Abstract
NMR magnetization transfer from water and ammonia-catalyzed exchange of the imino proton have been used to probe enhanced thermostability and conformational rearrangements induced by Mg2 + in two key activity fragments r(CACCUGGCGACAGGUG) and r(GGCCAAAAGCC) of the encephalomyocarditis virus (EMCV) picornavirus internal ribosome entry site (IRES). We have measured some of their r(G·C) base-pair lifetimes and dissociation constants under different MgCl2 conditions, and we compare them with those of other short RNA duplexes. The RNA fragment r(CACCUGGCGACAGGUG) adopts two topologies, a palindromic duplex with two conformations and a hairpin, whose equilibrium can be monitored: the duplex form is destabilized by Mg2 + and temperature, a delicate balance wherein the entropic contribution of the free energy helps populate the hairpin state. For both fragments, the opening rates of the r(G·C) pairs are lower in the presence of Mg2 + and their dissociation constants are smaller or comparable. Analysis of the results suggests that Mg2 + has a preferential and specific effect on the r(CACCUGGCGACAGGUG) hairpin in the region close to the r(G·C) closing pair of the GCGA tetraloop, and the ion moves diffusively around r(GGCCAAAAGCC), thereby differentiating the GNRA and RAAA hairpin motifs that are both involved in the biological regulation functions of the EMCV IRES.
Keywords: Base-pair kinetics, EMCV IRES, GNRA tetraloop, Imino proton exchange, Mg2 + , NMR, RAAA hairpin motif
Introduction
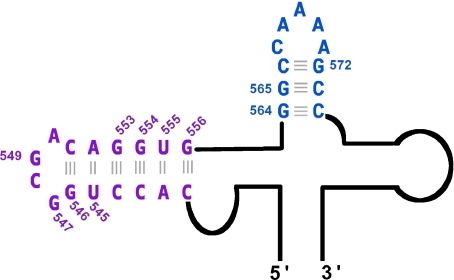
Picornaviruses are the leading cause of infectious morbidity in the world and replicate in the eukaryotic cell cytoplasm with the help of a genome segment called the internal ribosome entry site (IRES) [1–3]. Encephalomyocarditis virus (EMCV) is a member of the cardiovirus genus of the positive-sense picornaviruses. Its IRES is organized in five structural domains, termed 1 to 5 or G to L from the 5′ to the 3′ end [4]. Domain 3 contains a hammerhead region (Fig. 1) partly constituted by apical stem loops with two phylogenetically conserved motifs bearing the sequences GNRA and RAAA (N can be A, C, G or U; R is A or G). In the EMCV IRES, the GCGA tetraloop (in mauve, Fig. 1) and the AAAA submotif (in blue, Fig. 1) are essential for the activity of the IRES within mammalian cells [5, 6] and maintain the IRES three-dimensional structural integrity, including its dependence on ions. Specific binding of the GNRA hairpin to the central zone of domain 3 was also observed and may be stabilized by the RAAA hairpin local conformation [7]. 1H NMR and UV melting experiments [8] have shown that addition of Mg2 + induces selective changes to the chemical shifts of the imino protons and a dramatic increase in apparent melting temperature of fragments containing the GCGA tetraloop and the AAAA tetranucleotide, suggesting that Mg2 + may promote folding of the EMCV IRES and enhance its thermodynamic stability. Here, we present an experimental study of Mg2 + effects on the conformational equilibria of two EMCV IRES fragments, one containing a GNRA hairpin motif and the other an RAAA hairpin motif, and make a quantitative local analysis of the retardation of solvent exchange with r(G·C) base-pairs upon Mg2 + addition, interpreting it in terms of base-pair kinetics.
Fig. 1.
