Abstract
Capillary liquid chromatography coupled with electrospray ionization to a quadropole ion trap mass spectrometer was explored as a method for the analysis of polar anionic compounds in complex metabolome mixtures. A ternary mobile phase gradient, consisting of aqueous acidic, aqueous neutral and organic phases in combination with an aqueous compatible reversed-phase stationary phase allowed metabolites with a wide range of polarities to be resolved and detected. Detection limits in the full scan mode for glycolysis and tricarboxylic acid cycle intermediates were from 0.9 to 36 fmol. Using this system, 111 ± 9 (n = 3) metabolites were detected in Escherichia coli lysate samples. Reducing column I.D. from 50 μm to 25μm increased the number of metabolites detected to 156 ± 17 (n = 3). The improvement in number of metabolites detected was attributed to an increase in separation efficiency, an increase in sensitivity, and a decrease in adduct formation. Implementation of a second separation mode, strong anion exchange, to fractionate the sample prior to capillary RPLC increased the number of metabolites detected to 244 ± 21 (n = 3). This improvement was attributed to the increased peak capacity which decreased co-elution of molecules enabling more sensitive detection by mass spectrometry. This system was also applied to islets of Langerhans where more significant improvements in metabolite detection were observed. In islets, 391 ± 33 small molecules were detected using the two-dimensional separation. The results demonstrate that column miniaturization and use of two-dimensional separations can yield a significant improvement in the coverage of the metabolome.
1. Introduction
Analysis of small molecule pools present in cells holds the promise of elucidating novel biochemical pathways, functions of genes, drug effects and mechanisms of pathophysiological states. Coupling of separation methods such as capillary electrophoresis (CE) [1–3], gas chromatography (GC) [4,5], reversed-phase liquid chromatography (RPLC) [6,7], strong anion-exchange chromatography (SAX) [8] and hydrophilic interaction liquid chromatography (HILIC) [9]) with mass spectrometry (MS) has proven effective for detecting large numbers of metabolites in complex mixtures. Despite the resolving power of these methods, they generally only detect a fraction of the metabolites that are present. Several factors contribute to the difficulty of detecting all metabolites in a given sample. The many components demand high peak capacity of the separation to detect the complete metabolome. The huge variation of metabolite concentrations requires wide dynamic range of the measurement. Finally, the chemical heterogeneity of metabolites in terms of polarity, size, and charge suggest that not all components will be compatible with a single extraction and separation protocol. In this work, we examine how column miniaturization and use of two-dimensional (2D) separations impacts the coverage of the metabolome in complex mixtures by increasing peak capacity and sensitivity.
One approach to improving peak capacity is to improve efficiency by using smaller particles, as in ultra-high-pressure liquid chromatography (UHPLC). Application of UHPLC to metabolomic analysis has yielded impressive resolution of complex mixtures of metabolites [10]. It is also well known that using 2D separations can also improve peak capacity. While dual column GC-MS is becoming prevalent for analysis of some classes of metabolites [11,12], 2D condensed-phase separations for metabolites has received less attention. In one example, a combination of conventional RPLC with a fast 2nd dimension of carbon-coated zirconia RPLC was used to analyze plant extracts and allowed detection of 100 small molecules in 25 min [13]. An offline combination of LC with CE has been used to separate and detect 63 metabolite standards [14]. The use of absorbance detection likely limited both detection and identification of more metabolites in these cases. The relative lack of multi-dimensional analysis for metabolites is in contrast to proteomics where multi-dimensional separations have been widely developed and characterized [15,16].
In this work, we examined the effect that 2D separations would have on the numbers of metabolites detected in complex mixtures from extracts of both prokaryotes and eukaryotes. Polar anions were selected as a group of compounds to target because they include energy metabolites, central to any organism, and their related products. Therefore, a combination of SAX and RPLC for the two-dimensions of separation was used.
In addition to exploring the effect of 2D separations, we also examined the utility of decreasing column inner diameter (I.D.) for improving the number of metabolites detected in a complex mixture. Column miniaturization improves separation efficiency [17] and electrospray ionization efficiency [18]. Both of these effects are expected to aid in detection of more metabolites in a complex mixture. For these reasons, and because of the reduced sample consumption, capillary LC is commonly used in proteomics. In contrast, capillary LC has been relatively unexplored for metabolomic analysis (although, CE has seen some use for this application). This lack of application of capillary LC may be because most metabolomic studies have been directed towards samples which are relatively abundant, such as plants, microbes, and bodily fluids, thus negating the need to analyze small quantities. Nevertheless, miniaturization may be expected to improve sensitivity, and therefore the number of compounds detected. Furthermore, use of miniaturized columns should also enable analysis of more limited samples including specific mammalian tissues [19–22] such as embryos, neurons, and islets of Langerhans, which are too small to analyze with currently used methods.
2. Experimental
2.1 Reagents and Materials
All chemicals were purchased from Sigma-Aldrich (St Louis, MO USA) unless otherwise noted. All solvents were purchased from Burdick and Jackson (Muskegon, MI USA). Fused silica capillary was purchased from Polymicro Technologies (Phoenix, AZ USA). Reversed phase chromatographic stationary phase was purchased from Waters (Milford, MA USA). SAX chromatographic stationary phase was donated by Dionex (Sunnyvale, CA USA). Eppendorf Safelock Biopur tubes were purchased from Fisher Scientific (Fairland, NJ).
