Abstract
A senescent CD4+CD28− T cell subset develops with aging and in chronic inflammatory diseases like rheumatoid arthritis, and is implicated in plaque rupture and myocardial infarctions. This subset is pro-inflammatory, cytotoxic for endothelial cells, and aberrantly expresses genes like CD70, perforin and killer-cell immunoglobulin-like receptor (KIR) genes. Why CD4+CD28− cells overexpress these genes is unclear. We found that the CD70, perforin and KIR2DL4 promoters are demethylated in CD4+CD28− T cells, and that DNA methyltransferase 1 (Dnmt1) and Dnmt3a levels are decreased in this subset. siRNA “knockdown” of Dnmt1, but not Dnmt3a, in CD4+CD28+ T cells caused similar demethylation and overexpression of KIR2DL4, perforin and CD70, while simultaneous knockdown of Dnmt1 and Dnmt3a caused greater demethylation and overexpression of these genes than Dnmt1 alone. We conclude that decreased Dnmt1 and Dnmt3a causes demethylation and overexpression of these and perhaps other genes in CD4+CD28− cells, potentially contributing to pathologic functions by this subset.
Keywords: T cell, DNA methylation, aging, rheumatoid arthritis, senescence
INTRODUCTION
Chronic stimulation of CD4+ T cells results in the development of a “senescent” CD28− subset in vitro, in chronic inflammatory diseases like rheumatoid arthritis (RA), and with aging [1; 2; 3]. CD4+CD28− T cells aberrantly express genes not normally expressed by CD4+CD28+ T cells, such as members of the killer cell immunoglobulin-like receptor (KIR) gene family and perforin [4; 5], normally expressed by NK cells [6], and overexpress interferon–γ (IFNγ) [7], LFA-1 [8] and CD70 [9]. The CD4+CD28− T cells acquire cytolytic activity, lysing endothelial and other cells through mechanisms involving the aberrantly expressed KIR molecules and self class I MHC molecules, and have been isolated from ruptured atherosclerotic plaques of people dying from myocardial infarctions, implicating these cytotoxic and inflammatory functions in acute coronary events [4; 6]. The mechanisms causing aberrant overexpression of genes like KIR, perforin and CD70 in CD4+CD28− T cells are incompletely understood.
Our group and others have found that some of the genes aberrantly overexpressed in CD4+CD28− T cells, including KIR, perforin, IFNγ, LFA-1 and CD70, are suppressed in CD4+ T cells from young healthy people by DNA methylation, and that demethylation of their promoters is sufficient to increase their transcription in T cells [10; 11; 12; 13]. Total genomic T cell DNA is demethylated in RA and aging, although specific subsets have not been studied [14; 15]. Levels of T cell DNA methyltransferase 1 and 3a (Dnmt1 and Dnmt3a), which maintain DNA methylation patterns during mitosis and DNA repair [16], also decrease with aging [17], although again subsets have not been studied, the significance of the decreases has not been established, and the relative roles of Dnmt1 and Dnmt3a in maintaining T cell DNA methylation patterns during mitosis have not been determined.
We hypothesized that the aberrant expression of methylation sensitive genes in the CD4+CD28− T cell subset is due to demethylation of crucial promoter sequences, caused by decreased levels of Dnmt1 and/or Dnmt3a. We therefore compared expression as well as methylation status of crucial regulatory regions of the CD70, perforin and KIR2DL4 promoters in CD4+CD28+ and CD4+CD28− T cells, and compared the results with levels of Dnmt1 and Dnmt3a in these subsets. We also used siRNA “knockdowns” to determine the consequences of decreased Dnmt1 and Dnmt3a levels on the expression and methylation status of these genes in T cells.
MATERIALS AND METHODS
Subjects
Patients with RA were recruited from the outpatient rheumatology clinics at the University of Michigan. Older subjects (ages 50–85) were recruited from the Human Subjects Core of the University of Michigan Claude D. Pepper Older Americans Independence Center. Young healthy controls (ages 18–30) were recruited by advertising. All RA patients fulfilled the American College of Rheumatology criteria for the diagnosis of RA [18]. This protocol was approved by the University of Michigan Institutional Review Board. A small blood sample was obtained from each subject at the time of their initial recruitment and tested for the presence of CD4+CD28− T cells. Those with ≥ 10% CD4+CD28− T cells were invited back for a larger blood draw. In all 60 subjects were screened for the subset, and 27 (21 RA patients and 6 older subjects) met entry criteria. Clinical information regarding these subjects is shown in Table 1. No differences in methylation sensitive gene expression were observed between CD4+CD28− T cells isolated from the RA patients and the 6 elderly non-RA subjects, and no relationship was seen between medications received and gene expression in the RA subjects, as reported by others [19].
Table 1.