Schematic diagram showing the EMCV IRES hammerhead region containing the 16-mer (in mauve) and 11-mer (in blue) hairpins. The secondary structure base-pair numbering is based on the nomenclature proposed previously [2]. The four residues spanning 5′ -[G547-A550]-3′ form the GCGA tetraloop. The four residues spanning 5′ -[A568-A571]-3′ constitute the AAAA tetranucleotide motif
Base pairing disruption studies constitute a useful characterization and complement to nucleic acid structure determination by NMR, giving a relatively complete picture of the base-pair elementary dynamics and a comprehensive local description of the structure accessibility. Over the past two decades, base-pair kinetics has proven to be a very sensitive probe of nucleic acid structures [9, 10]. For example, d(G·C) base-pair lifetimes are increased by two orders of magnitude in Z DNA as compared to B DNA [11, 12]. To address the question of a possible EMCV IRES conformational rearrangement induced by Mg2 + , we have undertaken an NMR investigation of the imino proton exchange for three r(G·C) base-pairs in the two key activity fragments r(CACCUGGCGACAGGUG) (in mauve, Fig. 1) and r(GGCCAAAAGCC) (in blue, Fig. 1) under different MgCl2 conditions. The fragments r(CACCUGGCGACAGGUG) and r(GGCCAAAAGCC) respectively contain the GCGA tetraloop and the AAAA submotif sequences, and are henceforward designated as the 16-mer and the 11-mer, respectively, in the following text.
Materials and methods
Imino proton exchange theory
Imino proton exchange from a base-pair is a two-step process [13] requiring base-pair opening, followed by transfer to a proton acceptor such as NH3. The proton acceptor contribution to the exchange time is given by:
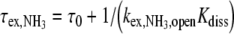 |
1 |
where τ0 is the base-pair lifetime, Kdiss the base-pair dissociation constant, and 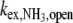 the proton transfer rate from the open pair. The imino proton transfer rate from the open pair
the proton transfer rate from the open pair. The imino proton transfer rate from the open pair 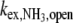 induced by NH3 is:
induced by NH3 is:
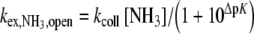 |
2 |
where kcoll is the collision rate, [NH3] is the ammonia concentration, and ΔpK is the pKa difference between the imino proton (pKG = 9.3, pKU = 9.2) and ammonia (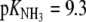 ).
).
Oligonucleotide synthesis
The fragments r(CACCUGGCGACAGGUG) and r(GGCCAAAAGCC) were synthesized using phenoxyacetyl  -cyanoethyl phosphoramidites on a 2-
-cyanoethyl phosphoramidites on a 2-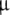 M scale and purified as previously described [10]. The RNA concentration was determined from the UV absorbance using the A 260 values computed according to a nearest neighbor model [14].
M scale and purified as previously described [10]. The RNA concentration was determined from the UV absorbance using the A 260 values computed according to a nearest neighbor model [14].
Sample preparation
The NMR samples contained 0.2 mM 2,2-dimethyl-2-silapentane-5-sulfonate, whose methyl peak was set to 0 ppm for chemical shift reference. The sample pH was measured at room temperature before and after each experiment and adjusted using 0.1 to 1 M HCl and NaOH solutions.
Polyacrylamide gel electrophoresis
The 16-mer and 11-mer RNA fragments were subjected to electrophoresis at 0°C for 45 min (35 V/cm) in a 15% non-denaturing polyacrylamide gel (acrylamide/bis-acrylamide, 19:1) in 1 × TAE, pH 8.0.
NMR methods
All the NMR experiments were performed on a 500 MHz Bruker Advance spectrometer. The imino protons were assigned by 2D NOESY (nuclear Overhauser enhancement spectroscopy) and spectra collected with mixing times of 90 and 250 ms. The 1H NMR titration of the 16-mer sample at 0°C, pH 6.0, was carried out by monitoring the integrals of the imino proton resonance peaks. Ammonia-catalyzed exchange was measured at pH 8.9, a value high enough to provide a fair proportion of NH3. Varying the pH from 6.0 to 8.9 had little effect on the chemical shifts. The ammonia was added to the samples from a stock solution whose concentration was 6.5 M. The ammonia-catalyzed exchange experiments were carried out as previously described [10].
