Abstract
Objectives
Mucosal inflammatory responses are orchestrated largely by proinflammatory chemokines. We aimed to evaluate the expression of the chemokine GCP-2 in clinically healthy vs. diseased gingival tissues and to explore possible correlations with clinical and microbiological markers of periodontitis.
Background
The chemokine granulocyte chemotactic protein 2 (GCP-2/CXCL6) is involved in neutrophil recruitment and migration. Previous studies showed that GCP-2 is upregulated in mucosal inflammation, e.g., in inflammatory bowel disease, similarly to the functionally and structurally related chemokine interleukin 8 (IL-8). Nevertheless, unlike IL-8, a role of GCP-2 in gingival inflammation has not yet been demonstrated.
Methods
Gene expression in 184 ‘diseased’ and 63 ‘healthy’ gingival tissue specimens from 90 periodontitis patients was analyzed using Affymetrix U133Plus2.0 arrays. GCP-2 expression was further confirmed by real time RT-PCR, western blotting and ELISA while the localization of GCP-2 protein in the gingival tissues was analyzed by immunohistochemistry. Plaque samples from the adjacent periodontal pockets were collected and evaluated for 11 periodontal species using checkerboard DNA-DNA hybridizations.
Results
Among all known chemokines, GCP-2 mRNA was the most expressed (3.8 fold, p<1.1×10−16) in ‘diseased’ vs. ‘healthy’ tissue, as compared to a 2.6 fold increased expression of IL-8 mRNA (p<1.2×10−15). Increased expression of GCP-2 correlated with higher levels of ‘red’ and ‘orange’ complex pathogens and with increased probing depth, but not with attachment loss. Immuno-histochemistry showed that GCP-2 was expressed in gingival vascular endothelium.
Conclusion
GCP-2 correlates with the severity of periodontitis and appears to act as a hitherto unrecognized functional adjunct to IL-8 in diseased gingival tissues.
Keywords: Periodontal disease, inflammatory mediator, Chemokines, Chemotaxis, Matrix metalloproteinase, Microarray
INTRODUCTION
Neutrophils play a pivotal role as a first line of cellular defense against pathogens in periodontal homeostasis, whilst defects of neutrophil function, but also hyperactive neutrophils are associated with severe periodontal disease (1). Neutrophils are recruited to an inflammatory site by a gradient of specific chemokines, which represent a family of chemotactic cytokines produced by different cell types including epithelial and endothelial cells in response to activation by microbial metabolic products or pro-inflammatory cytokines (2).
Human granulocyte chemotactic protein-2 (GCP-2, CXCL6) is a CXC chemokine with a conserved Glu-Leu-Arg (ELR+) motif (3). Similar to other ELR+ CXC chemokines such as IL-8 (CXCL8) and ENA-78 (CXCL5), it possesses potent chemotactic and proangiogenic properties (4). GCP-2, similar to IL-8, activates target cells by binding to CXC chemokine receptors (CXCR)-1 and (CXCR)-2 (5). Both receptors are expressed by neutrophil granulocytes, but not by other blood-derived cells, such as lymphocytes or monocytes .
IL-8 is consistently expressed in periodontal health, and mediates neutrophil recruitment to the gingival tissues adjacent to the periodontal crevice and into the gingival crevicular fluid, maintaining a subclinical inflammatory response to the ubiquitous microbiota of the dental plaque (6, 7). In periodontal disease, the expression of IL-8 is strongly up-regulated and correlates with disease activity (8, 9), whereas a regulated expression of the functionally related GCP-2 has not been described yet (10). However, a role for GCP-2 has been demonstrated in other mucosal chronic inflammatory conditions as inflammatory bowel disease (11, 12) or chronic rhinosinusitis (13, 14). In this study, we sought to evaluate the expression of GCP-2 in clinically healthy vs. diseased gingival tissues and to explore possible correlations with clinical and microbiological markers of periodontitis.
MATERIALS & METHODS
The design and procedures of the study were approved by the Columbia University Medical Center Institutional Review Board.
Subjects
A total of 90 subjects with moderate to severe periodontitis (63 with chronic and 27 with aggressive periodontitis) were recruited among the patients referred for periodontal therapy to the Clinic for Post-doctoral Periodontics, Columbia University College of Dental Medicine. Eligible patients were (i) at least 13 yr old; (ii) had a minimum of 24 teeth present; (iii) had no past history of systematic periodontal therapy other than occasional prophylaxis provided by the referring general dentist, (iv) had received no systemic antibiotics or anti-inflammatory drugs for at least 6 months, (v) harbored a minimum of 4 teeth with radiographic bone loss, (vi) did not suffer from diabetes mellitus, (vii) did not suffer from any of the systemic conditions or genetic disorders that entail a diagnosis of “Periodontitis as a manifestation of systemic diseases”, (viii) were not pregnant, and (ix) were not current users of tobacco products or of nicotine replacement medication. Signed informed consent was obtained prior to enrollment.