2.2 Capillary Column Fabrication
Capillary columns were prepared as previously described [23]. Briefly, macroporous photopolymerized frits were placed 5 cm from the end of 50 cm long by 50 μm or 25 μm I.D. capillaries. Using a P-2000 laser puller, tips were pulled 1 cm downstream of the frit, then etched for 30 seconds in 49% HF (v/v) and rinsed to create a ~3 μm opening at the outlet of the column unless stated otherwise. Capillaries were packed with 3 μm Atlantis C18 aqueous reversed phase particles (Waters) using an acetone-slurry packing method [17]. All columns had 30 cm packed bed length and were subsequently cut to 35 cm total length.
For the SAX capillary columns, frits were formed by tapping 10 μm diameter borosilicate particles (Duke Scientific Palo Alto, CA USA) into the orifice of a 50 cm long by 250 μm I.D. capillary to a distance < 2 mm. The glass particles were then sintered in place with a flame [17]. SAX particles (CarboPac PA-100, 8.5 μm diameter) were slurry-packed at 2,000 psi, using 1 M NaCl as the solvent, to a bed length of 40 cm.
2.3 Chromatography Conditions
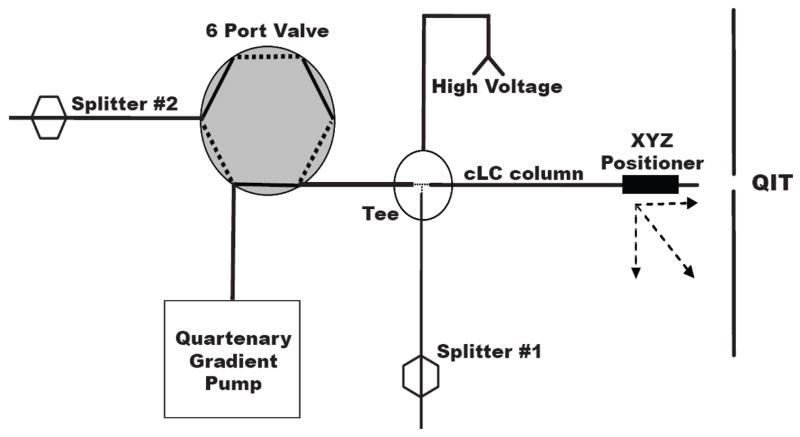
The RPLC system utilized two flow-splitters, one before the injection valve and one after the injection valve, to reduce flow into the system and control flow during injection (Figure 1). To avoid extra-column band broadening from the use of a conventional six port injection valve onto capillary columns, samples were loaded off-line. In the injection procedure, flow was diverted from the column to splitter 1 using the injection valve. The column was uncoupled from the system and sample (9 nL for 50 μm I.D. and 2.5 nL for 25 μm I.D.) pumped onto the column using gas pressure applied to a sample reservoir. The column was then reconnected to the splitting tee and the injection valve actuated to initiate flow through the column. With this procedure, the HPLC pump (Model 626, Waters) was able to maintain a constant pressure and initiated flow immediately after the valve actuation. Despite multiple re-connections, no air bubbles were observed during the course of these experiments. Three mobile phases were used to form the reversed phase gradient. Mobile phase A was 20 mM formic acid pH 2.7, mobile phase B was 20 mM ammonium acetate pH 6.5, and mobile phase C was acetonitrile. All gradient transitions were linear, with the following profile (time: %A, %B, %C); initial time 0 min: 100%, 0%, 0%; hold 5 min.; 10 min: 0%, 100%, 0%; 18 min: 0%, 80%, 20%; 23 min: 0%, 65% 35%; 60 min: 0%, 25%, 75%; 65 min: 100%, 0%, 0%. The column was re-equilibrated for 30 min with mobile phase A following gradient completion. Extended gradient consisted of 0 min: 100%, 0%, 0%; hold 5 min.; 10 min: 0%, 100%, 0%; 23 min: 0%, 80%, 20%; 34 min: 0%, 65% 35%; 60 min: 0%, 25%, 75%; 65 min: 100%, 0%, 0%.
Figure 1.
Block diagram of capillary LC system illustrating splitters and voltage application. For sample loading, the 6 port valve is set in “load” position (flow path represented by dashed line), the column is removed from the tee, sample loaded onto column using gas pressure sample reservoir (500 psi) and then reconnected to tee. The valve is actuated to inject position (solid lines) and gradient initiated. With this configuration, the pump is maintained at near constant pressure during the entire experiment.
The SAX chromatography system was operated in a conventional 6 port injection valve format. After injection of 10 μL of sample, a step gradient was used to elute anionic metabolites off the column. All gradient steps were 10 min in duration with mobile phase A: 5 mM ammonium formate pH 6.5 and mobile phase B: 700 mM ammonium formate (NH4HCO2) pH 6.5. Buffer was adjusted using ammonium hydroxide and formic acid as needed to reach the desired pH. The gradient profile was as follows (time: %A, %B): 0 min: 100%, 0%; 10 min: 86%, 14%; 20 min: 72%, 28%; 30 min: 58%, 42%; 40 min: 44%, 56%; 50 min: 30%, 70%; 60 min: 0%, 100%. Six fractions were collected from the capillary outlet in 10 minute intervals. Fractions were then evaporated to dryness using a vacuum centrifuge and reconstituted to 10 μL with 10 mM formic acid for further analysis using RPLC-MS.
2.4 Mass Spectrometry
Detection was accomplished using a LCQ Deca XP Plus quadrupole ion trap mass spectrometer (Thermo-Electron, San Jose, CA USA) operated in negative mode (spray voltage: -1.0 kV). Spectra were obtained in centroid mode with the mass spectrometer tuned for nucleotides (GTP) and the capillary temperature set to 150 °C. Experiments were performed in full scan mode with the maximum injection time of 25 ms and the scan range between 50–1500 m/z. The capillary column was interfaced to the mass spectrometer using a nanospray interface (Thermo-Electron).
