Abstract
Increased 14-3-3σ expression has been observed by immunohistochemistry in papillary and anaplastic tumors, but not follicular thyroid cancers. 14-3-3σ mRNA expression and methylation status was examined in tumor cell lines and primary thyroid tissues using real-time RT-PCR, bisulfite sequencing and methylation-specific PCR. Most of the 27 CpG’s in the gene’s CpG island were methylated in normal thyroid, TPC-1, NPA, FTC-238 and 2–7, which did not express 14-3-3σ. In contrast, they were unmethylated in KAK-1 and anaplastic lines KAT4 and DRO-90. 14-3-3σ expression was not increased in thyroid carcinomas, the majority of which had a methylated CpG island. In addition, 5-aza-dC treatment increased 14-3-3σ expression in the FTC-238 and NPA cell lines, which had low baseline expression. We conclude 14-3-3σ expression in thyroid carcinomas is regulated by CpG island hypermethylation.
Keywords: 14-3-3σ, Thyroid, Cancer, Hypermethylation
1. Introduction
14-3-3σ (stratifin or SFN) is a member of the 14-3-3 family of proteins which play critical roles in signal transduction pathways and cell cycle regulation [1]. 14-3-3σ was initially characterized as a human mammary epithelium-specific marker 1 (HME1) and functions as a negative regulator of cyclin-dependent kinases [2,3]. 14-3-3σ is the only isoform which is induced following DNA damage [4]. Increased 14-3-3σ levels lead to G2/M arrest via sequestration of cyclinB1-cdc2 complexes in the cytoplasm [2]. Furthermore, this protein may also be involved in G1/S transition via binding to CDK2 and CDK in vitro [3].
Previous work from our laboratory shows that 14-3-3σ expression is tissue specific [5]. Certain normal tissues such as skin, mouth, prostate, airway and breast express 14-3-3σ, whereas other tissues have undetectable levels of this protein. Experiments also demonstrated that 14-3-3σ expression in these tissues is linked to an unmethylated CpG island, whereas loss of 14-3-3σ expression is associated with its hypermethylation.
14-3-3σ also has an important role in carcinogenesis and is lost in several tumor types. Nacht et al. reported that 14-3-3σ expression was down-regulated in breast cancer cell lines and primary breast carcinomas [6,7]. These investigators also noted that loss of heterozygosity (LOH) and intragenic mutations were not major contributing mechanisms, rather 14-3-3σ was epigenetically silenced by hyper-methylation of a discrete CpG rich region. Demeth-ylation of the gene coincided with re-expression of 14-3-3σ mRNA in cell lines which had lost expression of this gene.
Loss of 14-3-3σ expression in association with hypermethylation has also been reported in several other cancers, including ovarian [8], hepatocellular [9], prostate [10], some lung [11], gastric [12] and endometrial carcinomas [13]. It is important to point out that this is not the only mechanism for 14-3-3σ down-regulation. In some breast cancer and prostate cancer cell lines, loss of 14-3-3σ has been shown to occur at the post-translational level and appears to be mediated by proteasome directed protein degradation [10,14].
Interestingly, in some tumor types, 14-3-3σ expression appears to be increased rather than lost. Higher 14-3-3σ expression (by immunohistochemistry) was noted in lung cancers when compared to normal lung tissues [15]. 14-3-3σ expression has also been reported to be increased in head and neck squamous cell carcinomas [16] and pancreatic cancers by microarray and real-time PCR studies [17], when compared to corresponding normal tissues. Perathoner et al. [18] also demonstrated strong 14-3-3σ expression by Western blot analysis in 4/8 colorectal carcinoma cell lines and 6/8 tumor tissue pairs. 14-3-3σ was also overexpressed by immuno-histochemistry in 39% of colorectal carcinomas and appeared to be associated with a worse prognosis. Overexpression in this tumor type also appeared to be epigenetically controlled by demethylation of the CpG island [19].
Many genes important in thyroid carcinogenesis have been shown to be inactivated by promoter hypermethylation, including p16 [20], thyroid-stimulating hormone receptor gene (TSH-R), SLC26A4 (Pendred syndrome gene) [21,22] to name a few. Exposure to external beam radiation is one of the most important risk factors for thyroid carcinoma development. It can therefore be hypothesized that 14-3-3σ, which is upregulated in response to DNA damage, has an important role in thyroid carcino-genesis. In fact, Ito et al. [23,24] using immunohistochemistry and Western blot analyses, have demonstrated that 14-3-3σ expression is increased in papillary and follicular variant of papillary thyroid tumors, however, the protein does not appear to be expressed in follicular thyroid cancers. Highest levels of expression were noted in anaplastic thyroid cancers [23]. The authors also hypothesized that 14-3-3σ may contribute to the dedifferentiation of thyroid carcinoma cells since increased protein expression was noted in higher stage papillary thyroid carcinomas and anaplastic thyroid cancers. The mechanism of this variable 14-3-3σ expression in thyroid tumors has not been previously studied. We hypothesized that 14-3-3σ expression in thyroid carcinoma is epigenetically regulated by aberrant methylation of its CpG-rich island.
2. Materials and methods
2.1. Cell culture and thyroid tissues
Thyroid tumor cell lines studied included KAK-1 (derived from a follicular adenoma), TPC-1, KAT10, NPA (Papillary carcinoma), FTC-133, FTC-236, FTC-238 (follicular carcinomas, the latter two represent lines derived from a lymph node metastasis and a lung metastasis from the same patient), XTC-1 (Hurthle cell carcinoma), 2–7 (poorly differentiated thyroid cancer), DRO-90 and KAT4 (both anaplastic carcinoma). The cell lines were maintained in Glutamine plus DME H-21:F-12 media (Fisher#MT-15-090-CM) containing 10% FBS,10 µg/ml Insulin, 0.01 U/ml TSH (Sigma# T-8931), 250 ng/ml Fungizone, 50 µg/ml Penicillin. DRO-90 was maintained in RPMI 1640 with L-glutamine. The cells were incubated to 80–90% confluence prior to total RNA extraction. Airway epithelial cells, which are known to express high levels of the 14-3-3σ transcript [5] were used as a positive control.
