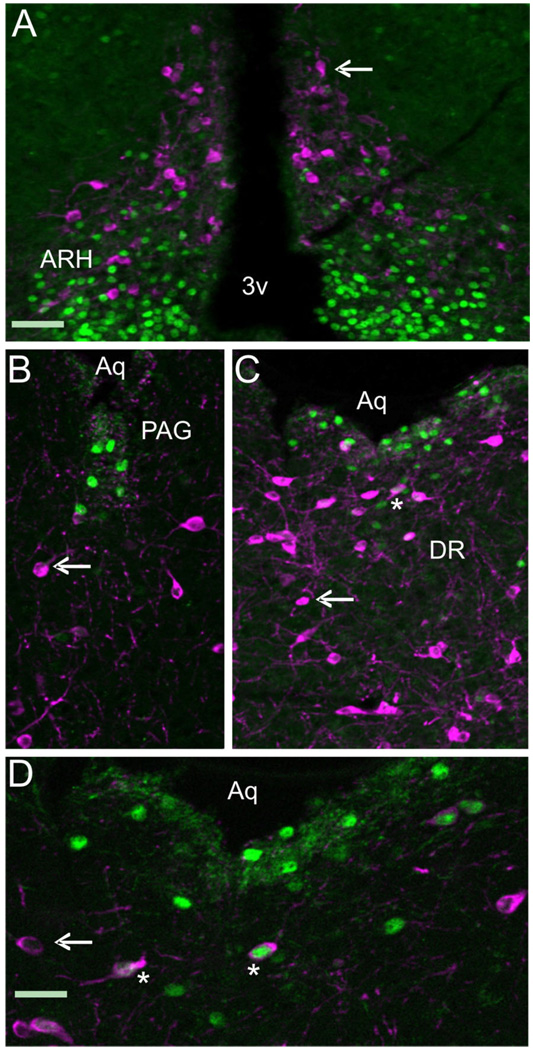
Figure 6.
A series of photomicrographs illustrating the coexpression of LepRb-Cre LacZ with pSTAT3. A–G: LepRb-Cre LacZ sections labeled for pSTAT3 (magenta) and LacZ (green). Coexpression was observed in (A) nucleus of the solitary tract (NTS), (B) in the dorsal raphe (DR) and periaqueductal gray (PAG), (C) in the DMH(d): dorsal medial hypothalamus dorsal part and DMH(v) dorsal medial hypothalamus ventral part, (D) in the arcuate nucleus (ARH), (E) in the lateral hypothalamus, (F) in the ventral premammilary nucleus (PMV), and (G) in the medial preoptic nuclei (MPO). Single-labeled cells appear green with uniform or punctuate cytoplasmic expression. Double-labeled cells have magenta/white nuclei with punctuate or uniform green cytoplasm. PMV double-labeled neurons are almost exclusively punctuate with respect to LacZ expression. Aq, aqueduct; 3v, third ventricle; cc, central canal; f, fornix. Scale bar =100 µm.
Brainstem and cerebellum
GFP immunoreactivity was evident in brainstem in areas previously described to present LepRb mRNA expression (Mercer et al., 1998a), including the lateral parabrachial nucleus, superior part (LPBS; Fig. 2I), the intermediate reticular nucleus, and the nucleus of the solitary tract (NTS), specifically in the medial, intermediate, and ventral subdivisions (Fig. 2J–K, Fig. 5, Fig. 6A). In addition, we also found that the superficial layer of the superior colliculus had a few GFP-IR cells and the deep gray matter subdivision of the superior colliculus was moderately stained. The deep mesencephalic nucleus, the rostral linear nucleus, and the Edinger-Westphal nucleus had low to moderate numbers of GFP-IR cells (Fig. 2F). We also found that the ventral tegmental area (VTA) showed GFP/LacZ labeling (Fig. 2F, Fig. 5E), while the substantia nigra (SN) showed little staining (Fig. 5F). Weakly expressing areas included the cuneiform nucleus, the ventrolateral tegmental area, and the oral part of the pontine reticular nucleus. Moderate expression was found in the periaqueductal gray matter (PAG) while the most robust expression in the pons localized to the dorsal raphe nucleus (DR, Fig. 2G,H, Fig. 6B). The dorsal motor nucleus of the vagus and the intercalated nucleus of the medulla also presented moderate GFP immunoreactivity (Fig. 2J,K). In the cerebellum we found virtually no GFP-IR cells.
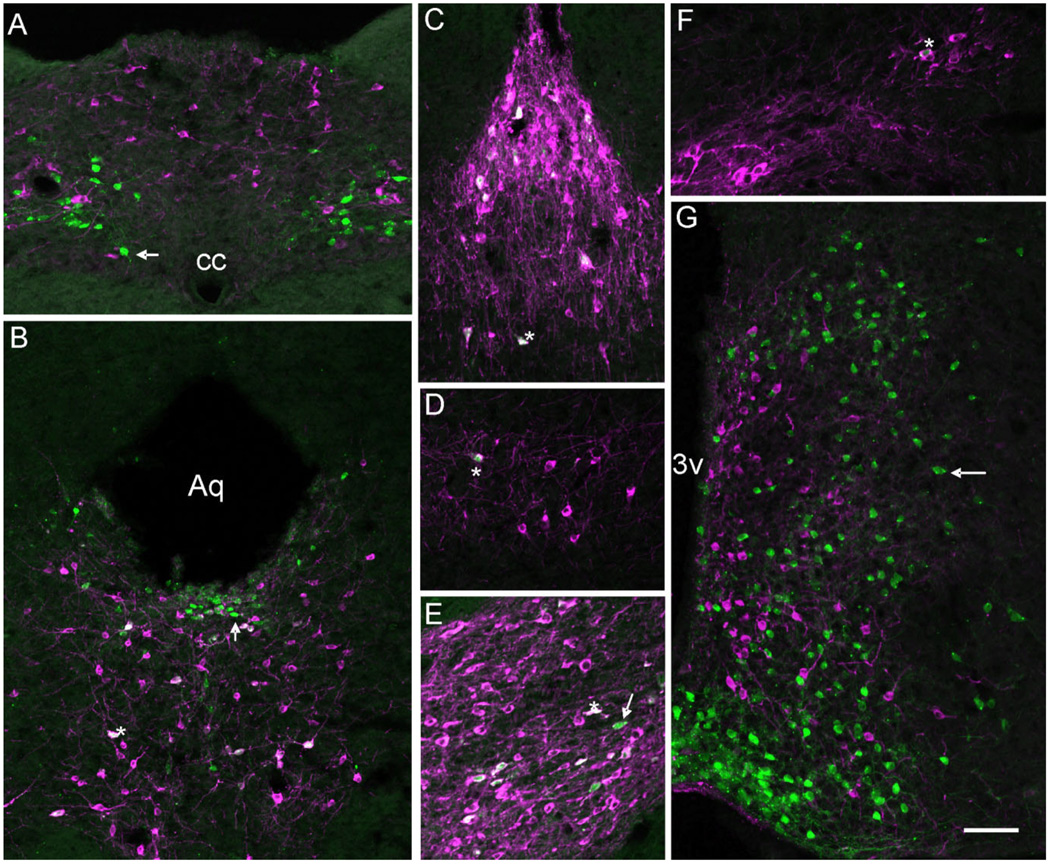
Figure 5.