Clinical examination and procedures
All participants underwent a full-mouth examination of the periodontal tissues at six sites per tooth, using a manual probe. The examination included assessments of presence/absence of dental plaque and bleeding on probing (BoP), and linear measurements of probing pocket depth (PPD) and clinical attachment level (CAL). Identification of donor sites and harvesting of gingival tissue samples was performed as earlier described (15). In brief, a ‘diseased’ interproximal papilla showed BoP, PPD ≥ 4mm, and CAL ≥ 3mm, whilst a ‘healthy’ papilla demonstrated no BoP, PPD ≤ 4mm and CAL ≤ 2mm. All tissue specimens were collected during periodontal surgery. Each patient contributed with one to three ‘diseased’ tissue samples (184 samples in total), 2/3 of all patients contributed with one ‘healthy’ tissue sample (63 in total).
Subgingival plaque samples were obtained from the adjacent periodontal pockets and analyzed for 11 periodontal species (Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Fusobacterium nucleatum, Prevotella intermedia, Campylobacter rectus, Micromonas micros, Eikenella corrodens, Veillonella parvula, and Actinomyces naeslundii) using checkerboard hybridizations as earlier described (16, 17).
mRNA quantification
Total RNA from 184 ‘diseased’ and 63 ‘healthy’ gingival tissue specimens was extracted, amplified, reverse transcribed, labeled, and hybridized with AffymetrixU133Plus2.0 arrays, as earlier described (15). Independent confirmation of the obtained microarray data was performed by quantitative real-time PCR in tissue samples from 5 patients who showed a strong differential expression for GCP-2 mRNA in ‘diseased’ gingival samples. Three patients contributed with a pair of ‘healthy’ and ‘diseased’ tissue samples each, while two patients contributed with ‘diseased” tissue samples only. The Taqman Gene Expression Assays Hs00237017_m1 and Hs99999905_m1 were used for GCP-2 and glyceraldehyd-3-phosphatedehydrogenase (GAPDH), respectively (Applied Biosystems, Foster City, CA). Three technical replicates per sample and gene were performed.
Immunoblot analysis
Total gingival tissue protein was prepared by homogenization and lysis of frozen biopsy tissues in modified RIPA buffer [50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.25% (w/v) sodium deoxycholate, 1% (v/v) NP-40, 0.1% (w/v) sodium dodecylsulfate, 1% (v/v) Triton X-100 (all from Sigma-Aldrich)] containing Protease Inhibitor Cocktail III (Calbiochem, San Diego, CA) on ice. Lysates were cleared by ultracentrifugation, and total protein concentration was determined using a Bradford assay (Bio-Rad, Hercules, CA). Samples were heat-denatured in 4× Laemmli buffer and size-separated by 12% SDS PAGE, blotted onto nitrocellulose membranes (Amersham Pharmacia Biotech, Arlington Heights, IL) and blocked with blocking buffer [PBS containing 0.05% (v/v) Tween-20, 5% (w/v) non-fat dry milk]. Blots were incubated with anti-human GCP-2 murine monoclonal antibody (MAB333, clone 60910, R&D Systems) at 1:250 overnight at 4°C. Immuno-detection was performed using a biotinylated rabbit anti-mouse antibody (Amersham Pharmacia Biotech), Streptavidin-HRP (Amersham Pharmacia Biotech) and enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech). Densitometric analysis was performed using the software imagej 1.38× (NIH, Bethesda, ML).
Immunohistochemistry
‘healthy’ and ‘diseased’ gingival tissue specimens, obtained as described above, were embedded in OCT Tissue Tek (Sakuma, Japan) and snap frozen in isopentane/liquid nitrogen. Five micrometer cryostat sections were prepared and dried on coated object slides (Superfrost plus, Walldorf, Germany) for 30 minutes at room temperature. Fixation of specimens was carried out with absolute acetone at −20°C for 10 minutes, endogenous peroxidase activity was blocked with 2% hydrogen peroxide in methanol. The slides were subsequently incubated with a murine monoclonal anti-human GCP-2 antibody (MAB333, clone 60910, R&D Systems, Minneapolis, MN) or the appropriate mouse isotype control (dilution: 1:100 in PBS) for 3 hours at room temperature. Immuno-reactivity was visualized using HRP-coupled secondary antibodies and diaminobenzidine as a chromogen. Sections were counterstained with hematoxylin and examined by light microscopy.
Measurement of GCP-2 by ELISA
The level of GCP-2 in gingival tissue lysates was determined using a Quantikine human CXCL6/GCP-2 ELISA kit (R&D Systems), according to the manufacturers protocol. One ‘healthy’ and one ‘diseased’ gingival tissue specimen from five consecutively recruited patients was homogenized and lysed as described above. The absolute amount of protein was quantified using a Bradford assay, was adjusted to a total protein concentration of 1 mg/ml, and was used undiluted for the assay. The assay was performed with two technical replicates.