Normal thyroid tissues and tumor samples were obtained from the Tissue Procurement Core at the University of Iowa Hospitals and Clinics. Tissues were obtained in accordance with a protocol approved by the Institutional Review Board for Human subjects. The tissues were harvested and snap frozen in liquid nitrogen at the time of thyroidectomy. The samples were stored at −80 °C until total RNA extraction. Diagnoses of banked specimens were confirmed by histological examination.
2.2. Drug treatment
Cell lines were grown for 5–7 days in the presence or absence of 1 µM of 5-aza-2′-deoxycytidine, a known DNA methyltransferase inhibitor (Sigma, St. Louis, MO), and then analyzed for 14-3-3σ mRNA expression by real-time PCR, as described below. The duration of drug treatment was selected based on the growth patterns of this cell line and previous studies from our lab using other thyroid cancer cell lines [25].
2.3. RNA extraction and cDNA synthesis
Total RNA was extracted after homogenization of cells and tissues using TRIzol (Invitrogen, Carlsbad, CA), followed by DNAase digestion (RNAase free DNA-ase set, Qiagen, Valencia, CA) and on-column clean up with the RNAeasy MinElute kit (Qiagen, Valencia, CA). Total RNA (2 µg) was reverse transcribed with the Superscript II RNAase H-reverse transcriptase kit (Invitrogen, Carlsbad, CA) for cDNA synthesis using random hexamer primers. The cDNA reaction was diluted to 1:10 for use as template for real-time RT-PCR.
2.4. Real-time RT-PCR
14-3-3σ primers and probes were obtained from Applied Biosystems (Assays on Demand kit®, HS00602835_s1 SFN Applied Biosystems, Foster city, CA). 18S ribosomal primers and probes (Applied Biosystems, Foster city, CA) were used as internal controls for the expression analyses. PCR amplifications were performed in a final reaction volume of 50 µl containing 25 µl of Taqman Universal PCR Master mix (Applied biosystems, Foster City, CA) and 2.5 µl of the primers and probe mix, as per the manufacturers’ instructions. All reactions were carried out in the ABI PRISM® 7700 Sequence detector Thermocycler (Applied Biosystems, Foster city, CA). The δCt for 14-3-3σ mRNA expression was calculated relative to the Ct (threshold cycle) of 18S ribosomal RNA. All reactions were performed in duplicate and relative mRNA expression was calculated using the formula 2(−δδCt), as previously described [26]. Appropriate positive and negative controls were used for PCR analyses.
2.5. Immunohistochemistry
Immunohistochemical staining was performed on formalin fixed paraffin embedded tissue sections using a polyclonal antibody against 14-3-3σ (Abcam, Cambridge, MA). Paraffin sections were deparafinized in xylenes, rehydrated using graded alcohols, and rinsed in distilled water. Antigen retrieval was performed using citrate buffer, pH 6.0, and two cycles of microwave heating. After slides were cooled to room temperature, endogenous biotin was quenched with 3% H2O2. Sections were covered with primary antisera (14-3-3σ, rabbit, polyclonal, 1:4000 dilution), incubated overnight at 4 °C, and rinsed. Sections were then covered with secondary antisera (anti-rabbit, Envision+System Labeled Polymer-HRP, Dako, Carpinteria, CA) and incubated 30 min at room temperature and rinsed. Visualization was performed with Dako Liquid DAB+substrate chromogen, and counterstaining was performed with Harris formula hematoxyln. Negative control slides were prepared by substituting non-immune rabbit IgG for the primary antibody. All the cases were evaluated by a single pathologist (JW) who was blinded to the results of the real-time RT-PCR. The scoring system was based on extent of staining, not intensity of staining:No staining = 0, < 25% cells positive = 1+, 25–75% cells positive = 2+ and >75% cells positive = 3+.
2.6. DNA extraction and methylation analysis
The methylation status of the 14-3-3σ CpG island was examined using sodium bisulfite genomic sequencing, as previously reported by our lab [5]. Briefly, genomic DNA was isolated using the DNAeasy Tissue kit (Qiagen, Valencia, CA) and modified with sodium bisulfite using EZ-DNA methylation kit (Zymo research, Orange, CA). Nested PCR was performed using previously published primers for the 14-3-3σ CpG island [5]. The first round of PCR was performed with 2.4 µL of modified DNA in a total reaction volume of 25 µL. PCR conditions were as follows: 95 °C for 5 min, 35 cycles of 92 °C for 2 min, 58 deg;C for 3 min, 72 °C for 2 min and a final extension of 72 °C for 5 min. In the second round of PCR, 2.5 µL of the first PCR product was amplified as indicated above, with the exception of an annealing temperature of 60 °C.
The PCR product was electrophoresed on a 2% agarose gel, followed by gel extraction and cloning into a TOPO TA vector according to the manufacturer’s instructions (TOPO TA Cloning Kit, Invitrogen, Carlsbad, CA, USA). Five to ten positive recombinants were isolated using a Qiaprep Spin Plasmid Miniprep kit (Qiagen, Valencia, CA) and sequenced on an ABI automated DNA sequencer. The methylation status of the individual CpG sites was determined by comparison of the sequence obtained with the known 14-3-3σ sequence. The number of methylated CpGs at a specific site was divided by the number of the clones analyzed to calculate the percentage methylation at each CpG site. For calculation of the total or overall promoter methylation frequency, the total of all the CpG sites that were methylated from all the clones was counted and divided by the total number of CpG sites. Bisulfite-treated DNA was also used for methylation-specific PCR using previously published primer sets [13] to distinguish between methylated and unmethylated DNA in both tumor cell lines and primary thyroid tissues and tumors.
