INTRODUCTION
Methylation of nucleotide bases is important for many biological processes(Bird, 1996 #5568; Jones, 2002 #5569; Robertson, 2005 #5570)(Ahmad, 1996 #5678)(Chela-Flores, 1990 #5682) from embryogenesis{Egger, 2004 #86} to facilitation of B-A and B-Z transitions.{Frederick, 1987 #87; Mooers, 1995 #88; Behe, 1981 #89; Fujii, 1982 #90} The HhaI system in particular is a restriction-modification system consisting of a methyltransferase (Mtase) and endonuclease, which together act as a defense mechanism in prokaryotic systems, protecting the cell from invasive DNA. Both proteins recognize the sequence [5′-G↓CGC-3′]2 (the arrow indicates cleavage and the underlined cytosine base is the methylation target). The Mtase DNA target forms a classic B-form double helix, providing no path to methylate the cytidine base; however, the crystal structure of the HhaI Mtase-cognate DNA ternary complex demonstrates that the cytosine is exo-helical and is surrounded in the binding pocket of the Mtase (Figure 1A).(ref Klimasauskas_1994) The physical removal from the stacked base in the helix explains how the methylation position of the base is accessible, however it is still unclear whether flipping of the target base occurs through an active (protein-induced) or passive (captured) process.(ref) Recent computational and mutational studies give evidence to support active flipping {Klimasauskas, 1998 #45}{Shieh, 2006 #71}{Zhou, 2007 #25}{Estabrook, 2004 #32}{Huang, 2003 #41}{Huang, 2004 #42} through the major groove pathway{Bouvier, 2007 #27}{Luo, 2005 #54}{Horton, 2004 #38} as well as passive flipping.{Chen, 2000 #28}{Dornberger, 1999 #79}
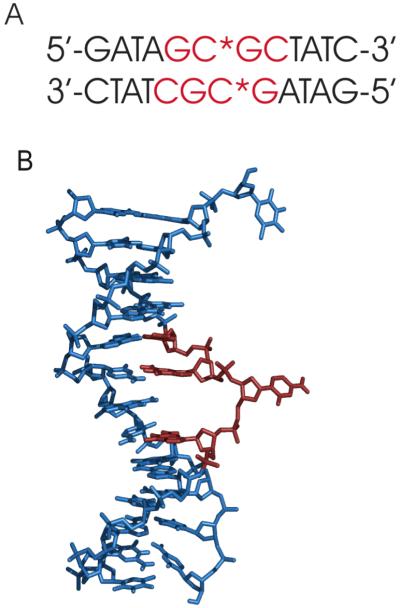
Figure 1.
(A). Structure of the HhaI target DNA in the protein-DNA complex (protein removed), showing the extrahelical deoxycytidine. The recognition sequence is shown in red. (B) Sequence of the target DNA, Residues highlighted in red are both labeled at the 5′/5” position and part of the recognition sequences.
Crystal structures of the DNA-HhaI Mtase complex, where the target C is altered to A, U or an abasic site to form a mismatched base-pair demonstrate that base flipping still occurs.{Kumar, 1997 #47}{O’Gara, 1998 #63} This strongly suggests that the primary interaction between the protein and the DNA that induces the base flipping mechanism would have to occur between the protein and the phosphodiester backbone of the DNA. The large-scale conformational changes of the target nucleotide required for interaction with the Mtase imply that it is reasonable to expect that some form of conformational flexibility of the phosphodiester backbone may characterize the native DNA binding site in order to overcome energetic barrier required for such torsion angle changes. A computational study by Banavali and co-workers found that the α, β, ε, and ζ torsional angles of the two phosphodiester groups surrounding the target base occupy many different states, supporting a more flexible phosphodiester group.{Banavali, 2006 #19}
The recognition process between the Mtase the methylation target has been amply studied,{Cheng, 2001 #29}{Daujotyte, 2004 #15}{Varnai, 2002 #11}{Youngblood, 2006 #13}{Estabrook, 2006 #16}{Zhou, 2007 #25} yet, so far there has been only a limited quantitative analysis of the effect of methylation on the backbone of the target site in this sequence{Meints, 2001 #57}{Banyay, 2002 #26} and very few studies on the recognition mechanism of the endonuclease.{Vardimon, 1984 #10} To date, there is no crystal structure of the endonuclease and target DNA. Recent analysis of the motion of the furanose ring by solid-state NMR of the nucleotides in and around the recognition sequence of the HhaI methyltransferase target DNA has shown that the 2′-deoxycytidine (dC) residues in the recognition sequence exhibit a much larger amplitude of motion and are more restricted to a single conformation than the other residues in and around the recognition sequence.(ref Meints JACS 2007)
We have previously suggested that motions specific to the HhaI target sequence facilitate base extrusion by facilitating overcome energetic barriers. In our present study, we are also interested in quantifying and understanding the degree to which methylation perturbs the local dynamics of the DNA sequence. We have extended our studies to investigate the range of motions of the phosphodiester backbone within the recognition site by reporting solid state NMR lineshape and relaxation data and investigate the angular trajectories of these motions by modeling the deuterium solid-state lineshapes of the 5′/5” deuterons in the protein recognition site. We observe increased conformational flexibility in the backbone of the target site. Interestingly this mobility is “quenched” and the direction of motion is altered upon methylation, suggesting how dynamics may assist the endonuclease in discerning between methylated and unmethylated DNA.