2.5 Sample Preparation: Escherichia coli (E. coli)
For cells cultured in minimal media, E. coli N99 was first grown in LB media on an orbital shaker overnight. The following morning, 25 μL of culture was added to 50 mL of minimal media (48 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 18.8 mM NH4Cl, 2.0 mM MgSO4, 0.1 mM CaCl2). Cells were grown on a shaker at 37 °C until OD600 = 1.5 (~20 h) when 15 mL of cultured E. coli was added to 35 mL of water at 4 °C. Afterwards, the sample was spun and rinsed with cold water.
After a second rinse-resuspension, the E. coli suspension was centrifuged, supernatant decanted and replaced with −20 °C 500 μL of 50:50 methanol-water (v/v). Cells were lysed using a sonic dismembrator (Fischer Scientific, model 60) set to power of 9 W for 10 s. The lysate was then mixed 50:50 (v/v) with chloroform and set to equilibrate on ice for 10 min. The final solvent composition was 25:25:50 (v/v/v) methanol: water: chloroform. The aqueous portion was then removed and centrifuged at 4 °C, 1200 g for 10 min. Supernatant was collected and vacuum centrifuged to near dryness (< 5 μL), then reconstituted with mobile phase A to 100 μL for analysis. Samples were stored at −80 °C until use (< 1 week).
2.6 Sample Preparation: Islets of Langerhans
Islets of Langerhans were isolated from CD-1 mice (20–30 g) as previously described [24] with slight modifications. Briefly, collagenase XI (0.5 mg/mL) was infused through the pancreatic duct. Pancreas was then removed and incubated in 5 mL of collagenase XI (0.5 mg/mL) at 37 °C for 7 min. Islets were isolated by passing digested pancreas through a 100 μm pore diameter nylon cell strainer (Fisher Scientific) with 10 mL of Krebs ringer buffer. This procedure collected islets that were greater than 100 μm in diameter. Islets were transferred to a Petri dish by inverting the filter which was rinsed with 5 mL of buffer. Islets were manually selected using a pipette and transferred to a Petri dish containing cell culture media (10 mM glucose RPMI media 1640 with L-glutamine) supplemented with 10% fetal bovine serum and 100 units/mL of penicillin and 100 μg/mL of streptomycin. Islets were incubated in this media at 37 °C, 5% CO2 for 2 to 7 days before use.
A total of 50 islets were transferred from media to a rinse solution of 10 mM glucose in water. Islets were rapidly removed in 15 μL of rinse solution, placed in a Biopure tube and tapped down to deposit islets at the bottom of the tube. Rinse solution was then removed to ~ 5 μL and 10 μL of 50:50 MeOH-H2O (v/v) at −20 °C was added. Islets were triturated using a pipette until they appeared shredded as imaged through an inverted microscope. The process between rinse and ending trituration took ~45 sec. After islets were lysed, 15 μL of chloroform was added to the lysate and allowed to equilibrate on ice for 10 min. Sample was then treated as E. coli samples, with a post evaporation reconstitution volume of 15 μL.
2.7 Data Processing
Peaks were discriminated from noise using MS Processor version 8.0 from Advanced Chemistry Development (Toronto, Canada). Detection of peaks, using a reconstructed ion chromatogram format, was performed using the following parameters: MCQ threshold = 0.85; smoothing window width = 3 scans; minimum full width at half maximum height = 3 scans; signal-to-noise = 5. Peaks were matched between runs when tr < 3% RSD and Δm/z < 0.5. Statistical comparison was performed using a two tailed student’s t-test.
3. Results and Discussion
3.1 Chromatography of Metabolite Standards
Our initial experiments focused on developing a capillary RPLC system for separation of polar metabolites. In order to ensure separation of a wide scope of small molecules, 24 metabolites from glycolysis and the tricarboxylic acid (TCA) cycle were targeted for separation and detection (see Table 1 for compounds used). These analytes were chosen because they cover a broad range of charges and polarities, from polar glucose-6-phosphate to nonpolar FAD. A silica-based reversed-phase particle designed to be compatible with 100% aqueous mobile phases (Atlantis) was used to facilitate retention and separation of highly polar compounds.
Table 1.
Figures of merit for metabolite standards. % RSDs were analyzed based on peak height for 5 μM n = 3. Lower limits of detection were calculated as the concentration that would give a S/N of 3 based on the average S/N ratio of peaks at sample concentrations of 5 μM, n = 3. Detection limits were determined using gradient separation conditions similar to those used in Figure 2.
