Abstract
We describe a detailed and widely applicable method for comprehensive proteomic profiling of the fission yeast Schizosaccharomyces pombe by 2-dimensional high performance liquid chromatography-electrospray ionization-tandem mass spectrometry that demonstrates high sensitivity and robust operation. Steps ranging from the preparation of total proteins, digestion of proteins to peptides, and separation of peptides by two-dimensional (1. strong cation exchange and 2. reversed-phase) high performance liquid chromatography followed by tandem mass spectrometry and data processing have been optimized for our instrumentation platform. Using this technology, we identify ca. 3400 proteins per sample and have identified an estimated 4600 proteins in vegetative cells (equal to ca. 90 % of the predicted S. pombe proteome) at a false discovery rate of ≤ 0.02. Considering the fact that ~500 genes are strongly induced during sexual differentiation, and sexual differentiation was not included in our experiments, the proteomic profiling technique affords what should be virtually complete coverage of the vegetative S. pombe proteome. In addition, these methods are widely applicable, having been used for proteomic profiling of several other organisms.
Keywords: Multidimensional liquid chromatography, tandem mass spectrometry, proteomic profiling, Schizosaccharomyces pombe, spectrum counting
1. Introduction
With the advent of many fully sequenced genomes, there is increased emphasis on a physiological understanding of the information encoded by the genome, much of which is proteins. Whereas large scale techniques such as mRNA expression profiling, genome-wide RNA interference studies, SNP mapping, and chromatin immunoprecipitation linked with comprehensive sequencing are providing the bulk of functional genomics information, it is widely held that a full understanding of the genomic information will also require comprehensive profiling of the cellular proteome.
A proteome is generally considered to be the total protein complement of cells or tissues. “Shotgun” proteomic methods were introduced as a multidimensional protein identification technology (MudPIT) platform by Yates and colleagues [1] and are used to identify and characterize proteins on a large scale. MudPIT, variations thereof, and additional mass spectrometry- (MS) based technologies have proven to be powerful tools for characterization of many different proteomes (reviewed in [2]). Technologies have been developed to examine specific subsets of proteins, for example, those containing post-translational modifications (PTM) such as phosphorylation of S, T and Y residues [3–6]. In addition, analysis of the total proteome may reveal changes in response to specific stimuli, developmental events, or during disease, which are of great interest to biomedical scientists because they may help to elucidate the mechanisms of these processes. Furthermore, the proteins undergoing consistent changes during these biological processes are potential biomarkers.
In this paper, we describe methods for comprehensive proteomic profiling of the fission yeast Schizosaccharomyces pombe by 2-dimensional (2D) high performance liquid chromatography- (HPLC) electrospray ionization- (ESI) tandem MS (MS/MS). This unicellular eukaryote, whose genome is predicted to encode 5,027 proteins [7], is a widely used model system for the study of cell cycle progression, cell division, DNA damage response and repair, stress responses, and mechanisms controlling cell morphology, among others.
We demonstrate a high degree of coverage of the S. pombe proteome (below), and the techniques described herein should also be applicable to proteins derived from many different sources. For example, we have applied the procedures to protein preparations from primary cultured mouse cells (nearly 9,000 protein identifications), whole and enriched fractions of mouse serum, and total protein extracts from the central nervous system of mice. In addition, samples containing decreased protein complexity, including immunoprecipitates, tandem affinity purifications and tissue culture supernatants have been successfully analyzed in our laboratory using the one-dimensional (1D) HPLC-MS/MS procedure comprising the reversed-phase (RP) dimension of peptide separation coupled directly to ESI-MS/MS. This same RP-ESI-MS/MS procedure is used on each fraction from strong cation exchange (SCX) chromatography. SCX is the first dimension of peptide separation of the total proteome of S. pombe, which is the primary focus of the procedures described in this article.
2. Experimental protocols for comprehensive total proteome analyses
2.1. Culture of S. pombe and preparation of total protein
A single colony of the desired S. pombe strain is cultured in sterile YES medium (5 g/l yeast extract (cat. no. BP1422; Fisher Scientific, Inc.), 30 g/l glucose (cat. no. D16–1, Fisher), 150 mg/l adenine (cat. no. A9126, Sigma-Aldrich Chemical Co., St. Louis, MO) and 75 mg/l uracil, (cat. No. U0570, Sigma-Aldrich)) to an absorbance at 260 nm of 0.8–1.0 [8]. The cells are pelleted by centrifugation, supernatant is removed, and cells are frozen immediately in liquid nitrogen and stored at −80 °C. For preparation of the total proteome of S. pombe, cells are kept on ice (0 °C), and 450 μl of ice-cold denaturing lysis buffer containing protease inhibitors is added (10 mM Tris pH 7.4, 0.55% CHAPS (detergent), 7.7 M urea, 2.2 M thiourea, 200 mM dithiothreitol, 5 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 1 mM phenylmethylsulfonylfluoride) [8]. Nitrile gloves, safety goggles, and a lab coat should be worn when handling the lysis buffer, its components and other solutions that include it. Cells are re-suspended by pipetting and transferred to a 1.5 ml microcentrifuge tube containing ca. 0.6 ml of 0.5 mm Zirconium/silica beads (Biospec Inc., Bartlesville, OK; Cat. No. 11079015z). Cell walls and membranes are disrupted in a Fastprep device (FastPrep FP120, Savant Inc., Holbrook, NY) for 40 seconds at setting 6 and the tube is immediately placed on ice. Pressure is released from the tube by loosening the cap, a hole is introduced into the bottom of the tube using a 21-gauge needle, and the tube is centrifuged inside a 15 ml conical bottomed tube (Corning, Inc., Corning, NY) at 5,000 rpm in a clinical centrifuge for 1 min at 4° C to collect the cell homogenate. The cell homogenate is transferred to a 2.0 ml microcentrifuge tube (Sarstedt Inc., Nümbrecht, Germany, part no. 72.694.005), centrifuged at 14,000 rpm, 4° C, and the supernatant (clear cell lysate) is collected. The pellet is extracted once more as described above, starting with the addition of 450 μl lysis buffer followed by the bead disruption and centrifugation steps. The lysates from each individual extraction are pooled. The protein concentration of the cell lysates is estimated using the Bio-Rad Protein Assay (Bio-Rad Inc., Hercules, CA) with lysis buffer as a blank.