Results and discussion
16-mer and 11-mer topologies
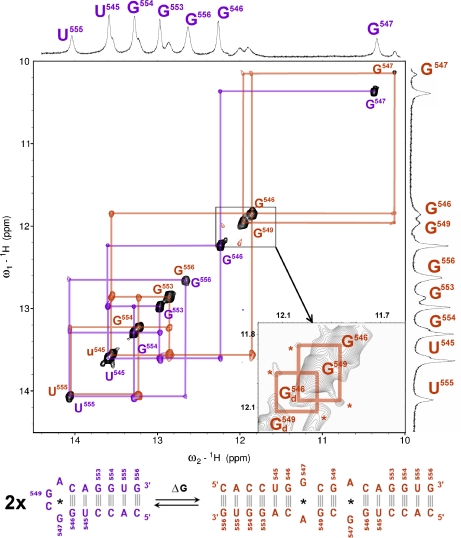
The 16-mer and 11-mer structures were mostly investigated by NOESY NMR. Figure 2 (top) displays the 2D 1H NOESY spectrum of the 16-mer which shows two families of exchangeable protons at the frequencies of the imino protons (10–15 ppm), demonstrating the presence of two different species. RNA is especially prone to adopting conformations other than the native state. NOESY methods allowed us to disentangle the resonance peaks whose assignments are colored in mauve and orange, corresponding to a monomer (the hairpin in mauve) and a palindromic duplex (orange) of the 16-mer (Fig. 2, bottom). The hairpin loop r(G549) imino proton is not visible and is in fast exchange with the solvent on the chemical shift scale. In contrast, the presence of the hairpin loop r(G547) imino proton resonance peak at 10.4 ppm and its weaker intensity indicate that it is partially involved in a non-canonical r(G·A) base-pair. As regards the 16-mer duplex, the duplex r(G549) imino proton resonance peak at 11.9 ppm shows that the additional Watson–Crick base-pairs r(G549·C) is formed. Additionally, the presence of the duplex r(G547) imino proton resonance peak at 10.1 ppm and its weaker intensity indicate that the duplex r(G547) imino proton is partially involved in a non-canonical r(G·A) base-pair. The hairpin and duplex r(U555) and r(G556) imino protons have identical chemical shifts (14.1 and 12.7 ppm, respectively).
Fig. 2.
Top: examples of imino proton NOESY cross-peaks of the monomer and the duplex of the 16-mer. The mauve color indicates the peaks of the monomer form, i.e., the 16-mer hairpin. The protons of the duplex form are colored orange. This color coding is conserved throughout the text. The lower inset shows an enlargement containing the 2D region between 11.8 and 12.1 ppm. r(G546) and r(G549) imino protons of the 16-mer duplex conformer with r(G547·A) in its closed state are termed G546 and G549; r(G546) and r(G549) imino protons of the 16-mer duplex conformer with r(G547·A) in its disrupted state are named G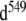 ; their respective cross-peaks are labeled with orange stars. Solution conditions: strand concentration 0.5 mM, 15°C, pH 6.0. Bottom: schematic of the equilibrium between the hairpin (mauve) and the duplex (orange) form of the 16-mer. Non-canonical r(G547·A) base-pairs are indicated by black stars
; their respective cross-peaks are labeled with orange stars. Solution conditions: strand concentration 0.5 mM, 15°C, pH 6.0. Bottom: schematic of the equilibrium between the hairpin (mauve) and the duplex (orange) form of the 16-mer. Non-canonical r(G547·A) base-pairs are indicated by black stars
Several features reveal an internal motion in the region of the duplex r(G547·A) base-pair. Two conformers of the 16-mer duplex are present: the first one has its non-canonical r(G547·A) base-pair formed and the second one has its r(G547·A) base-pair in a disrupted state. This 16-mer duplex conformational exchange is implied by cross-peaks (labeled with orange stars in the inset of Fig. 2) between the r(G546) and r(G549) imino protons (G546 at around 11.8 ppm and G549 at around 11.9 ppm in the inset of Fig. 2) of the duplex conformer with r(G547·A) in its closed state and the r(G546) and r(G549) imino protons (G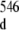 at around 12.0 ppm and G
at around 12.0 ppm and G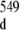 at around 12.1 ppm in the inset of Fig. 2) of the duplex conformer with r(G547·C) in its disrupted state. The broadening of r(G546) and of r(G549) imino proton resonance peaks (1D spectrum with orange labeling, Fig. 2) of the duplex conformer with r(G547·A), in its closed state, is due to exchange with water which is facilitated by the disruption of the adjacent r(G547·A) base-pair. The difference between the chemical shifts of G546 and of G
at around 12.1 ppm in the inset of Fig. 2) of the duplex conformer with r(G547·C) in its disrupted state. The broadening of r(G546) and of r(G549) imino proton resonance peaks (1D spectrum with orange labeling, Fig. 2) of the duplex conformer with r(G547·A), in its closed state, is due to exchange with water which is facilitated by the disruption of the adjacent r(G547·A) base-pair. The difference between the chemical shifts of G546 and of G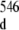 , or between the chemical shifts of G549 and of G
, or between the chemical shifts of G549 and of G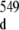 , approximately equals 100 Hz (0.2 ppm × 500 Hz ppm − 1). The frequency of r(G547·A) motion between r(G547·A) disrupted and closed states is then much less than 100 Hz, the corresponding constant rate being therefore much larger than 10 ms. This result, together with the fact that the 16-mer duplex r(G547) imino proton is not visible in the r(G547) disrupted state leads to the conclusion that the r(G547) imino proton in the r(G547) disrupted state is accessible and in fast exchange with the solvent on the chemical shift scale. The r(G547) disrupted state is radically different from the r(G547) open state described in the imino proton exchange theory (Section 2.1).