Subject Information
| Subject. | Sex | Age | CD28-% | Dx | Medications |
|---|---|---|---|---|---|
| 1 | M | 77 | 25 | RA | Pred, Plaq, MTX |
| 2 | M | 77 | 15.4 | RA | Pred, Plaq, MTX, α-TNF |
| 3 | F | 84 | 18.4 | RA | Doxy, LF, α-TNF |
| 4 | F | 65 | 10.1 | RA | MTX, Pred |
| 5 | F | 68 | 10.6 | RA | α-TNF |
| 6 | F | 70 | 11.3 | RA | MTX, α-TNF |
| 7 | F | 58 | 13.1 | RA | MTX, Pred, α-TNF |
| 8 | F | 54 | 10.1 | RA | Pred, Aza, Abat |
| 9 | M | 65 | 16 | RA | Pred, α-TNF |
| 10 | F | 85 | 27.4 | RA | MTX, Pred |
| 11 | F | 55 | 9.94 | RA | MTX, Plaq |
| 12 | M | 57 | 10.8 | RA | SSZ, MTX, Pred |
| 13 | F | 69 | 20.2 | RA | Pred, Plaq, LF |
| 14 | F | 50 | 12.5 | RA | LF, α-TNF |
| 15 | M | 72 | 16.2 | RA | Pred, α-TNF |
| 16 | M | 73 | 14.4 | RA | MTX, Plaq, Pred |
| 17 | M | 66 | 12.9 | RA | MTX, Pred |
| 18 | M | 65 | 11.7 | RA | Pred, Plaq, LF |
| 19 | F | 55 | 11.2 | RA | LF, Pred, α-TNF |
| 20 | M | 71 | 19.1 | RA | Pred, Plaq, MTX |
| 21 | F | 65 | 14.3 | RA | Pred, LF |
| 22 | M | 77 | 16.27 | OA | |
| 23 | M | 65 | 18.7 | OA | |
| 24 | F | 74 | 13.3 | Control | |
| 25 | M | 78 | 18.2 | Control | |
| 26 | F | 79 | 19.1 | Control | |
| 27 | M | 80 | 18.3 | Control |
Abbreviations: RA, rheumatoid arthritis; OA, osteoarthritis; Pred, prednisone; Plaq, plaquenil; MTX, methotrexate; α-TNF, TNF antagonist; doxy, doxycycline; LF, leflunomide, Aza, azathioprine, Abat, abatacept; SSZ, sulfasalazine.
Flow cytometric analysis
PBMC were isolated from peripheral blood then stained with fluorochrome conjugated monoclonal antibodies, fixed, and analyzed using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ) as previously described [10]. The following monoclonal antibodies were used: FITC-anti-CD3, FITC-anti-CD4, PE-anti-CD4, PE-anti-CD70, PE-anti-perforin, PE-mouse IgG1 and FITC-mouse-IgG1, PE-mouse-IgG2b and PE-mouse-IgG3 (all from BD PharMingen, San Diego, CA). Anti-CD158d-PE (anti-KIR2DL4) was obtained from R&D Systems. For CD28 staining, PE-conjugated anti-CD28 or a PE-conjugated isotype control and FITC-anti-CD4 (BD PharMingen) were used.
Cell separation
PBMC were isolated by density gradient centrifugation and CD4+ cells isolated using CD4 microbeads (Miltenyi, Auburn, CA) as previously described [10]. CD28+ cells were then isolated either by staining with FITC-conjugated anti-CD28 (BD Biosciences, Franklin Lakes, NJ) followed by depletion with anti-FITC MicroBeads (Miltenyi Biotech), or by using a CD28 MicroBead kit (Miltenyi Biotech). Purity is typically 95–98% by flow cytometry.
T cell siRNA transfection and culture
PBMC from young healthy donors were isolated by density gradient centrifugation and total T cells purified using a Pan T cell isolation kit II (Miltenyi). T cells were typically > 94% CD3+. The T cells were then cultured in RPMI 1640 (Thermo) containing 10% fetal bovine serum and 1% penicillin/streptomycin. T cells were transfected with 1.4 μg siRNA-DNMT1 and/or siRNA-DNMT3a (QIAGEN) on days one and two using the Amaxa (Gaithersburg, MD) human T cell nucleofector kit and program U-014. Controls included a nonspecific siRNA provided by the manufacturer. 6 hours after transfection the cells were stimulated with PHA, and 24 hours later Dnmt transcripts were measured relative to β-actin using real time RT-PCR, while methylation and expression of methylation sensitive genes was measured 72 hours after PHA stimulation. Where indicated, T cells were experimentally demethylated by stimulating PBMC with PHA for 16 hours then treating with 5 μM 5-azaC for another 72 hours [10].
Real-time quantitative RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Valencia, CA). The primers were as previously described for CD70 [20], perforin and β-actin [10], and KIR2DL4 by Uhrberg et al [21]. The Dnmt1, Dnmt3a and Dnmt3b primers were also as described [22]. Real-time RT-PCR was carried out using a Rotor-Gene 3000 (Corbett Robotics, San Francisco, CA) and a QuantiTect SYBR Green RT-PCR kit (QIAGEN) according to the manufacturer’s instructions. cDNAs synthesized from T cells were used to generate standard curves for β-actin, Dnmt1, Dnmt3a, Dnmt3b, CD70, perforin and KIR2DL4 transcripts. Each assay was repeated at least twice.
Bisulfite conversion and pyrosequencing
Genomic DNA was isolated from T cells using the DNeasy blood & tissue kit (QIAGEN), then bisulfite treated using the EZ DNA Methylation-Gold kit (ZYMO Research). The pyrosequencing primers used are shown in Table 2, and were designed and produced by EpigenDx (Worcester, MA). PCR was performed using HotStar Taq (QIAGEN) and the following conditions: initial incubation 95° C for 15 min, then 45 cycles of 95° C for 30 s, 60° C for 30 s and 72° C for 30 s for the CD70 promoter; 45 cycles of 95° C for 30 s, 53° C and 56° C for 30 s and 72° C for 30 s for the perforin promoter; and 45 cycles of 95° C for 30 s, 60° C for 30 s and 72° C for 30 s for the KIR2DL4 promoter. The PCR product of each gene was then used for the individual sequencing reactions (Table 2). Biotinylated PCR products were immobilized on streptavidin-coated sepharose beads (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). 40 μl of binding buffer including 2 μl streptavidin-coated sepharose beads was added to 40 μl of PCR product and mixed using a shaker, incubating at room temperature for 10 minutes while agitating constantly to keep the beads dispersed. The biotinylated amplicon was purified and denatured using a PyroMark Vacuum Prep Workstation (Biotage, Uppsala, Sweden). The DNA was resuspended in 12 μl annealing buffer including 0.5 μm sequencing primer in a 96-well PSQ96 plate (Biotage). The sequencing primers were allowed to anneal on a heat plate set to 85° C for 2 minutes then the samples were cooled to room temperature. After completion of primer annealing, sequencing was running on the Pyro Mark MD system (Biotage) with Pyro Gold Reagents (Biotage) according to the manufacturer’s instructions. Data were analyzed using Pyro Q-CpG Software (Biotage).