2.7. Statistical analysis
The SPSS software was used to perform the statistical analyses. All statistical tests were two-sided and performed at the 5% level of significance.
3. Results
3.1. 14-3-3σ mRNA expression in tumor cell lines and thyroid tissues
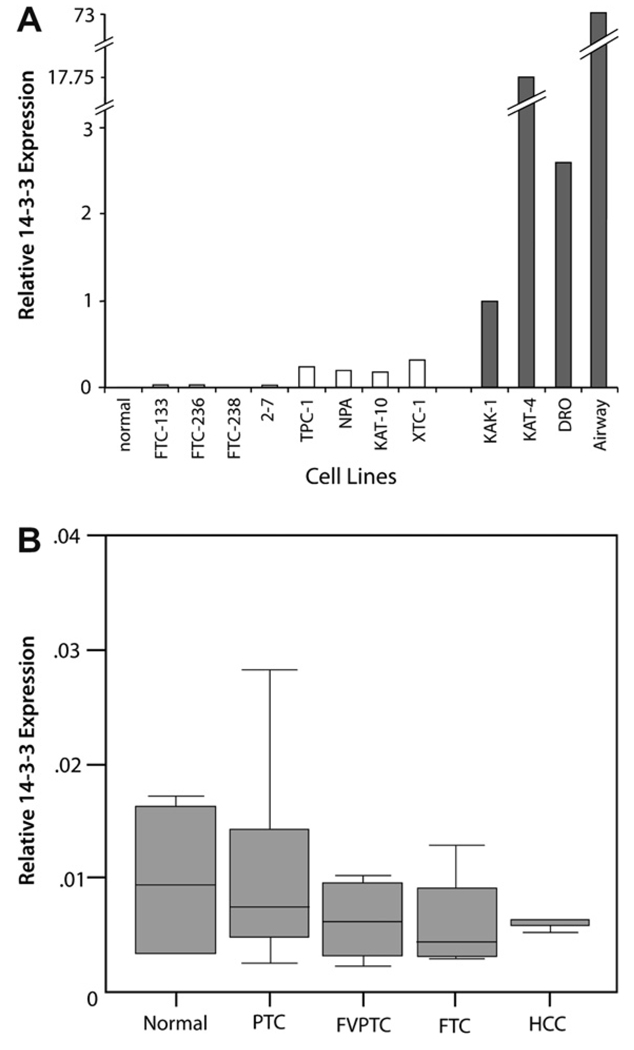
As displayed in Fig. 1(A), The 14-3-3σ transcript was undetectable by real-time RT-PCR in, all the follicular carcinoma-derived cell lines and the poorly differentiated cell lines 2–7. Very low levels of transcript were detectable in the papillary and well-differentiated cell lines (TPC-1, NPA and KAT-10) and the Hurthle cell cancer line XTC-1. Markedly increased 14-3-3σ mRNA levels were noted in KAK-1 and the anaplastic carcinoma cell lines KAT-4 and DRO-90. In total, 3/11 (28%) of cell lines demonstrated 14-3-3σ upregulation.
Fig. 1.
(A) 14-3-3σ expression in normal thyroid tissues (two samples) and thyroid tumor cell lines using real-time RT-PCR. The expression level in KAK-1 was designated as = 1. No expression was detected in normal thyroid tissues, all follicular (FTC) cell lines and the poorly differentiated cell lines 2–7. Very low expression levels were seen papillary tumor cell lines TPC, NPA, and KAT-10; and the Hurthle cell cancer line XTC-1. Markedly elevated 14-3-3σ levels were noted in KAK-1 and the anaplastic carcinoma cell lines KAT-4 and DRO-90. Airway cells were used as a positive control for 14-3-3σ expression. (B) Box plots of relative 14-3-3σ expression in normal thyroid tissues and primary thyroid cancers using real-time RT-PCR. The solid lines represent the median values and the boxes represent the interquartile range (25th to 75th percentile).
14-3-3σ expression was also examined in 36 primary thyroid tissues (4 normals, 14 papillary thyroid cancers, 10 follicular variant of papillary carcinomas, 4 follicular thyroid cancers, 3 Hurthle cell cancers and 1 anaplastic cancer) by real-time RT-PCR. The 14-3-3σ transcript was undetectable in all normal thyroid tissues and tumor types examined as shown in Fig. 1B.