MATERIALS AND METHODS
SYNTHESIS
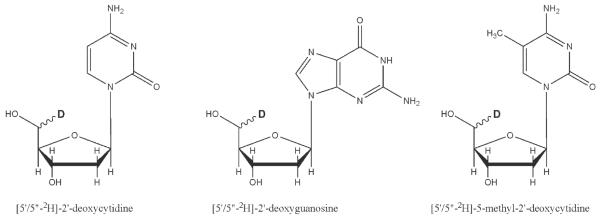
Deuterium labels were incorporated non-stereospecifically into the 5′ methylene group at positions dG5, dC6, dG7, dC8, and 5-methyl-dC6 in [d(G1A2T3A4G5C6G7C8T9A10T11C12)]2 (Figures 1B, and 2). [5′/5”-2H]-2′-dC and [5′/5”-2H]-2′-dG were synthesized by a method similar to that of Orban and Reid.{Orban, 1989 #17}[22] The starting material for the dC synthesis was N4-3′-O-acetyl-2′-dC, which was purchased from Sigma. The starting material for the dG synthesis was N2-isobutyryl-3′-O-acetyl-2′-dG, synthesized from N2-isobutyryl-5′- (dimethoxytrityl)-2′-dG. The 3′ hydroxyl group was acetylated with acetic anhydride and purified. N2-isobutyryl-5′- (dimethoxytrityl)-3′-O-acetyl-2′-dG was converted to N2-isobutyryl-3′-O-acetyl-2′-dG by deprotection of the 5′- dimethoxytrityl derivative in 0.25 M trichloroacetic acid in chloroform and purified by silica gel chromatography. The starting material for the 5-methyl-dC synthesis was [6-methyl-2H4]-2′-deoxythymidine which was prepared by enzymatically glycosylating [d6, methyl-2H4]-thymine, using 2′-deoxyadenosine as the pentosyl donor.{Krenitsky, 1986 #85} Deuterated nucleosides were converted to N-acyl-5′-O-( dimethoxytrityl)- 3′-O-(2-cyanoethyl-N,N-diisopropylphosphoramidite) as described previously[Gait, 1984 #131] and converted to N4-triazole derivatives using the procedure of Cowart et al.{Cowart, 1989 #81} Deprotection of the DNA in concentrated ammonia (2 days, 55 °C) converted the N4-protected thymidine derivative to [5′/5”-2H]-5-methyl-2′-deoxycytidine. Oligonucleotides were purified on Sephadex size exclusion resin, salted (10% NaCl by weight), packed into a 5mm solid-state NMR Kel-F sample chamber, and hydrated by vapor diffusion in a humidity chamber containing saturated salts in deuterium-depleted water (75% relative humidity at 20°C).[Weast, 1979 #24] Water content was quantified gravimetrically by the parameter W (number of water molecules per nucleotide base pair) and is accurate to ±1 waters per nucleotide base pair.
Figure 2.
Non-stereospecific labeling of 2′-dC, 2′-dG, and 5-methyl-2′-dC at the 5′/5” position.