Table 2.
Primers
| Pyrosequencing Primers | ||||
|---|---|---|---|---|
| Gene name | Primer name | Primer sequence 5′ -> 3′ | 5′ mod | PCR length |
| CD70 | ADS320FP | TGGATTATTTAAGGTTAGGAGTTTA | 269 bp | |
| ADS320RPB | TATACCCCTCTCCTACATTTTTT | biotin | ||
| Perforin-1 | ADS432FP | AGTTTTTGGTGAAGTTGGGATTA | 320 bp | |
| ADS432RPB | AATCACACTTTAAAACTCATCCCTTAC | biotin | ||
| Perforin-2 | ADS433FP | TGAGGTATAGTGAGGTTGAAGAAT | 184 bp | |
| ADS433RPT | TAAACAAACCAACAAAACCATCTC | tailed | ||
| KIR2DL4 | ADS549FP | GTGTAGGGGTAAGTGAGTTTGAGA | 311 bp | |
| ADS549RPB | CACCAAAACATACCAAAATAATAACC | biotin | ||
| Sequencing Primers | |||
|---|---|---|---|
| Gene name | Primer name | Primer sequence 5′ -> 3′ | CpGs No. |
| CD70 | ADS320FS1 | AAATTAGTTAGGTATGGTGG | 11 |
| Perforin-1 | ADS432FS2 | TAGAAGAGGGTGGGGCTATTG | 4 |
| Perforin-2 | ADS433FS | GAATTTTATTAGTTTATATTG | 7 |
| KIR2DL4-1 | ADS549FS1 | GGGGTAAGTGAGTTTGAGAT | 2 |
| KIR2DL4-2 | ADS549FS2 | TGATTGATTTATTATTTGAA | 1 |
| KIR2DL4-3 | ADS549FS3 | TTTTATATGTTGTGGCTAATG | 7 |
Statistical analysis was performed using Student’s t-test.
RESULTS
Methylation sensitive gene overexpression in CD4+CD28− T cells
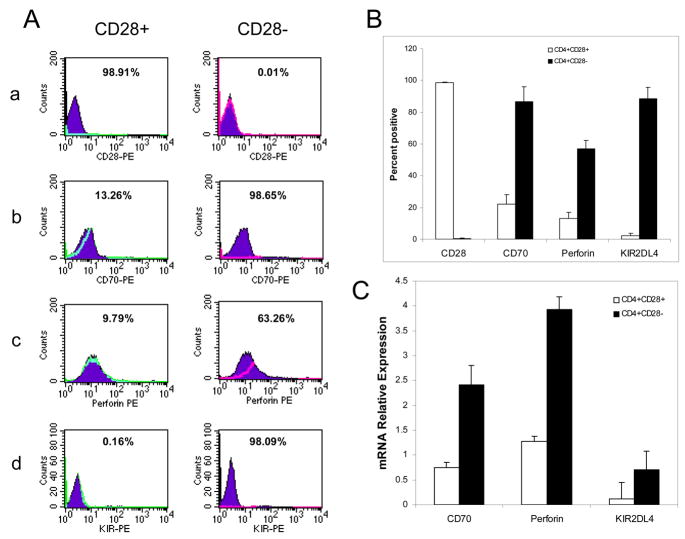
Initial studies confirmed overexpression of the methylation sensitive genes CD70, perforin and KIR2DL4 on CD4+CD28− T cells using flow cytometry. Figures 1a–d show representative histograms of CD70, perforin and KIR2DL4 in CD4+CD28+ and CD4+CD28− T cells from a patient with RA. Figure 1B shows the mean±SD of 3 independent experiments similarly comparing CD70, perforin and KIR2DL4 levels on CD4+CD28+ and CD4+CD28− subsets from 3 RA patients. Expression of all 3 proteins is significantly higher on the CD4+CD28− subset (p<0.001 for all genes).
Figure 1. Methylation sensitive gene expression in CD4+CD28+ and CD4+CD28− T cells.
A. PBMCs were isolated from RA patients with CD4+CD28− comprising ≥ 10% of the CD4+ population, then the CD4+CD28+ (left column) and CD4+CD28− (right column) subsets were separated and stained with a. Anti-CD28-PE, b. anti-CD70-PE, c. anti-perforin-PE or d. anti-KIR2DL4-PE. Filled curves represent staining with an IgG isotype control and open curves represent staining with the specific PE-conjugated antibodies. B. Mean+SD of three serial experiments similarly measuring CD28, CD70, perforin and KIR2DL4 on purified CD4+CD28+ (open bars) and CD4+CD28− (closed bars) cells (p<0.001, CD4+CD28+ vs CD4+CD28− for all). C. CD4+CD28− and CD4+CD28+ T cells were isolated from 5 additional subjects, then CD70, perforin and KIR2DL4 transcript levels measured relative to β-actin using real-time RT-PCR. Open bars represent the mean+SD of transcripts from CD4+CD28+ T cells, and closed bars the mean+SD of transcripts from CD4+CD28− T cells ( p<0.001, CD4+CD28+ vs CD4+CD28− for all).
These results were confirmed at the mRNA level. CD4+CD28+ and CD4+CD28− subsets were purified from the peripheral blood of 5 additional subjects (3 RA and 2 OA), and KIR2DL4, perforin and CD70 transcripts measured relative to β-actin. Figure 1C shows that KIR2DL4, perforin and CD70 mRNA levels are significantly (p<0.001) higher in CD4+CD28− T cells than in CD4+CD28+ T cells, and that KIR2DL4 is barely detectable in CD28+ T cells. No significant differences were seen in methylation sensitive T cell gene expression between RA patients and healthy elderly subjects with the CD4+CD28− subset.