To clarified lysate containing ca. 1.5-mg of protein, an equivalent volume of lysis buffer is added. As an optional step, to facilitate relative quantification, purified protein standards from a divergent species, such as bovine serum albumin (BSA) and β-casein (stocks containing 1 nmol/μl; Sigma-Aldrich) can be added as standards to the cell lysates. For example, BSA can be added at 1 nmol/mg yeast protein and β-casein can be added at 1, 2, 3, etc. nmol/mg S. pombe protein for the protein preparations from different treatments, or different strains of the S. pombe cultures. The tubes containing the clarified lysates are incubated at room temperature for 30 min. N, N-dimethylacrylamide (Sigma-Aldrich) is added to 0.5%. The mixture is incubated at room temperature for 30 min, and 20 mM dithiothreitol (DTT; Sigma-Aldrich) from a 2M stock solution is used to quench the reaction for 5 minutes at room temperature [8]. The mixture is centrifuged at 14,000 rpm for 15 minutes at room temperature, and the supernatant is transferred into a fresh 1.5 ml microcentrifuge tube (Sarstedt, part no. 72.692.005).
Methanol-chloroform precipitation of proteins is performed by adding 4 sample-volumes of methanol and vortexing at the maximum speed for 30 sec at room temperature. One sample-volume of chloroform is added and the vortexing is repeated. Three sample-volumes of sterile, MilliQ-purified water (Millipore, Inc., Billerica, MA) are added and the samples are split into two identical aliquots, which are placed in two 2 ml microcentrifuge tubes (Sarstedt). The tubes are centrifuged for 2 minutes at 14,000 rpm. The upper (aqueous) layer is carefully removed, because most of the protein is at the interface of the organic and aqueous phases. Two sample-volumes of methanol are added to each aliquot, the tubes are vortexed and the two aliquots of each sample are pooled in a 2 ml microcentrifuge tube (Sarstedt). Tubes are centrifuged for 2 minutes, 14,000 rpm, at room temperature. Supernatant is removed carefully, the pellet is dried at room temperature and stored at −80° C.
2.2 Digestion of proteins and desalting of peptides
Protein pellets (described above; ca. 1.0 mg) are re-suspended at room temperature in 1.9 ml of 50 mM NH4HCO3 buffer, pH 8, 1 M urea (Chromasolve plus, HPLC grade water (Sigma-Aldrich) is used for this buffer and all subsequent aqueous solutions in this paper) by vortexing at a medium setting (6, on a scale of 1–10) for 30 min. Tubes are sonicated in a water bath sonicator (Fisher, model FS30) for 20 min, vortexed for 30 min, sonicated for another 25 min and vortexed for another 45 min. Between steps, full-strength vortexing of the mixture is performed for 30 sec. Portions of the pellet resist re-suspension, and the mixture appears cloudy. However, following trypsin digestion, a clear suspension is obtained. For digestion, 40 μl (20 μg) of sequencing grade modified trypsin (Promega Inc., Madison, WI) is added and digests are placed on an orbital incubator (Thermomixer, Eppendorf, Enfield, CT) overnight at 37 °C, 700 rpm.
Peptides are desalted using Sep Pak Plus C18 cartridges (Waters Inc, Milford, MA; part # Wat020515) connected to a 10 ml luer-lock syringe. The syringe is washed twice with 10 ml of acetonitrile (JT Baker; Mallinckrodt Baker, Phillipsburg, NJ) and twice with 10 ml of 50% acetonitrile/0.05% formic acid (formic acid from EMD Biosciences, San Diego, CA). Nitrile gloves, safety goggles, and a lab coat should be worn when handling formic acid and acetonitrile, and these chemicals should be used in a fume hood. The cartridge is washed twice with 10 ml of 50% acetonitrile/0.05% formic acid and once with 10 ml of 0.1% formic acid at a flow rate of ca. 5 ml/min. Two ml of 0.1% formic acid, followed by the sample are drawn into the syringe and loaded onto the cartridge at a flow rate of ca. 2 ml/min. Peptides are desalted using 2 × 10 ml of 0.1% formic acid (5 ml/min) followed by elution into a 2 ml microcentrifuge tube (Sarstedt) using 1.8 ml of 50% acetonitrile/0.05% formic acid at ca. 2 ml/min. The needle (21 gauge) is used to poke two holes in the cap of the microcentrifuge tube to allow vapors to escape from the tube under vacuum and to minimize sample loss due to bumping (when samples can be dispersed as they dry). Samples are dried overnight at 35 °C in a speed vac (model UVS400 with model SPD111V vacuum centrifuge; Thermo Electron (now Thermo Fisher Scientific) Inc., San Jose, CA). Following drying, the tube containing the peptides is sealed with a new cap and stored at −80 °C until re-suspension prior to SCX separation.
2.3 Separation of peptides using SCX
The first dimension of peptide separation is via SCX. Peptides from ca. 1 mg of protein (above) are re-suspended in 200 μl of 95% solvent C/5% solvent D (Solvent C = 5% acetonitrile/0.1% formic acid in H2O; Solvent D = 25% acetonitrile/0.1% formic acid in H2O, containing 500 mM KCl (Sigma-Aldrich)). The peptide mixture is vortexed at maximum speed for 30 s, vortexed at setting 6 for 30 min at room temperature, vortexed at maximum speed for 30 s and sonicated for 20 min. The peptide mixture is centrifuged for 10 min at 14,000 rpm, room temperature. The supernatant containing soluble peptides is transferred to a deactivated, Qsert snap cap glass sample vial, which includes caps with PTFR/silicone septa (Waters, P/N 186001124DV), sealed and immediately stored at 4 °C. Peptides are separated using SCX as soon as possible after preparation, within 1 hr or less, as described below. A potential effect of storing the fully prepared peptides longer before SCX separation has not been investigated.