A series of photomicrographs illustrating the coexpression of tyrosine hydroxylase (TH) and LepRb. A–G: LepRb-Cre LacZ sections labeled for TH (magenta) and LacZ (green) in (A) nucleus of the solitary tract (NTS), (B) the dorsal raphe nucleus (DR) and periaqueductal gray (PAG), (C) in the caudal linear nucleus (Cli), (D) in the A8 nucleus, (E) in the ventral tegmental area (VTA), (F) in the substantia nigra, compact part (SNC), and (G) in the mediobasal hypothalamus. Single-labeled cells appear green or magenta (arrow) while coexpression (asterisk) is indicated by cells appearing white due to color overlay or magenta/white with green nuclei. Aq, aqueduct; 3v, third ventricle; cc, central canal. Scale bar =100 µm.
Cerebral cortex, hippocampal formation, and septum
Previous studies have shown LepRb expression in the cerebral cortex and hippocampal formation (Huang et al., 1996; Mercer et al., 1996a), but detailed anatomical localization has been lacking. We found only few GFP-IR neurons in many cerebral cortex areas including the prefrontal, cingulate, restrosplenial, and insular as well as the secondary auditory, ectorhinal, perirhinal, lateral entorhinal, and somatosensory cortical areas (Fig. 2A–E). Moderate numbers of GFP-IR cells were also detected in the claustrum (Fig. 2A). In the hippocampus, dense populations of GFP-IR cells were found throughout the granular layer of the dentate gyrus, while the CA3 region of Ammon’s horn showed sparse labeling. No GFP-immunoreactivity was observed in the CA1 or CA2 hippocampal regions. In addition, weak GFP staining was seen in the ventral and intermediate regions of the lateral septal nucleus (Fig. 2B), and inconsistently in the caudate-putamen.
Amygdala, bed nucleus of the stria terminalis, and thalamus
We found no GFP-IR cells in the amygdala and a few scattered immunoreactive cells were observed in the bed nucleus of the stria terminalis (BSTN, Fig. 2B). Thalamic GFP immunoreactivity was limited to a few cells in the reuniens nucleus (Fig. 2C,D). We also found inconsistent GFP immunoreactivity in the anterior paraventricular thalamic nucleus and in the anteromedial thalamic nucleus.
Distribution of long-form leptin receptor mRNA in the mouse brain
Coronal sections of the mouse brain were hybridized with a long-form-specific LepRb probe (Fig. 3). A standard RNase control was performed and determined the specificity of the LepRb probe. Distribution of the expression was assessed as silver grain density and compared with that observed using GFP immunoreactivity in LepRb-EYFP mouse (Table 1). All nomenclature is in accordance to that found in the mouse brain atlas of Paxinos and Franklin (2001).
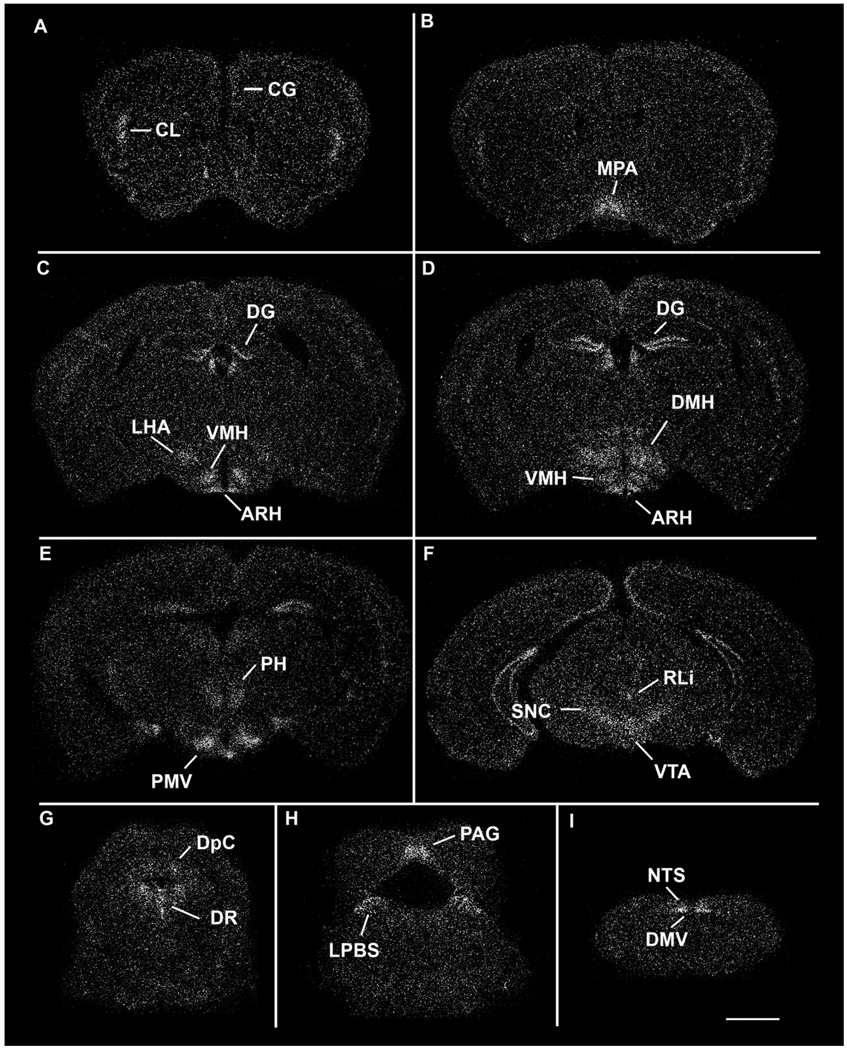
Figure 3.
A series of low-power darkfield autoradiographs illustrating the distribution of LepRb in the wildtype mouse brain. Sections are arranged in a rostral-to-caudal manner. ARH, arcuate nucleus; Cg, cingulate cortex; Cl, claustrum; DG, dentate gyrus; DMH, dorsomedial hypothalamic nucleus; DMV, dorsal motor nucleus of the vagus; DpC, deep gray layer of the superior colliculus; LPBS, superior lateral parabrachial subnucleus; NTS, nucleus of the solitary tract; PAG, periaqueductal gray; PH, posterior hypothalamic area; PMV, ventral premammillary nucleus; RLi, rostral linear nucleus; SNC, substantia nigra, compact part; VMH, ventromedial hypothalamic nucleus; VTA, ventral tegmental area. Scale bar = 1 mm.