Demethylation of CD70, perforin and KIR promoters in CD4+CD28− T cells
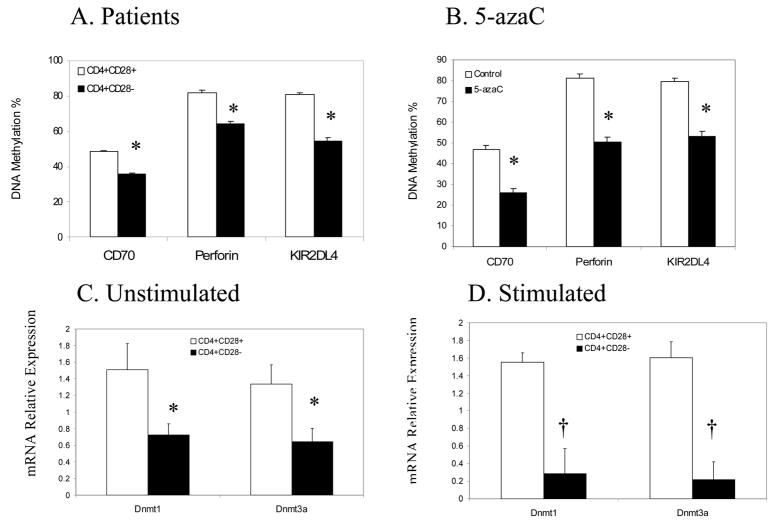
We next compared the methylation status of KIR2DL4, perforin and CD70 promoter regulatory elements in CD4+CD28− and CD4+CD28+ T cells from 3 RA patients using Pyrosequencing of bisulfite treated DNA. The sequences analyzed were previously reported to undergo transcriptionally relevant demethylation following treatment with 5-azacytidine, and included 11 CG pairs (−515 to −384) flanking the ITGAL (CD70) promoter [20], 9 CG pairs in the KIR2DL4 promoter (−250 to the transcription start site) [13], and 12 CG pairs in the PRF1 promoter and upstream enhancer [10]. Figure 2A shows that the promoters of these genes are significantly (p<0.001 for all) hypomethylated in CD4+CD28− T cells relative to CD4+CD28+ T cells. For reference, PBMC from 3 young healthy individuals were stimulated with PHA, treated with 5-azaC and 72 hours later ITGAL, KIR2DL4 and PRF1 promoter methylation was similarly compared in untreated and 5-azaC treated T cells by Pyrosequencing. Fig 2B shows a similar but slightly greater decrease in the methylation of the same sequences in these 5-azaC treated cells (p<0.001 for all 3 genes, untreated vs treated).
Figure 2. Promoter methylation and DNA methyltransferase expression.
A. PBMCs were isolated from 3 subjects with the CD4+CD28− subset then CD4+CD28+ and CD4+CD28− T cells were isolated as in figs 1 and 2. DNA was isolated from the cells and overall methylation of transcriptionally relevant promoter regions of the indicated genes measured by Pyrosequencing of bisulfite treated DNA. Open bars represent the mean overall methylation+SD of CD4+CD28+ T cells, and closed bars the mean+SD of CD4+CD28− T cells (* p<0.001, CD4+CD28+ vs CD4+CD28−). B. Freshly isolated PBMC from young healthy individuals were stimulated with PHA, treated or not with 5-azaC, and 72 hours later CD4+ T cells were purified, DNA isolated, and methylation of the CD70, perforin and KIR2DL4 promoters measured as in panel A. Open bars represent the mean+SD of the 3 determinations in untreated CD4+ T cells, and closed bars the mean+SD of the 5-azaC treated T cells (* p< 0.001). C. CD4+CD28+ (open bars) and CD4+CD28− (closed bars) T cells were isolated from PBMCs of 5 subjects, then Dnmt 1 and 3a transcripts measured relative to β-actin using RT-PCR. Results are presented as the mean+SD of the 5 determinations (* p<0.002, CD4+CD28+ vs CD4+CD28−). D. PBMCs from 3 additional subjects were stimulated with PHA for 18 hours, CD4+CD28+ (open bars) and CD4+CD28− (closed bars) subsets purified and Dnmt transcripts measured as in panel C. Results represent the mean+SD of the 3 determinations († p < 0.004).
DNA methyltransferase levels in CD4+CD28+ and CD4+CD28− T cells
Mechanisms potentially causing demethylation of these sequences were addressed by comparing Dnmt1, Dnmt3a and Dnmt3b transcripts in CD4+CD28− and CD4+CD28+ T cells from 5 subjects with the subset (3 RA and 2 healthy elderly subjects), using unstimulated and PHA stimulated cells. CD4+CD28+ and CD4+CD28− T cells were isolated from the peripheral blood of the subjects, and Dnmt 1, 3a and 3b transcripts measured relative to β-actin. Figure 2C shows that both Dnmt1 and Dnmt3a transcripts are significantly decreased in the CD4+CD28− subset (p<0.002). There was no appreciable difference between T cells from the RA patients and elderly subjects as before. As reported previously by our group [17], Dnmt3b levels were significantly lower than Dnmt1 and Dnmt3a in T cells, and no difference in Dnmt3b levels were seen between the CD4+CD28+ and CD4+CD28− subsets (not shown). Since T cell Dnmt levels increase following stimulation [23], PBMC from 3 additional RA patients were isolated, stimulated with PHA for 18 hours, then CD4+CD28+ and CD4+CD28− subsets purified and Dnmt transcripts similarly compared (Fig. 2D). Again, Dnmt1 and Dnmt3a transcripts are significantly lower in the CD28- subset.
