Abstract
JC virus (JCV) is a human polyomavirus that infects the majority of the human population worldwide. It is responsible for the fatal demyelinating disease Progressive Multifocal Leukoencephalopathy. JCV binds to cells using the serotonin receptor 5-HT2AR and α(2–6)- or α(2–3)-linked sialic acid. It enters cells using clathrin-dependent endocytosis and traffics to the early endosome and possibly to the endoplasmic reticulum. Viral DNA is then delivered to the nucleus where transcription, replication, and assembly of progeny occurs. We found that the early regulatory protein large T antigen accumulates in microdomains in the nucleus adjacent to ND-10 or PML domains. This observation prompted us to explore the role of these domains in JCV infection. We found that a reduction of nuclear PML enhanced virus infection and that an increase in nuclear PML reduced infection. Infection with JCV did not directly modulate nuclear levels of PML but our data indicate that a host response involving interferon-beta is likely to restrict virus infection by increasing nuclear PML.
Introduction
JC virus (JCV) is a human polyomavirus for that infects between 20 and 70% of the human population based on the most recent seorological evidence (Kean et al., 2009). JCV causes the fatal demyelinating disease Progressive Multifocal Leukoencephalopathy, that occurs following reactivation of latent JC virus (Padgett et al., 1971). Reactivation is triggered by immunosupression that allows the virus to migrate from peripheral sites into the central nervous system (CNS). In the CNS, JCV infects and lytically destroys the myelin-producing oligodendrocytes (Astrom, Mancall, and Richardson, 1958; Major et al., 1992; Richardson, 1961).
JCV is a small, non-enveloped, double stranded DNA virus with transcriptional units organized into early and late regions (Frisque, Bream, and Cannella, 1984). These regions are divided by the non-coding control region, containing a bidirectional promoter and the viral origin of replication (Danna and Nathans, 1972; Fareed, Garon, and Salzman, 1972). The early region contains small t, large T, T′135, T′136 and T′165. All of these products are produced from alternative splicing of the viral early mRNA (Trowbridge and Frisque, 1995). Large T (LT) antigen is a regulatory protein involved in deregulation of the cell cycle and DNA replication. LT is composed of several domains that interact with cellular factors to promote these actions (Krynska et al., 1997; Sullivan et al., 2000). After replication, JCV LT interacts with the late promoter and drives the expression of the viral capsid proteins Vp1, 2, and 3 (Sullivan and Pipas, 2002).
Upon entering the nucleus, DNA viruses can localize to distinct regions in the nuclear space where they regulate genome production and orchestrate efficient packaging of progeny virions. Nuclear domains are heterogeneous groups of proteins located within the nuclear matrix (Everett and Chelbi-Alix, 2007; Zimber, Nguyen, and Gespach, 2004). ND-10 or PML domains appear in a speckled pattern in the nucleus with immunofluorescent staining. However, the size and number of the domains varies by cell type. PML domains are mobile structures whose movements are ATP- and myosin-dependent. There are many proteins associated with this domain including Sp100, Daxx, and CREB binding protein (CBP). PML seems to provide the scaffolding for these proteins and allow for recruitment of other proteins. Daxx is a pro-apoptotic Fas-interacting protein, recruited to this domain through its C′ terminal region in association with SUMOylated PML (Ishov, Vladimirova, and Maul, 2004). Sp100 has a DNA binding domain and has been implicated in transcriptional modulation (Negorev, Ishov, and Maul, 2001). Many transcription factors, such as CBP, are also localized to ND-10. Cells can survive without the presence of PML, suggesting possible redundancies in its role (Zimber, Nguyen, and Gespach, 2004). The loss of PML is not without consequences: acute promyelocytic leukemia (APL) has been linked to a chromosomal translocation that causes a fusion between PML and the retinoic acid receptor, RARα. This fusion, in turn, causes PML to be re-localized to the cytoplasm, where it is unavailable to respond to cell stress (Zimber, Nguyen, and Gespach, 2004). PML protein can be modulated by arsenic and other heavy metals and stress. This modulation leads to changes in PML protein localization and levels that then influence the behavior of other domain components (Maul et al., 1995).
The PML domain is a common site of genome deposition by DNA viruses (Ishov and Maul, 1996). Adenovirus type 5 immediate protein pIX has been shown to form inclusion bodies in the nucleus that consist of PML protein surrounded by pIX as a way to modulate PML function and allow virus infection to proceed (Rosa-Calatrava et al., 2003). Herpesviruses also target the PML protein early in infection. Herpes simplex virus (HSV-1) immediate early protein ICP0 disrupts the ND-10 domain and causes PML to be degraded. Papillomaviruses, which are similar to polyomaviruses, have been shown to localize to ND-10 domains and this localization is dependent on the minor protein L2. Early during infection L2 deposits the genome in these domains and also accumulates there during viral assembly. However, this localization does not require the presence of the PML protein (Becker et al., 2004; Florin et al., 2002; Nakahara and Lambert, 2007). Other polyomavirus family members, simian virus 40 (SV40) and BKV, have been shown to replicate adjacent to PML nuclear bodies (Jul-Larsen et al., 2004). This replication is dependent on having a complete viral origin with LT binding sites and the expression of the LT protein itself (Leung et al., 2002). Additionally, JC virus minor proteins have been shown to accumulate in PML nuclear bodies. Assembled JC virus has been shown to reside in PML structures in post-mortem brains of patients with Progressive Multifocal Leukoencephalopathy (Shishido-Hara et al., 2008; Shishido-Hara et al., 2004).
In our study, we evaluated the relationship between JCV early regulatory protein large T and PML domains. We first observed that LT protein accumulates adjacent to PML domains. Surprisingly, elimination of PML protein enhanced JCV infection and the enhancement was manifest at the transcriptional level. Conversely, treatment of cells with IFN-β increased PML protein expression in the nucleus and led to a drastic inhibition of virus infection and early gene expression in a PML-dependent manner. PML protein was not disrupted by JCV over long-term infections, indicating that the virus is not degrading or relocalizing PML during infection. Taken together, these findings indicate that JCV infection is regulated by nuclear PML domains and their components.