Hypothalamus
The hypothalamus contained several high LepRb-expressing areas. In the preoptic division of the hypothalamus, we found robust expression in the MPA (Fig. 3B). In the anterior hypothalamus, weak hybridization was found in the supraoptic nucleus and the anterior hypothalamic area. Hybridization in the PVN was weak and inconsistent. In the tuberal hypothalamus, dense expression was distributed across the entire ARH (Fig. 3C–E). Additionally, moderate to high expression localized to the dorsomedial and central parts of the VMH, while no signal was observed in the ventrolateral division (Fig. 3C). Little to no expression was found in the rostral portion of the DMH, whereas the dorsal and ventral divisions in the caudal regions showed moderate to high expression levels (Fig. 3D). Weak signal was detected in the compact portion of the DMH. Finally, the LHA exhibited moderate hybridization (Fig. 3C). In the more caudal hypothalamic regions we saw a moderate signal in the supramammillary nucleus and the posterior hypothalamic area and robust expression in the PMV (Fig. 3E).
Brainstem and cerebellum
Substantial labeling was observed in various parts of the midbrain, pons, and medulla. The Edinger-Westphal nucleus contained moderate amounts of labeling while the compact part of the SN (Fig. 3F) and the VTA contained moderate to high expression. In contrast, the PAG, the deep gray matter of the superior colliculus, the deep mesencephalic nucleus, and the oral and caudal parts of the pontine reticular nucleus exhibited low to moderate levels of hybridization (Fig. 3F–H). Additionally, we saw hybridization in the superficial layer of the superior colliculus, although it was much less pronounced than that in the deep gray matter. The weak signal distributed across the PAG continued through the level of the pons, with the exception of the most caudal part of the dorsal PAG. We also found low levels of expression in the cuneiform nucleus and in the supratrigeminal nucleus. Moderate to high levels of expression were found in the rostral linear nucleus, dorsal raphe, the ventrolateral tegmental area, and the superior lateral parabrachial subnucleus (Fig. 3G,H). In the medulla, we found inconsistent hybridization in the ventral part of the medullary reticular nucleus as well as the hypoglossal nucleus. We saw moderate expression in the intermediate reticular nucleus and in the intercalated nucleus of the medulla. The dorsal motor nucleus of the vagus and the nucleus of the solitary tract both exhibited moderate to dense receptor expression (Fig. 3I). Within the NTS, hybridization localized to the dorsolateral, medial, ventral, ventrolateral, and intermediate parts (Herbert et al., 1990). In addition, we observed intermediate levels of LepRb signal in the granular and Purkinje layers of the cerebellum.
Cerebral cortex, hippocampal formation, and septum
Hybridization was observed in various regions of the cerebral cortex. Moderate expression was seen in prefrontal cingulate cortex (layer 2/3) and claustrum (Fig. 3A). Expression levels in layers 3, 4, and 6b of the granular and dysgranular divisions of the insular cortex, while not as robust as the claustrum, were classified as moderate. Weak hybridization was observed in the agranular and granular subdivision of the retrosplenial cortex (Fig. 3C,D), and in layers 3, 4, and 6b of the primary and the secondary motor cortices. The primary (hindlimb and forelimb subdivisions) and secondary somatosensory cortices (Fig. 3A–D), the temporal association cortex, the secondary auditory cortex, the ventral part as well as the ectorhinal, perirhinal and lateral entorhinal cortical areas had hybridization throughout layers 3, 4, and 6b. We found moderate expression in the dorsal endopiriform region and the piriform cortex had moderate hybridization signal (Fig. 3A–C). In the septum, we found very low levels of expression in the ventral and intermediate divisions of the lateral septal nucleus (Fig. 3A). The caudate putamen was inconsistently labeled, and in the cases where we found hybridization, it was weak. High levels of hippocampal expression were localized to the granular layer of the dentate gyrus (Fig. 3C,D). Substantial labeling was also seen in the CA2 and CA3 regions of Ammon’s horn (Fig. 3C,D). In contrast, CA1 expression was very weak (Fig. 3C,D).
Amygdala, bed nucleus of the stria terminalis, and thalamus
There was no LepRb expression in the amygdala. Weak expression in the BSTN was seen in the posterior region. Various regions of the thalamus expressed LepRb mRNA. The reuniens and the anteromedial thalamic nuclei were the thalamic regions with the densest expression (Fig. 3B). Additionally, low to moderate expression was found in the anterior paraventricular, throughout the lateral habenula (Fig. 3C), submedial, interanteromedial nuclei as well as the subparafascicular nucleus. Weak expression levels were seen in the central medial, centrolateral, and paracentral nuclei (Fig. 3D). In addition, we also observed an inconsistent labeling of the ventral medial, ventral posteromedial, and ventral posterolateral thalamic nuclei.
Colocalization of LepRb-Cre activated reporter and LepRb mRNA
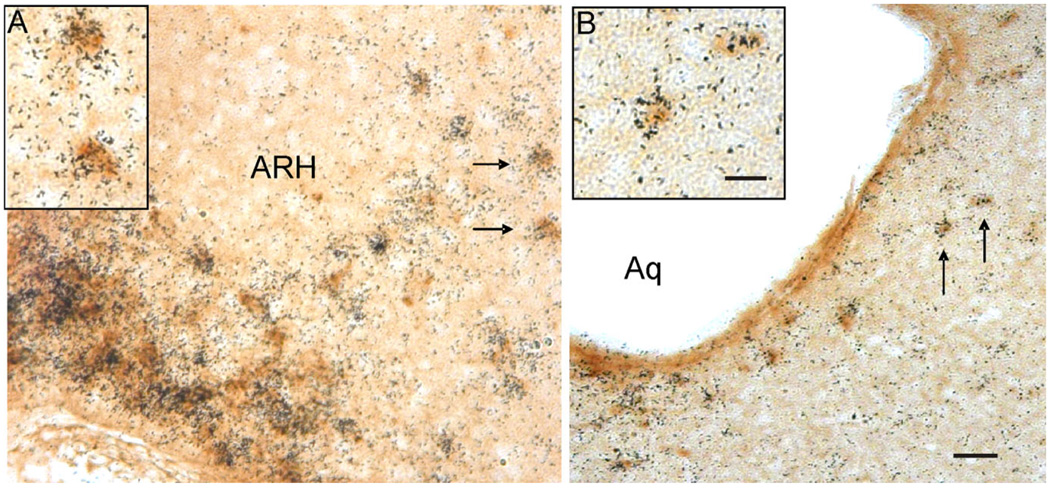
The experiments described above demonstrate that the LepRb-Cre mouse line activates reporter gene expression in an anatomical pattern similar to that of the endogenous LepRb gene. In order to ensure that the LepRb-Cre line was truly recapitulating the expression of LepRb, we performed dual ISHH for LepRb and IHC for the Cre-activated reporter LacZ and determined the extent of colocalization of the two labels throughout the brain. We found that nearly all LacZ-positive cells also expressed LepRb mRNA (Fig. 4, and data not shown) confirming that LepRb-Cre regulated LacZ is eutopically expressed in the mouse brain. Figure 4 illustrates examples of the hypothalamic areas that contained double-labeled LacZ and LepRb mRNA cells (ARH: Fig. 4A). Extrahypothalamic sites including the DR (Fig. 4B) also exhibited a high degree of colocalized LacZ and LepRb mRNA. The only exception to this finding was a small number of single-labeled LacZ cells that localized to the bed nucleus of the stria terminalis. The basis for this is not clear, although this anomaly may be due to low LepRb mRNA levels in the BSTN, which cannot be detected by ISHH assay but are detectable based on LacZ expression. In addition, some regions expressed significant levels of LepRb RNA but little or no LacZ, such as the VMH, the primary motor and somatosensory cortex, the subfascicular nucleus of thalamus, and the substantia nigra compact part. While the basis for these discrepancies is not known, this finding does not limit the utility of the LepRb-LacZ mice for studying those regions where LacZ is faithfully expressed.