, approximately equals 100 Hz (0.2 ppm × 500 Hz ppm − 1). The frequency of r(G547·A) motion between r(G547·A) disrupted and closed states is then much less than 100 Hz, the corresponding constant rate being therefore much larger than 10 ms. This result, together with the fact that the 16-mer duplex r(G547) imino proton is not visible in the r(G547) disrupted state leads to the conclusion that the r(G547) imino proton in the r(G547) disrupted state is accessible and in fast exchange with the solvent on the chemical shift scale. The r(G547) disrupted state is radically different from the r(G547) open state described in the imino proton exchange theory (Section 2.1).
We also observed that the 16-mer hairpin/duplex equilibrium is progressively displaced in favor of the duplex form. The 16-mer hairpin lifetime is around 3 months at 5°C, as monitored by real-time 1D 1H NMR. The 16-mer hairpin is trapped in a metastable state and evolves into the 16-mer duplex as a result of spontaneous random fluctuations. Conversely, the 11-mer sample exhibited only a monomer form (the 11-mer hairpin in blue, Fig. 1) as unraveled by 1D 1H spectra and 2D 1H NOESY spectra (not shown).
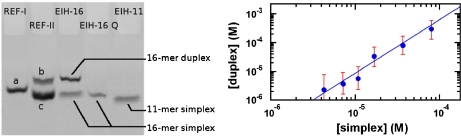
In order to confirm these observations, two types of experiments were carried out. Firstly, polyacrylamide gel electrophoresis corroborated that indeed the two 16-mer species migrate as a monomer and a duplex, whereas the 11-mer only exists as a monomer. Figure 3 (left) displays a polyacrylamide gel stained with ethidium bromide. In the REF-I and REF-II lanes, RNA linear fragments of known length were loaded: lane REF-I a refers to the band of the RNA duplex [r(CGCGAUCGCG)]2; lane REF-II b and c correspond respectively to the bands of the RNA duplex [r(GCGCGCGAUCGCGCGC)]2 and of the RNA monomer r(GCGCGCGAUCGCGCGC). Lane EIH-16 (EIH-16 stands for EMCV IRES Hammerhead 16-mer) shows two bands corresponding to the 16-mer monomer and duplex. Lane EIH-16 Q (EIH-16 Q stands for EMCV IRES Hammerhead 16-mer quenched) displays a unique band from a 16-mer sample heated at 90°C for 3 min and rapidly quenched in an ice-water bath. The 11-mer exists in a monomer form, as shown by lane EIH-11 (EIH-11 stands for EMCV IRES Hammerhead 11-mer).
Fig. 3.
Left: Polyacrylamide gel electrophoresis of the 16-mer and 11-mer. Samples in lanes REF-I and REF-II are references (see text). Samples in lanes EIH-16 and EIH-16 Q contained respectively, the 16-mer (2.5 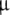 g) and the quenched 16-mer (1.0
g) and the quenched 16-mer (1.0 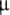 g). Lane EIH-11 displays the 11-mer (1.5
g). Lane EIH-11 displays the 11-mer (1.5  g). Right: NMR titration of the 16-mer duplex form vs. the hairpin concentration at 0°C, pH 6.0. The line of slope = 2 drawn through the data points confirms the formation of a duplex
g). Right: NMR titration of the 16-mer duplex form vs. the hairpin concentration at 0°C, pH 6.0. The line of slope = 2 drawn through the data points confirms the formation of a duplex
Secondly, the 1H NMR titration of the 16-mer sample at 0°C, pH 6.0, which proved that the concentration of the duplex increases as the second power of the monomer concentration (Fig. 3, right), defines the formation of the 16-mer duplex, as one expects for a stoichiometry of 2:
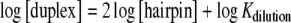 |
3 |
where Kdilution is the equilibrium dissociation constant of the duplex in the 16-mer hairpin/ duplex equilibrium at each dilution point. The 1H NMR titration of the 16-mer was also performed within the temperature range 5–40°C in steps of 5°C, pH 6.0, establishing the formation of the 16-mer duplex as well (not shown). The equilibrium between the two 16-mer forms (Fig. 2, below) is partly explained by the two supplementary r(G549·C) Watson–Crick hydrogen bonds in the 16-mer duplex form and the formation of the two non-canonical r(G547·A) base-pairs [15]. The 11-mer case does not reveal any advantageous hydrogen bond balance in favor of a duplex form, and the entropic driving force simply leads to the sole formation of a hairpin monomer.