Results
JC virus is capable of infecting cells in the absence of PML protein
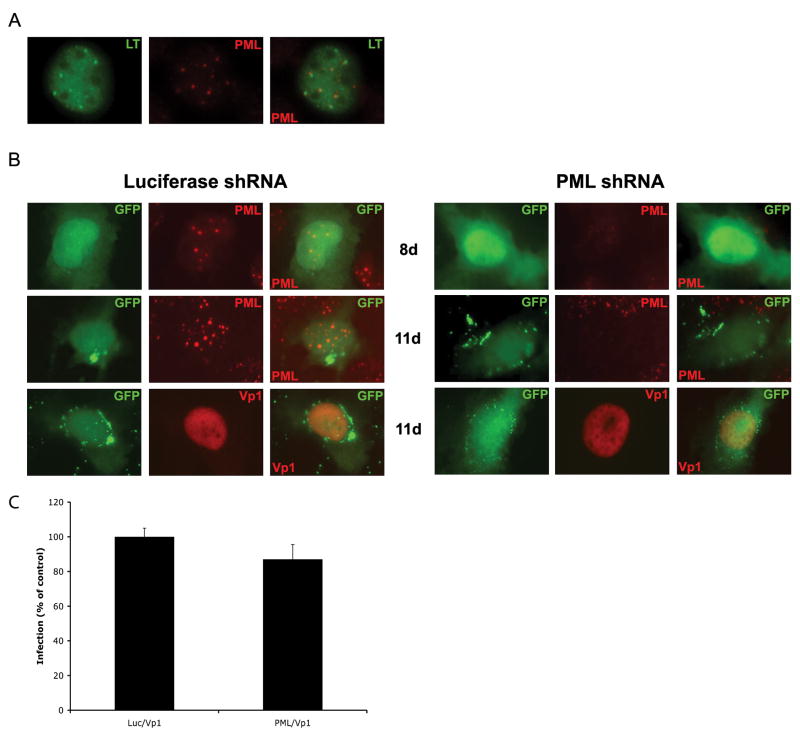
When assaying for JCV infection using an antibody that detected large T antigen (LT) we found that it localized in a pattern consistent with the expression pattern of PML containing nuclear domains. As other polyomaviruses are known to replicate adjacent to PML domains, we assayed whether the LT accumulations were associated with PML protein. We performed indirect immunofluorescence analysis of LT and PML protein expression at 48 hours post-infection on the human glial cell line, SVG-A. Both proteins exhibited a punctate, speckled pattern in the nucleus, and LT associated adjacent to many of the PML domains (Fig. 1A). The SVG-A cell line was derived by infecting primary human astrocytes with an origin defective SV40 expressing large T antigen. The SV40 LT expressed in these cells does not accumulate in a punctate pattern and does not localize to PML domains like SV40 LT expressed from infectious virus (Supplemental Fig. 1). This is likely due to the fact that localization to PML domains requires an intact origin of replication and this is lacking in SVG-A cells.
Figure 1. JC infection is not inhibited by shRNA knockdown of PML.
(A) Untreated SVG-A cells were infected with JCV and assayed 48h post-infection by indirect immunofluorescence analysis of JCV large T (LT) and PML protein expression. Samples were analyzed by epifluorescence microscopy using a magnification of 400. JCV LT is seen to accumulate in microdomains, which are adjacent to PML domains. (B) SVG-A cells were transfected with shRNA constructs directed at either a luciferase control (left panels) or the PML protein (right panels). The plasmids containing the hairpins also express GFP. After 8 days PML was knocked down in all GFP expressing samples and these transfected cells were infected with JCV. JCV infection was monitored 72 hrs later for the presence of JCV Vp1 protein or PML protein by indirect immunofluorescence in GFP-positive cells. Samples were analyzed by epifluorescence microscopy using a magnification of 400. (C) The number of double positive cells were quantified and represented as a percent of luciferase control.
To determine whether JC virus requires PML protein for infection, permissive SVG-A cells were transfected with shRNA constructs against PML protein or luciferase as a negative control. The ability of these constructs to eliminate PML was monitored with indirect immunofluorescence of the PML protein. The constructs expressing the shRNA also express GFP allowing for identification of shRNA expressing cells. After monitoring the levels of PML protein in GFP positive cells, it was determined there was complete knockdown of PML eight days post-transfection (Fig. 1B). After eight days, transfected cells were infected with JCV. JCV infectivity was monitored 72 hours post-infection. At that time, PML protein was still knocked down in the PML samples, but was unaffected in the samples that received the luciferase construct (Fig. 1B). JCV was able to infect both untreated and treated samples indicating that it does not require PML protein for a productive infection.
JCV LT accumulated in microdomains in the nucleus that are independent of PML
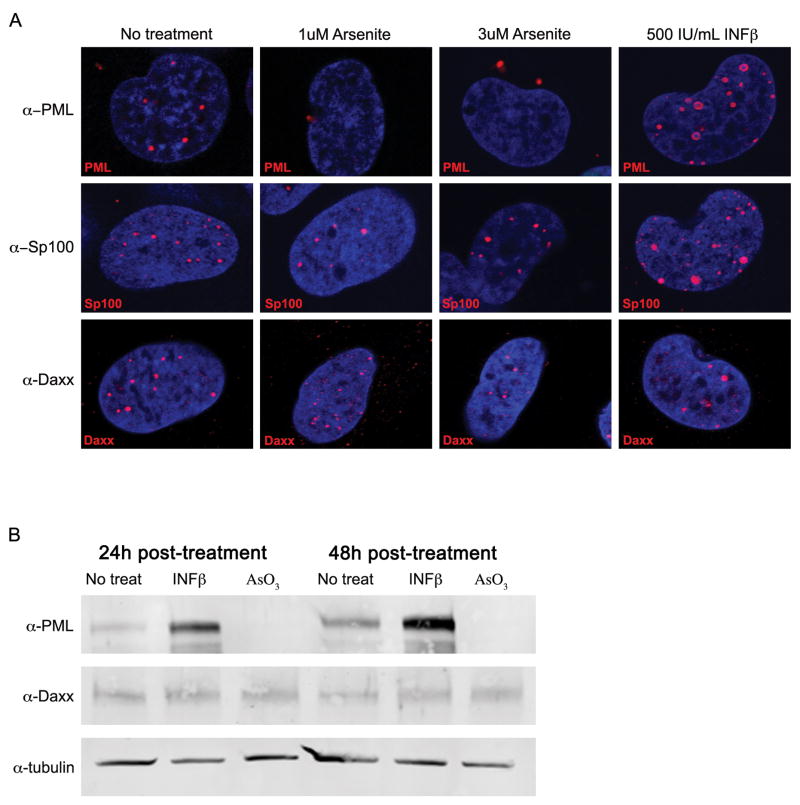
While shRNA indicated JCV is able to infect in the absence of PML, the percentage of cells receiving the knockdown was not sufficient to determine a global effect on infection. To see this global effect, ND-10 components were modulated with arsenite or interferon beta (INF-β) treatment. Arsenite is known to disrupt and degrade PML protein and INF-β is known to increase PML protein and transcript (Miller et al., 2002). The concentration of arsenite used is in the therapeutic range used to treat APL and did not cause toxicity in our cell line (data not shown) (Wang and Chen, 2008). Previous work in our lab determined that INF-β treatment was inhibitory to JCV infection (Dugan et al., 2008). Permissive SVG-A cells were treated with 1uM arsenite, 3uM arsenite, or 500IU/mL INF-β. After 48 hours of treatment, samples were stained by indirect immunofluorescence for PML, Sp100, and Daxx, and their cellular localization was evaluated using confocal microscopy. In the untreated samples, PML, Sp100, and Daxx all exhibit a pattern of expression with punctate domains speckled throughout the nucleus, that is the characteristic distribution of proteins in ND-10 (Fig. 2A). Upon arsenite treatment PML protein translocates from the nucleus to the cytosol (Fig. 2A). Arsenite had no effect on the nuclear localization of either SP100 or Daxx (Fig. 2A). Treatment of the cells with IFN-β led to an increase in both the number and size of PML and SP100 containing domains in the nucleus while Daxx expression was unaffected (Fig. 2A). To determine whether the overall protein levels of PML and Daxx were affected by arsenite and INF-β, SDS-PAGE and Western blot analyses were performed. After 24 or 48 hours of treatment, PML protein was eliminated from arsenite treated cells and was increased upon treatment with INF-β. Daxx protein levels remain constant over the time course (Fig. 2B). These results indicate that PML protein levels can be modulated using arsenite and IFN-β.