Figure 4.
Photomicrographs illustrating the colocalization of LacZ-immunoreactivity and LepRb mRNA in the hypothalamus and midbrain of the LepRb-Cre LacZ mouse. A: Cells in the arcuate nucleus of the hypothalamus (ARH). B: Cells in the dorsal raphe of LepRb-Cre LacZ tissue coexpressing LepRb mRNA and LacZ. Cells labeled with arrows denote examples of double-labeled cells presented at higher magnification in the panel inserts. Cells containing LepRb mRNA were labeled with silver grain clusters and LacZ-IR cells are labeled with a brown cytoplasmic precipitate. Scale bar: = 50 µm in A,B; 25 µm in inserts.
Chemical characterization of LepRb-expressing cells
The data presented in this report thoroughly describes the expression pattern of the long-form leptin receptor in the CNS. However, the neurochemical identity of cells expressing leptin receptor (outside of the hypothalamus) remains poorly described. Recent data suggest leptin, along with the orexigenic peptides orexin and ghrelin, may be directly acting on dopaminergic neurons of the VTA to affect energy homeostasis. Thus, to better understand the extent of leptin sensing by these and other dopaminergic neurons, we examined the coexpression of leptin receptor with a marker of noradrenergic and dopaminergic neurons, tyrosine hydroxylase. Four LepRb-IRES-Cre mice crossed with R26R-LacZ (LepRb-Cre LacZ) were used for these studies, with the data quantitated in Table 2. The greatest number of LacZ cells coexpressing TH were located in the VTA and adjacent caudal linear nucleus (Cli) of the midbrain (Fig. 5C,E). An average of 70% of the LacZ cells were also TH-positive in these two nuclei, with the greatest coexpression observed in the most caudal aspect of the VTA and most rostral part of the Cli. The dorsal raphe also demonstrated a significant number of LacZ cells expressing TH, although at a reduced frequency when compared to the VTA and Cli (Fig. 5B). Other nuclei demonstrating coexpression of TH and LacZ include the ventral and lateral periaqueductal gray (Fig. 5B), the rostral linear nucleus, A8 nucleus (Fig. 5D), and substantia nigra pars compacta (Fig. 5F). While the A8 and substantia nigra showed fewer lacZ/TH double positive cells when compared to the VTA or Cli, the percentage of coexpression was higher, as all observed LacZ-positive cells were also TH-positive. Interestingly, there was no coexpression of TH and lacZ noted outside of the midbrain. For example, there were no TH-positive cells of the hypothalamus and hindbrain that coexpressed LacZ (Fig. 5A,G).
Table 2.
Coexpression of LepR and TH in the CNS
| Nucleus | Number of cells TH/LacZ+ | Percent of LacZ cells that were TH/LacZ+ |
|---|---|---|
| A8 | 48 ± 15 | 100 |
| Caudal linear nucleus | 276 ± 40 | 70 ± 8 |
| Dorsal raphe | 169 ± 20 | 31 ± 8 |
| Periaqueductal gray ventral | 41 ± 15 | 14 ± 6 |
| Periaqueductal gray dorsal/lateral | 23 ±5 | 8± 1 |
| Rostral linear nucleus | 23 ±4 | 64 ± 20 |
| Substantia nigra compact part | 77 ± 10 | 100 |
| Ventral tegmental area | 485 ± 80 | 67 ± 5 |
Quantitation of TH and LepRb Cre-activated LacZ coexpression in the CNS. Limited nuclei showed coexpression of these two markers. Cell counts are presented as the number of double-positive cells per nuclei (average of 4 brains ± SEM). The percentage of all lacZ-positive cells within the nuclei that were also double-positive is presented in the right-most column.
CNS leptin receptor expression and peripheral leptin sensing
Another important question that we have addressed in our characterization of leptin receptor-expressing populations of neurons in the CNS is whether all leptin receptor-expressing cells can sense peripheral leptin. This point is then crucial to the prediction of whether specific populations of Lepr-positive neurons may be participating in the regulation of energy homeostasis. To answer the question of which CNS populations respond to peripheral leptin, we examined the ability of peripherally injected leptin in fasted mice to elicit phosphorylation of the signaling molecule STAT3. STAT3 phosphorylation has previously been shown to be downstream of leptin receptor activation in several tissues, including cells of the hypothalamus. Thus, we quantitated the coexpression of pSTAT3 with leptin receptor Cre-activated LacZ (Table 3). Mice were fasted overnight, then received either a bolus of recombinant leptin (5 mg/kg) IP or saline. Mice were sacrificed after 40 minutes, and brains were extracted and analyzed for STAT3 phosphorylation using a phosphoSTAT3-specific antibody. Saline controls exhibited similar minimal amounts of basal STAT phosphorylation in ventral arcuate cells as reported in Hosoi et al. (2002), while basal phosphorylation of STAT3 was not evident outside of the hypothalamus (data not shown). In the hindbrain, NTS LacZ-positive cells showed significant coexpression of pSTAT3 (Fig. 6A), with intercalate nucleus LacZ-positive cells showing low levels of coexpression. Coexpression of pSTAT3 and LacZ is indicated by the red nuclear pSTAT expression with punctuate green cytoplasmic (and occasionally green nuclear) LacZ expression. In the midbrain, few parabrachial lacZ-positive cells were also pSTAT3-positive, unlike that observed in ventral periaqueductal gray, where nearly all cells were positive for both lacZ and pSTAT3 (Table 3). Dorsal raphe lacZ cells also exhibited high levels of LacZ/pSTAT3 coexpression, with the majority of double-positive cells located close to the ventral surface of the aqueduct (Fig. 6B). In contrast, dorsal and lateral periaqueductal LacZ cells showed minimal coexpression of pSTAT3 (Table 3). Hypothalamic pSTAT3 expression was extensive, with most nuclei exhibiting high levels of LacZ/pSTAT3 coexpression. Ventral premammilary, arcuate, and ventromedial nuclei lacZ cells showed near complete overlap of pSTAT3 with LacZ expression (Fig. 6D,F). Unlike in the rest of the hypothalamus, a dichotomy of LacZ/pSTAT3 coexpression was seen in the dorsal medial nucleus. Ventral dorsal medial nucleus lacZ cells had high levels of pSTAT3 coexpression, while the dorsal portion of the dorsal medial nucleus had significantly fewer LacZ/pSTAT3 double-positive cells (Fig. 6C). A low frequency of LacZ and pSTAT3 coexpression was also observed in the lateral hypothalamus, with most double-labeled cells clustering around the fornix (Fig. 6E). Rostrally, medial preoptic area cells showed high levels of LacZ/pSTAT3 coexpression and were the only population of cells expressing pSTAT3 (Fig. 6G).