| Metabolite | % RSD | LLOD (fmol) | LLOD (μM) |
|---|---|---|---|
| G6P/F6P | 24 | 4 | 0.5 |
| F1,6P | 18 | 6 | 0.6 |
| Glyceraldehyde-3-Phosphate | 16 | 30 | 4 |
| 2/3 Phospho-glycerate | 26 | 30 | 4 |
| Bis-phospho-glycerate | 17 | 7 | 0.8 |
| Phosphoenol Pyruvate | 11 | 20 | 3 |
| Pyruvate | 33 | 20 | 2 |
| Acetyl CoA | 25 | 2 | 0.2 |
| Citrate | 31 | 10 | 1 |
| Isocitrate | 30 | 7 | 0.8 |
| α-Ketoglutarate | 18 | 10 | 1 |
| Succinyl CoA | 30 | 4 | 0.4 |
| Succinate | 10 | 9 | 1 |
| Fumarate | 27 | 10 | 2 |
| Malate | 19 | 3 | 0.3 |
| Oxaloacetate | 24 | 20 | 2 |
| ATP | 6 | 5 | 0.6 |
| ADP | 10 | 4 | 0.4 |
| NAD | 17 | 1 | 0.1 |
| GTP | 19 | 5 | 0.6 |
| GDP | 34 | 4 | 0.4 |
| FAD | 32 | 2 | 0.2 |
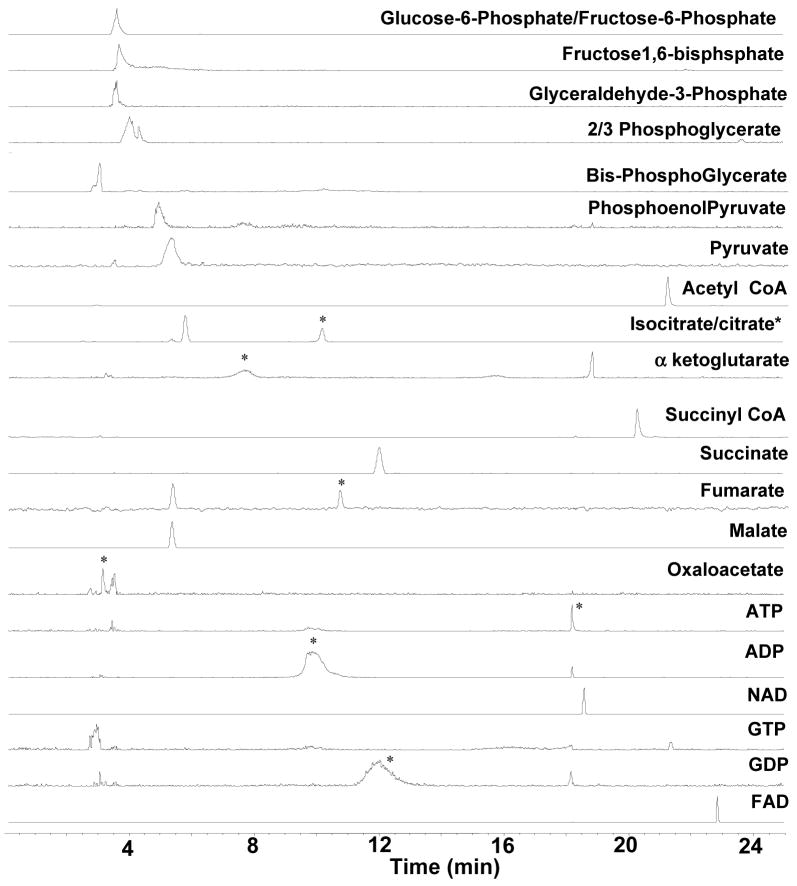
Work with this stationary phase revealed that manipulation of mobile phase pH was critical in allowing separation of the different types of analytes present in glycolysis and the TCA cycle. With aqueous mobile phases at pH < 3.0, polar organic acids were well-retained and resolved but, phosphorylated nucleotides yielded poor peak shape and peak height (See supporting information Figures S1 and S2). When the aqueous component of the mobile phase had pH > 6.5, the phosphorylated nucleotides were well resolved but, polar organic acids co-eluted near the column dead volume. Intermediate pH mobile phases (pH = 4.5) yielded poor resolution and peak shape for all classes of metabolite standards (data not shown). These data indicated that low pH mobile phase was necessary for separation of small organic acids while higher pH was required for separation of phosphorylated nucleotides. To resolve this problem, a gradient of formic acid pH 2.7 to ammonium formate pH 6.5 was used for the initial part of the separation followed by an increase in acetonitrile content to elute the less polar analytes. As seen in Figure 2, these conditions allowed separation of 23 of the 24 metabolite standards and were used for all further RPLC experiments.
Figure 2.
Reconstructed ion chromatogram illustrating resolution of 23 of 24 metabolite standards (10 μM of each metabolite) using ternary gradient of formic acid, ammonium formate and acetonitrile. Standards were mixed together before analysis. The LC column used was 30 cm long × 50 μm I.D. packed with Atlantis dC-18 3 μm particles and equipped with an integrated nanospray tip with 3 μm diameter. Gradient is given in the experimental section. Flow velocity was 100 nl/min. * indicate peak for the analyte when multiple peaks are detected.
For detection, negative mode ESI was used. Preliminary experiments revealed that negative mode ESI substantially decreased adduct formation and increased S/N when compared to positive mode ESI for these metabolites (data not shown). The use of capillary columns allowed for good sensitivity with lower detection limits of 0.3–12 fmol for glycolytic and TCA metabolite standards (see Table 1). These detection limits are a two to three order of magnitude improvement over previous reports even though the prior work used SRM for detection instead of full-scan mode [8,25]. For these measurements, only 9 nL of sample were consumed for the injection. Although the mass detection limit was considerably improved over previous results, the concentration detection limit was generally not improved over prior results. The use of full scan mode was chosen to facilitate an untargeted metabolomics approach, though this is likely the reason why cLODs were not improved over previous tandem MS modes. The good mass detection limits suggest that this method may have utility for sample limited metabolomic analysis; although that application was not explored in this work.
3.2 E. coli Metabolite Detection
When this method was applied to extracts of E. coli, 157 features were detected in the LC-MS data. We were interested in determining the number of actual endogenous small molecules detected; therefore, data were analyzed to eliminate false peaks and multiple peaks for single compounds. Peaks due to Na+ and formate adducts (as indicated by + 22 and + 46 m/z respectively and identical retention time to the base analyte), multiple charge states, and water loss, were eliminated from analyte count. Using the above criteria, a total of 111 ± 9 (n = 3) compounds were detected in the E. coli samples that were not present in background.