Figure 2. Arsenite treatment eliminates PML protein.
SVG-A cells were treated with media (no treatment), 1uM arsenite, 3uM arsenite, or 500IU/mL INF-β for 48 hrs. (A) Indirect immunofluorescence analysis of SVG-A cells and ND-10 components. Samples were mounted on slides with DAPI medium and analyzed by confocal microscopy using a 63X objective. Arsenite-treated samples showed PML re-localized from the nucleus to the cytoplasm, but Sp100 and Daxx samples were unaffected. Interferon beta treated samples of PML and Sp100 showed increases in domain size and number, but showed no effect on Daxx. (B) Analysis of protein levels of PML and Daxx. Whole cell lysates of SVG-A cells were resolved by SDS-PAGE and analyzed by Western blot at 24 and 48 hours post-treatment using antibodies specific for PML and Daxx proteins. PML protein was eliminated with arsenite treatment and increased with interferon beta treatment. Daxx protein levels remained unaffected. Tubulin was used as a loading control.
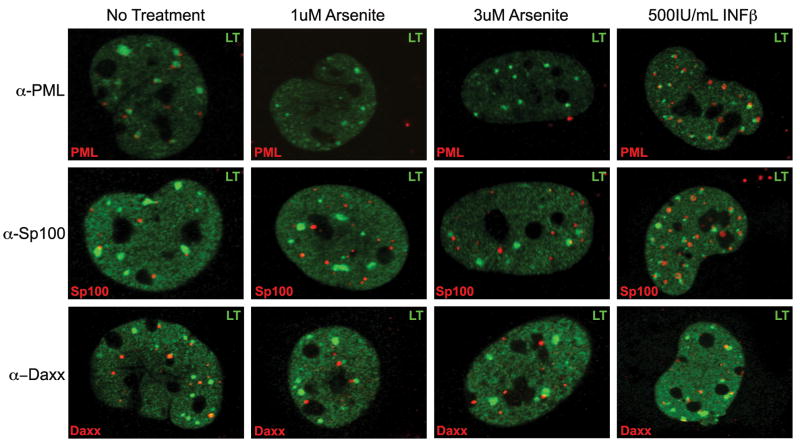
To determine whether viral proteins directly interact with ND-10 components, the localization of JCV LT protein and ND-10 proteins were evaluated 48 hours post infection using indirect immunofluorecence and confocal microscopy. JCV LT is expressed throughout the nucleus, but accumulates in larger speckled regions, or viral microdomains (Fig. 3). These accumulations are adjacent to PML, Sp100, and Daxx. The LT antigen containing domains are adjacent to the ND-10 domains and only partially overlap with ND-10 components (Fig. 3). Given that JCV LT accumulates in microdomains we wanted to determine whether the domains were dependent on the presence of PML. Upon arsenite treatment, when PML is removed from the nucleus, the LT microdomains are unaffected (Fig. 3). This indicates the virus does not use PML as a scaffold for its microdomains. When the samples are treated with INF-β and PML protein is increased a large percentage of LT microdomains associate with ND-10 components (Fig. 3). This increase in ND-10 association may partially explain the inhibitory effects seen with IFN-β treatment.
Figure 3. Large T accumulates in microdomains in the nucleus, which are independent of PML protein.
SVG-A cells were treated with media (no treatment), 1uM arsenite, 3uM arsenite, or 500IU/mL INF-β. Treated cells were infected with JCV and 48h post-infection samples were analyzed by indirect immunofluorescence and visualized using confocal microscopy with a 63X objective. LT was observed in the nucleus in accumulated microdomains. These accumulations were adjacent to the ND-10 components. When PML was removed using arsenite treatment, the microdomains were unaffected. When ND-10 domains components were upregulated with INF-β treatment, there was greater overlap of LT and the ND-10 proteins.
Arsenite treatment eliminates PML protein and enhances JCV infection
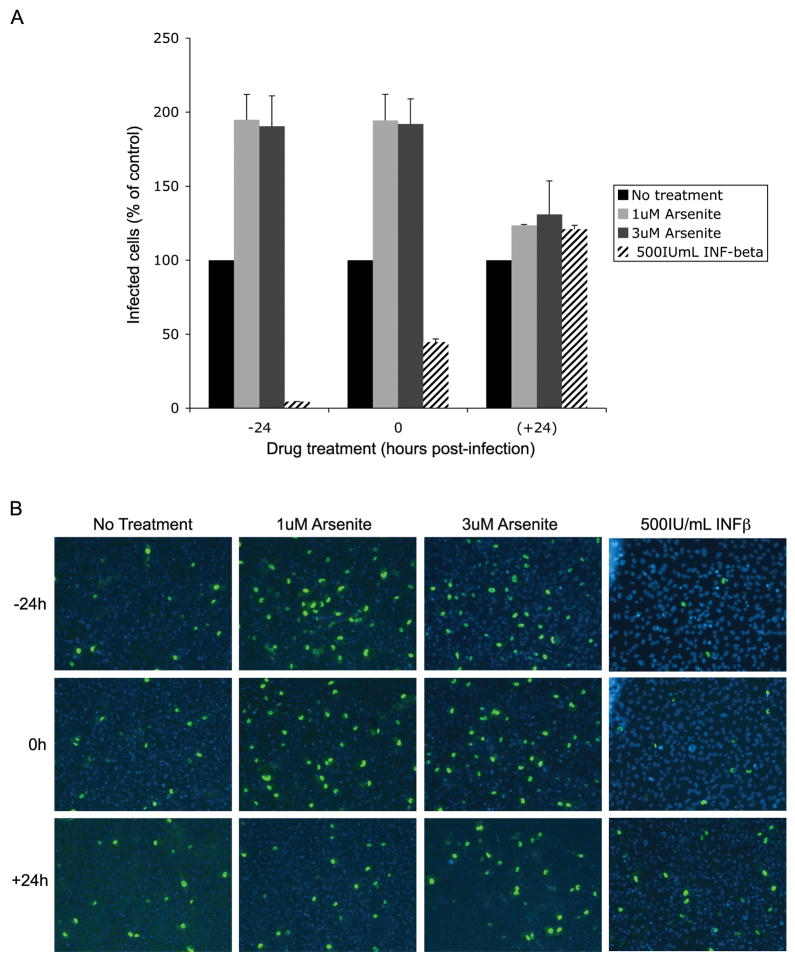
To determine the role of PML in JCV infection, we evaluated whether modulation of PML protein levels would affect JCV infection. SVG-A cells were either treated 24 hours before infection, at the time of infection, or 24 hours post-infection. Cells infected with JCV were scored 48 hours post-infection using an indirect immunofluorescence assay for LT protein (Fig. 4). The amount of JCV-positive cells were quantified and compared to those receiving no treatment. Cells pre-treated or treated at the time of infection with arsenite showed a 2-fold increase in infection. However, this increase was not seen when arsenite was added 24 hours post infection. Interferon beta treated cells showed the opposite trend: cells treated 24 hours prior to infection or at the time of infection show a 95–55% decrease in JCV LT positive cells (Fig. 4). Similarly, arsenite treatment led to an increase in viral protein expression and INF-β led to a decrease in viral protein expression when the late viral protein Vp1 was monitored (data not shown).