Effects of Dnmt suppression on methylation sensitive T cell gene expression
The functional significance of decreased Dnmt1 and Dnmt3a in CD4+CD28− cells was tested using siRNA transfection to “knock down” the transcripts in primary human T cells. Flow cytometric analysis confirmed that CD4+CD28− T cells are a minor subset of total CD4+ T cells in young healthy individuals (ages 18–30) (1.03±0.23%, mean±SEM, n=9). In initial studies, purified T cells from 4 healthy young individuals were transfected with Dnmt1 siRNA once, or two times on consecutive days. The cells were cultured for 6 hours following the final transfection, stimulated with PHA, and Dnmt1 transcripts measured in the cells 24 hours after the PHA stimulation. In cells transfected once, Dnmt1 levels decreased by 54±12% (mean±SD, n=4) relative to cells transfected without siRNA (“mock transfected”). However, a 70±2% decrease was observed when the cells were transfected twice with the Dnmt1 siRNA. This protocol was therefore used in subsequent studies.
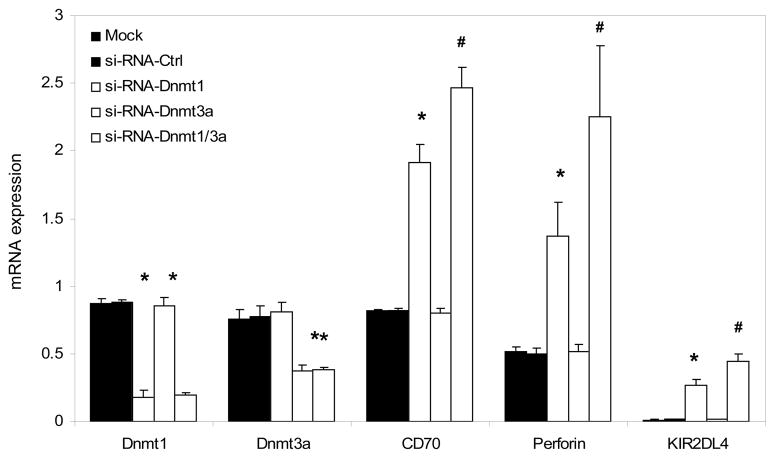
T cells from 3 young healthy controls were then transfected with siRNA specific for Dnmt1, Dnmt3a or both on days one and two, and 6 hours later stimulated with PHA. Controls included mock transfected T cells and T cells transfected with a nonspecific siRNA provided by the manufacturer. Twenty-four hours later Dnmt1 and Dnmt3a transcripts were measured relative to β-actin using real time RT-PCR. Changes in the methylation and expression of methylation sensitive genes (CD70, perforin and KIR2DL4) were measured 72 hours following the PHA stimulation. Figure 3 shows that Dnmt1 transcripts were suppressed by transfection with Dnmt1 with or without Dnmt3a siRNA, but not affected by Dnmt3a siRNA or the mock and no siRNA control transfections. Similarly Dnmt3a transcripts were suppressed by transfection with Dnmt3a siRNA with or without Dnmt1 siRNA, but not by Dnmt1 siRNA alone or the controls. These experiments confirm specificity of the siRNA’s for Dnmt1 and Dnmt3a. Figure 3 also shows that transfection with Dnmt1 siRNA increased levels of CD70, perforin and KIR2DL4 transcripts, and a further increase was seen with cotransfection of the Dnmt1 and Dnmt3a siRNAs. However, no effect was seen when the cells were transfected with the Dnmt3a siRNA alone, or with the control transfections.
Figure 3. Effect of Dnmt1 and Dnmt3a suppression on methylation sensitive T cell gene mRNA levels.
T cells were purified from the PBMC of 3 healthy subjects then transfected twice without siRNA (mock transfected, black bars), a control siRNA (dark dotted bars), Dnmt1-siRNA (open bars), Dnmt3a siRNA (crosshatched bars) or Dnmt1+Dnmt3a siRNA (light dotted bars). Six hours later the cells were stimulated with PHA. Dnmt1 and Dnmt3a transcripts were measured 24 hours after stimulation, and CD70, perforin and KIR2DL4 transcripts measured 72 hours after stimulation. Results are presented as the mean+SD of the three independent experiments. (* p<0.01, siRNA-Dnmt1 vs mock or negative control, and # p<0.03, siRNA-Dnmt1 vs siRNA-Dnmt1+Dnmt3a).
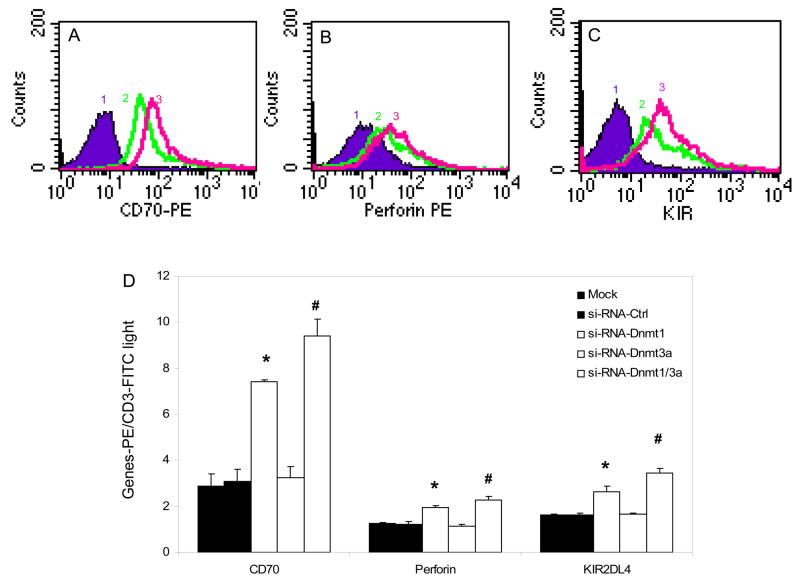
These results were confirmed at the protein level using flow cytometry. T cells from 3 additional young healthy controls were similarly transfected with siRNA specific for Dnmt1, Dnmt3a or both, and controls again included mock transfected T cells and T cells transfected with a nonspecific siRNA. Figure 4A shows a representative histogram comparing CD70 expression on mock transfected cells, cells transfected with Dnmt1 siRNA alone, or cells transfected with Dnmt1 and Dnmt3a siRNA, standardized to CD3 expression. Dnmt1 suppression causes an increase in CD70, and co-transfection with siRNA for Dnmt1 and Dnmt3a show a further increase. Controls confirmed no effect of the transfections on CD3 expression (not shown). Similar increases are seen in the expression of perforin (Fig. 4B) and KIR2DL4 (Fig. 4C). Figure 4D shows the mean±SD of the 3 experiments. As was seen at the RNA level, Dnmt1 suppression increases expression of all 3 proteins, and suppression of Dnmt1 and Dnmt3a gives a further increase. In contrast, methylation sensitive gene expression is the same in cells transfected with the controls and the Dnmt3a siRNA. These results closely parallel those observed at the mRNA level shown in fig 3.