Statistical analysis
For gene expression analyses, R version 2.3.1 (Linux OS) or SAS for PC version 9.1 (SAS Institute, Cary, NC) were used. Expression data were first normalized and summarized using the log scale robust multi-array analysis (RMA) (18) with default settings. Differential expression was assayed using a standard mixed-effects linear model approach, in which patients were conditioned as random effects to account for the within-mouth correlation of GCP-2 levels resulting from multiple gingival tissue samples being collected from each patient. Using this approach, we explored the association between GCP-2 mRNA levels (dependent variable) and the following independent variables (fixed effects): gingival tissue status (‘healthy’ vs. ‘diseased’ as described above), PPD, CAL, bacterial colonization level with each of the aforementioned eleven species, age, gender and race/ethnicity. All results reported herein stem from univariate models with the exception of PPD and CAL which were modeled simultaneously to obtain the independent association between either PPD or CAL and GCP-2 expression. GCP-2 expression fold change was computed by dividing the average raw expression values among the comparison group of the independent variable by the average expression in the reference group (i.e., average expression in ‘diseased’ tissue samples divided by average expression among ‘healthy’ samples). Therefore, fold change values represent relative differences in RNA levels.
For all experiments not involving microarrays, statistical analyses were performed using GraphPad Prism 5 (GraphPad, San Diego, CA) for unpaired, two-tailed t-tests. Differences between chemokine concentrations or Realtime PCR cycles were considered significant if p-values were < 0.05. Analysis of GCP-2 protein expression using western blotting and immuno-histochemistry was performed at least in triplicate with similar results.
RESULTS
Patients were on average 42 years old (range 13−76 years), 50% female, 76% Hispanic, 15% Black and 6% White (15). According to the criteria of the 1999 International Workshop, 70% of the patients had chronic and 30% aggressive periodontitis.
mRNA for GCP-2 is upregulated in periodontitis
Microarray data demonstrated that, among all chemokines, GCP-2 mRNA was the most expressed (3.8 fold, p<1.1×10−16) in ‘diseased’ vs. ‘healthy’ gingival tissues. In comparison a 2.6 fold increased expression of IL-8 mRNA (p<1.2×10−15) in ‘diseased’ vs. ‘healthy’ gingival tissues was observed. Table 1 summarizes microarray-based mRNA expression data for all chemokines, chemokine receptors and selected functionally related genes with a p-value below the Bonferroni threshold of 9.14×10−07.
Table 1.
Microarray analysis of chemokine and chemokine receptor mRNA expression in periodontal disease. Data are shown for all chemokines, chemokine receptors and selected functionally related genes with a p-value below the Bonferroni threshold of 9.14×10−07.1
| Probe | Gene | Description | Fold Change | p-value |
|---|---|---|---|---|
| 206336_at | CXCL6 | chemokine (C-X-C motif) ligand 6 | 3.85 | 1.1E-16 |
| 217028_at | CXCR4 | chemokine (C-X-C motif) receptor 4 | 3.56 | 1.1E-16 |
| 204470_at | CXCL1 | chemokine (C-X-C motif) ligand 1 | 3.45 | 1.1E-16 |
| 209201_x_at | CXCR4 | chemokine (C-X-C motif) receptor 4 | 3.07 | 1.1E-16 |
| 211919_s_at | CXCR4 | chemokine (C-X-C motif) receptor 4 | 2.90 | 1.1E-16 |
| 202859_x_at | IL8 | interleukin 8 | 2.57 | 1.2E-15 |
| 205242_at | CXCL13 | chemokine (C-X-C motif) ligand 13 | 2.42 | 7.9E-14 |