SOLID-STATE NMR SPECTROSCOPY
All experiments were performed on a home-built NMR spectrometer, operating at a deuterium Larmor frequency of 76.776 MHz, corresponding to a magnetic field strength of 11.74 Tesla. A quadrupolar echo pulse sequence with an eight-step phase cycling scheme was implemented with a delay of 40 μs between 90° pulses (typically 2.6 μs in duration) and a dwell time of 200 ns during acquisition. Data acquisition was initiated prior to the echo maximum. The time domain data were left-shifted and apodized with 3000 Hz Lorentzian line broadening prior to Fourier transformation. Partially-relaxed lineshapes and spin-lattice relaxation times were determined using an inversion recovery pulse sequence, which incorporated a 180° composite pulse to ensure broadband excitation.[Tycko, 1983 #25] To obtain powder-averaged Zeeman spin-lattice relaxation times, <T1Z>, the integrated intensity of the powder spectrum was monitored as a function of recovery time and analyzed using a non-linear least squares fitting routine.[deFontaine, 1975 #26] The relaxation and lineshape measurements reported in this section are for DNA samples equilibrated at 75-80% relative humidity, which correspond to hydration levels of W=10-11.8 moles of water per mole of nucleotide base pairs. As mentioned in Synthesis section the absolute uncertainly in W is ±1 water per nucleotide base pair or a relative uncertainly of about 9-10% in the range studied. For crystalline DNA in the B form the hydration level is approximately W=10. Therefore the relaxation and lineshape data presented here are characteristic of a DNA in a B form helix. In all experimental spectra, the center isotropic peak is due to residual HDO.
SIMULATIONS
Simulated line shapes were calculated from parameters describing the global and local motions (Figure 3) using the program MXET1 developed by the Vold group.{Vold, 1991 #77} To simulate the deuterium line shape, a form of the potential energy surface/landscape, U, (through which the C-D bond is traversing) must be chosen. A number of simple jump models have been used to simulate deuterium lineshape data acquired from the 5′ methylene groups of the phosphodiester backbones of several DNA sequences. The local motion of the 5′ methylene group has been modeled as a three-site gauche-trans isomerization,[Meints, #157] as a two-site jump, and a biaxial four-site jump.[Alam, 1991 #36] [Hatcher, 1998 #18]. As described in the aforementioned papers, these simple models have only approximated the experimental line shapes acquired for the 5′ methylene deuterons in DNA.
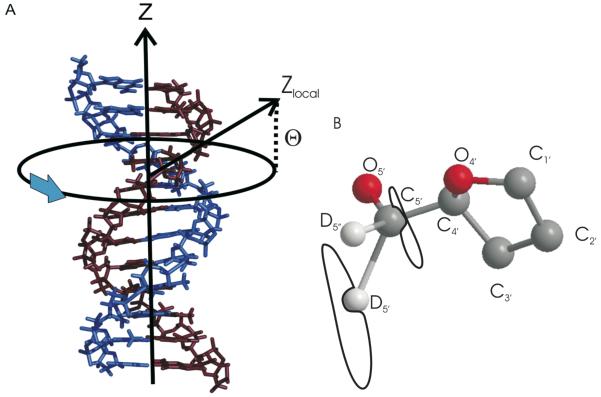
Figure 3.
The dynamic axes and coordinate systems used in the studies of backbone dynamics. (A) Lineshape simulations include two independent motions. A slow (rate of 104 Hz) uniform rotation of the DNA molecule occurs about the helical symmetry axis, labeled Z. Local motion of the backbone is referenced to a local coordinated system, where the z axis is indicated by the vector Zlocal defined by the angle Θ. (B) The elliptical trajectories for C5′ and D5′/5”. (Need to create a new part A)
General Formalism for 2H Line Shape Calculations
Most models proposed to simulate phosphodiester motions in nucleic acids assume segments of the backbone exchange between discrete conformations. Exchanges between discrete structural conformers adequately describe internal molecular reorientations when energy barriers exceed 10kBT. However, the barriers between conformational isomers of the phosphodiester backbone are likely much lower than 10kBT so a more realistic model of motion must be invoked. More realistic models of phoshodiester conformational dynamics model the motions of the constituent bonds as Brownian diffusors that move over energy barriers that intervene between low energy conformers. A great deal of work has been done on the Brownian dynamics of polymethylene chains, mostly in the context amino acid side chain motions in proteins and polymethylene chain motions in lipid (Wittebort, Olejniczak, Griffin, 1987). Solution of the relevant Fokker-Planck equations can be achieved by a discretization scheme as described by Nadler & Schulten (1986). The application of these methods to describing the dynamics of furnaose rings has been described in the literature and has succeeded in providing excellent fits to experimental 2H solid state NMR line shapes (Meints & Drobny, 2001; Meints et al 2007). Because this formalism has been adequately described in the literature, we only outline the method briefly.