3.2. 14-3-3σ methylation status in thyroid cell lines and tissues
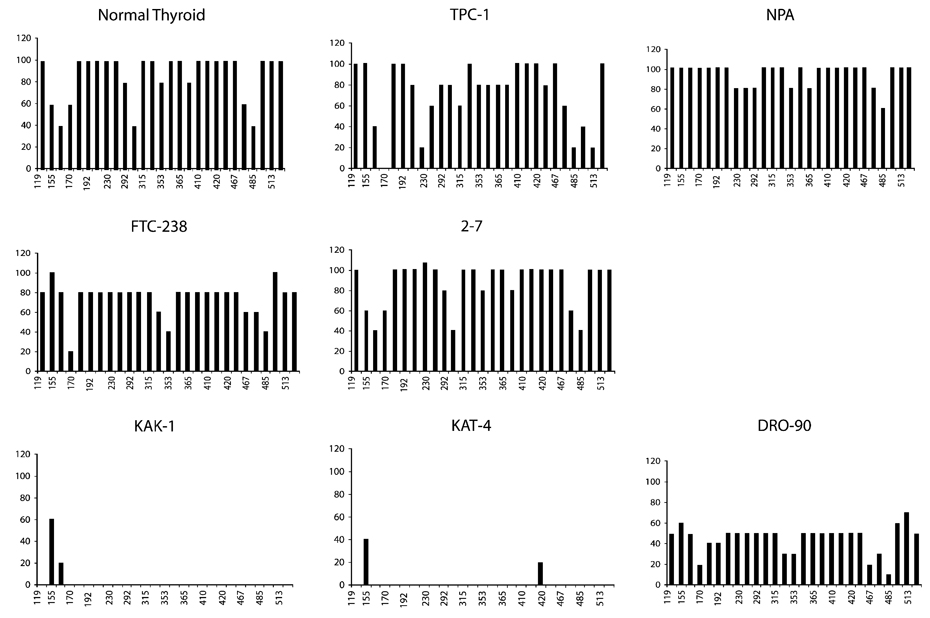
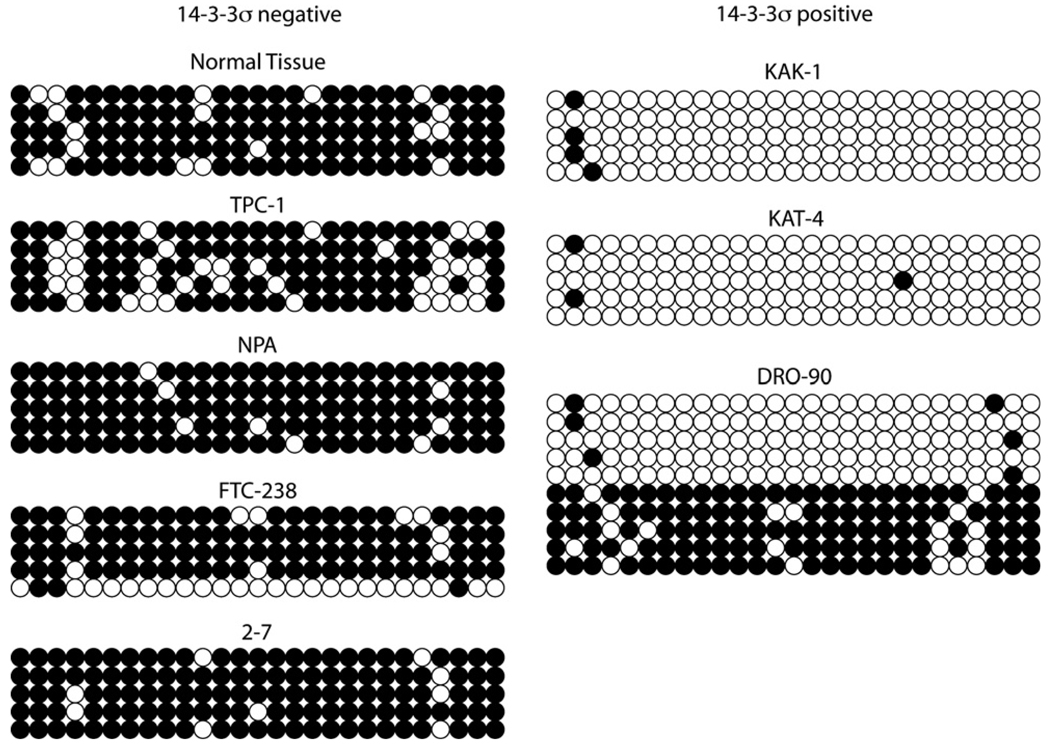
Bisulfite sequencing was performed in 7 of the 11 studied cell lines, in addition to normal thyroid tissues. Fig. 2 and Fig. 3 illustrate the methylation frequency and distribution at each of the CpG sites among the sequenced amplicons. The majority of the 27 CpGs investigated were methylated in normal thyroid tissues and in TPC-1, NPA, FTC-238 and 2–7, all of which did not express 14-3-3σ. In contrast, the CpGs were largely unmethylated in KAK-1 and KAT4; and DRO-90 appeared to contain a mixed population of cells with 4/10 methylated clones. All of these cell lines expressed abundant 14-3-3σ transcript. Overall methylation frequency of the 14-3-3σ CpG island in the various cell lines is depicted in Table 1. The total methylation frequency in cell lines expressing 14-3-3σ ranged from 2% to 34% and from 73% to 94% in cell lines expressing little to no transcript. This difference was statistically significant, (t-test, p < 0.01).
Fig. 2.
Methylation frequency at the 27 CpG sites in the 14-3-3σ gene. The X-axis represents the nucleotide position relative to the transcription start site (as previously described) [20], and the Y-axis represents the % methylation at that site. The upper two panels represent 14-3-3σ non-expressers, whereas the lower panel depicts the cell lines expressing 14-3-3σ mRNA.
Fig. 3.
Bisulfite sequencing of the 14-3-3σ promoter in normal thyroid tissues and cell lines. The bubble charts show the methylation status of each of the 27 CpGs in the CpG island at the positions indicated in Fig. 2. Dark circles represent methylated cytosines, whereas open circles represent unmethylated cytosines. Each row represents a single sequenced amplicon.
Table 1.
Relationship between 14-3-3σ expression and promoter methyl-ation frequency
| Cell line | 14-3-3σ expression | Methylation frequency (%) |
|---|---|---|
| Normal | − | 87 |
| KAK-1 | + | 3 |
| TPC-1 | − | 73 |
| NPA | − | 94 |
| KAT-10 | − | n.d. |
| FTC-133 | − | n.d. |
| FTC-236 | − | n.d. |
| FTC-238 | − | 74 |
| XTC-1 | − | n.d. |
| 2–7 | − | 93 |
| KAT-4 | + | 2 |
| DRO-90 | + | 34 |
n.d, not done.