Mg2 + control of the 16-mer hairpin/duplex equilibrium
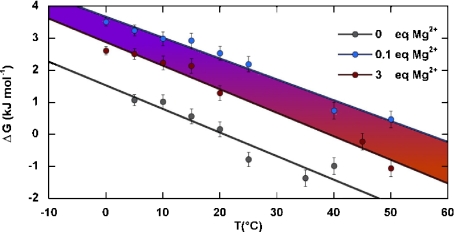
The analysis of such energetic properties would arguably suggest that the 16-mer hairpin/ duplex equilibrium could be monitored by distinguishing the electrostatic contribution of both topologies. Figure 4 demonstrates that Mg2 + favors the 16-mer hairpin formation and provides evidence that 16-mer hairpin enhanced stability arises from Mg2 + . Also, we did not observe any changes in the chemical shifts and the integrals of the 16-mer imino proton resonance peaks upon addition of NaCl (1, 3, and 6 molar equivalents), of KCl (1, 3, and 6 molar equivalents), of MnCl2 (3 molar equivalents), or of CoCl2 (3 molar equivalents). Therefore, a simple increase in the ionic strength did not produce the same effect as the addition of Mg2 + . This observation seems to indicate the specificity of Mg2 + for the 16-mer hairpin. The temperature dependence of the Gibbs free energy ΔG is provided by the equilibrium ratio between the two 16-mer species:
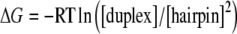 |
4 |
where RT is the thermal energy. The thermodynamic parameters were derived for the equilibrium between the hairpin and the duplex: ΔH increases from 22 to 24 kJ mol − 1 as Mg2 + is added, and ΔS takes an approximately constant value of 74 J mol − 1 K − 1. The derivative of the Gibbs free energy ΔG with respect to the number of added Mg2 + equivalents provides the variation of the chemical potential 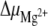 :
:
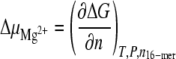 |
5 |
where n is the number of added Mg2 + equivalents, n16 − mer is the number of moles of 16-mers, and T and P are respectively the temperature and the pressure. 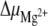 does not change much with temperature and varies between 330 J mol − 1 eq − 1 at 0°C and 510 J mol − 1 eq − 1 at 50°C.
does not change much with temperature and varies between 330 J mol − 1 eq − 1 at 0°C and 510 J mol − 1 eq − 1 at 50°C.
Fig. 4.
The temperature dependence of the Gibbs free energy, ΔG, of the equilibrium between the hairpin and duplex forms of the 16-mer, parameterized by Mg2 + concentration from 0.1 to 3 molar equivalents. For the sake of clarity, the experimental points are only displayed for 0 (gray), 0.1 (blue), and 3 (red) molar equivalents of Mg2 + . A color gradient from orange to mauve symbolizes displacement of the equilibrium between the hairpin (mauve) and the duplex form (orange) of the 16-mer
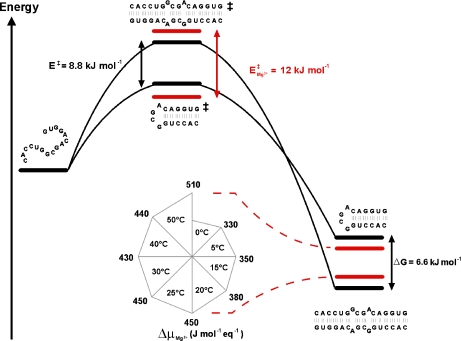
Figure 5 schematically presents a plausible energetic path for the RNA 16-mer sequence, incorporating the various data obtained from the evolution of the 16-mer hairpin with temperature and upon Mg2 + addition. The single-stranded 16-mer random conformation was observed at 90°C; its energy level is therefore placed above those of the 16-mer hairpin and duplex in the diagram. The temperature variation results help in positioning plausible 16-mer transition state energy levels. The 16-mer hairpin/duplex equilibrium Gibbs free-energy differences ΔG and the activation energies 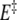 and
and 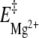 were obtained from the previously mentioned and other 1H NMR titrations of the 16-mer performed within a range of temperatures 0–50°C, with and without Mg2 + . ΔG is around 6.6 kJ mol − 1 and