Table 3.
Coexpression of LepR and Phospho-STAT3 in the CNS
| Nucleus | Number of cells pSTAT3/LacZ+ | Percent of LacZ cells that were pSTAT3/LacZ+ |
|---|---|---|
| Intercalate nucleus | 37 ± 12 | 26 ± 5 |
| Nucleus of the solitary tract | 378 ± 70 | 72 ± 12 |
| Parabrachial nucleus | 122 ± 45 | 20 ± 10 |
| Dorsal raphe | 305 ± 125 | 75 ± 13 |
| Periaqueductal gray ventral | 323 ± 45 | 97 ± 4 |
| Periaqueductal gray dorsal/lateral | 125 ± 90 | 15 ± 8 |
| Periaqueductal gray | 142 ± 15 | 42 ± 10 |
| Posterior hypothalamic area | 12 ±2 | 97 ± 5 |
| Ventral premammilary nucleus | 1200 ± 145 | 94 ± 3 |
| Arcuate nucleus | 5360 ± 200 | 98 ± 1 |
| Ventral medial nucleus | 185 ± 30 | 94 ± 5 |
| Dorsal medial nucleus ventral | 778 ± 100 | 94 ± 3 |
| Dorsal medial nucleus dorsal | 218 ± 40 | 23 ± 6 |
| Lateral hypothalamic area | 195 ± 100 | 29 ± 11 |
| Periventricular nucleus | 20 ±1 | 90 ± 14 |
| Medial preoptic area | 613 ± 115 | 86 ± 5 |
Quantitation of pSTAT3 and LepRb Cre-activated lacZ coexpression in the CNS. Limited nuclei demonstrated coexpression of both markers. Cell counts were presented as the number of double-positive cells per nuclei (average of 3 brains ± SEM). The percentage of all LacZ-positive cells within the nuclei that were also double-positive is presented in the right-most column.
Peripheral leptin sensing by midbrain dopaminergic neurons
Figure 5 and Figure 6 demonstrate that a population of TH + dopaminergic neurons in the midbrain coexpress LepRb, while a subset of those LepRb neurons were also shown to sense peripheral leptin as evidenced by STAT3 phosphorylation. To determine whether the TH-positive neurons were also able to phosphorylate STAT3 upon peripheral leptin administration, we performed double IHC on serial sections from mice used to generate data for Table 3 and Figure 6. As expected, no TH neurons in the hypothalamus exhibited pSTAT3 expression (Fig. 7A). Interestingly, few TH-expressing neurons express pSTAT3 in the ventral PAG (Fig. 7B,C) and dorsal raphe (Fig. 7C,D) in the areas that exhibited STAT3 phosphorylation in the previous experiment. Thus, while many TH neurons express LepRb, very few cells displayed phosphorylation of STAT3 following peripheral leptin administration. The neurochemical identity of the STAT3+/TH– neurons in the midbrain remains to be established.
Figure 7.
A series of photomicrographs illustrating the coexpression of TH with pSTAT3. A–D: LepRb-Cre LacZ sections labeled for TH (magenta) and pSTAT3 (green) in (A) arcuate nucleus (ARH), (B) periaqueductal gray (PAG), (C,D) dorsal raphe (DR). Single-labeled cells are magenta or green (arrow), while double-labeled cells have a green/white nucleus surrounded by magenta cytoplasm (asterisk). 3v: third ventricle, Aq: aqueduct. Scale bars = 100 µm for A–C; 50 µm in D.
DISCUSSION
It is well established that the CNS is the principal target of leptin action (Schwartz et al., 2000; Spiegelman and Flier, 2001; Niswender and Schwartz, 2003; Benoit et al., 2004). This conclusion is supported by the great potency of leptin when administered centrally (Campfield et al., 1995; Seeley et al., 1996) and the fact that selective removal of neuronal LepRb recapitulates the phenotype of leptin deficiency (Cohen et al., 2001). However, the complete set of leptin’s neural targets has not been established. The studies presented in this report validate a novel mouse model that now provides a means for studying the anatomic and functional characteristics of neuons that express LepRb.
Several groups have discovered that the LepRb transcript has been found to be expressed at low levels in many brain regions, thus making its detection technically challenging. Visualization of the EYFP protein as a marker for LepRb-expressing cells using standard IHC is a rapid and robust method of detection, provided that the GFP marker protein can be shown to be eutopically expressed. Consistent with previous descriptions of the central LepRb mRNA distribution, we found GFP immunoreactivity in regions of the hypothalamus and brainstem that have previously been described as expressing LepRb mRNA in the rat brain. We also used the LepRb-EYFP mice to identify GFP-IR cells in the MPA, SNC, DR, Edinger-Westphal nucleus, rostral linear nucleus, and several other regions not previously known to express LepRb in the mouse.
Distribution of LepRb mRNA in the wildtype mouse brain
Prior descriptions of LepRb expression in the mouse have focused primarily on the hypothalamic and brainstem populatios (Huang et al., 1996; Mercer et al., 1996; Fei et al., 1997; Elmquist et al., 1998; Shioda et al., 1998). While our results correlate well with earlier work in the mouse, we expand on these studies by comprehensively examining LepRb expression throughout the mouse brain. Consistent with previous studies, we found dense hybridization in various hypothalamic nuclei, with the highest expression in the ARH. We found little to no LepRb mRNA-expressing cells in the PVN as has been previously reported in the mouse (Mercer et al., 1996). Interestingly, we saw substantial MPA staining, whereas earlier studies in both rat and mouse reported no visible hybridization in this region (Mercer et al., 1996; Elmquist et al., 1998). Our observation that MPA LepRb-IRES-Cre LacZ cells exhibited significant pSTAT3 expression is further evidence of the presence of functional long-form leptin receptor in this region.
Similar to the rat, we found cells containing LepRb mRNA throughout the cortex. Many of the cortical regions found to express LepRb were similar across rat and mouse, but there were a few areas that differed. For example, hybridization was observed in layer V of the rat neocortex (Shioda et al., 1998), whereas in the mouse LepRb mRNA is restricted to layers 3, 4, and 6b. Consistent with previous descriptions, we find LepRb mRNA in the dentate gyrus (DG) (Shioda et al., 1998), but we also found hybridization in the CA2 and CA3 regions of the hippocampus. Additionally, we found that mouse and rat had common cerebellum expression including hybridization in the granular and Purkinje layers. Many extrahypothalamic areas including the DR, LPBS, and regions of the dorsal vagal complex previously found to express LepRb in the rat similarly express LepRb message in mouse (see also Mercer et al., 1998a). Prior studies along with our analysis of pSTAT3 expression suggest that there are significant populations of leptin-responsive neurons in some of these extrahypothalamic areas (Fernandez-Galaz et al., 2002; Hosoi et al., 2002; Munzberg et al., 2003). These findings underscore the importance of establishing the physiological relevance and requirement for extrahypothalamic leptin signaling in the regulation of metabolic homeostasis. Investigation of the role of leptin in extrahypothalamic CNS sites may be accomplished using Cre/loxP technology to selectively delete leptin receptor from select nuclei (Cohen et al., 2001). As well, using a Flp recombinase/frt system similar to Coppari et al. (2005), the necessity of leptin signaling in extrahypothalamic CNS nuclei could be analyzed through the restricted reactivation of leptin receptor expression in an otherwise leptin receptor null mouse.