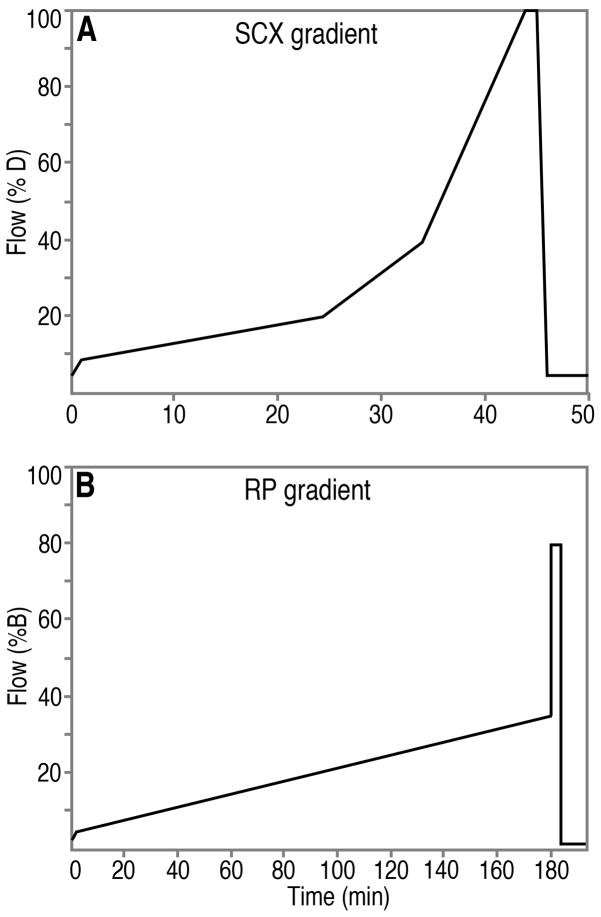
Beginning approximately 2 hr before peptides are re-suspended, preparation for SCX separation is initiated using automated Paradigm Multi-Dimensional Liquid Chromatography (MDLC) instrumentation, which includes an autosampler/fraction collector, HPLC and UV detector (Fig. 1; Michrom Bioresources, Inc., Auburn, CA). Xcalibur v. 2.0.6 software (Thermo Electron) with custom modifications (Michrom) controls all of the instrument functions. The SCX column (polysulfoethyl A, 5 μM beads, 200 Å pore size; 2 × 150 mm, PolyLC, Inc., Columbia, MD) is prepared for the SCX separation by washing with 40:40:20 isopropanol:acetonitrile:H20 (40:40:20 wash solvent) for 50 min at a flow rate of 200 μl/min. Subsequently, 4 blank SCX gradients are performed using solvents C and D, (at 200 μl/min). The SCX gradient proceeds from 5% (solvent) D to 9% D from 0 to 1.0 min, 9% D to 20% D from 1.1 to 24.0 min, 20% D to 40% D from 24.1 to 34.0 min, from 40% D to 100% D from 34.1 to 44.0 min, 100%D from 44.1 to 45.0 min, 100% D to 5% D from 45.1 to 46.0 min and 5% D from 46.1 to 50.0 min (Fig. 2A). Immediately following completion of the blank gradients, 10 μl of 95% solvent C/5% solvent D is drawn into the syringe (pre-sample solvent to facilitate complete sample introduction into the sample loop) followed by 400 μg (80 μl) of the re-suspended peptides (in 95% solvent C/5% solvent D; above). This sample (90 μl) is introduced into the sample injection port of the autosampler (Fig. 1A) and pushed into the sample loop, through PEEKsil tubing, via the force generated by the syringe (Fig. 3A). (Remaining sample is stored at −80 °C, and could be re-injected, but has not been used because of the reliability of the SCX separation.) After sample introduction, HPLC valve 1 switches, the sample is loaded onto the SCX column via the flow from pumps C and D (Fig. 3B), and the peptides are separated using the SCX gradient (Fig. 2A). Offline collection of 24 × 400 μl fractions is performed using 24 × 750 μl vials (Sun-sri, Inc., www.sun-sri.com, part number 501 307; Fig. 1B, 3B). Each SCX fraction (400 μl, 2 min/fraction) is manually capped (Sun-sri, part number 501 318) within several minutes of collection and stored in an autosampler/fraction collector drawer (Fig. 1B, 1C, 1D) at 4 °C. An example of a typical trace of an SCX separation, in which absorbance at 214 nm (A214) was recorded, is shown in Fig. 4A. The procedures result in reproducible A214 profiles, with only small variations. The A214 values throughout the SCX separation are an approximate gauge of the quantity of peptides in the SCX fractions during the corresponding time ranges. Good separation of the population of peptides is obtained using this protocol, as demonstrated below.
Figure 1.
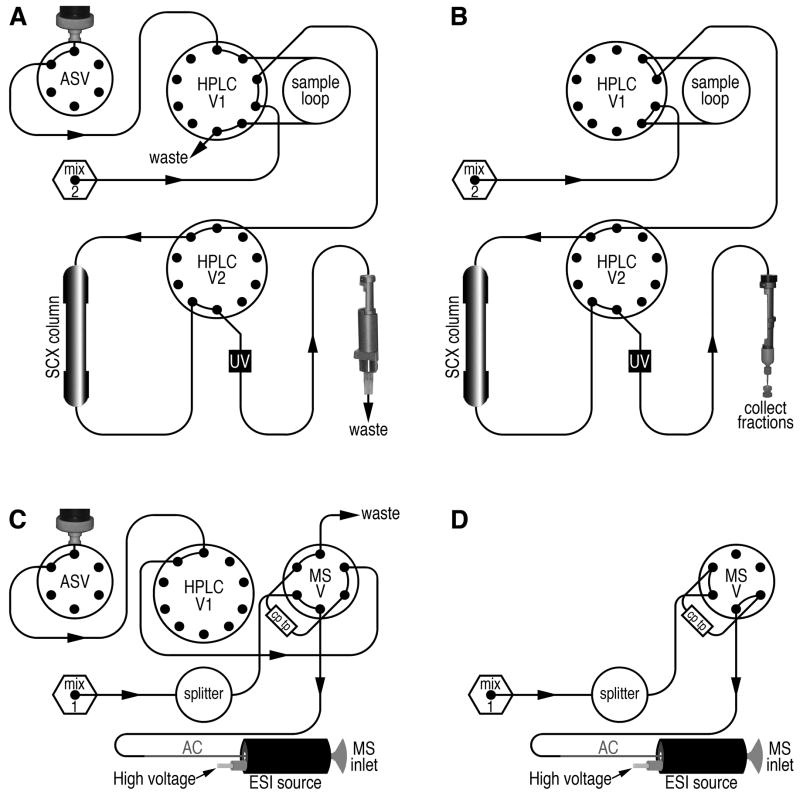
Instrumentation platform used in the proteomic analyses. (A) Sample injection, from the syringe mounted on the autosampler, into the sample injection port (red arrow). Samples for SCX and RP separations are introduced into the HPLC at this point. (B) Offline collection of SCX fractions using the fraction collection tool (blue and tan), which is magnetically suspended from the autosampler arm (black cylinder). The SCX eluate drips into open vials (red arrow). (C) Syringe drawing up a portion of a SCX fraction (black arrow) for subsequent re-injection into the sample injection port (shown in panel A), for RP HPLC-MS/MS analysis. Scale bars represent 2.5 cm (A–C). (D) Composite instrumentation platform, including the UV detector (UV); solvent bottle holder on top of the module controlling gas flow and choice of 40:40:20 isoproponal:acetonitrile:water (40:40:20 wash solvent) or HPLC solvents (solv.); HPLC pumps/mixers/splitter for solvents A/B and solvents C/D (pumps A/B use a splitter but C/D do not) and 2 valves (HPLC) with the necessary plumbing (PEEK and PEEKsil tubing), SCX and immobilized metal affinity chromatography (IMAC) columns attached (the IMAC application for phosphopeptide enrichment will be described elsewhere); the sample loop mounted on HPLC valve 1 (loop); the sample injection port (port); polymeric trap for peptide capture and desalting (RP-pep. capture) on the valve that is mounted on the mass spectrometer; the RP analytical column (RP column) mounted in the ESI source (ESI); the autosampler/fraction collector containing refrigerated drawers for reagent, sample and SCX fraction storage; and the hybrid linear ion trap- (ion trap) OrbitrapXL (Orbitrap) mass spectrometer; scale bar represents 10 cm.