Figure 4. Removal of PML protein enhances JCV infection.
SVG-A cells were treated with media (no treatment), 1uM arsenite, 3uM arsenite, or 500IU/mL INF-β at indicated time points. Treated cells were infected with JCV and infection was scored 48 hrs later using indirect immunofluorescence of JCV LT. (A) Positive cells were quantified and the data represent an average of three infections compared to no treatment (set at 100%). There is a drastic increase in infection when samples are pre-treated with arsenite. Conversely, there is a decrease in infection when samples are treated with INF-β. There is no effect when samples are treated 24 hrs post-infection. Error bars represent the standard deviation of the samples. (B) Representative indirect immunofluorescence images of JCV LT 48 hrs post-infection. Samples were mounted on slides with DAPI medium.
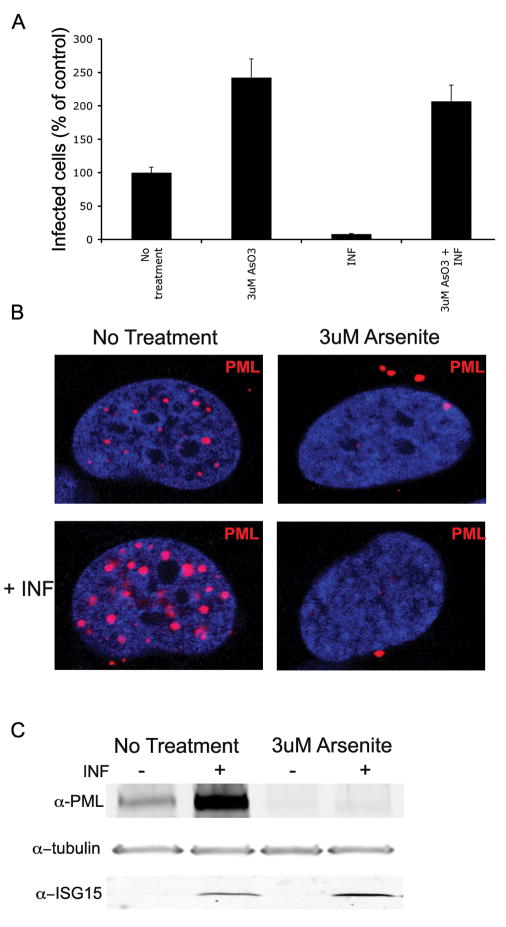
Treatment of cells with IFN-β or arsenite may have effects on other cellular processes. To determine whether the effect of interferon on JCV was mediated through PML, SVG-A cells were pre-treated 24 hours prior to JCV infection with arsenite to eliminate PML. The SVG-A cells were infected and INF-β was added at the time of infection. Infection was evaluated 48 hours later with an indirect immunofluorescence assay of LT. JCV infection was no longer inhibited by INF-β when PML was removed with arsenite (Fig 5A). To confirm the PML protein status in cells treated with both arsenite and interferon, uninfected SVG-A cells were pre-treated with arsenite for 24 hours and then treated with INF-β. These cells were analyzed by confocal microscopy or SDS-PAGE and Western blot analysis (Fig 5B&C). Both assays showed that PML was eliminated in the samples pre-treated with arsenite and that the addition of INF-β did not induce PML expression. To verify that INF-β was still active in cells treated with arsenite, cell lysates were evaluated for ISG-15. ISG-15 is an interferon responsive gene and is still upregulated by interferon-β even in the presence of arsenite (Fig 5C). These results indicate that changes in PML protein levels have dramatic effects on viral gene expression and infection. Consistent with our previous shRNA findings, JCV is not only able to infect cells in the absence of PML, its infectivity is drastically increased when PML is removed. When PML protein is upregulated with interferon, JCV infection is drastically decreased. Additionally, INF-β decreases JCV infection in a PML dependent manner.
Figure 5. Interferon-β requires PML to inhibit JCV infection.
(A) SVG-A cells were pre-treated 24 hours prior to infection with media (no treatment) or 3uM arsenite. Cells were infected with JCV and interferon-β (INF) was added at the time of infection. Arsenite and interferon-b were both maintained during the course of the infection. Infection was scored 48 hrs later using indirect immunofluorescence of JCV LT. Positive cells were quantified and the data represent an average of three infections compared to no treatment (set at 100%). There is a drastic increase in infection when samples are pre-treated with arsenite. Conversely, there is a decrease in infection when samples are treated with INF-β. However, IFN-β is no longer able to inhibit infection in samples were PML was eliminated with arsenite. Error bars represent the standard deviation of the samples. (B) SVG-A cells were pre-treated 24 hours with media (no treatment) or 3uM arsenite. After 24 hour pre-treatment IFN-β was added to cells. Treated cells were visualized using confocal microscopy with a 63X objective 24 hours post-interferon treatment. Samples treated with INF-β alone showed increase number and size of PML speckles. However, when SVG-A cells were treated with INF after arsenite treatment, PML was no longer present. (C) SVG-A cells were pre-treated 24 hours with media (no treatment) or 3uM arsenite. After 24 hour pre-treatment IFN-β was added to cells. Whole cell extracts were collected 24 hours later. Samples treated with INF-β alone showed an increased amount of PML protein but this increase is not seen when cells have been pre-treated with 3uM arsenite. INF-β is still active in arsenite treated samples as shown by ISG15 induction. Tubulin is shown a loading control.
Arsenite has no effect on JCV entry but does increase viral transcripts
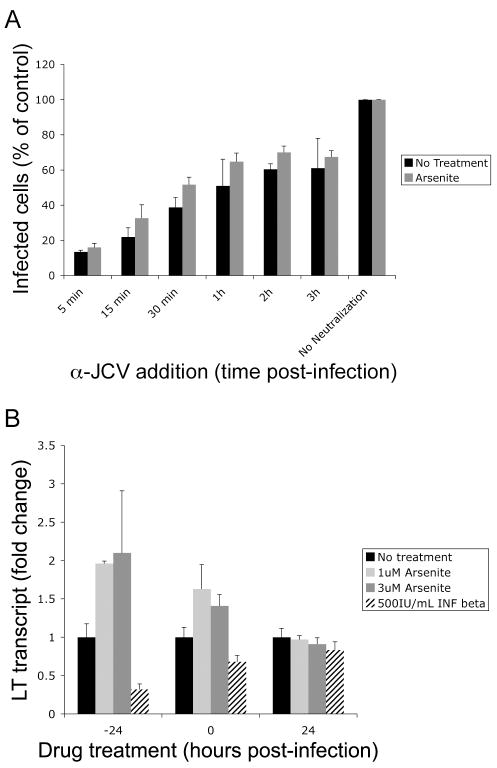
Upon arsenite treatment, the number of cells infected with JCV increases substantially indicating that arsenite induces a global change in the cells that makes them more permissive to viral infection. To evaluate which step of the viral lifecycle was enhanced by arsenite, we began by analyzing viral entry. Effects on entry were evaluated by infecting cells that were either pre-treated with arsenite or treated with media alone. JCV infection was neutralized at regular intervals post-infection using neutralizing anti-JCV rabbit serum. Infection was scored 48 hours later using an indirect immunofluorescence assay of LT (Fig. 6A). Both untreated and arsenite treated cells exhibited the same rate of neutralization, suggesting that the arsenite dependent enhancement in JCV infection is not due to an increase in viral entry.