Figure 4. Effect of Dnmt1 and Dnmt3a suppression on methylation sensitive T cell gene protein levels.
Panels A–C show representative histograms of T cells transfected twice without siRNA or siRNA negative control (curve 1), with Dnmt1-siRNA (curve 2) or with Dnmt1-siRNA+Dnmt3a-si-RNA (curve 3), stimulated with PHA, and 72 hours later stained with anti-CD70-PE (A), anti-perforin-PE (B) or anti-KIR2DL4-PE (C). D. T cells from three healthy controls were similarly transfected without siRNA (mock transfected, black bars), a control siRNA (dark dotted bars), Dnmt1-siRNA (open bars), Dnmt3a siRNA (crosshatched bars) or Dnmt1+Dnmt3a siRNA (light dotted bars) then stimulated with PHA as described in fig 3. 72 hours later CD70, perforin and KIR2DL4 expression were measured relative to CD3 by flow cytometry. Results are presented as the mean+SD of three independent experiments measuring CD70-PE, perforin-PE or KIR2DL4-PE MFI relative to CD3-FITC MFI. (* p≤0.003, Mock or negative control vs siRNA-Dnmt1, # p≤0.04, siRNA-Dnmt1 vs siRNA-Dnmt1+Dnmt3a).
Effects of Dnmt suppression on T cell gene methylation
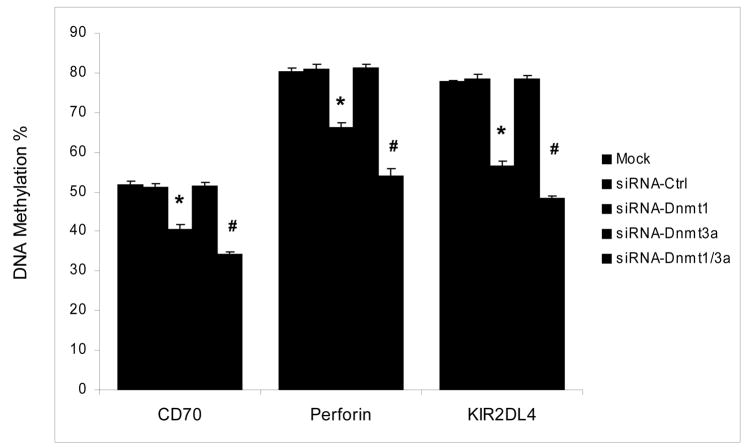
These results suggest that Dnmt1 is the enzyme primarily responsible for maintaining DNA methylation patterns, and that Dnmt3a may have an additive effect, but suppression of Dnmt3a is not sufficient to demethylate these genes. We therefore compared methylation of the CD70, perforin and KIR2DL4 promoters in T cells from 3 young healthy control subjects transfected with siRNA specific for Dnmt1, Dnmt3a or both using the same protocols as before. Controls again included mock transfected T cells and T cells transfected with a nonspecific siRNA. DNA was then isolated and methylation of the CD70, perforin and KIR2DL4 promoters compared by Pyrosequencing of bisulfite treated DNA as in figure 2. Figure 5 shows that Dnmt1 suppression demethylates the promoters of all 3 genes, and suppression of both Dnmt1 and Dnmt3a has a small additive effect, paralleling the effects on gene expression shown in figs 3 and 4. Again, transfection with the Dnmt3a siRNA or controls had no effect on promoter methylation. Taken together, our data strongly suggest that Dnmt1 plays a key role in the human T cell DNA methylation, and Dnmt3a may act as an accessory to support the function of Dnmt1. The results also suggest that decreases in Dnmt1, and possibly Dnmt3a, contribute to the aberrant overexpression of some genes in the CD4+CD28− subset.
Figure 5. Effect of Dnmt1 and Dnmt3a suppression on CD70, perforin and KIR2DL4 promoter methylation.
T cells from three healthy controls were transfected without siRNA (mock transfected, black bars), a control siRNA (dark dotted bars), Dnmt1-siRNA (open bars), Dnmt3a siRNA (crosshatched bars) or Dnmt1+Dnmt3a siRNA (light dotted bars) then stimulated with PHA as described in fig 4. 72 hours later CD70, perforin and KIR2DL4 promoter methylation were measured by Pyrosequencing. The results represent the mean+SD of the three independent experiments. (* p<0.001, mock or negative control vs siRNA-Dnmt1, and # p≤0.002, siRNA-Dnmt1 vs siRNA-Dnmt1+Dnmt3a).
DISCUSSION
T cells undergoing replicative stress in RA, aging and chronic infections [1; 2; 3; 24] become senescent. Characteristic changes include altered secretion of a variety of molecules including proteases, cytokines and growth factors, telomere shortening with eventual proliferative arrest, and resistance to apoptosis [25; 26]. The mechanisms causing senescence are multiple, and include oxidative damage [27; 28], decreased clearance of aberrant proteins [29], and changes in DNA including telomere shortening, translocations, deletions and mutations [30]. Why senescent CD4+ T cells aberrantly express genes like perforin and KIR, normally confined to cytotoxic lymphocytes, is not explained by these mechanisms. However, since these cells are implicated in plaque rupture and myocardial infarctions understanding the mechanisms involved may lead to ways to prevent these complications associated with T cell senescence.