Fitting the experimental 2H solid state NMR line shape using the discretization scheme of Nadler and Schulten (1986) requires construction of an exchange matrix π with elements defined as
| (1) |
where , Ui is the value of the potential at the ith discrete site along the reorientational trajectory, and . The correlation time τ is related to the coefficient of reorientational diffusion D by δ2 = 2Dτ where δ is the unit angular displacement, and to the jump rate k between adjacent sites .
With the jump matrix constructed according to equation (1), the 2H line shape is obtained as outlined in Wittebort et al (1987), Vold & Vold (1995), and Meints et al (2007). The 2H NMR lineshape I(ω,τ1,τ2,ΩCL) obtained by application of the quadrupolar echo pulse sequence (i.e. two ninety degree pulses separated by a time τ1, with acquisition of the free induction decay commenced after a time τ2 following the second pulse) to a polycrystalline sample is the Fourier transform if the time domain response m(t):
| (2) |
where the time domain response is
| (3) |
and where λ and λ* are eigenvalues of the matrices A = iω+π and A* = -iω+π, respectively, i.e. T-1AT = λ and (T *)-1A*T* = λ *. The matrix ω is diagonal with elements
| (4) |
where the solid angle relates the principal axis system (P) of the (uniaxial) electric field gradient tensor of the deuteron to the jth site along the reorientation trajectory of the C-D bond which is defined relatic=ve to a molecular-fixed frame C. The second angle ΩCL relates the molecular frame C to the lab frame L, and is averaged randomly in a polycrystalline sample.
In summary, to obtain a 2H simulated line shape, the ω and π matrices must be determined. The ω matrix includes the set of angles that define the sequence of solid angles that orient the C-D bond in the C frame. The matrix ω may therefore be associated with the pathway or trajectory that the C-D bond follows. The matrix π includes information on the nature of the energy surface that the C-D encounters as it travels along the trajectory defined by ω as well as the diffusion coefficient that defines the rate at which a C-D bonds executes a mean displacement in the course of its travels along the trajectory. The structures of the two matrices ω and π used to obtain optimal fits to the experimental 2H line shapes will be separately discussed below.
Description of Motions of the Phosphodiester Backbonen
The 5′ methylene group moves in part as a result of rotation about the C4′-C5′ bond, which is quantified in turn by the torsion angle γ. The torsional state of this bond affects the position of the phosphate group relative to the base and furanose ring, and is therefore an important determinant of nucleic acid structure. Motions about this bond at nucleotide C6 and in the adjacent nucleotides of the GCGC moiety would exert a profound effect on the local structure of the phosphodiester backbone with which the methyltranferase protein must interact.
The orientational state of the C4′-C5′ bond has been -studied in isolated nucleotides and in nucleic acids by X-ray crystallography, solution NMR spectroscopy, and by theoretical modeling (Saenger, section 4.8). On the basis of these studies, three rotational isomers produced by rotations about C4′-C5′ and defined in terms of the torsion angles O5′-C5′-C4′-O4′ and O5′-C5′-C4′-C3′are observed: +sc, where the C5′-O5′ bond is gauche to both C4′-O4′ and C4′-C3′, ap, where the C5′-O5′ bond is gauche to C4′-O4′ and trans to C4′-C3′, and -sc where the C5′-O5′ bond is trans to C4′-O4′ and gauche to C4′-C3′. Dynamic modulation of the D5′/5” line shape might therefore result from rotations about the C4′-C5′ axis, resulting in an exchange between the +sc, ap, and -sc rotational. isomeric states. Because of the large angular excursions that the C5′-D5′/5” bonds would experience as a result of such an exchange, such a mechanism would exert a profound effect on the D5′/5” line shape were it to occur at a rate k>104 Hz. Although this model has produced some agreement with D5′/5” line shape obtained from the T nucleotides in a DNA dodecamer containing the Eco R1 restriction site [d(CGCGAATTCGCG)]2 only qualitative agreement was obtained when this model was applied to [d(G1A2T3A4G5C6G7C8T9A10T11C12)]2 enriched with 2′-deoxycytidine-2H5′/5” at nucleotide C6 (Meints & Drobny, 2001).