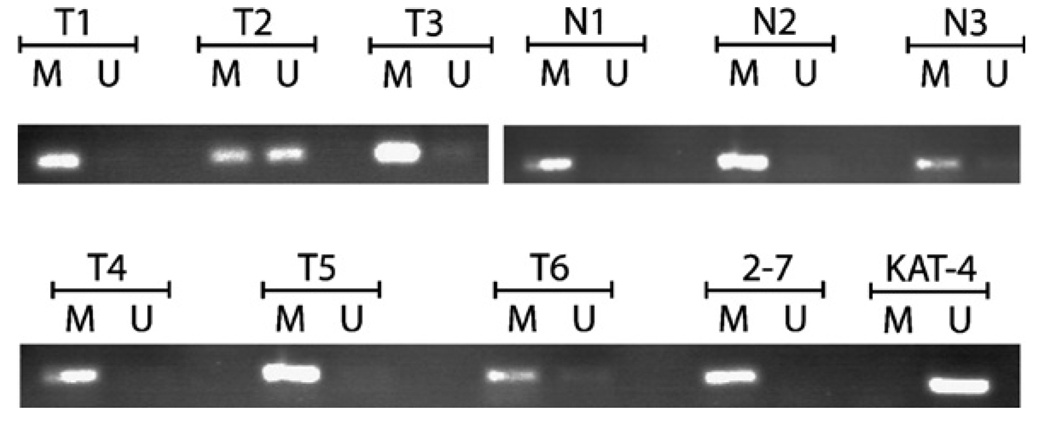
Methylation-specific PCR was performed on DNA derived from the tissues examined by real-time RT-PCR, as shown in Fig. 4. All 4/4 (100%) normal thyroid tissues were methylated and did not express 14-3-3σ. Of the 32 tumors, bright unmethylated bands were noted in 2 Papillary cancers and 3 Follicular variant of papillary cancers, for an overall methylation rate of 27/32 or 84%. There was no statistical difference in the methylation rate between tumors and normal tissues (Fisher’s exact test). The 5 tumors with the unmethylated transcript also had undetectable 14-3-3σ expression.
Fig. 4.
Methylation-specific PCR analysis of various thyroid tumor cell lines and representative primary tumors and normal thyroid tissues. Bisulfite-treated DNA was used as template for methylation-specific PCR. All the normal tissues were methylated. T1 = anaplastic cancer, T2 and T3 = papillary cancer T4 and T6 = follicular cancers, T5 = Hurthle cell cancer. Note that T2 is unmethylated. M = primers specific to methylated template DNA, U = primers specific to unmethylated template DNA, T = tumor, N = normal.
3.3. 5-aza-dC treatment and 14-3-3σ reactivation
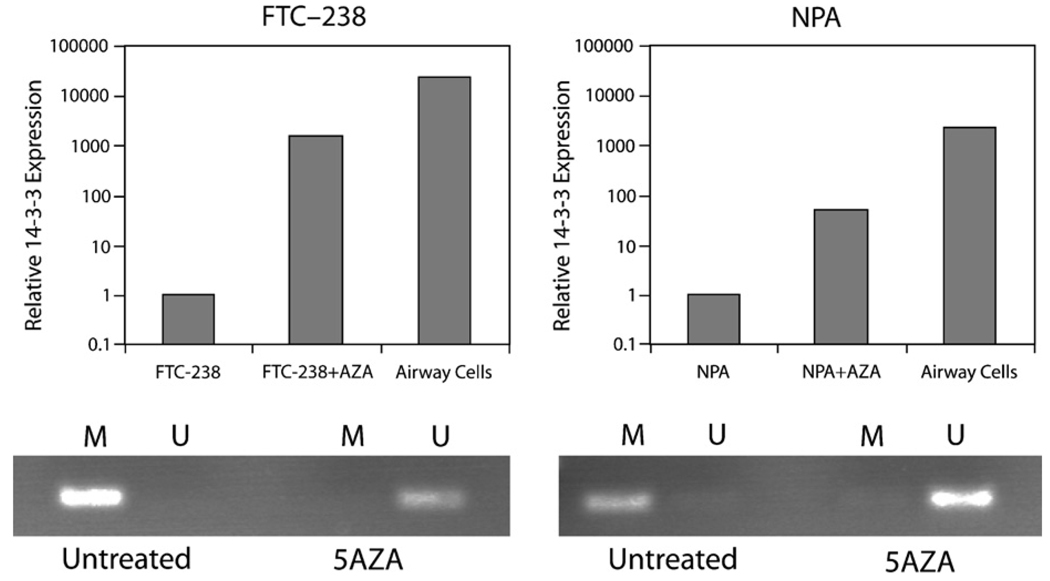
The real-time RT-PCR and sequencing data suggest an association between cytosine methylation and 14-3-3σ expression. To test whether CpG methylation in 14-3-3σ was functionally linked to its silencing in thyroid cell lines, we attempted to induce 14-3-3σ expression in the 14-3-3σ negative FTC-238 and NPA cell lines, by treatment with 5-aza-dC. This treatment resulted in significantly increased 14-3-3σ mRNA expression, as shown in Fig. 5. This was accompanied by demethylation of the CpG island, as examined by methylation-specific PCR.
Fig. 5.
Effect of 5-aza-dC treatment on 14-3-3σ mRNA expression. Treatment of FTC-238 and NPA which do not express 14-3-3σ at baseline, led to a marked increase in expression levels, along with demethylation of the CpG island, as shown in the lower panels. Airway cells were used as positive control for mRNA expression.