The present studies demonstrate that the aberrant overexpression of KIR, perforin, and CD70 in CD4+CD28− T cells is associated with demethylation of regulatory regions and decreased levels of Dnmt1 and Dnmt3a relative to CD4+CD28+ T cells, and the siRNA studies indicate that decreases in Dnmt1 and Dnmt3a are likely responsible. KIR genes are typically expressed by NK cells, and perforin by cytotoxic CD8+ T cells and NK cells, but neither gene is normally expressed by CD4+ T cells [6]. Similarly, CD70 is expressed on some but not all CD4+ T cells [31]. We previously reported that pharmacologic inhibition of DNA methylation in CD4+ T cells is sufficient to demethylate the same regulatory regions of KIR2DL4, perforin and CD70 found to be demethylated in CD4+CD28− cells, causes overexpression of all three genes, and that methylation of the relevant regulatory sequences in reporter constructs suppresses promoter function [10; 13; 20]. This suggests that the requisite transcription factors are present in CD4+ T cells, and that KIR, perforin and CD70 expression is suppressed primarily by DNA methylation and a repressive chromatin configuration. These experiments also exclude unanticipated effects of the pharmacologic agents used previously. It should be noted that others have very recently reported that the KIR2DL4 promoter is partially demethylated in CD4+CD28− T cells [32], although transformed lines were primarily studied in this report, and transformed lines are characterized by abnormal DNA methylation [33] limiting interpretation of the results.
The present studies also demonstrate that CD4+CD28− T cells express lower levels of Dnmt1 and Dnmt3a than CD4+CD28+ T cells. Dnmt1 preferentially methylates hemimethylated DNA and is primarily involved in maintaining DNA methylation patterns, while Dnmt3a and Dnmt3b can methylate unmethylated as well as hemimethylated DNA, and can function as de novo as well as maintenance methyltransferases [34]. Complete inhibition of DNA methylation in human, but not necessarily mouse, cells requires suppression of both maintenance and de novo DNA methyltransferases [35]. We have previously reported that Dnmt1 and Dnmt3a levels decrease with age in unfractionated T cells, and that T cells express little if any Dnmt3b [17]. The present studies confirm the low levels of Dnmt3b in T cells, and indicate that the decreases in Dnmt1 and Dnmt3a are greater in CD4+CD28− than in CD4+CD28+ T cells.
The relative contributions of Dnmt1 and Dnmt3a to maintenance T cell DNA methylation has not been established previously. The present results demonstrate that decreasing Dnmt1 levels alone is sufficient to cause demethylation and overexpression of the three methylation sensitive genes studied in CD4+CD28+ T cells, while decreasing Dnmt3a levels alone has no effect, and suppression of both methyltransferases had an additive effect beyond that of Dnmt1 alone. Since CD4+CD28− T cells have lower levels of these transcripts, it is reasonable to propose that the decreases in the methyltransferases are responsible for the DNA demethylation observed in the CD28- subset. This is consistent with observations that inhibition of both maintenance and de novo DNA methylation is required to demethylate CpG islands in transformed cells. However, these reports indicate a more prominent role for Dnmt3b than Dnmt3a [35; 36]. Why T cells preferentially express Dnmt3a rather than Dnmt3b is unclear.
The reason for the decrease in Dnmt1 and Dnmt3a in these cells is unknown, but important to establish because correcting the abnormality may suggest ways to prevent or correct aberrant gene expression in this subset. Dnmt1 is regulated by signals through the ERK and JNK pathways [23; 37; 38], and Dnmt3a by signaling through the ERK pathway [23]. Others have reported that signaling through these pathways is diminished in aged T cells [39; 40]. Our group has reported that inhibiting ERK pathway signaling with a MEK inhibitor is sufficient to cause DNA demethylation and overexpression of CD70 in T cells [20], so decreased ERK pathway signaling could contribute. This raises the possibility that decreased JNK signaling may have an additive effect on methylation sensitive genes in CD4+CD28− T cells. These studies are in progress.
Acknowledgments
The authors thank Ms. Cindy Bourke for her expert secretarial assistance. This work was supported by PHS grants AG25877, AR42525, ES015214, the University of Michigan Pepper Center grant AG024824, and a Merit grant from the Veterans Administration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goronzy JJ, Weyand CM. Rheumatoid arthritis. Immunol Rev. 2005;204:55–73. doi: 10.1111/j.0105-2896.2005.00245.x. [DOI] [PubMed] [Google Scholar]
- 2.Koetz K, Bryl E, Spickschen K, O’Fallon WM, Goronzy JJ, Weyand CM. T cell homeostasis in patients with rheumatoid arthritis. Proc Natl Acad Sci U S A. 2000;97:9203–8. doi: 10.1073/pnas.97.16.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schonland SO, Lopez C, Widmann T, Zimmer J, Bryl E, Goronzy JJ, Weyand CM. Premature telomeric loss in rheumatoid arthritis is genetically determined and involves both myeloid and lymphoid cell lineages. Proc Natl Acad Sci U S A. 2003;100:13471–6. doi: 10.1073/pnas.2233561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yen JH, Moore BE, Nakajima T, Scholl D, Schaid DJ, Weyand CM, Goronzy JJ. Major histocompatibility complex class I-recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193:1159–67. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namekawa T, Wagner UG, Goronzy JJ, Weyand CM. Functional subsets of CD4 T cells in rheumatoid synovitis. Arthritis Rheum. 1998;41:2108–16. doi: 10.1002/1529-0131(199812)41:12<2108::AID-ART5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Nakajima T, Schulte S, Warrington KJ, Kopecky SL, Frye RL, Goronzy JJ, Weyand CM. T-cell-mediated lysis of endothelial cells in acute coronary syndromes. Circulation. 2002;105:570–5. doi: 10.1161/hc0502.103348. [DOI] [PubMed] [Google Scholar]