Because 120 degree angular excursions between two or three rotational isomers failed to simulate the 2H solid state NMR line shape, motional models with more limited amplitudes were developed. In addition, the 4′ carbon is a component of the furanose ring whose displacements may couple, to an extent, to the 5′ methylene group. Therefore the reorientation of the C5′-D5” bond may not be described by a simple rotation about a single bond axis. To reflect coupling to the furanose ring motion, a lateral displacement of the 5′ carbon was incorporated into the dynamic model. Table 1 shows a list of trajectories for the C5′-D5′/5” as a function of the displacement of the 5′ carbon. The trajectory of the C5′-D5” bond is described in Table 1by a set of azimuthal/equatorial angles (θi,φi) where i labels the site along the trajectory. Typically ten sites were used in each simulation. Discretizing the trajectory into more than 10 sites did not change in the line shape markedly.
Table 1.
| C5′-D5′/5” Trajectory (θ,ϕ) as a Function of Lateral Displacement of C5′ | |||
|---|---|---|---|
| Trajectory Site | 0.30 Å | 0.32 Å | 0.37 Å |
| 1 | (78.2, 333.7) | (78.0, 334.2) | (77.5, 335.7) |
| 2 | (82.8, 345.0) | (83.1, 346.1) | (83.6, 348.5) |
| 3 | (87.8, 345.4) | (88.4, 346.4) | (90.0, 348.8) |
| 3 | (88.8, 341.5) | (89.4, 342.6) | (91.0, 345.2) |
| 5 | (86.6, 330.1) | (86.9, 330.6) | (87.8, 331.9) |
| 6 | (83.8, 314.3) | (83.9, 313.6) | (84.2, 311.8) |
| 7 | (82.1, 303.3) | (82.3, 301.8) | (82.9, 298.1) |
| 8 | (79.3, 302.4) | (79.4, 300.9) | (79.9, 297.2) |
| 9 | (75.0, 310.8) | (74.7, 309.8) | (74.1, 307.3) |
| 10 | (73.1, 325.8) | (72.5, 325.9) | (70.9, 326.2) |
In addition to the localize motion of nucleotides in DNA, previous studies (Meints et al, 2001, 2007) show that a global rotation of the entire dodecamer about the helix axis is required to fit 2H line shapes acquired at hydration levels greater than or equal to W=10. The global motion is described by six site jump, diffusion on a cone model. It was previously determined{Alam, 1991 #2} that for hydration levels of W=10 and above, the helical rate of rotation is 104 Hz. The half angle of the cone, which also describes the orientation of the local dynamic axis of the C5′-D bond with respect to the longitudinal helix axis, has previously been set to 20°, but for these studies of the more flexible phosphodiester backbone, it was varied from 20° - 60°.
Potentials
Most studies of the geometry of the DNA phosphodiester backbone assume that as the C-D bond reorients along its trajectory, it traverses a multi-well -potential for the 5′ methylene group.(ref. Saenger) In general such potentials can be represented as combinations of sine and cosine functions,(ref) with the the simplest, approximation to a n well potential being
| (5) |
where U0 is the barrier height between wells (Figure 4A), , N is the number of sites (i.e. typically N=10), and j labels the site number With the potential defined by equation 5 and the correlation time defined in terms of the diffusion coefficient D the exchange matrix π is completely defined .
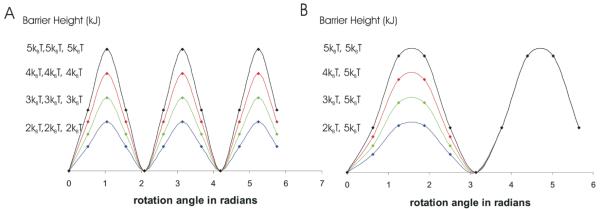
Figure 4.
(A) Varying barrier height in three-well potential (B) Varying barrier height in two-well potentials
Lineshapes simulated with a three-well potential could not fit the experimental data; however, a simple periodic two-well potential (Figure 4B)
similar to those used to fit some of the furanose ring data reproduced the experimental data, likely due to the coupled motion between the phosphodiester backbone and furanose rings.