Figure 6. Arsenite does not enhance JCV entry but increases viral transcripts.
(A) SVG-A cells untreated or pre-treated with arsenite were infected with JCV. The infection was neutralized at indicated times post-infection with an anti-JCV rabbit serum. JCV positive cells were scored 48 hrs post-infection using indirect immunofluorescence of JCV LT. The average of three trials is plotted compared to no neutralization controls set as 100%. Error bars represent the standard deviation of the samples. (B) Cellular RNA was harvested from SVG-A cells treated with media (no treat), 1uM arsenite, 3uM arsenite, or 500IU/mL interferon beta at indicated times. Viral transcripts were detected using qPCR with JCV specific LT primers and probes. All samples were performed in triplicate. An average of three experiments is represented. Error bars indicate the standard deviation of the samples. Similar to infection results, arsenite increased viral transcripts while INF-β decreased viral transcripts. There was no effect when treatment was performed 24 hrs post-infection.
ND-10 domains contain a variety of transcriptional regulators whose localization and activity are modulated by other domain components. To determine if viral transcripts were increased during infection, SVG-A cells were treated with arsenite and INF-β. Cells were either pre-treated, treated at the time of infection or 24 hours post-infection. Cells were harvested 48 hours post-infection and viral transcript levels were assayed using qPCR with viral specific primers and probes to LT. We found that arsenite treatment significantly enhanced virus specific transcription and that IFN-β treatment significantly inhibited viral transcripts (Fig 6B). These data demonstrate that the effect of arsenite and IFN-β on JCV infection is at the level of transcription and suggest that ND-10 domain components repress viral transcription.
JCV does not eliminate PML during infection
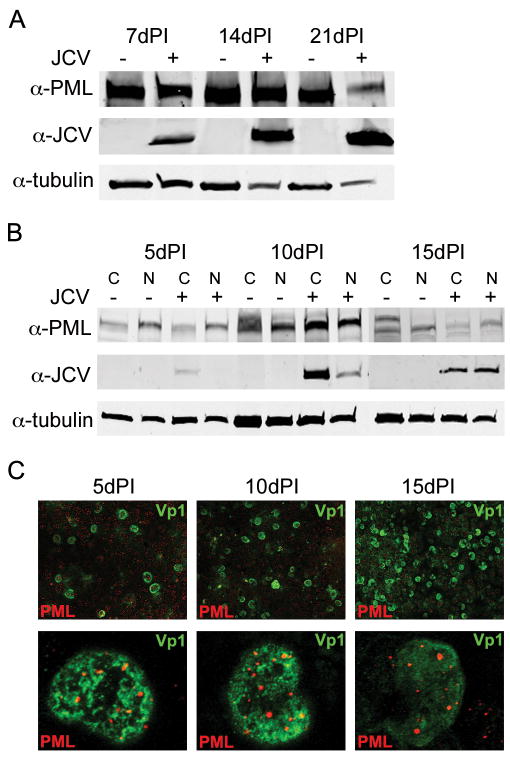
Many viruses interact with PML and modulate its expression during infection. To evaluate PML protein during JCV infection, permissive SVG-A cells were infected with JC virus. PML and Vp1 protein levels were monitored over a long time course of infection. Whole cell extracts were collected 7, 14, and 21 days post-infection and their PML and viral protein levels were monitored by Western blot analysis (Fig. 7A). Tubulin levels were also evaluated as a control. JCV protein levels increase over time, but the presence of PML protein remains throughout the infection. The decrease in PML at 21 days post-infection is due to cytopathic effects (CPE) of the virus. When quantified by infrared scanning of proteins, PML is expressed at the same level relative to tubulin as the no infection control (data not shown). To determine whether PML is re-localized during infection, cytoplasmic and nuclear extracts were collected at 5, 10, and 15 days post-infection, all days prior to the development of CPE (Fig. 7B). Cells collected at these time points were also stained for the presence of JCV Vp1 and PML and evaluated by confocal microscopy (Fig. 7C). Over these time points, JCV infection spreads throughout the culture. However, there is no effect on the nuclear localization of the PML protein, since it is still observed in all samples. PML also maintains its characteristic nuclear speckled pattern over the time of infection. This is also confirmed through immunoblot analysis of the cytoplasmic and nuclear extracts (Fig. 7B). PML has many isoforms and post-translational modifications, which can be modulated by the cell cycle or other cellular proteins. We observed multiple PML reactive bands in the cytoplasmic fractions over time. This is probably due to increased numbers of cells in different phases of the cell cycle. Although other viruses are able to eliminate PML or sequester it during infection, we demonstrate JCV is unable to remove or modulate PML during infection.
Figure 7. JCV does not eliminate PML protein during infection.
(A) SDS-PAGE and Western blot analysis of whole cell lysates from uninfected and JCV infected SVG-A cells 7, 14, and 21 days post-infection (dPI). JCV infection spreads over time, but PML protein levels remained constant throughout the infection. (B) Cytoplasmic (C) and nuclear (N) extracts from uninfected and JCV infected cells at 5, 10, and 15 days post-infection (dPI). There was no change in the cellular localization of PML during the course of JCV infection. (C) Representative indirect immunofluorescence images of JCV VP1 and PML 5, 10, and 15 days post-infection (dPI). Top panels represent 20X field of JCV infection, which spreads throughout the culture over the course of infection. Bottom panels represent 63X images of Vp1 positive nuclei showing no change in the appearance of PML domains during the infection.
Discussion
DNA viruses deliver their genomes to the nucleus of the host cell. Within the nucleus they are able to modulate host cell proteins and activities in order to transcribe and replicate their genomes. Polyomaviruses use their early regulatory protein LT to interact with p53 and Rb, thereby regulating the cell cycle and allowing for viral replication (Stubdal et al., 1997; Sullivan et al., 2000; Zalvide, Stubdal, and DeCaprio, 1998). Other DNA viruses interact with nuclear domain components such as PML. Herpesviruses cause PML to be degraded through a proteasome-dependent pathway to efficiently complete their lifecycle (Everett et al., 2006; Maul, Guldner, and Spivack, 1993), while adenovirus sequesters PML to remove PML’s repressive effects on its lifecycle (Rosa-Calatrava et al., 2003). In the current study we explored the relationship between JC virus regulatory proteins and nuclear domain components. Previous studies have shown viral capsid proteins Vp2 and Vp3 localize adjacent to PML domains. It is hypothesized that this localization is necessary for packaging and assembly of JCV virions (Shishido-Hara et al., 2008; Shishido-Hara et al., 2004). Using shRNA, we were able to show that PML protein is not required for JC virus to produce its early and late proteins as determined by indirect immunofluorescence.