3.4. 14-3-3σ expression by immunohistochemistry
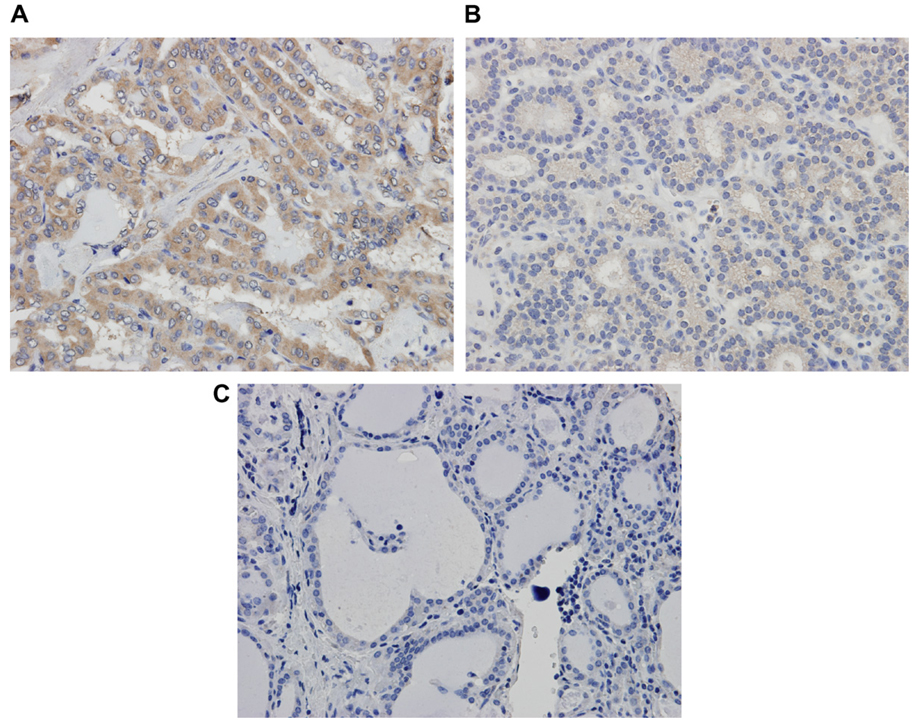
Due to the discrepancy in our real-time RT-PCR data and published expression data [23] in primary thyroid tumors, 14-3-3σ expression was also examined by immu-nohistochemistry in 21 available archived tumors corresponding to the above specimens. The results are shown in Table 2 and Fig. 6. Squamous cell carcinoma was used as a positive control. 14-3-3σ staining was observed primarily in the cytoplasm. None of the normal tissues showed 14-3-3σ expression. Staining was noted in majority of papillary cancers and follicular variant of papillary cancers. Faint staining was noted in 1 follicular cancer, 1 Hurthle cell cancer, and the only anaplastic cancer that was analyzed. However, the pattern of staining was highly variable between the tumors.
Table 2.
14-3-3σ IHC results by tumor type
| Tissues | 0 | 1+ | 2+ | 3+ | Total |
|---|---|---|---|---|---|
| PTC | 1 | 4 | 3 | 1 | 8/9 (89%) |
| FVPTC | 2 | – | 1 | 2 | 3/5 (60%) |
| FC | 2 | 1 | – | – | 1/3 (33%) |
| HCC | 2 | 1 | – | – | 1/3 (33%) |
| Anaplastic | – | 1 | – | – | 1/1 (100%) |
Fig. 6.
Immunohistochemistry for 14-3-3σ. (A) Papillary thyroid cancer with intense staining, (B) Follicular cancer with minimal staining and (C) non-neoplastic thyroid tissue with no staining.
4. Discussion
Although 14-3-3σ expression has been previously examined in thyroid tumors and normal tissues using immunohistochemistry,[23] the mechanism of its regulation has not been studied. In the present study, we have used a panel of thyroid tumor cell lines and primary tumors to determine if aberrant methylation of the gene’s CpG island is responsible for the regulation of 14-3-3σ expression. Our experiments confirm that normal thyroid tissues do not express 14-3-3σ, as detected by real-time RT-PCR. In the previous studies of thyroid tumors [23,27], 14-3-3σ expression was shown to be increased, rather than lost, in papillary thyroid and follicular variant of papillary carcinomas, with highest expression levels in poorly differentiated and ana-plastic tumors. Interestingly, primary follicular thyroid carcinomas did not appear to overexpress 14-3-3σ [23,27]. Our work in cell lines also reflects this to some extent, in that low levels of 14-3-3σ transcript were detected in well-differentiated cell lines, with highest levels seen in the anaplastic cancer-derived cell lines. Furthermore, as in the previous work, the follicular cancer-derived cell lines showed negligible 14-3-3σ expression. The increased 14-3-3σ expression seen in the KAK-1 cell line is intriguing, given that it is derived from a follicular adenoma. A review of the literature demonstrates that KAK-1 appears to be morphologically similar to other thyroid carcinoma-derived cell lines such as KAT-4 and KAT-10 [28]. In addition, it seems to have lost expression of several genes, similar to other thyroid carcinoma cell lines [25,29–31]. In this regard, KAK-1 may behave more like a malignant rather than a benign tumor-derived cell line in culture.
Per our experiments, increased 14-3-3σ expression does not appear to be a frequent event in primary thyroid cancers. The 14-3-3σ transcript was undetectable in all the 4 normal and 32 primary thyroid cancers examined, including papillary and ana-plastic tumors. This is in marked contrast to previously published data [23] in which 82/82 papillary cancers and 21/23 anaplastic cancers were reported to express 14-3-3σ by immunohistochemistry. Studies have shown that real-time RT-PCR is a reliable technique for the quantification of gene expression [32,33]. In general, the use of immuno-histochemistry as a test of gene expression tends to be significantly affected by the sensitivity and specificity of the antibodies used (polyclonal vs. monoclonal) and the type of tissues examined (frozen vs. formalin fixed). In addition, immunohistochemistry results can vary depending upon different scoring and interpretive criteria used to evaluate the cases [34]. In order to minimize these effects, our immunohistochemistry analyses were performed in a single lab, with the results being scored independently by a single pathologist (JW). Antibody to 14-3-3σ is commercially available and we have used a rabbit polyclonal antibody, similar to the published studies. Staining was observed in the majority of papillary cancers and follicular variant of papillary cancers, with little staining in follicular cancers. This is similar to previous work, however, the results were discrepant with those obtained by real-time RT-PCR. It is important to note that polyclonal antibodies are not always optimal for immunohistochemical analyses. The pattern of staining observed also suggests that these results are more likely to represent non-specific binding, and that the true expression is reflected by the real-time RT-PCR results.