Assuming a given potential, lineshapes are simulated as a function of U0, D, and q. In almost all data sets a sharp peak appears in the center of the spectrum, which is due to residual deuterons in the deuterium-depleted water used to hydrate the DNA. This water peak is simulated by adding a Lorentzian peak to the center of the simulated lineshape.
RESULTS
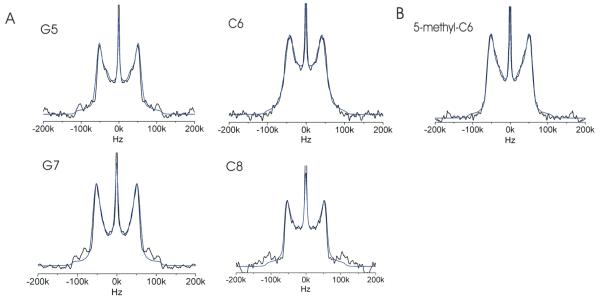
In order to probe local mobility of the backbone in the d(G5C6G7C8) site, deuterium lineshapes and spin-lattice relaxation rates were obtained for the 5′/5” deuterons of dG5, dC6, dG7, dC8, and 5mdC6. Experimental lineshapes and lineshape simulations are shown in Figure 5A and 5B. Motions ranging from ps-us are can be determined from the spin-lattice relaxation rates, which are calculated from partially relaxed lineshapes (Figure 6). Simulations can elucidate the detailed nature of local molecular motions including the direction and amplitude of reorientational motion.
Figure 5.
(A) The four deuterium lineshapes (black) for each of the labeled sites in the non-methylated DNA dodecamer with the simulation (blue) of each overlayed. Simulation parameters are described in Table 2. (B) The experimental solid state deuterium lineshape obtained for D5′/5” in the backbone of methylated C6 (black) and simulated lineshape (blue) (C) Spectrum of classical Pake doublet. The width between the horns is ¾ of the quadrupolar coupling constant, which is 140-220 Hz for deuterium.
Deuterium spin interactions are dominated by the modulation of the electric quadrupole moment with the surrounding electric field gradients, and are not largely influenced by CSA or dipolar couplings. In contrast to relaxation studies of spin ½ nuclei (where dynamic properties can only be compared from the relaxation of magnetically similar spins) the relative dynamics of deuterons can be obtained on a qualitative level and to a good approximation by straightforward comparison of relaxation and lineshape data regardless of structural site. Knowledge of the electric field gradient (EFG) tensor is still critical for evaluation of relaxation and lineshape data, but fortunately tensor variations are small for deuterons bonded to aliphatic carbons, allowing for direct comparisons between different sites.
Qualitative Analysis of the Lineshape and Partially Relaxed Lineshape Data
Static lineshape experiments probe motions ranging from ns-ms timescale. Motions with rates occurring in this range produce “intermediately-averaged” lineshapes, which differ from the classical Pake doublet (Figure 5C) most often by raising of the center of the lineshape, narrowing of the overall lineshape and a loss of definition of the perpendicular edges, “the horns,” of the type shown in Figures 5A and 5B with quantification of the lineshape features in Table 1. Qualitatively, the extent of motion is manifested by the amount of deviation from the Pake doublet form.
The quadrupolar echo lineshape of dG5 (Figure 5A) shows a narrowed classical Pake doublet with slightly broadened horns, a slightly raised center, while the inner edge of the horns becomes jagged; The dG7 lineshape is quite similar and of all the residues in the recognition site, deviates the least from the classical Pake doublet form, with only a slight rise in the center of the lineshape, and less narrowing of the overall lineshape and broadening in the horns. The dC6 and dC8 lineshapes deviate substantially from the classic pake doublet form. Here, both the centers are markedly raised and the horns are broadened, and beginning to lost definition. In addition these lineshapes are significantly more narrowed that either the dG5 or dG7. The lineshape results indicate the dC6 and dC8 are the most mobile on the ns-ms timescale, followed by dG5 and dG7.
The <T1z> values (Table 2) also suggest that dC6 (46 ± 4 ms) is the most mobile site. Interestingly, both dG7 (53 ± 3 ms) and dC8 (51 ± 4 ms) have comparable spin-lattice relaxation times, (only slightly less than dC6) implying that they have the same degree of motion. Here, the <T1z> value of dG5 (68 ± 7 ms), is significantly longer than the others indicating less mobility. These results are not contrary, but highlight the differences in timescales that these two experiments probe. While dG6 and dC8 appear to be able to exhibit motions over a large timescale (ps-ms), both dG5 and dG7 exhibit mobility over a smaller range of timescales. dG5 appears to exhibit motions slow motions (us-ms) whereas dG7 experiences fast motions (ps-ns). (Add chart?)