| 211506_s_at | IL8 | interleukin 8 | 2.26 | 1.9E-12 |
| 209924_at | CCL18 | chemokine (C-C motif) ligand 18 | 2.18 | 1.1E-16 |
| 214146_s_at | PPBP | chemokine (C-X-C motif) ligand 7 | 2.11 | 7.0E-14 |
| 32128_at | CCL18 | chemokine (C-C motif) ligand 18 | 2.08 | 2.4E-15 |
| 203666_at | CXCL12 | chemokine (C-X-C motif) ligand 12 | 2.05 | 1.1E-16 |
| 209774_x_at | CXCL2 | chemokine (C-X-C motif) ligand 2 | 1.90 | 2.6E-12 |
| 203936_s_at | MMP9 | matrix metallopeptidase 9 | 1.81 | 1.8E-13 |
| 210072_at | CCL19 | chemokine (C-C motif) ligand 19 | 1.78 | 5.6E-10 |
| 209687_at | CXCL12 | chemokine (C-X-C motif) ligand 12 | 1.76 | 1.4E-15 |
| 208335_s_at | DARC | Duffy blood group, chemokine receptor | 1.71 | 1.1E-16 |
| 205098_at | CCR1 | chemokine (C-C motif) receptor 1 | 1.66 | 1.1E-16 |
| 207850_at | CXCL3 | chemokine (C-X-C motif) ligand 3 | 1.61 | 6.1E-11 |
| 214974_x_at | CXCL5 | chemokine (C-X-C motif) ligand 5 | 1.60 | 8.8E-07 |
| 1405_i_at | CCL5 | chemokine (C-C motif) ligand 5 | 1.59 | 6.4E-11 |
| 206337_at | CCR7 | chemokine (C-C motif) receptor 7 | 1.59 | 9.0E-12 |
| 1555759_a_at | CCL5 | chemokine (C-C motif) ligand 5 | 1.56 | 1.3E-12 |
| 205099_s_at | CCR1 | chemokine (C-C motif) receptor 1 | 1.48 | 5.2E-14 |
| 205114_s_at | CCL3 | chemokine (C-C motif) ligand 3 | 1.48 | 7.2E-09 |
| 206390_x_at | PF4 | chemokine (C-X-C motif) ligand 4 | 1.45 | 3.4E-13 |
| 206366_x_at | XCL1 | chemokine (C motif) ligand 1 | 1.44 | 1.1E-16 |
| 214567_s_at | XCL1 | chemokine (C motif) ligand 1 | 1.42 | 4.4E-13 |
| 204655_at | CCL5 | chemokine (C-C motif) ligand 5 | 1.41 | 1.1E-08 |
| 205392_s_at | CCL14 | chemokine (C-C motif) ligand 14 | 1.40 | 2.9E-09 |
| 204103_at | CCL4 | chemokine (C-C motif) ligand 4 | 1.39 | 1.6E-10 |
| 214038_at | CCL8 | chemokine (C-C motif) ligand 8 | 1.35 | 4.1E-07 |
| 207794_at | CCR2 | chemokine (C-C motif) receptor 2 | 1.31 | 3.6E-12 |
| 220565_at | CCR10 | chemokine (C-C motif) receptor 10 | 1.30 | 1.4E-11 |
| 219161_s_at | CKLF | chemokine-like factor | 1.28 | 2.1E-11 |
| 223451_s_at | CKLF | chemokine-like factor | 1.24 | 5.3E-11 |
| 211434_s_at | CCRL2 | chemokine (C-C motif) receptor-like 2 | 1.23 | 2.5E-11 |
| 206126_at | BLR1 | chemokine (C-X-C motif) receptor 5 | 1.15 | 3.9E-07 |
| 210133_at | CCL11 | chemokine (C-C motif) ligand 11 | 1.15 | 6.8E-07 |
| 210548_at | CCL23 | chemokine (C-C motif) ligand 23 | 1.13 | 1.1E-07 |
| 224027_at | CCL28 | chemokine (C-C motif) ligand 28 | 0.90 | 8.7E-08 |
| 220351_at | CCRL1 | chemokine (C-C motif) receptor-like 1 | 0.71 | 4.4E-16 |
| 222484_s_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 0.59 | 3.7E-14 |
| 218002_s_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 0.56 | 5.1E-15 |
| 237038_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 0.55 | 1.1E-16 |
Multiple Affymetrix probes may map to the same gene. Fold change is defined as the ratio of gene expression in disease over expression in health. Thus, a fold change < 1.0 indicates a down-regulation in disease vs. health.
Confirmatory realtime RT-PCR showed a mean difference of 5.64 cycles between ‘healthy’ and ‘diseased’ samples, resulting into a 25.64 = 49.8 fold increased expression of GCP-2 mRNA (Figure 1).
Figure 1. Realtime RT-PCR analysis of GCP-2 mRNA in ‘healthy’ and ‘diseased’ gingival tissues.
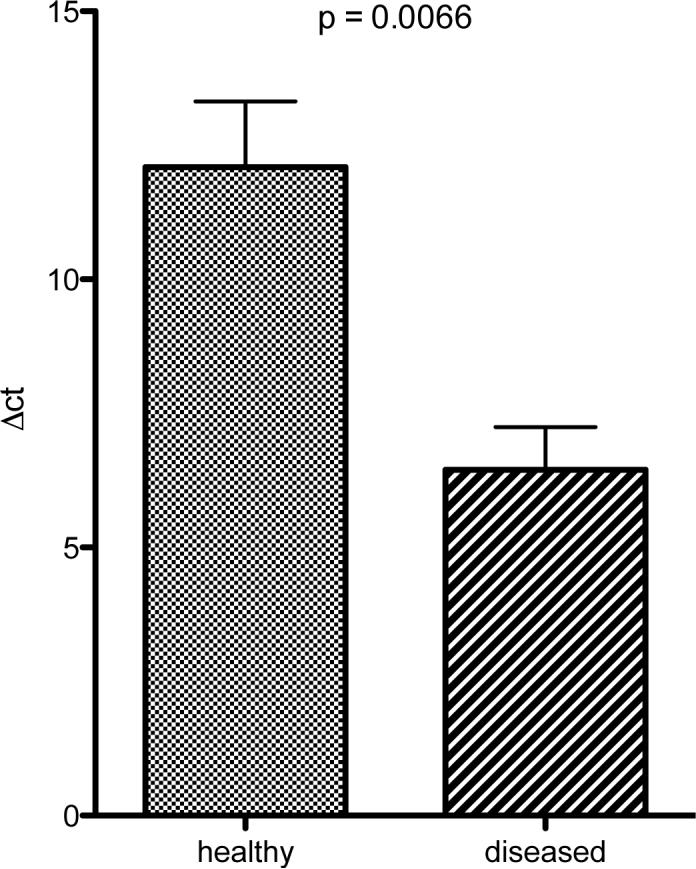
Confirmation of microarray expression data for GCP-2 mRNA in five patients, each contributing with two gingival specimens. The observed mean difference of 5.62 cycles corresponds to a mean up-regulation in ‘diseased’ vs. ‘healthy’ gingival tissue of 49.8 fold.