Our studies also show that the expression of 14-3-3σ in thyroid tumor cell lines and primary thyroid tissues and tumors is linked to the methylation status of the 14-3-3σ CpG island. As judged by bisulfite sequencing, high overall methylation frequencies were seen in cell lines that had low levels of 14-3-3σ expression. In contrast, cell lines showing appreciable expression had low overall methylation frequencies in the 2–3% range. DRO-90 demonstrated a mixed population of cells with an overall methylation frequency of 34%, i.e. the CpG islands were largely unmethylated in this cell line. With respect to primary thyroid tissues, the methylated form of the 14-3-3σ CpG island was detected in all tumor and normal samples. This is not an unexpected finding as the tissue samples were obtained macroscopically rather than microdissected, and thus can be expected to contain both normal and neoplastic tissue. However, a bright band corresponding to the unmethylated product was also observed in 5 (15%) tumors which did not express the transcript. This may be due to the fact that out specimens were not microdissected. Alternatively, it suggests that 14-3-3σ expression in a small proportion of these tumors may be regulated by a different mechanism.
This association between gene expression and methylation status was further confirmed by the induction of 14-3-3σ expression in the FTC-238 and NPA cell lines after treatment with the DNA methyltransferase inhibitor 5-aza-dC. Demethylation of the CpG island as evidenced by MS-PCR was accompanied by upregulation of 14-3-3σ expression by real-time RT-PCR.
The exact function and biological significance of altered 14-3-3σ expression in thyroid cancer cells has not thus far been elucidated. Previous studies in other tumors suggest that 14-3-3σ may be a prognostic marker, and high grade, advanced stage endometrial carcinomas over expressed 14-3-3σ when compared to low-grade lesions [13]. In these tumors, increased 14-3-3σ expression was correlated with hypomethylation of the CpG island. Similar findings have been reported for colorectal [19] and pancreatic cancers [35] and it has been postulated that 14-3-3σ over expression may be responsible for promoting proliferation and/or preventing apoptotic signal transduction [18]. Elevated 14-3-3σ expression has been reported as contributing to drug resistance in breast cancer cell lines [36], and its increased expression in the anaplastic cancer cell lines in the current study, may be able to explain, at least in part, the widespread resistance to chemo-therapeutic agents noted in this aggressive malignancy [37].
To summarize, we have shown that transcriptional activation of 14-3-3σ in thyroid cancer cells is mediated by altered methylation of its CpG island. These findings are not only confirmatory but also extend work by other investigators highlighting the epigenetic control of 14-3-3σ in both normal and neoplastic tissues. Additional studies are needed to further characterize the role of epigenetic mechanisms in regulating 14-3-3σ expression in both well-differentiated and undifferentiated primary thyroid carcinomas.
Acknowledgements
The authors wish to thank Dr. Orlo Clark and the Endocrine Surgery Laboratory at the University of California, San Francisco (for the gift of the TPC, FTC and XTC cell lines), Dr. Kenneth Ain (for the KAK-1, KAT4 and KAT10 cell lines), Dr. Guy J. Juillard (for the DRO-90 cell line), Dr. Bhuvanesh Singh (for the NPA and 2–7 cell lines).
Footnotes
This work was supported in part by a Holden Comprehensive Cancer Center seed grant (GL), and R01 CA73612 (FED).
References
- 1.Mhawech P. P 14-3-3 proteins-an update. Cell Res. 2005;15:228–236. doi: 10.1038/sj.cr.7290291. [DOI] [PubMed] [Google Scholar]
- 2.Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 3.Laronga C, Yang HY, Neal C, Lee MH. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J. Biol. Chem. 2000;275:23106–23112. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- 4.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cells. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 5.Oshiro MM, Futscher BW, Lisberg A, Wozniak RJ, Klimecki WT, Domann FE, Cress AE. Epigenetic regulation of the cell type-specific gene 14-3-3sigma. Neoplasia. 2005;7:799–808. doi: 10.1593/neo.05274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nacht M, Ferguson AT, Zhang W, Petroziello JM, Cook BP, Gao YH, Maguire S, Riley D, Coppola G, Landes GM, Madden SL, Sukumar S. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59:5464–5470. [PubMed] [Google Scholar]
- 7.Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, Sukumar S. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. USA. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akahira J, Sugihashi Y, Suzuki T, Ito K, Niikura H, Moriya T, Nitta M, Okamura H, Inoue S, Sasano H, Okamura K, Yaegashi N. Decreased expression of 14-3-3 sigma is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin. Cancer Res. 2004;10:2687–2693. doi: 10.1158/1078-0432.ccr-03-0510. [DOI] [PubMed] [Google Scholar]
- 9.Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T, Kaneto H, Iku S, Ozeki I, Karino Y, Satoh T, Toyota J, et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298–5302. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 10.Urano T, Takahashi S, Suzuki T, Fujimura T, Fujita M, Kumagai J, Horie-Inoue K, Sasano H, Kitamura T, Ouchi Y, Inoue S. 14-3-3sigma is down-regulated in human prostate cancer. Biochem. Biophys. Res. Commun. 2004;319:795–800. doi: 10.1016/j.bbrc.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 11.Osada H, Tatematsu Y, Yatabe Y, Nakagawa T, Konishi H, Harano T, Tezel E, Takada M, Takahashi T. Frequent and histological type-specific inactivation of 14-3-3sigmain human lung cancers. Oncogene. 2002;21:2418–2424. doi: 10.1038/sj.onc.1205303. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14-3-3 sigma gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 2000;60:4353–4357. [PubMed] [Google Scholar]