Using the average peak width for the compounds detected and the retention time window, a peak capacity of 110, based on unity resolution, was calculated for this separation. In calculating peak capacity, we determined the peak capacity separately for the two different gradients (a pH gradient and the organic phase gradient), because the peaks had different widths in these gradients, and added them together. Detection of more small molecules than indicated by the calculated peak capacity is due to the use of the mass spectrometer as the detector, which substantially increases the peak capacity.
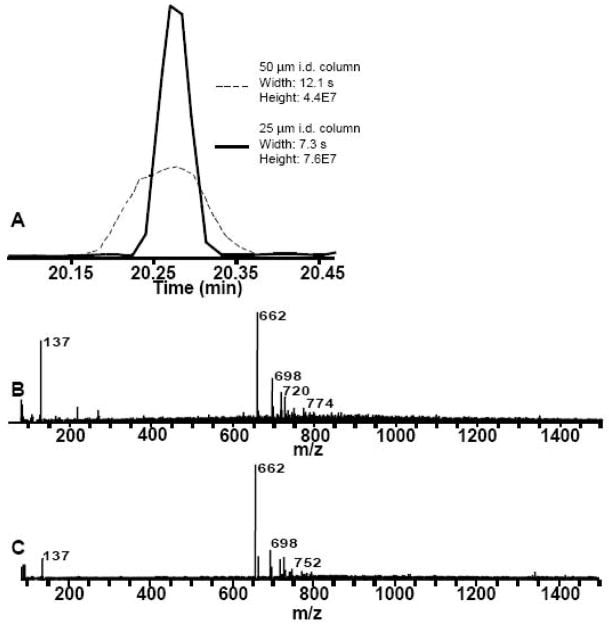
To evaluate the effect of column dimension on detection of metabolites in E. coli, the capillary column I.D. was reduced from 50 μm to 25 μm. For these experiments, the gradient conditions and mobile phase velocity were kept the same but the volume injected was decreased by 4 in proportion to the column volume. The ESI tip I.D. was decreased from 3.0 to 1.5 μm and the volumetric flow rate from 104 to 25 nL/min. Decreasing the column I.D. is expected to improve separation efficiency primarily by decreasing the A term of the Van Deemter equation [17]. Furthermore, the lower flow rate and smaller emitter tips may be expected to improve ionization efficiency and therefore sensitivity [26, 27]. Both of these effects are seen in the comparison of the reconstructed ion chromatograms for m/z 662, chosen as a typical analyte, shown in Figure 3. On average, peak width decreased by 25 ±14% (n = 4) while the signal increased by 88 ± 21% (n = 4) for decreasing the column I.D. by 2-fold. The signal increased even though 4-fold less sample was injected. The improvement in signal intensity cannot be solely attributed to improved peak shape as peak area was also increased (23% greater) in the 25 μm column.
Figure 3.
Effect of decreasing column I.D. on peak signal and mass spectrum.
A. Reconstructed ion chromatogram of 662 m/z from 50 μm I.D. and 25 μm I.D. column. B. Average mass spectrum over width of 662 m/z peak from 50 μm I.D. column. C. Average mass spectrum over width of 662 m/z peak from 25 μm I.D. column. Comparison of B and C illustrates fewer adducts to the 662 m/z peak and a 73% increase in signal intensity.
An added benefit of the decrease in column I.D. was a decrease in adduct formation. As shown in Figure 3B and C, peaks 698 m/z and 720 m/z, adducts to 662 were reduced from ~40% of base height to ~20%. The increase in signal intensity discussed above can be in large part attributed to the reduced adduct formation and increased ionization efficiency [18].
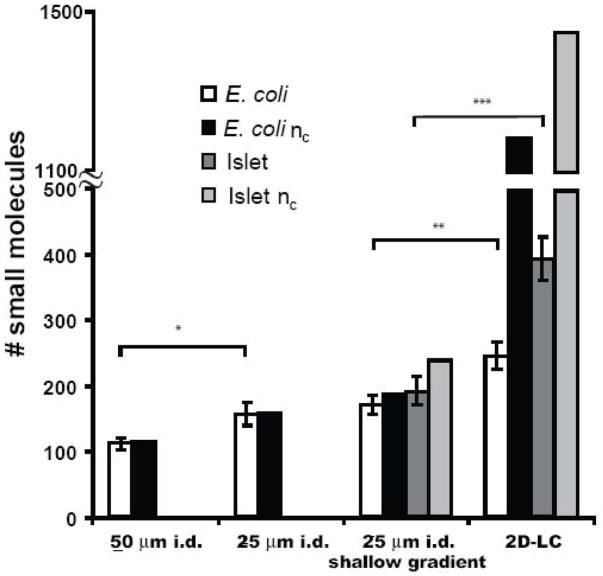
As summarized in Figure 4, the decrease in peak width associated with decreasing the column bore increased peak capacity from 114 to 168. The actual number of peaks detected in E. coli extracts increased in proportion to the peak capacity from 111 ± 9 to 156 ± 17 (n = 3). These results illustrate a multi-faceted improvement in decreasing column I.D. for complex metabolite detection. We also examined the effect of extending the gradient time from 11 to 24 min. This manipulation had a modest, but not statistically significant, improvement in peak capacity and number of metabolites detected (see Figure 4).
Figure 4.
Effect of chromatographic changes on the peak capacity and number of small molecules detected in complex mixtures. Error bars represent 1 standard deviation. Statistical significance between samples is as follows: * p < 0.05; ** p < 0.01; *** p < 0.005.