Table 2.
Quantitative values of key features of the 5 experimental lineshapes in Figure 2
| Label Site | Horn Width (kHz) | QCCeff (kHz) | Λ | Center Height (% of total height) | Full Width at Half Max. (kHz) |
|---|---|---|---|---|---|
| G5 | 102 ± 2 | 136 ± 2 | 0.78 | 47% | 118 ± 2 |
| C6 | 90 ± 2 | 120 ± 2 | 0.69 | 57% | 114 ± 2 |
| G7 | 104 ± 2 | 139 ± 2 | 0.79 | 38% | 108 ± 2 |
| C8 | 106 ± 2 | 141 ± 2 | 0.81 | 56% | 121 ± 2 |
| 5mC6 | 105 ± 2 | 140 ± 2 | 0.80 | 46% | 125 ± 2 |
There is a significant change in the lineshape of [5′/5”-D]-dC6 upon methylation (Figure 5A, 5B), as the horns expand and “unbend” approaching the classical Pake doublet lineshape indicating that this site becomes more rigid upon methylation. Additionally, the <T1z> value increases (46 ± 4 ms to 71 ± 6 ms) from the unmethylated to methylated dC6. Though the extent of the dynamics of dC6 is not clear from the lineshapes and relaxation data, what does remain clear is the dramatic loss of mobility associated with the methylation of dC6.
The backbone dynamics can also be discussed through an analysis of the quadrupolar coupling constant:
The static QCC for the 5′/5”-methylene deuteron is QCCstatic =175 ± 1 kHz.{Alam, 1991 #2} For deuterium powder patterns that retain a Pake pattern, the amplitude reduction factor (ARF) Λ, can be used to assess motional averaging of the QCC. The ARF is defined as
and can be roughly equated to an order parameter. For dC6, the value of 120 ± 2 kHz for QCCeff gives a value of Λ of 0.69 (Table 1). For 5-methyl-dC6, QCCeff is 140 ± 2 kHz, and Λ is 0.80 (Table 1), which is on the same order as the QCCeff and Λ values for the non-target bases in the recognition sequence (Table 1). This implies differential motional averaging in the methylated versus unmethylated DNAs, with motional averaging of the phosphodiester backbone being significantly diminished in the methylated DNA.
QUANTITATIVE ANALYSIS
The parameters describing the best-fit lineshapes are shown in Table 3. The dG5 lineshape was simulated best by a two-well periodic potential with unequal barriers of 2kBT and 4.5kBT. The amplitude used was 0.3 Å with a jump rate of 108 Hz where the zlocal is 50° above the plane of the helical rotation. The dG7 lineshape was fit with a two-well potential with higher barriers of 4kBT and 8kBT. The amplitude and jump rate were 0.32 Å and 108 Hz as before, and the orientation of zlocal at this site was 40° above the plane of the helical rotation. The dC8 lineshape was fit by a two-well potential with unequal barriers of 1kBT and 3kBT, an amplitude of 0.32 Å and a jump rate of 108 Hz with the orientation of zlocal at this site was 30° above the plane of the helical rotation.
Table 3.
Spin-lattice relaxation time for [5′/5”-2H]-dG5 through dC8 from the DNA sequence [d(GATAG5C6G7C8TATC)]2
| Sample (from [d(GATAG5C6G7C8TATC)]2) | Hydration (waters/nucleotide) | <T1Z> (ms) |
|---|---|---|
| [5′/5”-2H]-dG5 | W = 11.3 ± 1 | 68 ± 7 |
| [5′/5”-2H]-dC6 | W = 11.8 ± 1 | 46 ± 4 |
| [5′/5”-2H]-dG7 | W= 11.0 ± 1 | 53 ± 3 |
| [5′/5”-2H]-dC8 | W = 11.0 ± 1 | 51 ± 4 |
| [5′/5”-2H]-5-methyl-dC6 | W = 11.3 ± 1 | 71 ± 6 |
The unmethylated dC6 lineshape differed significantly from the non-target sites and was best using a simple equal barrier two well-potential with barriers of 4.5 kBT. Both the amplitude (0.37 Å) and jump rate (109 Hz) were larger than the other sites, and the zlocal orientation was the largest (60°) This indicates that the dC6 5′/5” deuterons are equally likely to be in the two most favored conformations and that they are moving at a greater rate through a larger volume in space than the neighboring sites.