Association between demographic, clinical or bacteriological characteristics and GCP-2 mRNA expression
Age, race and gender were not related to GCP-2 expression levels. No significant difference in expression levels of the chemokine could be detected between subjects with aggressive and chronic periodontitis. A one standard deviation increase of either P. gingivalis, T. forthythia, C. rectus or P. intermedia was associated with an approximate 1.55 fold increase in GCP-2 expression (all p-values<0.001) while T. denticola (p<0.0001) and M. micros (p<0.01) were each associated with an approximate 1.4 fold increase in expression. A one mm increase in PPD was associated with a 1.33 fold increase in GCP-2 expression independent of CAL. Conversely, CAL was not associated with GCP-2 expression after accounting for PPD level.
GCP-2 protein expression is increased in periodontitis
Immunoblot analysis (Figure 2) showed stronger expression of GCP-2 in ‘diseased’ than in ‘healthy’ gingival tissue samples. The higher expression of GCP-2 protein in periodontitis was also confirmed by ELISA measurements (Figure 3).
Figure 2. Western blot analysis of GCP-2 protein in ‘healthy’ and ‘diseased’ tissue samples.
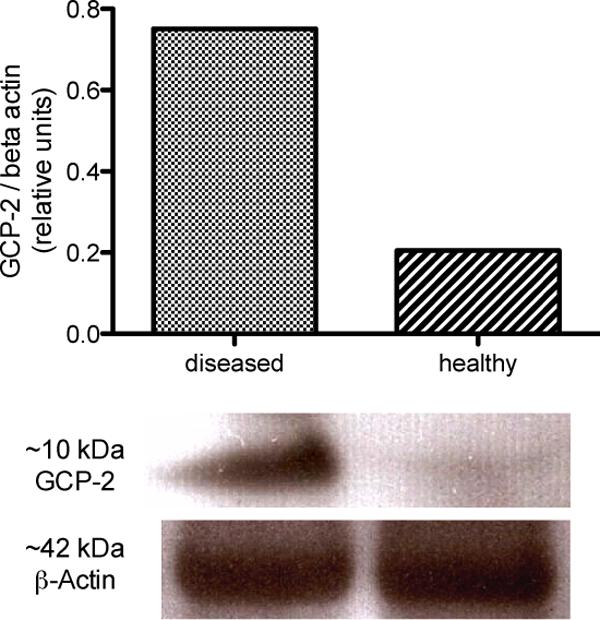
Stronger GCP-2 protein expression in ‘diseased’ gingival tissues. The band for GCP-2 was detected at approx. 10 kDa while equal loading was demonstrated by β-actin probing. Densitometric analysis revealed a 3.6 fold up-regulation of GCP-2 protein in ‘diseased’ tissue. The analysis was performed in 3 consecutively recruited patients each contributing with one ‘diseased’ and one ‘healthy’ gingival tissue specimen.
Figure 3. ELISA analysis of GCP-2 protein in homogenized in ‘healthy’ and ‘diseased’ tissue samples.