3.3 Two-Dimensional Capillary Chromatography
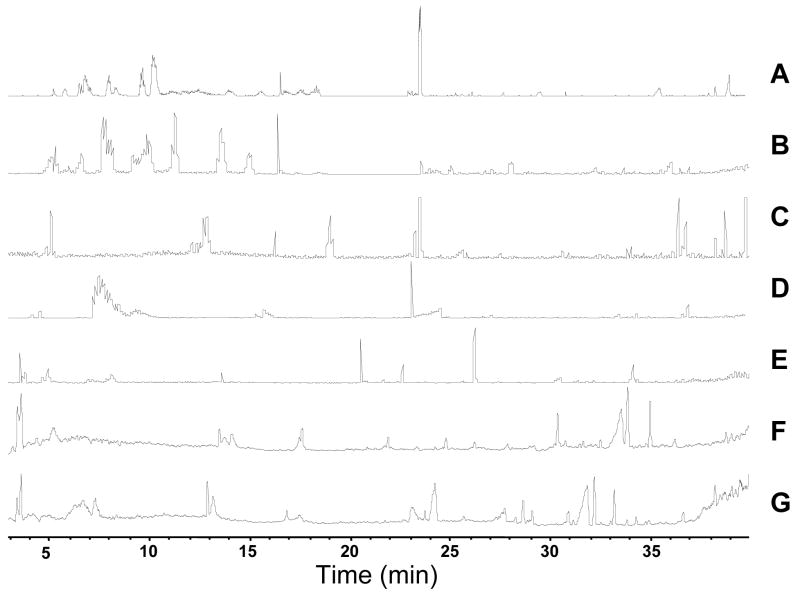
After establishing the peak capacity and number of components detected on the reversed phase column, the effect of pre-fractionation using a SAX column was evaluated. For this investigation, six fractions resulting from step gradients of 100 mM increments of ammonium formate were collected from the SAX column off-line and stored for a second separation on the reversed phase column. The SAX-RPLC-MS method was applied to extracts of E. coli lysate. Figure 5B–G shows the base peak chromatogram of six SAX fractions injected onto a capillary RPLC column. The use of a 2D separation resulted in detection of 244 ± 21 compounds compared to 170 ± 14 (n = 3) for the single dimension RPLC with the same RPLC gradient. As the SAX eluent was dried and then reconstituted to the same volume as initially injected (Figure 5B–G), dilution effects of the first dimension should not be present. The peak capacity of the separation is estimated as 970 (6 SAX fractions × 161 peaks for RPLC).
Figure 5.
Base peak chromatograms from 1D and 2D separation of small molecules from E. coli. RPLC was performed using 30 cm × 25 μm I.D. column. 2D was performed by off-line fraction collection in 10 minute step intervals from SAX capillary column. [Brackets] indicate the average number of small molecules detected. A:1-D RPLC-MS [170] B-G 2-D SAX-RPLC B: 5 mM NH4HCO2 fraction [79] C. 100 mM NH4HCO2 fraction [46] D. 200 mM NH4HCO2 fraction [45] E. 300 mM NH4HCO2 fraction [38] F. 400 mM NH4HCO2 fraction [19] G. 500 mM NH4HCO2 fraction [16]
Carryover, defined as detection of a given compound in more than 1 SAX fraction, was found for approximately 14% of the small molecules detected. The majority of the carryover was present between the first two fractions. This indicated that those compounds which were weakly retained on the SAX column were overloaded, whereas those compounds of higher affinity were well retained and preconcentrated in the 1st dimension.
In principle the MS adds a significant resolving component making it unnecessary to use extremely high peak capacity separations; however, the above results illustrate that the improved separation resolution increased the coverage of the metabolome. At least part of this effect is due to signal enhancement associated with reducing the number of co-eluting components. Signal enhancement was quantified by comparing signal intensity from co-eluting compounds in the 1-D RPLC with those same compounds which were separated into different fractions by SAX and analyzed by RPLC. Seven sets of peaks were found to co-elute in the 1D analysis which were resolved into different fractions by SAX. These peaks increased signal intensity on average by 47% (± 29%). Presumably this improvement is due to reduction of competing ionization and other matrix effects.
3.4 Islet Analysis
The majority of metabolomic analyses have involved plants and prokaryotes; however, metabolomic analysis of specific tissues is mammalian systems is emerging as an important application [23,28]. We therefore also evaluated the effectiveness of these methods on islets of Langerhans. Islets are microorgans dispersed throughout the pancreas that contain about 3000 cells each. Islets secrete insulin in a process driven by glucose metabolism. Metabolic disturbances of islets are a possible root cause of type 2 diabetes, therefore considerable benefit may emerge from metabolomic analysis of these cells. Only ~70 islets can be isolated from a single mouse and the smallest reasonable sample volume for extracting this amount of tissue is ~ 20 μL; therefore, minimizing the sample volume required for analysis is of interest. The use of capillary columns is well-suited for analysis of such samples.
Figure 6A shows the base peak chromatogram from a capillary RPLC-MS analysis of lysate from 50 islets in 15 μL. Average peak width for these samples (n = 3) was 8 s, which is ~ 2 s less than those found with E. coli samples. The decrease in average peak width was due to detecting a larger number of hydrophobic compounds, which tend to elute as narrower zones in this gradient. As a result of the decrease in peak width, the peak capacity calculated for the separation was increased to 240 from 161 for the E. coli sample (Figure 4). Using the criteria described above for the E. coli analysis for distinguishing peaks from compounds, an average of 191 ± 14 (n = 3) compounds were detected in the islet samples.