Figure 2.
HPLC gradient profiles for the SCX separation (A) and RP separation (B) of peptides are shown. For the SCX separation, solvents C and D are used, and for the RP separation, solvents A and B are used. Solvent composition is described in the text.
Figure 3.
Simplified schematic diagram of the plumbing for liquid flow through the HPLC, autosampler, UV detector flow cell, SCX and RP columns. The flow paths that are used for the separations described in this article are all shown. All tubing is 1/16” outer diameter (OD) PEEK, with varied inner diameters, with the exception that PEEKsil tubing (also 1/16” OD) is used for all sample flow paths. Tubing was obtained primarily from Michrom and also from Upchurch Scientific (Seattle, WA). (A) Flow paths for sample introduction and SCX column stabilization, immediately prior to SCX separation are shown. Abbreviations: ASV, autosampler valve; HPLC V1, HPLC valve 1; mix 2, mixer 2 (for solvents C and D); HPLC V2, HPLC valve 2; UV, UV detector flow cell. Two flow paths, marked by arrows, are active at this stage: 1) Flow of the sample, from the syringe, for sample introduction into the (100 μl) sample loop, and 2) flow from pumps C and D (not shown for clarity), through mixer 2, for stabilization of solvent flow and pressure through the SCX column prior to the SCX separation. (B) Flow path during the SCX separation, which no longer involves sample introduction. To enter this stage following that shown in panel A, HPLC valve 1 switches, resulting in reversal of the sample flow back out of the sample loop and onto the SCX column. Concurrent to the switching of valve 1, the autosampler automatically suspends the fraction collection tool above the vials (Fig. 1B), in order to collect the eluate from the SCX column. The fraction collection tool is automatically moved to a fresh vial every 2.0 min, resulting in collection of 24 × 400 μl fractions. The SCX gradient, consisting of solvents C and D (see the text and Fig. 2A), is applied, resulting in peptide separation throughout the gradient. (C) Flow paths for sample introduction immediately prior to reversed phase (RP) separation; abbreviations: MSV, valve mounted on the mass spectrometer; mix 1, mixer 1 (for solvents A and B); cp tp, capillary peptide trap (polymeric); and AC, C18 reversed-phase, capillary-scale (1/16″ OD) analytical column. Two flow paths, marked by arrows, are active at this stage: 1) Flow of the sample, from the syringe, for sample introduction onto the peptide trap (cp tp) followed by desalting of the captured peptides, and 2) flow from pumps A and B (not shown for clarity), through mixer 1, for stabilization of solvent flow and pressure through the C18 analytical column (AC). The high voltage (1.8 kV) is manually activated at the start of this stage using the tune page, which is part of the control software for the mass spectrometer. (D) Flow path during the reversed-phase separation, which no longer involves sample introduction. To enter this stage following that shown in panel C, the valve mounted on the mass spectrometer switches, resulting in reversal of the solvent flow back through the peptide trap, gradient elution of the peptides off of the peptide trap, their separation in the analytical column, elution, and ionization in the ESI source, just prior to introduction into the mass spectrometer.
Figure 4.