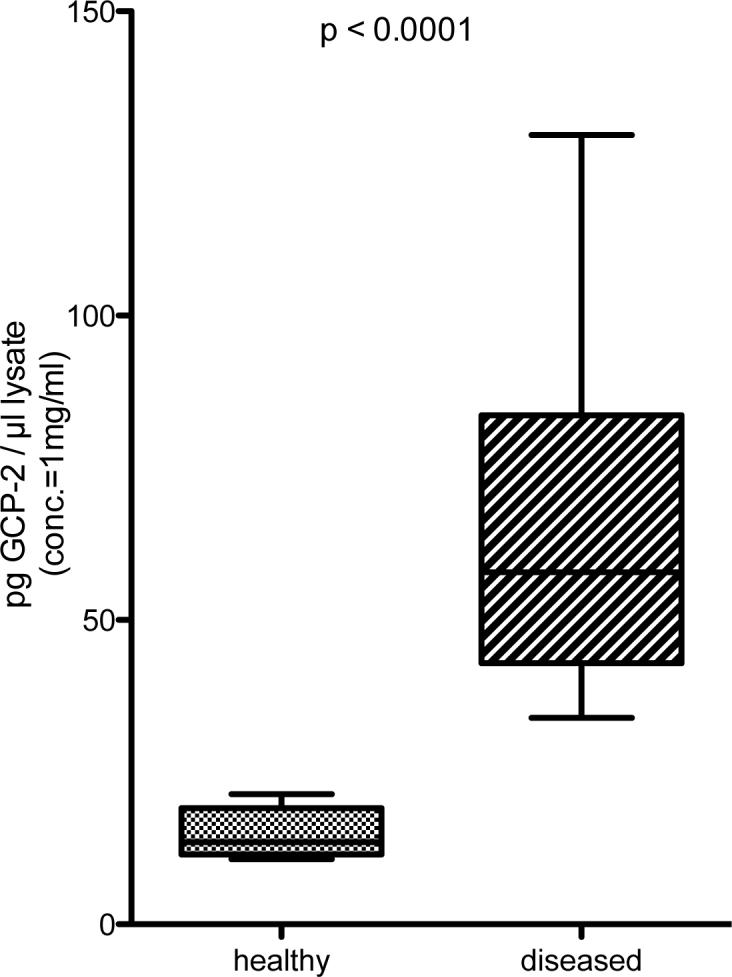
Up-regulation of GCP-2 protein in ‘diseased’ gingival tissue by ELISA (n=5 consecutively recruited patients, each contributing with one ‘diseased’ and one ‘healthy’ gingival tissue specimen).
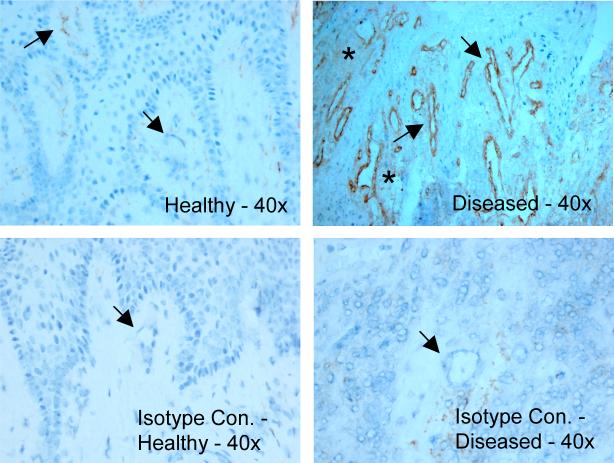
In frozen sections of gingival tissue, a pronounced immunoreactivity for GCP-2 was observed in endothelium in ‘diseased’ gingival tissue, whilst only mild staining was observed in ‘healthy’ tissue (Figure 4). In a similar manner, sections from ‘diseased’ tissues showed a stronger diffuse staining for the secreted chemokine than ‘healthy’ tissue sections (Figure 4).
Figure 4. Immunostaining of GCP-2 protein in frozen tissue sections.
GCP-2 expression is enhanced in ‘diseased’ gingival tissues. GCP-2 immunoreactivity is predominant in endothelial cells of the gingival microvasculature (arrows). Furthermore, a diffuse positive staining of the connective tissue can be detected primarily in ‘diseased’ samples as a typical sign of chemokine secretion (asterisk).
DISCUSSION
Our data are the first to report a differential expression of the ELR+ CXC chemokine GCP-2 in periodontally ‘healthy’ and ‘diseased’ gingival tissues and in fact demonstrate that GCP-2 in the mostly up-regulated among all known chemokines in periodontitis. The levels of GCP-2 expression in the gingival tissues correlated positively with clinical and microbiological markers of periodontitis, such as PPD and levels of red/orange complex periodontal pathogens. In analyses accounting for PPD, a correlation between GCP-2 and CAL was not found, suggesting that GCP-2 expression reflects current periodontal inflammatory status, rather than cumulative history of periodontitis.
Whilst IL-8 has been shown to be expressed in periodontal pocket epithelium in both periodontal health and disease in order to maintain a continuous neutrophil migration into the sulcus/pocket via interaction with CXC neutrophil receptors (7), our data suggest that GCP-2 expression originates from the microvascular endothelium of inflamed gingival tissue. The observed expression pattern of GCP-2 in periodontal inflammation corroborates earlier findings by Gijsbers et. al (11), who detected pronounced GCP-2 expression in intestinal microvascular endothelium exclusively adjacent to regions with ulcerated or eroded epithelium in specimens from patients with inflammatory bowel disease (11). Since periodontitis is also characterized by loss of epithelial integrity in the pocket (19), the observed up-regulation of GCP-2 in the gingival microvascular endothelium appears to reflect a similar, supplementary neutrophil recruitment mechanism to the site of tissue injury. It is plausible to suggest that, in situations of gingival health or incipient periodontal infection (i.e., in relatively shallow pockets colonized by a low periodontal pathogen burden) microbial metabolic products stimulate junctional and pocket epithelial cells to release IL-8 and recruit a steady stream of neutrophils. In states of more severe disease, characterized by deeper pockets and higher levels of virulent, invading pathogens, the theater of inflammatory warfare shifts from the pocket epithelium to the gingival connective tissue and the vascular endothelium, forming a second line of defense (20). Hence, the enhanced recruitment of neutrophils from the bloodstream to the connective tissue seems to be primarily mediated by GCP-2, as indicated by the increased GCP-2 expression relative to IL-8 expression. Our data therefore suggest that, with increasing severity of the periodontal lesion, GCP-2 neutrophil recruitment in the vascular endothelium is complementary to the IL-8 activity in the pocket epithelium.