were obtained from the previously mentioned and other 1H NMR titrations of the 16-mer performed within a range of temperatures 0–50°C, with and without Mg2 + . ΔG is around 6.6 kJ mol − 1 and 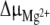 per se, whose average over this temperature range roughly equals 420 J mol − 1 eq − 1, has little influence on the 16-mer hairpin/duplex equilibrium, as corroborated by the coexistence of the two species in solution. An input of thermal energy combined with a Mg2 + addition has a larger activation effect in favor of the 16-mer hairpin and agrees with the explanation that the 16-mer hairpin is trapped in a metastable state:
per se, whose average over this temperature range roughly equals 420 J mol − 1 eq − 1, has little influence on the 16-mer hairpin/duplex equilibrium, as corroborated by the coexistence of the two species in solution. An input of thermal energy combined with a Mg2 + addition has a larger activation effect in favor of the 16-mer hairpin and agrees with the explanation that the 16-mer hairpin is trapped in a metastable state: 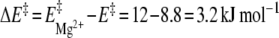 . The 16-mer hairpin lifetime is then around 6 months at 5°C, as monitored by real-time 1D 1H NMR. This duplex-to-hairpin transition induced by Mg2 + is surprising, since one might have expected the reverse switch to occur at increasing polyelectrolyte concentrations: given the salt screening of the electrostatic repulsions between the negatively charged phosphate groups, the 16-mer hairpin/duplex equilibrium should have been even more displaced in favor of the duplex form. An explanation for the greater stability of the hairpin may be the following. The favorable stacking energy and the adverse conformational entropy are the two largest factors that determine RNA stability [16]. In most models, the thermodynamic stability of an RNA hairpin is ascribed to a cooperative two-state (all-or-none) process, where ΔG of the tetraloop is assumed to be purely entropic and sequence-independent. The directionality of the GNRA backbone is reversed to reveal the two antiparallel strands of the stem and provokes steric repulsion. In compensation, the GNRA stability comes from intraloop interactions such as non-canonical r(G·A) base pairing and cation-mediated interactions, which cause a sequence dependence and an enthalpic contribution to ΔG of the tetraloop. The 16-mer hairpin r(G546) site belongs to the closing base-pair between the loop and the stem (Fig. 1, in mauve), and constitutes a strong candidate for binding a Mg2 + ion [8, 17]. Its immediate surroundings in the 16-mer duplex form may be perturbed such that the hairpin/duplex equilibrium is displaced into a state with a larger 16-mer hairpin proportion. The chemical shift of the 16-mer hairpin r(G547) increases by around 0.3 ppm after incremental addition of 3 molar equivalents of Mg2 + [8], whereas the chemical shift of the 16-mer duplex r(G547) varies by less than 0.01 ppm. The Mg2 + -induced local reorganization observed for the 16-mer hairpin may not be possible in the 16-mer duplex, where the addition of Mg2 + may hamper the internal motions of its non-canonical r(G547·A) base-pairs, implying an uncompensated loss of conformational entropy. The conformationally restricted 16-mer r(G546) region helps populate the hairpin conformer at the expense of the competing duplex.