While the distribution of LepRb expression in the rat and the mouse was largely similar, some differences were observed. In the thalamus, LepRb expression was observed in rat and mouse, but this expression localized to separate subregions in these two species. Additionally, the rat, but not the mouse, shows expression in the substantia nigra, reticular part, and area postrema. The finding that LepRb is not expressed in the mouse AP is consistent with a previous report (Mercer et al., 1998). The physiological role of the differential expression in mouse and rat is not yet known, but such differences highlight the importance of studies of central leptin action in both species.
Colocalization of LacZ and LepRb mRNA
To document the fidelity of the LepRb-Cre line, we performed a dual ISHH/IHC for LacZ and LepRb mRNA. We found coexpression of LepRb message in nearly all of the LepRb-Cre-activated LacZ cells throughout the brain. However, in the BSTN and PVN we found LacZ-IR cells that did not coexpress LepRb mRNA. These areas showed inconsistent labeling above background levels in the wildtype brain. Therefore, we attributed this discrepancy across studies to the LepRb mRNA levels being below the detection threshold of our ISHH technique. Additionally, the sparse lacZ expression in these areas, which generally correlates with low hybridization levels, indicates that, in these areas, LepRb mRNA is at levels subthreshold to the detection methodology used here. It is also possible that leptin receptor expression in BNST and PVN occurs during a restricted period of development, causing Cre-dependent activation of LacZ expression that is observed in the adult in the absence of adult leptin receptor expression. In other regions, we found LepRb-expressing cells with no detectable LacZ immunoreactivity. These were regions of low to moderate LepRb levels as detected by ISHH. This result may be due to the inability of the Cre recombinase, which is likely translated at a much reduced level (as a result of the IRES sequences employed) than that of the endogenous leptin receptor transcript to induce recombination in areas of low leptin receptor/Cre transcript abundance. Alternatively, suppression of the IRES used to drive Cre expression or even inhibition of expression from the ROSA26 locus in specific cell types could also lead to false negatives.
These few discrepancies aside, however, the LepRb-Cre mice permit rapid and straightforward identification of LepRb neurons, despite the very low levels LepRb mRNA in most neurons.
Characterization of LepRb expression in catecholaminergic neurons
We used the LepRb-LacZ mice, shown to recapitulate the endogenous expression pattern of LepRb, to chemically characterize LepRb-containing cell types. Previous reports support a role for catecholaminergic neurons in food intake regulation (Schwartz et al., 2000), although the mechanism of dopamine’s regulation of energy homeostasis remains unclear. Notably, Szczypka et al. (2000) illustrated the requirement for a functional domaninergic system for hyperphagia in the ob/ob mouse. Moreover, recent studies have shown that dopaminergic neurons in the VTA are targets of leptin (Fulton et al., 2006; Hommel et al., 2006). In the present study, we used the LepRb-LacZ model to elucidate the extent of neuroanatomical connectivity of LepRb and TH-containing neurons in the mouse brain. Similar to results in the rat (Figlewicz et al., 2003) and the recent reports in mice (Fulton et al., 2006; Hommel et al., 2006), we found that VTA LacZ-IR cells coexpressed TH protein. The extent of coexpression of TH with LacZ, however, was greater than previously suggested (Fulton et al., 2006, Hommel et al., 2006). In addition, we observed several other locations within the midbrain that demonstrated TH/LacZ coexpression, in the DR, SNC, Rli, Cli, and A8 nuclei. Outside of the midbrain, LepRb and TH cell populations are also present in the ARH and the NTS; however, we found no colocalization in these areas, suggesting that leptin’s direct action on the TH system is likely limited to dopaminergic neurons in the midbrain.
The potential role of leptin signaling in midbrain dopaminergic neurons remains poorly understood. The recent report from Hommel et al. (2006) suggests that leptin acts to directly inhibit dopaminergic neurons, with a knockdown of leptin receptor in the VTA enhancing food intake and nocturnal locomotion. Action at other midbrain dopaminergic loci, however, has yet to be investigated. This task is made more difficult by the fact that these nuclei, such as the Cli, have poorly described roles in the control of CNS function. Furthermore, the significance of leptin receptor expression in Rli, SNc, and A8 cell groups might also be questioned, as few TH/leptin receptor doubly expressing neurons were observed. Lastly, few TH neurons of the midbrain demonstrated STAT3 phosphorylation, suggesting that these cells do not sense peripheral leptin or that leptin elicits the activation of an alternate signaling cascade.
CNS sensing of peripheral leptin
Our results demonstrate that numerous nuclei within the CNS express long-form leptin receptor, suggesting that these cell groups may be capable of sensing changes in this peripherally derived metabolic signal. Phosphorylation of the signaling molecule STAT3 has been used previously as a direct measure of leptin receptor activation (Ghilardi et al., 1996). While neuronal deletion of STAT3 reproduces the metabolic phenotype of the LepR null animal (Gao et al., 2004), deletion of the STAT3 binding site on the long-form leptin receptor produced a phenotype similar to the leptin receptor null mouse (Bates et al., 2003). Interestingly, few of the CNS locations expressing leptin receptor show STAT3 phosphorylation upon peripheral administration of leptin. This is in agreement with a prior study that looked at STAT3 phosphorylation (in the absence of leptin receptor detection) in the hindbrain (Hosoi et al., 2002). The few regions of the CNS that showed pSTAT3 and leptin Cre-LacZ colocalization included select regions of the NTS, PAG, hypothalamus, and medial preoptic nuclei (MPO). Within these nuclei, cells were occasionally observed to be pSTAT3 +/LacZ–, suggesting that not all LepRb cells coexpressed LacZ. As well, mosaicism of pSTAT3/LacZ coexpression was seen within many nuclei expressing pSTAT3, with the exception of the arcuate, PVN, and VMH nuclei of the hypothalamus. Within the dorsal part of the DMV, the NTS, the MPO, and the LH, variable numbers of LacZ LepRb cells showed an absence of STAT3 phosphorylation. In general, areas that demonstrated pSTAT3 were those proximal to cerebral ventricles or the aqueduct, suggesting leptin delivery from the periphery to the CSF via the choroid plexus is the main CNS source of this peripheral hormone. Indeed, Zlokovic et al. (2000) have shown high affinity transport of leptin from the periphery into the hypothalamus and CSF via the choroid plexus. A reduced rate of transport across the blood−brain barrier may explain the lack of effect of peripheral leptin on STAT phosphorylation in areas such as the VTA and hippocampus, where the acute dose of leptin administered in this study had little effect. However, increasing the dose of administered leptin to achieve sensing of leptin (as measured by STAT3 phosphorylation) by these nuclei would likely take the injection of supraphysiological levels of leptin. It is therefore unlikely that areas such as the VTA and dentate gyrus of the hippocampus undergo leptin-mediated STAT3 phosphorylation. Thus, if leptin is acting in a physiologically relevant manner in these nuclei expressing LepRb, it may be via a JAKSTAT-independent mechanism.