PML domains are sites where many proteins, including a variety of transcription factors, are localized depending on their post-translational modification status. Treatment of cells with arsenite has been shown to cause changes in the SUMOylation and phosphorylation status of PML (Lallemand-Breitenbach et al., 2001; Zhu et al., 1997). These changes affect the activities of transcriptional regulators within the domain (Everett et al., 2007). Arsenite has been used in the treatment of acute promyelocytic leukemia (APL) since the 1990’s, the concentrations that were used in our experiments are in the same therapeutic range as used to treat APL. Arsenite was found to be more effective at causing remission than the all-trans retinoic acid treatment used previously. It is also used to treat APL that is resistant to retinoic acid treatment (Wang and Chen, 2008).
In our experiments, altering PML protein had drastic effects on the capacity of JCV to infect SVG-A cells. Upon arsenite treatment, PML is eliminated and infection is enhanced. The opposite response is seen when PML is upregulated with interferon-β, thereby inhibiting infection. The inhibition by interferon-β was dependent on the presence of PML since it is no longer able to be repressive in arsenite treated cells where PML is eliminated. We did not observe any specific viral transcription factors activated from the cytoplasm in response to changes in PML (data not shown). However, there are many other regulators within the ND-10 domains, which can be repressed by PML or other domain components. This is supported by the increase in viral transcript levels seen with arsenite treatment.
To understand whether any JCV proteins interact with the ND-10 domain or regulate its localization during infection, we evaluated LT microdomains in association with ND-10 components, as well as levels and localization of PML itself during a long-term infection. When the early viral protein large T was evaluated, we noted it accumulated in microdomains in the nucleus. These viral domains were localized adjacent to ND-10 components PML, Sp100, and Daxx. When PML was eliminated with arsenite treatment, these viral domains were unaffected, suggesting they do not require PML as a scaffold protein. Upon interferon treatment, however, the viral domains became smaller and more closely associated with ND-10 components. This suggests the antiviral nature of PML domains could be through sequestering viral proteins or transcription factors within the PML domains. JCV did not alter the expression of PML during infection: PML was seen at constant levels throughout JCV infection. PML was also not re-localized during infection suggesting PML removal is not necessary to achieve infection by JCV.
It is beneficial for viruses to localize their processes to distinct regions, since it makes the process of viral replication and packaging most efficient. Why would polyomaviruses choose to localize close to domains known to have antiviral properties? PML domains are also sites of many regulators, including cell cycle regulators and transcription factors. Polyomaviruses are small viruses that require many cellular factors to efficiently complete their lifecycles. Cell cycle regulators such as p53 are associated with PML domains and are also critical for LT induction of S-phase in order to allow for viral replication. SV40 and BKV require localization close to PML domains for their replication. Overall these domains are very complex, tightly localized, and have many properties that would be valuable for viruses to exploit.
Materials and methods
Cells, viruses, and antibodies
SVG-A cells are a subclone of the original SVG human glial cell line established by transformation of human fetal glial cells by an origin-defective SV40 mutant (Aksamit et al., 1985). SVG-A cells were maintained in a humidi ed 37°C CO2 incubator in Eagle’s minimal essential medium (EMEM) (Mediatech Inc., Herndon, Va.), supplemented with 10% heat-inactivated fetal bovine serum (Mediatech Inc.). The PAB597 hybridoma produces a monoclonal antibody against the SV40 major capsid protein VP1 and was a generous gift from Ed Harlow. This antibody has previously been shown to cross-react with JCV VP1 (Atwood et al., 1995). The PAB962 hybridoma produces a monoclonal antibody against the JCV large T antigen, which was a generous gift from the Tevethia lab. The Mad-1/SVEΔ strain of JCV was used in these experiments as it has enhanced growth kinetics over wild type strains of the virus. The coding sequences of the Mad-1/SVEΔ strain is entirely from the Mad-1 strain. The regulatory region has elements of both Mad-1 and SV40(Liu and Atwood, 2001; Liu, Hope, and Atwood, 1998). Rabbit serum containing anti-JCV neutralizing antibody was used to neutralize infection. Other antibodies used were directed against PML H-238 and PGM3 (these detect all isoforms of PML) (Santa Cruz Biotech, Santa Cruz, CA), Sp100 (H-60), ISG15 (H-150) (Santa Cruz Biotech, Santa Cruz, CA), tubulin (Santa Cruz Biotech, Santa Cruz, CA) and Daxx (Abcam, Cambridge, MA),
Indirect immunofluorescence
SVG-A cells were grown to 50% confluence on coverslips. Cells were either treated with 1uM arsenite, 3uM arsenite, or 500IU/mL interferon beta 24h prior to infection. Drugs were removed during infection and the cells were incubated with JCV at 37°C for 1.5 hrs. At the end of the incubation, cells were rinsed in EMEM then fed with EMEM (drugs were replaced and maintained for the duration of the experiment). For 24h post-infection drug treatment, medium was removed and replaced with medium containing drug concentrations described above. JCV infection was assayed 48 hrs post-infection for LT or 72 hrs for Vp1. Cells were fixed in 2% paraformaldehyde (PFA) for 20 mins and washed several times in phosphate-buffered saline (PBS) (137 mM NaCl, 2.682 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4, pH 7.2). Cells were permeabilized in 0.5% Triton-X 100 at room temperature for 15 mins, then incubated with a 1:10 dilution of the PAB597 or PAB962 monoclonal antibody in PBS at 37°C for 1h. Cells were washed three times in PBS, incubated with a 1:500 dilution of goat anti-mouse Alexa Fluor 488 (Molecular Probes, Carlsbad, CA) at 37°C for 45 min, and rinsed in PBS. Coverslips were mounted onto slides with mounting medium containing DAPI (Vector Labs, Burlingame, Calif.). JCV-positive cells were visualized on a Nikon epifluorescence microscope Eclipse E800 (Nikon Inc., Melville, N.Y.) and scored by counting. At least ten visual fields were counted for each sample.
Confocal microscopy
SVG-A cells were grown to 50% confluence on coverslips. Cells were treated either with 1uM arsenite, 3uM arsenite, or 500IU/mL interferon beta 24 hrs prior to infection. Drugs were removed during infection and the cells were incubated with JCV at 37°C for 1.5 hrs. At the end of the incubation, cells were rinsed in EMEM and fed with EMEM (drugs were replaced and maintained for the duration of the experiment). At 48 hrs post-infection, cells were fixed in 2% PFA for 20 mins, washed several times in PBS, permeabilized in 1% Triton-X 100 at RT for five mins, blocked in 5% bovine serum albumin (BSA) at RT for 1h, and incubated with PAB962 (1:10) and either PML, Sp100, or Daxx (all 1:50) at RT for 1h. Cells were then washed four times in PBS with 0.5% BSA and 0.05% Tween-20 (PBS-BT) and incubated with a 1:1,000 dilution of goat anti-mouse Alexa Fluor 488 (Molecular Probes, Carlsbad, CA) and goat anti-rabbit Alexa Fluor 594 (Molecular Probes, Carlsbad, CA) at 37°C for 45 min. Cells were washed four times in PBS-BT, one time in PBS, and one time in diH2O. Coverslips were mounted onto slides with mounting medium containing DAPI (Vector Labs, Burlingame, Calif.). Cells were visualized on a Zeiss LSM 510 meta laser-scanning confocal microscope (Carl Zeiss, New York, NY) using a 63X objective.