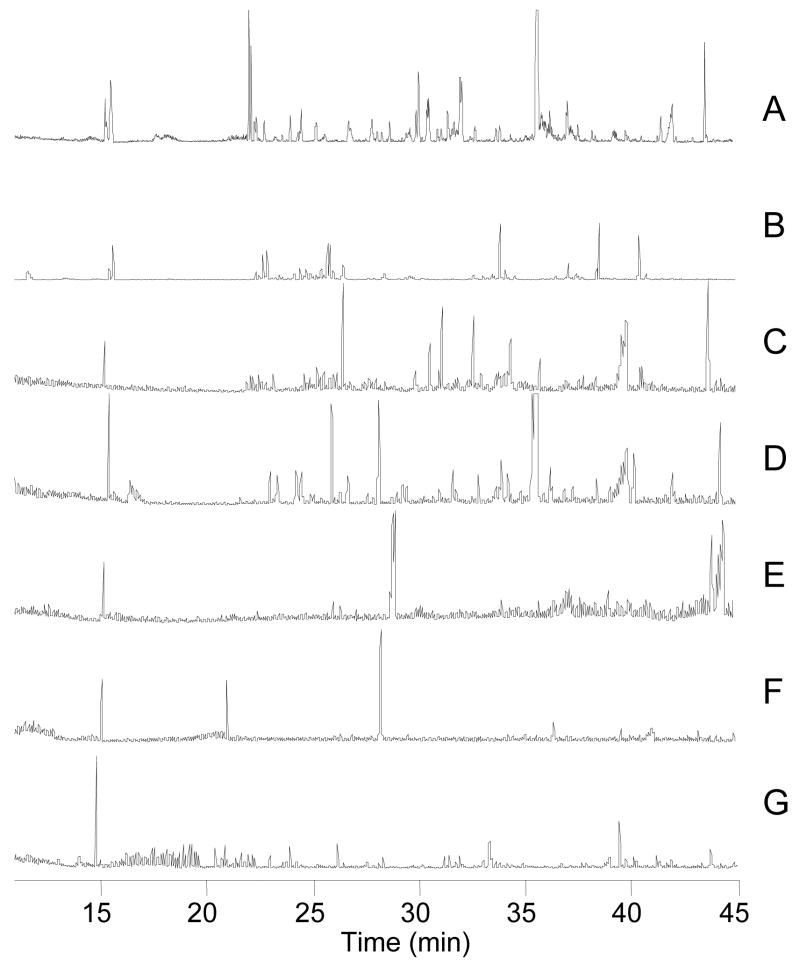
Figure 6.
Base peak chromatograms from 1D and 2D separation of small molecules from 50 islets of Langerhans. Experiment performed as in Figure 6. [Brackets] indicate the average number of small molecules detected. A:1-D RPLC-MS [191] B-G 2-D SAX-RPLC B: 5 mM NH4HCO2 fraction [155] C. 100 mM NH4HCO2 fraction [70] D. 200 mM NH4HCO2 fraction [73] E. 300 mM NH4HCO2 fraction [41] F. 400 mM NH4HCO2 fraction [36] G. 500 mM NH4HCO2 fraction [18].
As with the E. coli samples, use of the 2D separation significantly increased the coverage of the metabolome (Figures 4 and 6B–G). Whereas the single dimension RPLC yielded on average 191 small molecules, 2D-LC allowed for detection of 391 ± 33 peaks with a peak capacity of 1200. Figure 6 illustrates that the number of peaks detected decreased over the course of the step gradient, with the majority found in the first three fractions. The limited number of compounds found in the last three steps suggests that metabolites with a high degree of charge are relatively rare in islets.
Interestingly, islets yielded significantly more metabolites than the E. coli (Figure 4) even though the islet samples were about 1% as concentrated on a per gram of tissue basis. Many of these metabolites were fairly large (m/z > 600) and non-polar (i.e., eluted during the organic phase of the gradient). The greater complexity of the metabolome is likely due to in part to the media in which the cells are cultured. Islets were incubated in media containing glucose, amino acids, vitamins and fetal bovine serum, which would allow for synthesis and uptake of more metabolites than the media containing only glucose as a fuel as was the case for E. coli. The differences in metabolites detected may also reflect inherent differences in the complexity of the metabolomes of the different tissue types. Because of the greater complexity of the islet metabolome, the use of 2D separation was even more important for increasing the number of detected metabolites in these samples than for the E. coli samples (compare increase in detected metabolites in Figure 4).
Our experiments have addressed the potential impact of column dimensions and multidimensionality on metabolite separations. Many other factors are important in determining the quality of metabolomic data including sample extraction, quantification, and peak identification. Recent data has suggested for example that the use of water rinses on bacteria samples, such as those used here, prior to extraction may result in disturbance of metabolite levels [29]. Therefore, development of metabolomic methods for quantification will require consideration of those variables as well.
4. Conclusion
The work presented here demonstrates the utility of an aqueous-compatible stationary phase in combination with a ternary gradient to separate and detect metabolites with a wide range of polarities and charge, including those in glycolysis and the TCA cycle. Reduction of column I.D. led to superior peak shape, reduction of adducts, and higher signal intensity. The use of multiple dimensions improved peak capacity as expected. Combined, these effects more than doubled the number of metabolites detected from a complex metabolomic samples as summarized in Figure 4. A comparison of the number of metabolites in the E. coli and islet samples reveals detection of approximately 60% more in the islets. Furthermore, the islet samples had more high molecular weight molecules. These differences may reflect either an inherently more complex metabolome or effects of incubation media or both. Regardless, these results illustrate the potential for high sensitivity and high resolution for metabolomic analysis by multi-dimensional separations on capillary columns.
Supplementary Material
Figure S1
Separation of metabolite standards under acidic (pH 2.7) conditions with ACN. Gradient was 0–20% ACN over 20 min. Metabolite concentrations were 10 μM each.
Figure S2
Separation of metabolite standards under neutral (pH 6.5) conditions with ACN gradient. Gradient was 0–20% ACN over 20 min. Top 5 traces are reconstructed ion chromatograms (RIC) for mass of each compound indicated. Bottom trace is total ion chromatogram (TIC). Metabolite concentrations were 10 μM each.