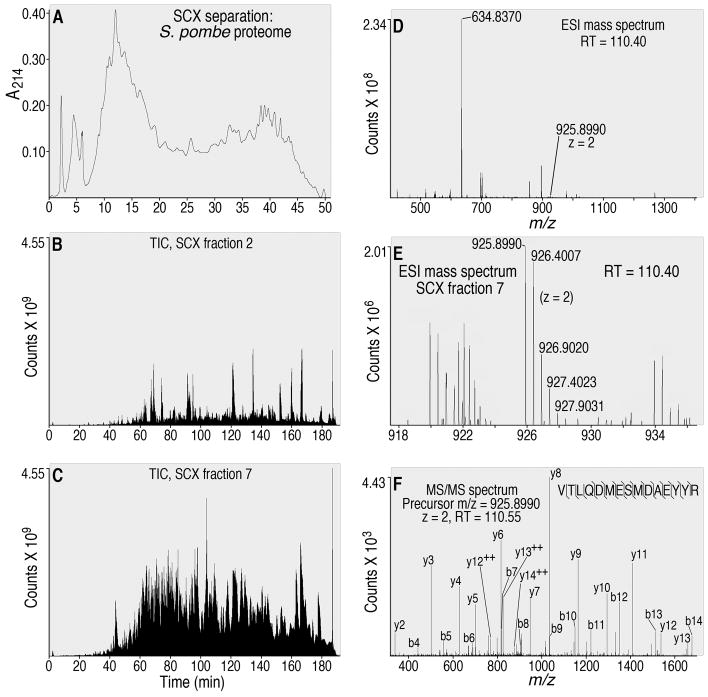
Representative results of a 2D HPLC-MS/MS analysis of the S. pombe proteome. (A) Typical trace of a SCX separation of tryptic peptides; A214 = absorbance at 214 nm. (B) Total ion chromatogram (TIC) resulting from re-injection of 10% of SCX fraction 2 and (C) SCX fraction 7 for RP HPLC-MS/MS analysis. The number of counts recorded by the mass spectrometer, in arbitrary units (Thermo Fisher Scientific), is shown. (D) The entire m/z range (the instrument is programmed to scan from m/z 300 to 2,000, but most scans contain a portion of this range) of an ESI mass spectrum resulting from re-injection of SCX fraction 7, at an RP retention time of 110.40 min. (E) The m/z range from 918 to 936 of the same ESI mass spectrum as in (D). Note the doubly charged ion with a measured monoisotopic m/z of 925.8990 and a theoretical monoisotopic m/z of 925.9007, which demonstrated 1.8 ppm mass accuracy in this scan for this ion. The isotopes of this same ion, with an increased mass due to the natural incorporation of 1, 2, 3 or 4 heavy atoms are also labeled. (F) MS/MS spectrum resulting from collision-induced dissociation (CID) of the precursor ion with an m/z of 925.8990 (shown in panel E). This MS/MS spectrum, when searched against the S. pombe proteome, demonstrated that the precursor ion (measured m/z of 925.8990) was derived from the pub1 ubiquitin protein ligase E3 (accession number SPAC11G7.02). b- and y product ions obtained upon CID are labeled, as are sites in the peptide sequence that were detectably fragmented via CID.
2.4 Reversed-phase HPLC-tandem mass spectrometry
An LTQ OrbitrapXL mass spectrometer (Fig. 1D; Thermo Fisher Scientific) is used as the primary analytical instrument in this analysis. Caution should be exercised to avoid contact with the high voltage cable of the mass spectrometer during operation, to prevent electric shock hazards. To optimize performance, calibration of the mass spectrometer is performed monthly using positive ion mode calibration mix, and mass calibration of the Orbitrap is performed every three days, according to the manufacturer (Thermo Fisher Scientific). Although a variety of standards can be used for Orbitrap mass calibration, including the positive ion mode calibration mix, we use MALDI peptide standards II (Bruker Daltonics, Inc., Billerica, MA; part number 222570). The peptide standards are re-suspended in 125 μl of 0.1% formic acid. Caffeine (5 ng/ml final concentration; Sigma-Aldrich) is added to the peptide standards, and the final volume of the standards mixture (peptides plus caffeine) is 2.5 ml in 50% methanol/0.05% formic acid. For mass calibration, the standard mixture is infused at 2 μl/min through an ADVANCE ionization source (Michrom). Tuning is also performed every time the Orbitrap mass accuracy is calibrated. Tuning is on the 2+ ion from Substance P (peptide sequence RPKPQQFFG, with a mass to charge ratio (m/z) of 674.37134, although other ions may be chosen) for optimum sensitivity, and only increases in signal are saved in the tune file that is used with the MS/MS method (described below). Tuning is repeated (typically 3–4 times) until no further gains in signal are obtained.
For sample introduction into the mass spectrometer, the autosampler opens a refrigerated drawer (internal temperature of 4 °C; Fig. 1D) containing the specified SCX fraction, draws up a sample of the peptide mixture from the fraction into the syringe (40 of the 400 μl; Fig. 1C), closes the refrigerated drawer, introduces the peptides into the injection port (Fig. 1A, 1D, 3C), where they are pushed to and captured by a polymeric peptide trap with retention characteristics of C8 RP media (Michrom; Fig. 1D, 3C).
SCX fractions later in the gradient contain more acetonitrile than early fractions, because they contain more solvent D, which contains 25% acetonitrile, in contrast with solvent C, which contains 5% acetonitrile (see Fig. 2A). Thus, hydrophilic peptides in later SCX fractions could be captured inefficiently by the peptide trap due to the presence of an increased concentration of acetonitrile. To address this possibility, we partially dried the later SCX fractions to ca. 50% of their initial volume, which could deplete the more volatile acetonitrile. These SCX fractions were returned to their original volume with 0.1% formic acid in water. Although different samples could behave differently, we did not detect an effect of partial dry-down on the number of peptide and protein identifications, nor on the total ion chromatogram (TIC) profile (described below), consistent with the proposal that peptides from the later SCX fractions were not lost due to more acetonitrile in Solvent D than in Solvent C.
To desalt the peptides on the C8 trap, the autosampler injects 50 μl of a mixture of 98% solvent A/2% solvent B (Fig. 3C), which flows through the C8 peptide trap to desalt the peptides that are bound to the C8 trap. (Solvent A = 0.1% formic acid in water and solvent B = 100% acetonitrile). The remainder (360 μl) of each SCX fraction can be stored at 4 °C for several months, and longer-term storage is at −80 °C. After thawing of the fractions, a minor decrease in the number of protein identifications (the identifications are via peptides derived from the proteins) has occurred with some of the SCX fractions, so it is advisable to promptly run replicate runs, if required.
Following sample loading and desalting on the peptide trap, the valve on the mass spectrometer is switched to place the trap inline with the analytical column (150 mm X 0.2 mm Magic C18, 3 um particles, 200 Å pores; Michrom; Fig. 3D). An RP gradient, consisting of 2–5% solvent B from 0 to 2.0 min, 5%–35% B from 2.1 to 180.0 min, 80% B from 180.1 to 184.0 min and 2% B from 184.1 to 194.1 min, is applied (Fig. 2B). Peptide carryover and fouling of the RP media is minimized by washing the peptide trap and analytical column with 40:40:20 wash solvent and RP blanks, in which 40:40:20 wash solvent is injected in place of sample and the 180 min gradient is compressed to 15 min in order to reserve the majority of the instrument time for data collection. An ADVANCE ESI source (Michrom) is used to ionize the peptides as they elute from the RP column and are introduced into the mass spectrometer (Fig. 3D).