shRNA knockdown
shRNA constructs were generated using siSTRIKE plasmid (Promega, Madison, WI) and primers: [(PML): 5′accgAGATGCAGCTGTATCCAAGTTCAAGAGACTTGGATACAGCTGCATCTTTTTTTc3′; (PML2): 5′tgcagAAAAAAAGATGCAGCTGTATCCAAGTCTCTTGAACTTGGATACAGCTGCATCT-3′; (Luc): 5′accgGTGCGTTGCTAGTACCAACTTCAAGA GAGTTGGTACTAGCAACGCACTTTTTTc; (Luc2): 5′tgcagAAAAAAGTGCGTTGCTAGTACCAACTCTCTTGAAGTTGGTACTAGCAACGCAC]. SVG-A cells at 50% confluence were transfected with 1ug of either PML plasmids or control luciferase plasmid using FuGene per manufacturers directions. Knockdown was monitored using indirect immunofluorescence of PML at 24-hr intervals as described. Cells were fixed in 2% PFA for 20 mins, washed several times in PBS, and permeabilized in 0.5% Triton-X 100 at RT for 15 mins. Cells were then incubated with PML antibody (1:50) at 37°C for 1 hr, washed three times in PBS, incubated with goat anti-rabbit Alexa Fluor 594 (1:500) (Molecular Probes) at 37°C for 45 min, and rinsed in PBS. Coverslips were mounted onto slides with 100% glycerol. Eight days post-transfection, transfected cells were infected with JCV at 37°C for 1.5 hrs. At the end of the incubation, cells were rinsed in EMEM then fed with EMEM. JCV infection was assayed 72 hrs for Vp1. Cells were fixed in 2% PFA for 20 mins and washed several times in PBS. Cells were permeabilized in 0.5% Triton-X 100 a RT for 15 mins, then incubated with a 1:10 dilution of the PAB597 monoclonal antibody in PBS at 37°C for 1h. Cells were washed three times in PBS, incubated with a 1:500 dilution of goat anti-mouse Alexa Fluor 594 (Molecular Probes, Carlsbad, CA) at 37°C for 45 min, and rinsed in PBS. Coverslips were mounted on slides with 100% glycerol. Cells were visualized on a Nikon epifluorescence microscope (Eclipse E800; Nikon Inc., Melville, N.Y.).
Western blot analysis
SVG-A cells were grown to 70% confluence in 75-cm2 flasks and treated with 3uM arsenite or 500IU/mL interferon beta in EMEM. Cells were then harvested 24 or 48 hrs post-treatment by washing twice in cold PBS and lysing in ice cold radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris HCl, pH 7.4, 0150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Sigma-Aldrich], 1 mM sodium orthovanadate) for 30 mins on ice. Samples were collected by pelleting membranes by centrifugation at 13,000rpm for 10 mins and then removing the protein-containing supernatant. Protein samples were loaded onto a 4–15% Tris-HCl polyacrylamide gel (Bio-Rad, Hercules, CA), run at 30mA, transferred to nitrocellulose membranes using a mini-trans blot apparatus (Bio-Rad), and blocked with 5% milk in PBS containing 0.05% Tween 20 (PBS-T). Blots were probed with the respective antibodies all diluted in 5% milk in PBS-T, washed in PBS-T, and then incubated with goat anti-rabbit Alexa Fluor 680 (Molecular Probes) antibody diluted 1:5,000 in 5% milk in PBS-T. This was followed by further washes with PBS-Tween 20 and one in PBS. Blots were viewed using an infrared scanner (LI-COR, Lincoln, NE) and analyzed using Odyssey software (LI-COR).
Nuclear and cytoplasmic extracts
SVG-A cells were grown to 70% confluence in 150-cm2 flasks and treated with 3uM arsenite or 500IU/mL interferon beta in EMEM. Cells were then harvested 1, 4, and 24 hrs post-treatment by scraping. Cell pellets were washed two times in cold PBS and cells were pelleted at 2,000rpm at 4°C. Pellets were resuspended in five times cell pellet volume of pre-lysis buffer (100mM HEPES pH 7.9; 15mM MgCl2; 100mM KCl, 0.01M DTT and protease inhibitors) on ice for 15 mins. Cells were pelleted at 2,000rpm for five minutes and supernatant was removed. Pellet was resuspended in twice the pellet volume of lysis buffer and a syringe was used to disrupt the membrane. The samples were centrifuged at 10,500rpm for 20 mins and the cytosolic containing supernatant was frozen at −20°C. The remaining pellet was resuspended in 2/3 cell pellet volume of extraction buffer (20mM HEPES H 7.9; 1.5mM MgCl2; 0.42M NaCl; 0.2mM EDTA, 25% (v/v) glycerol, 0.01M DTT and protease inhibitors) and a syringe was used to disrupt the nuclear membrane. The samples were then incubated on a rocking platform at 4°C for 30 mins. Samples were collected by centrifugation at 10,000rpm for 15 mins and the nuclear-containing supernatant was frozen at −20°C. Samples were then subjected to western blot analysis as previously described.
Neutralization of JCV
50% confluent SVG-A cells were pre-treated with EMEM or EMEM containing 3uM arsenite for 24 hrs. SVG-A cells were then infected with JCV for the indicated time at 37°C. Cells were washed in EMEM and media containing anti-JCV antiserum was added to the cells. Viral infection was assayed 72 hrs later for Vp1 as previously described.
qPCR of JCV transcripts
SVG-A cells were grown to 50% confluence on 6-well plates in triplicate. Cells were either treated with 1uM arsenite, 3uM arsenite, or 500IU/mL interferon beta 24 hrs prior to infection. Drugs were removed during infection and the cells were incubated with JCV at 37°C for 1.5 hrs. At the end of the incubation, cells were rinsed in EMEM and fed in EMEM (drugs were replaced and maintained for the duration of the experiment). For 24h post-infection drug treatment, medium was removed and replaced with medium containing above described drug concentrations. 48h post-infection cells were washed with cold PBS and detached with trypsin. Cells were pelleted at 2,000rpm for five mins and washed two times with cold PBS. RNA was extracted using RNeasy Kit (Qiagen, Germantown, MD). Sample concentrations were determined and 1ug of RNA was used for one-step PCR with probes (BioRad). Primers used (LT): ttcttcatggcaaaacaggtctt and ttccaccaggattcccattc; Probe used: ccacttctcattaaatg as previously described (Applied Biosystems, Foster City, CA) (McNees et al., 2005). GAPDH was used as an internal control. GAPDH primers and probes were obtained from Applied Biosystems. All samples were performed in triplicate.