For example, leptin is capable of activating other metabolically relevant signal transduction cascades, such as mammalian target of rapamycin (mTOR) (Cota et al., 2006), AMP-activated kinase (AMPK) (Minokoshi et al., 2004), or phosphoinositol-3-kinase (PI3K) (Niswender et al., 2001; Hill et al., 2008) in a STAT3-independent manner. Currently, the requirement for STAT3 phosphorylation and participation in these signaling cascades is unknown (Robertson et al., 2008). Alternatively, leptin may be acting during neuronal development, as has been shown previously (Bouret et al., 2004; Udagawa et al., 2006). Expression of leptin receptor in the adult cell that does not show STAT3 phosphorylation may then be a vestige of earlier expression, at a time of leptin action in the embryo or early postnatal animal. Finally, leptin receptor expression in LacZ+/pSTAT3– cells may act in a leptin-independent manner, modulating other receptor or membrane protein function.
Obviously, the functional significance of an inability of peripheral leptin to induce STAT3 phosphorylation has yet to be investigated and remains an important question in the characterization of the action of leptin in the CNS.
In summary, we have systematically defined the distribution of LepRb mRNA expression in the mouse brain. We have further characterized and validated a mouse model that expresses EYFP selectively in LepRb-containing cells. Finally, we assessed the extent of colocalization of LepRb-LacZ and TH throughout the mouse brain and investigated the responsiveness of LepRb cells to peripheral leptin. The LepRb-EYFP/LacZ mouse represents the first mouse strain that facilitates the precise and selective identification of LepRb expressing neurons and its use in studies of in vitro slices will enable electrophysiological, imaging, and neurochemical studies specifically in LepRb neurons.
Acknowledgments
Grant sponsor: National Institutes of Heath; Grant numbers: DK056116, MH061583, DK053301 (to J.K.E.), DK062656 to (J.L.L.).
LITERATURE CITED
- Ahima RS, Saper CB, Flier JS, Elmquist JK. Leptin regulation of neuroendocrine systems. Front Neuroendocrinol. 2000;21:263–307. doi: 10.1006/frne.2000.0197. [DOI] [PubMed] [Google Scholar]
- Badman MK, Flier JS. The gut and energy balance: visceral allies in the obesity wars. Science. 2005;307:1909–1914. doi: 10.1126/science.1109951. [DOI] [PubMed] [Google Scholar]
- Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC, Jr, Elmquist JK, Lowell BB. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AW, Wang Y, Banks AS, Lavery HJ, Haq AK, Maratos-Flier E, Neel BG, Schwartz MW, Myers MG., Jr STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003;421:856–859. doi: 10.1038/nature01388. [DOI] [PubMed] [Google Scholar]
- Benoit SC, Clegg DJ, Seeley RJ, Woods SC. Insulin and leptin as adiposity signals. Recent Prog Horm Res. 2004;59:267–285. doi: 10.1210/rp.59.1.267. [DOI] [PubMed] [Google Scholar]
- Bjorbaek C, Kahn BB. Leptin signaling in the central nervous system and the periphery. Recent Prog Horm Res. 2004;59:305–331. doi: 10.1210/rp.59.1.305. [DOI] [PubMed] [Google Scholar]
- Bouret SG, Draper SJ, Simerly RB. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science. 2004;304:108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- Campfield LA, Smith FJ, Guisez Y, Devos R, Burn P. Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- Cohen P, Zhao C, Cai X, Montez JM, Rohani SC, Feinstein P, Mombaerts P, Friedman JM. Selective deletion of leptin receptor in neurons leads to obesity. J Clin Invest. 2001;108:1113–1121. doi: 10.1172/JCI13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppari R, Ichinose M, Lee CE, Pullen AE, Kenny CD, McGovern RA, Tang V, Liu SM, Ludwig T, Chua SC, Jr, Lowell BB, Elmquist JK. The hypothalamic arcuate nucleus: A key site for mediating leptin’s effects on glucose homeostasis and locomotor activity. Cell Metab. 2005;1:63–72. doi: 10.1016/j.cmet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Dahlstroem A, Fuxe K. Evidence for the existence of monoamine-containing neurons in the central nervous system. I. Demonstration of monoamines in the cell bodies of brain stem neurons. Acta Physiol Scand Suppl. 1964;232:231–255. [PubMed] [Google Scholar]
- DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Elias CF, Saper CB, Maratos-Flier E, Tritos NA, Lee C, Kelly J, Tatro JB, Hoffman GE, Ollmann MM, Barsh GS, Sakurai T, Yanagisawa M, Elmquist JK. Chemically defined projections linking the mediobasal hypothalamus and the lateral hypothalamic area. J Comp Neurol. 1998;402:442–459. [PubMed] [Google Scholar]
- Elmquist JK, Saper CB. Activation of neurons projecting to the paraventricular hypothalamic nucleus by intravenous lipopolysaccharide. J Comp Neurol. 1996;374:315–331. doi: 10.1002/(SICI)1096-9861(19961021)374:3<315::AID-CNE1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Elmquist JK, Bjorbaek C, Ahima RS, Flier JS, Saper CB. Distributions of leptin receptor mRNA isoforms in the rat brain. J Comp Neurol. 1998;395:535–547. [PubMed] [Google Scholar]
- Fei H, Okano HJ, Li C, Lee GH, Zhao C, Darnell R, Friedman JM. Anatomic localization of alternatively spliced leptin receptors (Ob-R) in mouse brain and other tissues. Proc Natl Acad Sci U S A. 1997;94:7001–7005. doi: 10.1073/pnas.94.13.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Galaz MC, Diano S, Horvath TL, Garcia-Segura LM. Leptin uptake by serotonergic neurones of the dorsal raphe. J Neuroendocrinol. 2002;14:429–434. doi: 10.1046/j.1365-2826.2002.00783.x. [DOI] [PubMed] [Google Scholar]
- Figlewicz DP, Evans SB, Murphy J, Hoen M, Baskin DG. Expression of receptors for insulin and leptin in the ventral tegmental area/ substantia nigra (VTA/SN) of the rat. Brain Res. 2003;964:107–115. doi: 10.1016/s0006-8993(02)04087-8. [DOI] [PubMed] [Google Scholar]
- Gao Q, Wolfgang MJ, Neschen S, Morino K, Horvath TL, Shulman GI, Fu XY. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc Natl Acad Sci U S A. 2004;101:4661–4666. doi: 10.1073/pnas.0303992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim MH, Skoda RC. Defective STAT signaling by the leptin receptor in diabetic mice. Proc Natl Acad Sci U S A. 1996;93:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RB. Leptin—much more than a satiety signal. Annu Rev Nutr. 2000;20:45–75. doi: 10.1146/annurev.nutr.20.1.45. [DOI] [PubMed] [Google Scholar]