The LTQ OrbitrapXL is programmed to scan the precursor ions, with an m/z ratio of 300–2000, in the Orbitrap at a resolution of 60,000 in the profile mode. Use of the profile mode enables use of the data with label-free quantification software such as DeCyder MS (GE Healthcare, Chalfont St. Giles, United Kingdom) and SIEVE (Thermo Fisher Scientific). A top-4 data-dependent MS/MS method is used. Data-dependence means that the most abundant ions from each precursor ion scan (4 for this method) are individually selected for collision-induced dissociation (CID) and MS/MS scanning. Precursor charge state screening, monoisotopic precursor selection and dynamic exclusion are enabled. Dynamic exclusion specifies a repeat count of 2 and exclusion duration of 2.0 min. The amount of signal obtained from injection of samples contained in SCX fractions, monitored as the TIC, is relatively low in the first 3 SCX fractions (e.g. Fig. 4B) and the final 3 SCX fractions (data not shown), and is higher in the rest of the SCX fractions throughout the remainder of the SCX separation (e.g. SCX fraction 7, Fig. 4C). When signal from the mass spectrometer is monitored as a basal ion chromatogram, the profiles are similar to the TIC profiles (data not shown).
Throughout the RP gradient, approximately 14,000 spectra are recorded by the mass spectrometer. For example, an ESI mass spectrum (Fig. 4D) contained a doubly charged ion with a measured m/z of 925.8990 (1.8 ppm mass accuracy; Fig. 4E) that was ca. 100-fold less intense than the highest peak (i.e. base peak) in this spectrum (m/z 634.8370). When the doubly charged ion with a measured m/z of 925.8990 was fragmented using CID in the linear ion trap, the MS/MS scan, also recorded by the linear ion trap, revealed a mixture of fragment ions that could be clearly assigned to a specific peptide from a ubiquitin ligase (Fig. 4F). Thousands of additional MS/MS spectra are similarly assigned to peptides contained in the SCX fractions, using database searching and post-search processing.
2.5 Searching the raw MS data and post-search processing
The raw data is searched against an S. pombe protein database (http://www.sanger.ac.uk/Projects/S_pombe/protein_download.shtml) using Sorcerer™-SEQUEST® (SageN Research, Inc., San Jose, CA). The peptide mass tolerance is set to 10.0 ppm, fragment mass type to monoisotopic, and static Cys carboxymethylation (+ 58.005478 atomic mass units (amu)) as well as differential Met oxidation (+ 15.99492 amu) are specified. Although there are dozens of additional modifications to amino acid residues that can be specified, we have found that differential modification of Lys with tags from ubiquitination (GG, + 114.042931 amu and LRGG, + 382.228088 amu; [9]), and Ser, Thr and Tyr phosphorylation (+ 79.966331 amu) can be specified with minimal effects on the number of peptide/protein identifications. However, specification of additional differential modifications requires additional database search time. Moreover, the precise masses of the modifications are used because the accuracy of the Orbitrap mass analyzer is typically within ±2 ppm, and Sorcerer™-SEQUEST® easily accommodates these precise masses.
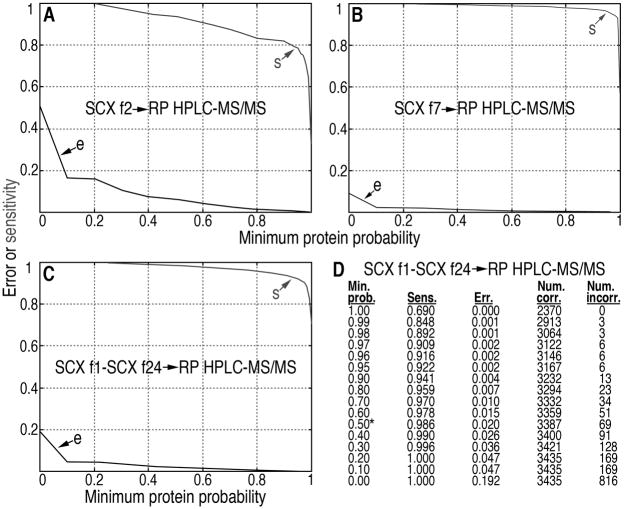
Post-search analysis is via the Trans-Proteomic Pipeline (TPP; Institute for Systems Biology, Seattle, WA). TPP is accessed from Sorcerer-SEQUEST. The results of the searches, including probabilities for correct assignments, are viewed using the pepXML Viewer with a focus on peptides, and by using the ProteinProphet® protXML Viewer with a focus on the peptides and the proteins that contain these peptides. The PepXML Viewer and the ProteinProphet protXML Viewer are tools of the TPP. Results from the PepXML viewer and ProteinProphet ProtXML viewer are both exported to Excel spreadsheets for archiving and further analyses. To export very large lists of peptides and proteins to Excel Spreadsheets, Qtools, which is a collection of computational tools that we have developed, are used. QTools are available from the authors upon request. In addition, a link is provided by the ProteinProphet ProtXML viewer for viewing graphs (from the TPP), which display the predicted sensitivity (fraction of all correct proteins with probabilities ≥ the minimum protein probability) and error (also known as the false discovery rate (FDR), which is the fraction of all proteins with probabilities ≥ the minimum protein probability that are incorrect) as defined by the TPP. Representative results are shown for the analyses of SCX fractions 2 and 7 (Fig. 5A, B, showing the same SCX fractions analyzed in Fig 4). For SCX fraction 2, modest decreases in sensitivity, and decreases in error (FDR) occur as the minimum protein probability increases (Fig. 5A). For SCX fraction 7, minimal decreases in sensitivity, and minimal decreases in error (FDR) occur as the minimum protein probability increases (Fig. 5B). Patterns similar to those in Fig. 5A are observed for early and late SCX fractions containing a relatively small number of peptides, potentially due to a relatively high proportion of noisy MS/MS spectra during analysis of the early and late SCX fractions. In contrast, SCX fractions containing relatively large numbers of peptides (the rest of the SCX fractions) typically show patterns similar to that in Fig. 5B. For a detailed description of parameters and methods used to calculate peptide and protein probabilities, see [10, 11].
Figure 5.