Supplementary Material
Acknowledgments
We thank all the members of the Atwood Laboratory for critical discussion during the course of this work. We thank the LeDuc Bioimaging Facility at Brown for help with microscopy and the Center for Genomics and Proteomics for help with real time PCR. Work in our laboratory is supported by grants from the National Cancer Institute (CA71878) and from the National Institute of Neurological Diseases and Stroke (NS43097) to WA. Megan L. Gasparovic was supported by a Ruth L. Kirschstein Predoctoral Fellowship #1F31NS053340-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Aksamit AJ, Mourrain P, Sever JL, Major EO. Progressive multifocal leukoencephalopathy: investigation of three cases using in situ hybridization with JC virus biotinylated DNA probe. Ann Neurol. 1985;18:490–496. doi: 10.1002/ana.410180412. [DOI] [PubMed] [Google Scholar]
- Astrom K, Mancall E, Richardson EP., Jr Progressive multifocal leukoencephalopathy. Brain. 1958;81:93–127. doi: 10.1093/brain/81.1.93. [DOI] [PubMed] [Google Scholar]
- Atwood WJ, Wang L, Durham LC, Amemiya K, Traub RG, Major EO. Evaluation of the role of cytokine activation in the multiplication of JC virus (JCV) in human fetal glial cells. J Neurovirol. 1995;1:40–49. doi: 10.3109/13550289509111009. [DOI] [PubMed] [Google Scholar]
- Becker KA, Florin L, Sapp C, Maul GG, Sapp M. Nuclear localization but not PML protein is required for incorporation of the papillomavirus minor capsid protein L2 into virus-like particles. J Virol. 2004;78(3):1121–8. doi: 10.1128/JVI.78.3.1121-1128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna KJ, Nathans D. Bidirectional replication of Simian Virus 40 DNA. Proc Natl Acad Sci U S A. 1972;69(11):3097–100. doi: 10.1073/pnas.69.11.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan AS, Maginnis MS, Jordan JA, Gasparovic ML, Manley K, Page R, Williams G, Porter E, O’Hara BA, Atwood WJ. Human alpha-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J Biol Chem. 2008;283(45):31125–32. doi: 10.1074/jbc.M805902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89(6–7):819–30. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Everett RD, Murray J, Orr A, Preston CM. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol. 2007;81(20):10991–1004. doi: 10.1128/JVI.00705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80(16):7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareed GC, Garon GF, Salzman NP. Origin and direction of simian virus 40 deoxyribonucleic acid replication. J Virol. 1972;10(3):484–91. doi: 10.1128/jvi.10.3.484-491.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Schafer F, Sotlar K, Streeck RE, Sapp M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology. 2002;295(1):97–107. doi: 10.1006/viro.2002.1360. [DOI] [PubMed] [Google Scholar]
- Frisque R, Bream G, Cannella M. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134(4):815–26. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci. 2004;117(Pt 17):3807–20. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- Jul-Larsen A, Visted T, Karlsen BO, Rinaldo CH, Bjerkvig R, Lonning PE, Boe SO. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp Cell Res. 2004;298(1):58–73. doi: 10.1016/j.yexcr.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5(3):e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krynska B, Gordon J, Otte J, Franks R, Knobler R, DeLuca A, Giordano A, Khalili K. Role of cell cycle regulators in tumor formation in transgenic mice expressing the human neurotropic virus, JCV, early protein. J Cell Biochem. 1997;67(2):223–30. doi: 10.1002/(sici)1097-4644(19971101)67:2<223::aid-jcb7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193(12):1361–71. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AY, Chan M, Tang SC, Liang R, Kwong YL. Real-time quantitative analysis of polyoma BK viremia and viruria in renal allograft recipients. J Virol Methods. 2002;103(1):51–6. doi: 10.1016/s0166-0934(01)00447-5. [DOI] [PubMed] [Google Scholar]
- Liu CK, Atwood WJ. Propagation and assay of the JC virus. Methods Mol Biol. 2001;165:9–17. doi: 10.1385/1-59259-117-5:9. [DOI] [PubMed] [Google Scholar]
- Liu CK, Hope AP, Atwood WJ. The human polyomavirus, JCV, does not share receptor specificity with SV40 on human glial cells. J Neurovirol. 1998;4:49–58. doi: 10.3109/13550289809113481. [DOI] [PubMed] [Google Scholar]
- Major EO, Amemiya K, Tornatore CS, Houff SA, Berger JR. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Guldner HH, Spivack JG. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74 (Pt 12):2679–90. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- Maul GG, Yu E, Ishov AM, Epstein AL. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59(4):498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- McNees AL, White ZS, Zanwar P, Vilchez RA, Butel JS. Specific and quantitative detection of human polyomaviruses BKV, JCV, and SV40 by real time PCR. J Clin Virol. 2005;34(1):52–62. doi: 10.1016/j.jcv.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Miller WH, Jr, Schipper HM, Lee JS, Singer J, Waxman S. Mechanisms of action of arsenic trioxide. Cancer Res. 2002;62(14):3893–903. [PubMed] [Google Scholar]
- Nakahara T, Lambert PF. Induction of promyelocytic leukemia (PML) oncogenic domains (PODs) by papillomavirus. Virology. 2007;366(2):316–29. doi: 10.1016/j.virol.2007.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negorev D, Ishov AM, Maul GG. Evidence for separate ND10-binding and homo-oligomerization domains of Sp100. J Cell Sci. 2001;114(Pt 1):59–68. doi: 10.1242/jcs.114.1.59. [DOI] [PubMed] [Google Scholar]
- Padgett B, ZuRhein G, Walker D, Echroade R, Dessel B. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet. 1971;I:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- Richardson EP., Jr Progressive multifocal leukoencephalopathy. N Engl J Med. 1961;265:815–823. doi: 10.1056/NEJM196110262651701. [DOI] [PubMed] [Google Scholar]
- Rosa-Calatrava M, Puvion-Dutilleul F, Lutz P, Dreyer D, de The H, Chatton B, Kedinger C. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 2003;4(10):969–75. doi: 10.1038/sj.embor.embor943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido-Hara Y, Higuchi K, Ohara S, Duyckaerts C, Hauw JJ, Uchihara T. Promyelocytic leukemia nuclear bodies provide a scaffold for human polyomavirus JC replication and are disrupted after development of viral inclusions in progressive multifocal leukoencephalopathy. J Neuropathol Exp Neurol. 2008;67(4):299–308. doi: 10.1097/NEN.0b013e31816a1dd3. [DOI] [PubMed] [Google Scholar]
- Shishido-Hara Y, Ichinose S, Higuchi K, Hara Y, Yasui K. Major and minor capsid proteins of human polyomavirus JC cooperatively accumulate to nuclear domain 10 for assembly into virions. J Virol. 2004;78(18):9890–903. doi: 10.1128/JVI.78.18.9890-9903.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubdal H, Zalvide J, Campbell KS, Schweitzer C, Roberts TM, DeCaprio JA. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17(9):4979–90. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Pipas JM. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol Mol Biol Rev. 2002;66(2):179–202. doi: 10.1128/MMBR.66.2.179-202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Tremblay JD, Fewell SW, Lewis JA, Brodsky JL, Pipas JM. Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol Cell Biol. 2000;20(15):5749–57. doi: 10.1128/mcb.20.15.5749-5757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge PW, Frisque RJ. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J Neurovirol. 1995;1(2):195–206. doi: 10.3109/13550289509113966. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111(5):2505–15. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- Zalvide J, Stubdal H, DeCaprio JA. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18(3):1408–15. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Koken MH, Quignon F, Chelbi-Alix MK, Degos L, Wang ZY, Chen Z, de The H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci U S A. 1997;94(8):3978–83. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber A, Nguyen QD, Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal. 2004;16(10):1085–104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.