Upon methylation, the dC6 lineshape is best modeled by parameters that are similar to the non-target sites with an amplitude that decreased to 0.32 Å and a slower jump rate (108 Hz). A two-well potential of unequal barriers of 2.5 kBT and 4.5 kBT (similar to dG5) and an orientation where zlocal decreases to 40° describes the lineshape the best. Each of these parameters support the conclusions drawn from the qualitative analysis, namely, that there is a dramatic loss of conformational flexibility upon methylation of dC6, and demonstrate that upon methylation, the dynamics of the target nucleotide become similar to the non-target sites in the recognition sequence.
DISCUSSION
Previously, this group has investigated the effects of methylation in the Dickerson dodecamer and the HhaI recognition sequence and demonstrated that motion in the target site is quenched upon methylation.(Geahigan_2000, Hatcher_1998, Hatcher_2001, Meints_2001) A computational study by Banavali and co-workers supports a more flexible phosphodiester backbone surrounding the target cytidine of the HhaI recognition sequence.{Banavali, 2006 #19} However, to date, there has been little to no analysis of the possible effect of methylation on recognition and cleavage by the endonuclease.
Here we have measured the lineshapes and <T1z> for the backbone of residues in the recognition site and the methylated dC6. A qualitative analysis of both the lineshapes and the relaxation data indicates that dC6 is the most mobile position in the recognition sequence. The lineshapes of the other sites in the recognition sequence and the methylated dC6 indicate some motion on the millisecond to nanosecond timescale, while the <T1z> values indicate that dG7 and dC8 experience additional dynamics on the nanosecond to picosecond timescale.
Our simulations demonstrate the dC6 site diffuses through the two favored conformations (equal barrier heights) whereas the dG5, dG7, and dC8 sites are primarily restricted to a single conformation, (unequal barrier heights) The difference in the barriers for dG7 is approximately twice that of the other non-target bases, indicating that dG7 must be more strongly restricted to a single conformation in order for dC6 to be able to move so freely. Upon methylation, we see dC6 become restricted mainly to a single conformation as the other bases are.
The most important conclusion from this work arises from the comparison of the dynamics between methylated and unmethylated dC6. While both deoxycytidine residues display dynamic behavior within their furanose rings,(Meints, 2007) only the dC6 position has significant motional averaging in its backbone methylene moiety. Neither of the deoxyguanosine residues displays any significant dynamic averaging at either position. This suggests that the dynamics observed at this position, which is the target site of the HhaI methyltransferase, might play some sort of role in the restriction-modification mechanism
The local dynamics play an important role, with the target site showing more local conformational flexibility than any other position within the binding site. Even more interestingly, based upon these results we suggest a model for how the endonuclease recognizes the methylated site. Our simulations show that in addition to a decrease in the amplitude of the local motion, seen both in the furanose ring and the backbone, the angle between the 5′/5” deuteron of dC6 and the plane of the helical rotation decreases by 20° upon methylation. This change in conformation of the backbone of the DNA may play a role in endonuclease recognition.
Figure 7.
Inversion Recovery Data to be added.
Table 4.
Model potentials and parameters used to fit each of the 5 simulated lineshapes in Figure 2. (Remove U(ø) column since all are the same, maybe put in caption)
| Label Site | U(ϕ) (J) | U0 (0≤ϕ<π) (J) | U0 (π ≤ϕ<2π) (J) | Amplitude (Å) | Jump Rate (Hz) | θ |
|---|---|---|---|---|---|---|
| G5 | 2kBT | 4.5kBT | 0.30 | 108 | 50 | |
| C6 | 4.5kBT | 4.5kBT | 0.37 | 109 | 60 | |
| G7 | 4kBT | 8kBT | 0.32 | 108 | 40 | |
| C8 | 1kBT | 3kBT | 0.32 | 108 | 30 | |
| 5mC6 | 2.5kBT | 4.5kBT | 0.32 | 108 | 40 |