Error, sensitivity and number of proteins are shown for protein identification vs. minimum protein probability, as predicted by ProteinProphet®, for the MS/MS data resulting from analysis of a typical sample of the total proteome of S. pombe. (A) Error (e) and sensitivity (s) are shown for injection of 10% of SCX fraction 2 onto RP media, separation using the RP gradient, coupled to ESI-MS/MS (SCX f2->RP HPLC-MS/MS); (B) the same analysis using SCX fraction 7 (SCX f7->RP HPLC-MS/MS) or the same analysis using the data from SCX fractions 1–24 (all fractions) searched together in a composite search of all the data from the sample (C; SCX f1-SCX f24->RP HPLC-MS/MS). (D) The ProteinProphet® Sensitivity/Error Info shows the protein identification statistics from a database search of the composite dataset (re-injection of SCX fractions 1–24) for the sample as a function of minimum protein probability (Min. prob.). The sensitivity of protein identification (Sens.) is the number of proteins identified divided by the total predicted number of correct proteins (3435 for this data set); error (Err.) is the estimated protein FDR at the given minimum protein probabilities; number correct (Num. corr.) = estimated number of correct protein identifications at the given minimum protein probabilities and number incorrect (Num. incorr.) = estimated number of incorrect protein identifications at the given minimum protein probabilities. At a minimum protein probability of 0.50 (0.50*), the FDR is estimated at 0.020 for this data set.
ProteinProphet allows user specification of the false discovery rate (FDR), as implied by Fig. 5. The FDR is set deliberately, within a reasonable range. We have seen that values of ca. 0.001 to 0.05 are often specified, largely depending on the goals of the experiment, and no widely accepted standard FDR seems to exist currently. In order to obtain high proteome coverage at a relatively low FDR, we accept protein identifications with a FDR of 0.02 or less in whole proteome profiling studies. In order to monitor the performance of the 2D HPLC-MS/MS procedure, the ProteinProphet statistics for all of the individual SCX fraction re-injections, at a FDR ≤ 0.02, are monitored. A representative set of these statistics is shown in Table 1. SCX fractions 1–3 as well as 23–24 yield a lower number of peptide (and hence protein) identifications, whereas the rest of the SCX fractions have larger numbers of identifications. The peak number of protein identifications is typically observed in SCX fraction 8 (1341 in the example shown in Table 1, which is from the same analysis as the results presented in Fig. 4 and 5). Because most of the SCX fractions yield a large number of identifications, the SCX gradient is proposed to effectively separate the complex population of peptides from the S. pombe proteome. These results also suggest that the RP HPLC-MS/MS platform effectively identifies large numbers of peptides (and hence proteins).
Table 1.
Summary of the results of a 2D-HPLC MS/MS analysis of the S. pombe proteome*
| Frac. | # of entries | Min. Prob. | Sensit | Error | #corr. | #incorr. |
|---|---|---|---|---|---|---|
| 1 | 332 | 0.50 | 0.984 | 0.016 | 327 | 5 |
| 2 | 173 | 0.90 | 0.795 | 0.014 | 171 | 2 |
| 3 | 288 | 0.90 | 0.800 | 0.019 | 283 | 5 |
| 4 | 499 | 0.60 | 0.980 | 0.017 | 490 | 8 |
| 5 | 1009 | 0.10 | 1.000 | 0.013 | 996 | 13 |
| 6 | 1220 | 0.10 | 1.000 | 0.020 | 1196 | 24 |
| 7 | 1255 | 0.50 | 0.995 | 0.018 | 1233 | 23 |
| 8 | 1341 | 0.60 | 0.980 | 0.017 | 1317 | 23 |
| 9 | 1323 | 0.70 | 0.962 | 0.017 | 1302 | 23 |
| 10 | 1257 | 0.80 | 0.934 | 0.015 | 1238 | 19 |
| 11 | 1113 | 0.80 | 0.939 | 0.017 | 1095 | 19 |
| 12 | 991 | 0.70 | 0.959 | 0.020 | 971 | 20 |
| 13 | 1022 | 0.70 | 0.969 | 0.017 | 1005 | 17 |
| 14 | 1164 | 0.60 | 0.983 | 0.017 | 1145 | 20 |
| 15 | 1235 | 0.60 | 0.978 | 0.019 | 1212 | 23 |
| 16 | 1273 | 0.60 | 0.982 | 0.019 | 1249 | 24 |
| 17 | 1252 | 0.60 | 0.976 | 0.017 | 1231 | 21 |
| 18 | 1152 | 0.70 | 0.959 | 0.019 | 1130 | 22 |
| 19 | 976 | 0.70 | 0.941 | 0.020 | 957 | 20 |
| 20 | 918 | 0.70 | 0.946 | 0.020 | 899 | 18 |
| 21 | 929 | 0.70 | 0.960 | 0.019 | 912 | 18 |
| 22 | 723 | 0.80 | 0.912 | 0.016 | 712 | 12 |
| 23 | 525 | 0.80 | 0.899 | 0.019 | 515 | 10 |
| 24 | 302 | 0.80 | 0.924 | 0.020 | 296 | 6 |
Abbreviations and definitions: Frac., SCX fraction that was re-injected to RP HPLC-MS/MS; # of entries, number of protein database entries (proteins) identified; Min. Prob., minimum probability that a protein identification is correct at a protein FDR of 0.02 or less; Sensit, sensitivity of identification of proteins at a protein FDR of 0.02 or less, i.e. the number of proteins identified divided by the total predicted number of correct entries; Error = estimated protein FDR (0.02 or less, and dependent on the minimum protein probability, which in turn is specified in increments of 0.01 to 0.1, depending on the range; see Fig. 5D); #corr, number correct = estimated number of correct protein identifications at the given FDR (0.02 or less); #incorr., number incorrect = estimated number of incorrect protein identifications at the given FDR (0.02 or less)
The total protein number identified from the sample is obtained from a composite search of the data, which includes all 24 SCX fraction re-injections for RP HPLC-MS/MS analysis, and a FDR of 0.02 or less is also specified (Fig. 5C, D). In this example, which is typical, 3,387 S. pombe proteins are required to explain the peptide identifications at the specified FDR. Because ProteinProphet groups the peptide data into the minimal number of proteins that are expected to be present, but some of the peptide sequences that are present belong to 2 or more proteins [11], the analysis also revealed that 4,249 of the predicted 5,027 protein entries (84.5%) from the S. pombe protein database were identified in this analysis. The sensitivity of this composite analysis was high and the error was low. Although the analysis of the data is ongoing, we estimate that by combining data from four repeat runs of S. pombe protein lysate, we may detect a total of approximately 4600 S. pombe proteins, which corresponds to approximately 92% of the predicted proteome. Since only the vegetative cell state was sampled in our experiments, this is likely to represent virtually complete proteome coverage. A recent study detected 4,399 proteins from Saccharomyces cerevisiae (budding yeast) with high confidence and similarly concluded that this number covered essentially the entire proteome of log phase cells [12].