Similarly to IL-8, GCP-2 triggers the degranulation of gelatinase B/matrix metalloproteinase (MMP)-9 in neutrophils (3), a proteinase that cleaves IL-8, ENA-78 and GCP-2 at the N-terminal end. This cleavage results in a potentiation of IL-8, and thus a positive feedback (21), generates a biologically inactive form of ENA-78 (22), but does not affect the biological activity of GCP-2, suggesting that GCP-2 can act as a potent chemoattractant for neutrophils even in a MMP-9 rich environment. Our data confirm the merely limited up-regulation of gelatinase B mRNA in periodontally diseased tissue recently reported by another group (23). This is likely due to the fact that neutrophils store gelatinase B in secondary secretory granules for rapid degranulation in an acute phase of inflammation rather than produce this proteinase in the connective tissue (24, 25).
Recently, a causative role of continuous and uncontrolled excessive neutrophil recruitment and activation, resulting in “neutrophil-mediated tissue injury”, has been suggested for localized aggressive periodontitis (LAP) (26, 27). Interestingly, we were unable to demonstrate a significant difference in GCP-2 expression between clinically distinct phenotypes of periodontal disease.
In summary, our data demonstrate a supplementary role for GCP-2 to the established one of IL-8 in enhancing neutrophil recruitment in established periodontitis lesions.
ACKNOWLEDGEMENT
Supported by a NIH grant DE015649 to Dr. Papapanou. Dr. Kebschull was partly supported by a stipend by Neue Gruppe Wissenschaftsfond, Wangen, Germany.
We acknowledge helpful discussions with Dr. Evie Lalla and the kind assistance of Dr. Vicky Woo with the micrographs.
REFERENCES
- 1.Del Fabbro M, Francetti L, Pizzoni L, Weinstein RL. [Congenital neutrophil defects and periodontal diseases]. Minerva Stomatol. 2000;49:293–311. [PubMed] [Google Scholar]
- 2.Kornman KS, Page RC, Tonetti MS. The host response to the microbial challenge in periodontitis: assembling the players. Periodontol 2000. 1997;14:33–53. doi: 10.1111/j.1600-0757.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 3.Proost P, Wuyts A, Conings R, et al. Human and bovine granulocyte chemotactic protein-2: complete amino acid sequence and functional characterization as chemokines. Biochemistry. 1993;32:10170–10177. doi: 10.1021/bi00089a037. [DOI] [PubMed] [Google Scholar]
- 4.Van Damme J, Wuyts A, Froyen G, et al. Granulocyte chemotactic protein-2 and related CXC chemokines: from gene regulation to receptor usage. J Leukoc Biol. 1997;62:563–569. doi: 10.1002/jlb.62.5.563. [DOI] [PubMed] [Google Scholar]
- 5.Wuyts A, Proost P, Lenaerts JP, Ben-Baruch A, Van Damme J, Wang JM. Differential usage of the CXC chemokine receptors 1 and 2 by interleukin-8, granulocyte chemotactic protein-2 and epithelial-cell-derived neutrophil attractant-78. Eur J Biochem. 1998;255:67–73. doi: 10.1046/j.1432-1327.1998.2550067.x. [DOI] [PubMed] [Google Scholar]
- 6.Tonetti MS, Freiburghaus K, Lang NP, Bickel M. Detection of interleukin-8 and matrix metalloproteinases transcripts in healthy and diseased gingival biopsies by RNA/PCR. J Periodontal Res. 1993;28:511–513. doi: 10.1111/j.1600-0765.1993.tb02114.x. [DOI] [PubMed] [Google Scholar]
- 7.Tonetti MS, Imboden MA, Lang NP. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J Periodontol. 1998;69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- 8.Mathur A, Michalowicz B, Castillo M, Aeppli D. Interleukin-1 alpha, interleukin-8 and interferon-alpha levels in gingival crevicular fluid. J Periodontal Res. 1996;31:489–495. doi: 10.1111/j.1600-0765.1996.tb01414.x. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald JE, Kreutzer DL. Localization of interleukin-8 in human gingival tissues. Oral Microbiol Immunol. 1995;10:297–303. doi: 10.1111/j.1399-302x.1995.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 10.Silva TA, Garlet GP, Fukada SY, Silva JS, Cunha FQ. Chemokines in oral inflammatory diseases: apical periodontitis and periodontal disease. J Dent Res. 2007;86:306–319. doi: 10.1177/154405910708600403. [DOI] [PubMed] [Google Scholar]