- Haycock JW, Waymire JC. Activating antibodies to tyrosine hydroxylase. J Biol Chem. 1982;257:9416–9423. [PubMed] [Google Scholar]
- Herbert H, Moga MM, Saper CB. Connections of the parabrachial nucleus with the nucleus of the solitary tract and the medullary reticular formation in the rat. J Comp Neurol. 1990;293:540–580. doi: 10.1002/cne.902930404. [DOI] [PubMed] [Google Scholar]
- Higuchi H, Yang HY, Sabol SL. Rat neuropeptide Y precursor gene expression. mRNA structure, tissue distribution, and regulation by glucocorticoids, cyclic AMP, and phorbol ester. J Biol Chem. 1988;263:6288–6295. [PubMed] [Google Scholar]
- Hill JW, Williams KW, Ye C, Luo J, Balthasar N, Coppari R, Cowley MA, Cantley LC, Lowell BB, Elmquist JK. Acute effects of leptin require PI3K signaling in hypothalamic proopiomelanocortin neurons in mice. J Clin Invest. 2008;118:1796–1805. doi: 10.1172/JCI32964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi T, Kawagishi T, Okuma Y, Tanaka J, Nomura Y. Brain stem is a direct target for leptin’s action in the central nervous system. Endocrinology. 2002;143:3498–3504. doi: 10.1210/en.2002-220077. [DOI] [PubMed] [Google Scholar]
- Huang XF, Koutcherov I, Lin S, Wang HQ, Storlien L. Localization of leptin receptor mRNA expression in mouse brain. Neuroreport. 1996;7:2635–2638. doi: 10.1097/00001756-199611040-00045. [DOI] [PubMed] [Google Scholar]
- Liu H, Kishi T, Roseberry AG, Cai X, Lee CE, Montez JM, Friedman JM, Elmquist JK. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003;23:7143–7154. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer JG, Hoggard N, Williams LM, Lawrence CB, Hannah LT, Trayhurn P. Localization of leptin receptor mRNA and the long form splice variant (Ob-Rb) in mouse hypothalamus and adjacent brain regions by in situ hybridization. FEBS Lett. 1996;387:113–116. doi: 10.1016/0014-5793(96)00473-5. [DOI] [PubMed] [Google Scholar]
- Mercer JG, Moar KM, Hoggard N. Localization of leptin receptor (Ob-R) messenger ribonucleic acid in the rodent hindbrain. Endocrinology. 1998;139:29–34. doi: 10.1210/endo.139.1.5685. [DOI] [PubMed] [Google Scholar]
- Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, Mu J, Foufelle F, Ferre P, Birnbaum MJ, Stuck BJ, Kahn BB. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature. 2004;428:569–574. doi: 10.1038/nature02440. [DOI] [PubMed] [Google Scholar]
- Morton GJ, Niswender KD, Rhodes CJ, Myers MG, Jr, Blevins JE, Baskin DG, Schwartz MW. Arcuate nucleus-specific leptin receptor gene therapy attenuates the obesity phenotype of Koletsky (fa(k)/fa(k)) rats. Endocrinology. 2003;144:2016–2024. doi: 10.1210/en.2002-0115. [DOI] [PubMed] [Google Scholar]
- Morton GJ, Blevins JE, Williams DL, Niswender KD, Gelling RW, Rhodes CJ, Baskin DG, Schwartz MW. Leptin action in the forebrain regulates the hindbrain response to satiety signals. J Clin Invest. 2005;115:703–710. doi: 10.1172/JCI200522081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzberg H, Huo L, Nillni EA, Hollenberg AN, Bjorbaek C. Role of signal transducer and activator of transcription 3 in regulation of hypothalamic proopiomelanocortin gene expression by leptin. Endocrinology. 2003;144:2121–2131. doi: 10.1210/en.2002-221037. [DOI] [PubMed] [Google Scholar]
- Niswender KD, Schwartz MW. Insulin and leptin revisited: adiposity signals with overlapping physiological and intracellular signaling capabilities. Front Neuroendocrinol. 2003;24:1–10. doi: 10.1016/s0091-3022(02)00105-x. [DOI] [PubMed] [Google Scholar]
- Niswender KD, Morton GJ, Stearns WH, Rhodes CJ, Myers MG, Jr, Schwartz MW. Intracellular signalling. Key enzyme in leptin-induced anorexia. Nature. 2001;413:794–795. doi: 10.1038/35101657. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KB. The mouse brain in stereotaxic coordinates. 2nd ed. San Diego: Academic Press; 2001. [Google Scholar]
- Robertson SA, Leinninger GM, Myers MG., Jr Molecular and neural mediators of leptin action. Physiol Behav. 2008;94:637–642. doi: 10.1016/j.physbeh.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Seeley RJ, van Dijk G, Campfield LA, Smith FJ, Burn P, Nelligan JA, Bell SM, Baskin DG, Woods SC, Schwartz MW. Intraventricular leptin reduces food intake and body weight of lean rats but not obese Zucker rats. Horm Metab Res. 1996;28:664–668. doi: 10.1055/s-2007-979874. [DOI] [PubMed] [Google Scholar]
- Shioda S, Funahashi H, Nakajo S, Yada T, Maruta O, Nakai Y. Immunohistochemical localization of leptin receptor in the rat brain. Neurosci Lett. 1998;243:41–44. doi: 10.1016/s0304-3940(98)00082-2. [DOI] [PubMed] [Google Scholar]
- Simmons DM, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabelled single stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Rainey MA, Palmiter RD. Dopamine is required for hyperphagia in Lep(ob/ob) mice. Nat Genet. 2000;25:102–104. doi: 10.1038/75484. [DOI] [PubMed] [Google Scholar]
- Udagawa J, Hashimoto R, Hioki K, Otani H. The role of leptin in the development of the cortical neuron in mouse embryos. Brain Res. 2006;1120:74–82. doi: 10.1016/j.brainres.2006.08.116. [DOI] [PubMed] [Google Scholar]
- van de Wall E, Leshan R, Xu AW, Balthasar N, Coppari R, Liu SM, Jo YH, MacKenzie RG, Allison DB, Dun NJ, Elmquist J, Lowell BB, Barsh GS, de Luca C, Myers MG, Jr, Schwartz GJ, Chua SC., Jr Collective and individual functions of leptin receptor modulated neurons controlling metabolism and ingestion. Endocrinology. 2008;149:1773–1785. doi: 10.1210/en.2007-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- Zigman JM, Elmquist JK. Minireview: from anorexia to obesity—the yin and yang of body weight control. Endocrinology. 2003;144:3749–3756. doi: 10.1210/en.2003-0241. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV, Jovanovic S, Miao W, Samara S, Verma S, Farrell CL. Differential regulation of leptin transport by the choroid plexus and blood-brain barrier and high affinity transport systems for entry into hypothalamus and across the blood-cerebrospinal fluid barrier. Endocrinology. 2000;141:1434–1441. doi: 10.1210/endo.141.4.7435. [DOI] [PubMed] [Google Scholar]