- 13.Nakayama H, Sano T, Motegi A, Oyama T, Nakajima T. Increasing 14-3-3 sigma expression with declining estrogen receptor alpha and estrogen-responsive finger protein expression defines malignant progression of endometrial carcinoma. Pathol. Int. 2005;55:707–715. doi: 10.1111/j.1440-1827.2005.01900.x. [DOI] [PubMed] [Google Scholar]
- 14.Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi K, Hashizume S, Kato M, Honjoh T, Setoguchi Y, Yasumoto K. Elevated expression levels of the 14-3-3 family of proteins in lung cancer tissues. Hum Antibodies. 1997;8:189–194. [PubMed] [Google Scholar]
- 16.Villaret DB, Wang T, Dillon D, Xu J, Sivam D, Cheever MA, Reed SG. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. Laryngoscope. 2000;110:374–381. doi: 10.1097/00005537-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 18.Perathoner A, Pirkebner D, Brandacher G, Spizzo G, Stadlmann S, Obrist P, Margreiter R, Amberger A. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin. Cancer Res. 2005;11:3274–3279. doi: 10.1158/1078-0432.CCR-04-2207. [DOI] [PubMed] [Google Scholar]
- 19.Ide M, Nakajima T, Asao T, Kuwano H. Inactivation of 14-3-3sigma by hypermethylation is a rare event in colorectal cancers and its expression may correlate with cell cycle maintenance at the invasion front. Cancer Lett. 2004;207:241–249. doi: 10.1016/j.canlet.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Boltze C, Zack S, Quednow C, Bettge S, Roessner A, Schneider-Stock R. Hypermethylation of the CDKN2/ p16INK4A promotor in thyroid carcinogenesis. Pathol. Res. Pract. 2003;199:399–404. doi: 10.1078/0344-0338-00436. [DOI] [PubMed] [Google Scholar]
- 21.Xing M, Usadel H, Cohen Y, Tokumaru Y, Guo Z, Westra WB, Tong BC, Tallini G, Udelsman R, Califano JA, Ladenson PW, Sidransky D. Methylation of the thyroid-stimulating hormone receptor gene in epithelial thyroid tumors: a marker of malignancy and a cause of gene silencing. Cancer Res. 2003;63:2316–2321. [PubMed] [Google Scholar]
- 22.Xing M, Tokumaru Y, Wu G, Westra WB, Ladenson PW, Sidransky D. Hypermethylation of the Pendred syndrome gene SLC26A4 is an early event in thyroid tumorigenesis. Cancer Res. 2003;63:2312–2315. [PubMed] [Google Scholar]
- 23.Ito Y, Miyoshi E, Uda E, Yoshida H, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Matsuura N, Kakudo K, Kuma K, et al. 14-3-3 sigma possibly plays a constitutive role in papillary carcinoma, but not in follicular tumor of the thyroid. Cancer Lett. 2003;200:161–166. doi: 10.1016/s0304-3835(03)00282-9. [DOI] [PubMed] [Google Scholar]
- 24.Ito K, Suzuki T, Akahira J, Makuma S, Saitou S, Okamoto S, Niikura H, Okamura K, Yaegashi N, Sasano H, Inoue S. 14-3-3sigma in endometrial cancer-a possible prognostic marker in early-stage cancer. Clin. Cancer Res. 2005;11:7384–7391. doi: 10.1158/1078-0432.CCR-05-0187. [DOI] [PubMed] [Google Scholar]
- 25.Lal G, Padmanabha L, Smith BJ, Nicholson RM, Howe JR, O’Dorisio MS, Domann FE., Jr RIZ1 is epigenetically inactivated by promoter hypermethylation in thyroid carcinoma. Cancer. 2006;107:2752–2759. doi: 10.1002/cncr.22325. [DOI] [PubMed] [Google Scholar]
- 26.Relative quantification of gene expression User Bulletin #2 ABI Prism 7700 Sequence Detection syst 1997.
- 27.Ito Y, Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Nakamura Y, Kakudo K, Kuma K, Miyauchi A. Caveolin-1 and 14-3-3 sigma expression in follicular variant of thyroid papillary carcinoma. Pathol. Res. Pract. 2005;201:545–549. doi: 10.1016/j.prp.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Rocha AS, De Wever O, Moreira S, Costa MJ, Vandekerckhove J, Mareel M, Soares P. Mutated E-cadherin: genomic and functional characterization in thyroid cells from the KAT family. Thyroid. 2004;14:902–909. doi: 10.1089/thy.2004.14.902. [DOI] [PubMed] [Google Scholar]
- 29.Ain KB, Taylor KD, Tofiq SG. Venkataraman, Somatostatin receptor subtype expression in human thyroid and thyroid carcinoma cell lines. J. Clin. Endocrinol. Metab. 1997;82:1857–1862. doi: 10.1210/jcem.82.6.4013. [DOI] [PubMed] [Google Scholar]
- 30.Venkataraman GM, Yatin M, Marcinek R, Ain KB. Restoration of iodide uptake in dedifferentiated thyroid carcinoma: relationship to human Nasymporter gene meth-ylation status. J. Clin. Endocrinol. Metab. 1999;84:2449–2457. doi: 10.1210/jcem.84.7.5815. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, Quiros RM, Maxhimer JB, Jiang P, Marcinek R, Ain KB, Platt JL, Shen J, Gattuso P, Prinz RA. Inverse correlation between heparan sulfate composition and heparanase-1 gene expression in thyroid papillary carcinomas: a potential role in tumor metastasis. Clin. Cancer Res. 2003;9:5968–5979. [PubMed] [Google Scholar]
- 32.Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 2002;30:503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- 33.Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FM. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol. Med. 2003;9:189–195. doi: 10.1016/s1471-4914(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 34.Nuciforo PG, Pellegrini C, Fasani R, Maggioni M, Coggi G, Parafioriti A, Bosari S. Molecular and immunohistochemical analysis of HER2/neu oncogene in synovial sarcoma. Hum. Pathol. 2003;34:639–645. doi: 10.1016/s0046-8177(03)00238-7. [DOI] [PubMed] [Google Scholar]
- 35.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes over-expressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 36.Liu Y, Liu H, Han B, Zhang JT. Identification of 14-3-3sigma as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res. 2006;66:3248–3255. doi: 10.1158/0008-5472.CAN-05-3801. [DOI] [PubMed] [Google Scholar]
- 37.Green LD, Mack L, Pasieka JL. Anaplastic thyroid cancer and primary thyroid lymphoma: a review of these rare thyroid malignancies. J. Surg. Oncol. 2006;94:725–736. doi: 10.1002/jso.20691. [DOI] [PubMed] [Google Scholar]