- 11.Gijsbers K, Van Assche G, Joossens S, et al. CXCR1-binding chemokines in inflammatory bowel diseases: down-regulated IL-8/CXCL8 production by leukocytes in Crohn's disease and selective GCP-2/CXCL6 expression in inflamed intestinal tissue. Eur J Immunol. 2004;34:1992–2000. doi: 10.1002/eji.200324807. [DOI] [PubMed] [Google Scholar]
- 12.Gijsbers K, Gouwy M, Struyf S, et al. GCP-2/CXCL6 synergizes with other endothelial cell-derived chemokines in neutrophil mobilization and is associated with angiogenesis in gastrointestinal tumors. Exp Cell Res. 2005;303:331–342. doi: 10.1016/j.yexcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 13.Rudack C, Maune S, Eble J, Schroeder JM. The primary role in biologic activity of the neutrophil chemokines IL-8 and GRO-alpha in cultured nasal epithelial cells. J Interferon Cytokine Res. 2003;23:113–123. doi: 10.1089/107999003321455507. [DOI] [PubMed] [Google Scholar]
- 14.Rudack C, Hermann W, Eble J, Schroeder JM. Neutrophil chemokines in cultured nasal fibroblasts. Allergy. 2002;57:1159–1164. doi: 10.1034/j.1398-9995.2002.23748.x. [DOI] [PubMed] [Google Scholar]
- 15.Demmer R, Behle JH, Wolf DL, et al. Transcriptomes in Healthy and Diseased Gingival Tissues. J Periodontol. 2008 doi: 10.1902/jop.2008.080139. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papapanou PN, Neiderud AM, Sandros J, Dahlen G. Checkerboard assessments of serum antibodies to oral microbiota as surrogate markers of clinical periodontal status. J Clin Periodontol. 2001;28:103–106. doi: 10.1034/j.1600-051x.2001.280116.x. [DOI] [PubMed] [Google Scholar]
- 17.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. “Checkerboard” DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Liu RK, Cao CF, Meng HX, Gao Y. Polymorphonuclear neutrophils and their mediators in gingival tissues from generalized aggressive periodontitis. J Periodontol. 2001;72:1545–1553. doi: 10.1902/jop.2001.72.11.1545. [DOI] [PubMed] [Google Scholar]
- 20.Heidemann J, Domschke W, Kucharzik T, Maaser C. Intestinal microvascular endothelium and innate immunity in inflammatory bowel disease: a second line of defense? Infect Immun. 2006;74:5425–5432. doi: 10.1128/IAI.00248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van den Steen PE, Proost P, Wuyts A, Van Damme J, Opdenakker G. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-alpha and leaves RANTES and MCP-2 intact. Blood. 2000;96:2673–2681. [PubMed] [Google Scholar]
- 22.Van Den Steen PE, Wuyts A, Husson SJ, Proost P, Van Damme J, Opdenakker G. Gelatinase B/MMP-9 and neutrophil collagenase/MMP-8 process the chemokines human GCP-2/CXCL6, ENA-78/CXCL5 and mouse GCP-2/LIX and modulate their physiological activities. Eur J Biochem. 2003;270:3739–3749. doi: 10.1046/j.1432-1033.2003.03760.x. [DOI] [PubMed] [Google Scholar]
- 23.Kubota T, Itagaki M, Hoshino C, et al. Altered gene expression levels of matrix metalloproteinases and their inhibitors in periodontitis-affected gingival tissue. J Periodontol. 2008;79:166–173. doi: 10.1902/jop.2008.070159. [DOI] [PubMed] [Google Scholar]
- 24.Bainton DF, Ullyot JL, Farquhar MG. The development of neutrophilic polymorphonuclear leukocytes in human bone marrow. J Exp Med. 1971;134:907–934. doi: 10.1084/jem.134.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birkedal-Hansen H. Role of matrix metalloproteinases in human periodontal diseases. J Periodontol. 1993;64:474–484. doi: 10.1902/jop.1993.64.5s.474. [DOI] [PubMed] [Google Scholar]
- 26.Kantarci A, Oyaizu K, Van Dyke TE. Neutrophil-mediated tissue injury in periodontal disease pathogenesis: findings from localized aggressive periodontitis. J Periodontol. 2003;74:66–75. doi: 10.1902/jop.2003.74.1.66. [DOI] [PubMed] [Google Scholar]
- 27.Kantarci A, Van Dyke TE. Lipoxin signaling in neutrophils and their role in periodontal disease. Prostaglandins Leukot Essent Fatty Acids. 2005;73:289–299. doi: 10.1016/j.plefa.2005.05.019. [DOI] [PubMed] [Google Scholar]