- 7.Weyand CM, Fulbright JW, Goronzy JJ. Immunosenescence, autoimmunity, and rheumatoid arthritis. Exp Gerontol. 2003;38:833–41. doi: 10.1016/s0531-5565(03)00090-1. [DOI] [PubMed] [Google Scholar]
- 8.Komocsi A, Lamprecht P, Csernok E, Mueller A, Holl-Ulrich K, Seitzer U, Moosig F, Schnabel A, Gross WL. Peripheral blood and granuloma CD4(+)CD28(−) T cells are a major source of interferon-gamma and tumor necrosis factor-alpha in Wegener’s granulomatosis. Am J Pathol. 2002;160:1717–24. doi: 10.1016/s0002-9440(10)61118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee WW, Yang ZZ, Li G, Weyand CM, Goronzy JJ. Unchecked CD70 expression on T cells lowers threshold for T cell activation in rheumatoid arthritis. J Immunol. 2007;179:2609–15. doi: 10.4049/jimmunol.179.4.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, Pipkin M, Lichtenheld M, Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression. J Immunol. 2003;170:5124–32. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- 11.Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL. Effect of promoter methylation on the regulation of IFN-gamma gene during in vitro differentiation of human peripheral blood T cells into a Th2 population. J Immunol. 2003;171:2510–6. doi: 10.4049/jimmunol.171.5.2510. [DOI] [PubMed] [Google Scholar]
- 12.Lu Q, Ray D, Gutsch D, Richardson B. Effect of DNA methylation and chromatin structure on ITGAL expression. Blood. 2002;99:4503–8. doi: 10.1182/blood.v99.12.4503. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2008 doi: 10.1016/j.clim.2008.08.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20:1869–72. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- 15.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–73. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 16.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–91. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Deng C, Lu Q, Richardson B. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev. 2002;123:1257–68. doi: 10.1016/s0047-6374(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 18.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 19.Thewissen M, Somers V, Hellings N, Fraussen J, Damoiseaux J, Stinissen P. CD4+CD28null T cells in autoimmune disease: pathogenic features and decreased susceptibility to immunoregulation. J Immunol. 2007;179:6514–23. doi: 10.4049/jimmunol.179.10.6514. [DOI] [PubMed] [Google Scholar]
- 20.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–9. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 21.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 22.Attwood J, Richardson B. Relative quantitation of DNA methyltransferase mRNA by real-time RT-PCR assay. Methods Mol Biol. 2004;287:273–83. doi: 10.1385/1-59259-828-5:273. [DOI] [PubMed] [Google Scholar]
- 23.Deng C, Lu Q, Zhang Z, Rao T, Attwood J, Yung R, Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–56. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 24.Koch S, Solana R, Dela Rosa O, Pawelec G. Human cytomegalovirus infection and T cell immunosenescence: a mini review. Mech Ageing Dev. 2006;127:538–43. doi: 10.1016/j.mad.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Campisi J. Replicative senescence: an old lives’ tale? Cell. 1996;84:497–500. doi: 10.1016/s0092-8674(00)81023-5. [DOI] [PubMed] [Google Scholar]
- 26.Campisi J. From cells to organisms: can we learn about aging from cells in culture? Exp Gerontol. 2001;36:607–18. doi: 10.1016/s0531-5565(00)00230-8. [DOI] [PubMed] [Google Scholar]
- 27.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–4. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 28.Van Remmen H, Hamilton ML, Richardson A. Oxidative damage to DNA and aging. Exerc Sport Sci Rev. 2003;31:149–53. doi: 10.1097/00003677-200307000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Macario AJ, Conway de Macario E. Sick chaperones and ageing: a perspective. Ageing Res Rev. 2002;1:295–311. doi: 10.1016/s1568-1637(01)00005-8. [DOI] [PubMed] [Google Scholar]
- 30.Guarente L. Do changes in chromosomes cause aging? Cell. 1996;86:9–12. doi: 10.1016/s0092-8674(00)80072-0. [DOI] [PubMed] [Google Scholar]
- 31.Lens SM, Baars PA, Hooibrink B, van Oers MH, van Lier RA. Antigen-presenting cell-derived signals determine expression levels of CD70 on primed T cells. Immunology. 1997;90:38–45. doi: 10.1046/j.1365-2567.1997.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, Weyand CM, Goronzy JJ. Epigenetic mechanisms of age-dependent KIR2DL4 expression in T cells. J Leukoc Biol. 2008;84:824–34. doi: 10.1189/jlb.0807583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrlich M. DNA methylation and cancer-associated genetic instability. Adv Exp Med Biol. 2005;570:363–92. doi: 10.1007/1-4020-3764-3_13. [DOI] [PubMed] [Google Scholar]
- 34.Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59:241–57. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 36.Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu JC, Yan PS, Huang TH. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res. 2003;63:6110–5. [PubMed] [Google Scholar]
- 37.MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–37. doi: 10.1074/jbc.270.19.11327. [DOI] [PubMed] [Google Scholar]
- 38.Rouleau J, MacLeod AR, Szyf M. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J Biol Chem. 1995;270:1595–601. doi: 10.1074/jbc.270.4.1595. [DOI] [PubMed] [Google Scholar]
- 39.Kirk CJ, Freilich AM, Miller RA. Age-related decline in activation of JNK by TCR- and CD28-mediated signals in murine T-lymphocytes. Cell Immunol. 1999;197:75–82. doi: 10.1006/cimm.1999.1567. [DOI] [PubMed] [Google Scholar]
- 40.Miller RA, Garcia G, Kirk CJ, Witkowski JM. Early activation defects in T lymphocytes from aged mice. Immunol Rev. 1997;160:79–90. doi: 10.1111/j.1600-065x.1997.tb01029.x. [DOI] [PubMed] [Google Scholar]