Acknowledgments
This work was supported by NIH R37 DK 46960.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards JL, Chisolm CN, Shackman JG, Kennedy RT. J Chromatogr A. 2006;1106:80. doi: 10.1016/j.chroma.2005.08.082. [DOI] [PubMed] [Google Scholar]
- 2.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. J Proteome Res. 2003;2:488. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 3.Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T. Anal Chem. 2002;74:2233. doi: 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- 4.Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Nat Biotech. 2000;18:1157. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson P, Gullberg J, Nordstrom A, Kusano M, Kowalczyk M, Sjostrom M, Moritz T. Anal Chem. 2004;76:1738. doi: 10.1021/ac0352427. [DOI] [PubMed] [Google Scholar]
- 6.Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. Anal Chem. 2003;75:6737. doi: 10.1021/ac034716z. [DOI] [PubMed] [Google Scholar]
- 7.Lu WJ, Kimball E, Rabinowitz JD. J Am Soc Mass Spectrom. 2006;17:37. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Mashego MR, Wu L, van Dam JC, Ras C, Vinkle JL, van Winden WA, van Gulik WM, Heijen JJ. Biotech Bioeng. 2004;85:620. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- 9.Lafaye A, Laberre J, Tabet JC, Ezan E, Junot C. Anal Chem. 2005;77:2026. doi: 10.1021/ac048657g. [DOI] [PubMed] [Google Scholar]
- 10.Wilson ID, Nicholson JK, Castro-Perez J, Granger JH, Johnson KA, Smith BW, Plumb RS. J Prot Res. 2005;4:2201. doi: 10.1021/pr049769r. [DOI] [PubMed] [Google Scholar]
- 11.Mohler RE, Dombek KM, Hoggard JC, Young ET, Synovec RE. Anal Chem. 2006;78:2700. doi: 10.1021/ac052106o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welthagen W, Shellie RA, Spranger J, Ristow M, Zimmerman R, Fiehn O. Metabolomics. 2005;1:65. doi: 10.1016/j.chroma.2005.05.088. [DOI] [PubMed] [Google Scholar]
- 13.Stoll DR, Cohen JD, Carr PW. J Chromatogr A. 2006;1122:123. doi: 10.1016/j.chroma.2006.04.058. [DOI] [PubMed] [Google Scholar]
- 14.Jia L, Tanaka N, Terabe S. Electrophoresis. 2005;26:3468. doi: 10.1002/elps.200500308. [DOI] [PubMed] [Google Scholar]
- 15.Washburn MP, Wolters D, Yates JR. Nat Biotechnol. 2001;19:242. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 16.Opiteck GJ, Lewis KC, Jorgenson JW, Anderegg RJ. Anal Chem. 1997;69:1518. doi: 10.1021/ac961155l. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy RT, Jorgenson JW. Anal Chem. 1989;61:1128. doi: 10.1021/ac00180a012. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt A, Karas M, Dulcks T. J Am Soc Mass Spectrom. 2003;10:300. doi: 10.1016/S1044-0305(98)00157-3. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy RT, Oates MD, Cooper BR, Nickerson B, Jorgenson JW. Science. 1989;246:57. doi: 10.1126/science.2675314. [DOI] [PubMed] [Google Scholar]
- 20.Woods LA, Powell PR, Paxon TL, Ewing AG. Electroanalysis. 2005;17:1192. doi: 10.1002/elan.200403240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolters AM, Jayawickrama DA, Sweedler JV. J Nat Prod. 2005;68:162. doi: 10.1021/np030302p. [DOI] [PubMed] [Google Scholar]
- 22.Harwood MM, Christians ES, Fazal MA, Dovichi NJ. J Chromatogr A. 2006;1130:190. doi: 10.1016/j.chroma.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 23.Haskins WE, Wang ZQ, Watson CJ, Rostand RR, Witowski SR, Powell DH, Kennedy RT. Anal Chem. 2001;73:5005. doi: 10.1021/ac010774d. [DOI] [PubMed] [Google Scholar]
- 24.Edwards JL, Kennedy RT. Anal Chem. 2005;77:2201. doi: 10.1021/ac048323r. [DOI] [PubMed] [Google Scholar]
- 25.Bajad SU, Lu WY, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. J Chromatogr A. 2006;1125:76. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Korner R, Wilm M, Morand K, Schubert M, Mann M. J Am Soc Mass Spectrom. 1996;7:150. doi: 10.1016/1044-0305(95)00626-5. [DOI] [PubMed] [Google Scholar]
- 27.Andren PE, Caprioli RM. J Mass Spectrom. 1995;30:817. doi: 10.1002/(SICI)1096-9888(199904)34:4<377::AID-JMS778>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 28.Andren PE, Emmett MR, Caprioli RM. J Am Soc Mass Spectrom. 1994;5:867. doi: 10.1016/1044-0305(94)87010-1. [DOI] [PubMed] [Google Scholar]
- 29.Kimball E, Rabinowitz JD. Anal Biochem. 2006;358:273. doi: 10.1016/j.ab.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Separation of metabolite standards under acidic (pH 2.7) conditions with ACN. Gradient was 0–20% ACN over 20 min. Metabolite concentrations were 10 μM each.
Figure S2
Separation of metabolite standards under neutral (pH 6.5) conditions with ACN gradient. Gradient was 0–20% ACN over 20 min. Top 5 traces are reconstructed ion chromatograms (RIC) for mass of each compound indicated. Bottom trace is total ion chromatogram (TIC). Metabolite concentrations were 10 μM each.