2.6 Semi-quantitative analyses of the S. pombe proteome using spectral counting
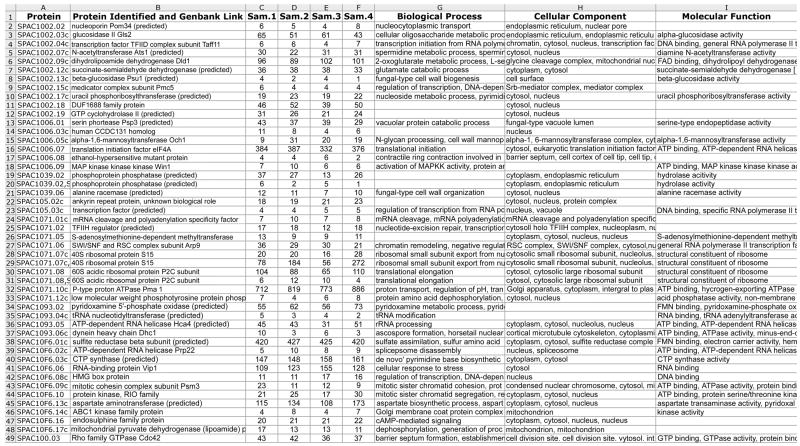
The data acquired by RP HPLC-MS/MS also contains quantitative information about protein abundance. For example, spectral counts are the number of times an ionized peptide from an ESI mass spectrum is selected by the mass spectrometer (in a data-dependent fashion) for fragmentation and scanning as an MS/MS scan. Relative numbers of spectral counts for individual proteins (derived from the proteins’ peptides) among samples are a semi-quantitative estimate of the relative abundance of these proteins [13]. Spectral counts are listed in ProteinProphet output, and QTools are also used to compute spectral counts of the proteins from the S. pombe samples using Peptide Prophet output. These QTools are Visual Basic macros for automated spectral count analysis, and include assignment of functions of the proteins using the Gene Ontology (http://www.geneontology.org/index.shtml) and other computational proteomic data analysis tools. A representative sample of spectrum counts measured in 4 different total protein samples from S. pombe, from the combined, composite analysis of all 24 SCX fractions that were analyzed using RP HPLC-MS/MS, is shown in Fig. 6. Those proteins with a high number of spectral counts are typically highly abundant, whereas proteins with fewer spectral counts are typically at lower abundance. It is difficult to state the degree of difference in spectrum counts among samples that reflects a significant difference in the quantity of the protein, but recent studies have reported statistical tools with an improved ability to assess the statistical significance of potential differences in spectral counts of proteins among samples [14]. We also found that spectral counts for proteins with a spectral count of 10 or greater are similar when these samples are analyzed in replicate analyses.
Figure 6.
Representative view of QTools output showing spectral counts of S. pombe proteins from 4 different samples in an experiment, each of which was subjected to 2D HPLC-MS/MS analyses. The protein identifier and description were derived from the S. pombe protein database (described in the text), and the numbers in the sample (Sam.) columns are the number of spectral counts from all of the peptides belonging to the identified proteins. The biological processes, cellular components and molecular functions, when known, are also shown.
3. Concluding remarks
Large-scale proteomic analyses require high quality sample preparation and analysis using sophisticated HPLC and mass spectrometry instrumentation. In this paper, we have described an analytical platform, including the methods that we routinely use for profiling of the S. pombe proteome, which reliably result in identification of the majority of the predicted S. pombe proteins from total proteome samples. These methods employ offline SCX separation of a highly complex mixture of peptides from the entire S. pombe proteome, to simplify the mixture of peptides contained in each fraction, followed by RP HPLC-ESI-MS/MS and database searches to identify the peptides contained in the SCX fractions. The peptides are, in turn, assigned to the proteins from which they originated, demonstrating that this is a “bottom up” proteomic approach. A FDR of ≤ 0.02 is specified in order to obtain high proteome coverage at what we consider to be an acceptable accuracy of identification. Moreover, spectral counts from the MS/MS data are used for semi-quantitative assessment, in order to identify potentially differentially expressed proteins. In conclusion, our study highlights the applicability of SCX and highly sensitive RP HPLC-ESI-MS/MS to comprehensive profiling of a eukaryotic proteome.
Acknowledgments
This work was funded by NIH grant R01 GM059780 to D.A.W., by the Burnham Institute for Medical Research NCI Cancer Center Support Grant 5 P30 CA30199-28 and by the La Jolla Interdisciplinary Neuroscience Center Cores Grant 5 P30 NS057096 from NINDS. We thank Kerry Nugent, Peter Kent, Lori Ann Upton and Dave Mintline for custom configuration of the HPLC hardware and system software, Patrick Chu and David Chang for assistance with Sorcerer-SEQUEST applications, and Dr. Ali Iranli for his efforts in developing QTools.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 2.Cravatt BF, Simon GM, Yates JR., 3rd Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 3.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Nat Methods. 2007;4:231–7. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 4.Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC. Anal Chem. 2004;76:2763–72. doi: 10.1021/ac035352d. [DOI] [PubMed] [Google Scholar]
- 5.Ficarro SB, Salomon AR, Brill LM, Mason DE, Stettler-Gill M, Brock A, Peters EC. Rapid Commun Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 6.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Mol Cell Proteomics. 2005;4:310–27. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Nature. 2008;453:1239–43. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt MW, Houseman A, Ivanov AR, Wolf DA. Mol Syst Biol. 2007;3:79. doi: 10.1038/msb4100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Proteomics. 2007;7:868–74. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- 10.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 11.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 12.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Nature. 2008;455:1251–4. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 14.Choi H, Fermin D, Nesvizhskii AI. Mol Cell Proteomics. 2008;7:2373–85. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]