. The 16-mer hairpin lifetime is then around 6 months at 5°C, as monitored by real-time 1D 1H NMR. This duplex-to-hairpin transition induced by Mg2 + is surprising, since one might have expected the reverse switch to occur at increasing polyelectrolyte concentrations: given the salt screening of the electrostatic repulsions between the negatively charged phosphate groups, the 16-mer hairpin/duplex equilibrium should have been even more displaced in favor of the duplex form. An explanation for the greater stability of the hairpin may be the following. The favorable stacking energy and the adverse conformational entropy are the two largest factors that determine RNA stability [16]. In most models, the thermodynamic stability of an RNA hairpin is ascribed to a cooperative two-state (all-or-none) process, where ΔG of the tetraloop is assumed to be purely entropic and sequence-independent. The directionality of the GNRA backbone is reversed to reveal the two antiparallel strands of the stem and provokes steric repulsion. In compensation, the GNRA stability comes from intraloop interactions such as non-canonical r(G·A) base pairing and cation-mediated interactions, which cause a sequence dependence and an enthalpic contribution to ΔG of the tetraloop. The 16-mer hairpin r(G546) site belongs to the closing base-pair between the loop and the stem (Fig. 1, in mauve), and constitutes a strong candidate for binding a Mg2 + ion [8, 17]. Its immediate surroundings in the 16-mer duplex form may be perturbed such that the hairpin/duplex equilibrium is displaced into a state with a larger 16-mer hairpin proportion. The chemical shift of the 16-mer hairpin r(G547) increases by around 0.3 ppm after incremental addition of 3 molar equivalents of Mg2 + [8], whereas the chemical shift of the 16-mer duplex r(G547) varies by less than 0.01 ppm. The Mg2 + -induced local reorganization observed for the 16-mer hairpin may not be possible in the 16-mer duplex, where the addition of Mg2 + may hamper the internal motions of its non-canonical r(G547·A) base-pairs, implying an uncompensated loss of conformational entropy. The conformationally restricted 16-mer r(G546) region helps populate the hairpin conformer at the expense of the competing duplex.
Fig. 5.
Energy diagram summarizing the Mg2 + effects on the equilibrium between the hairpin and duplex forms of the 16-mer. The dagger indicates transition states and their corresponding activation energy variations in the presence and absence of Mg2 + . Variations of the chemical potential 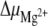 due to the addition of Mg2 + are displayed as a function of temperature
due to the addition of Mg2 + are displayed as a function of temperature
Mg2 + -parameterized ammonia-catalyzed r(G·C) imino proton exchange
Motivated by the importance of quantifying the Mg2 + -mediated changes, we focused on how much the Mg2 + cation modifies the rate constants of the base-pair opening–closing mechanisms and thereby helps stabilize the 16-mer hairpin GCGA tetraloop and the 11-mer simplex AAAA tetranucleotide. In this study, we characterized the 16-mer hairpin base-pairs r(G546·C) and r(G553·C), and the 11-mer simplex base-pair r(G572·C) as a first probing of the Mg2 + local effects on the base-pair opening–closing kinetics. The base-pairs r(G546·C) and r(G572·C) are both the closing base-pairs between the loop and the stem of the 16-mer and the 11-mer, and may give information about motional changes at these structural junctions upon Mg2 + addition. On the other hand, r(G553·C) is an inner-stem base-pair and may help for the analysis with respect to the previously studied RNA duplexes [10]. Using 1D 1H NMR magnetization transfer from water and ammonia-catalyzed exchange of the imino proton, we measured their base-pair lifetimes and their dissociation constants in the presence and in the absence of Mg2 + . NH3 remains the best choice for imino proton exchange studies due to its small size, high pKa, good solubility, neutral character, and lack of detectable proton on the 1H NMR spectrum at alkaline pH. The presence of the equilibrium between the two 16-mer species could have complicated the evaluation of the base-pair kinetic parameters. Since we were primarily interested in the base-pair kinetics of the 16-mer hairpin, the 16-mer sample was heated to 90°C for 3 min and rapidly cooled in an ice-water bath before conducting the measurements.
As predicted by the two-state (closed/open) model of the base-pair [13], the plot of 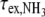 vs. the inverse of ammonia concentration is a straight line whose extrapolation to infinite catalyst concentration yields the base-pair lifetime
vs. the inverse of ammonia concentration is a straight line whose extrapolation to infinite catalyst concentration yields the base-pair lifetime 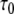 . The apparent dissociation constants Kdiss were computed according to Eq. 1 from the ratio of the rates of exchange catalysis measured in the RNA fragments and for the isolated nucleoside representing the base in the open pair situation. The apparent lifetime of the open pair, τopen, is equal to the product
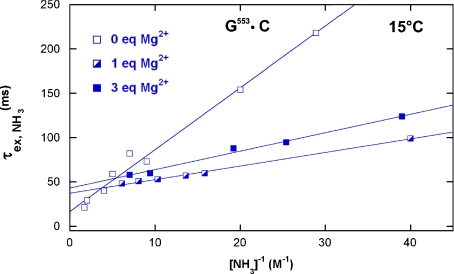
. The apparent dissociation constants Kdiss were computed according to Eq. 1 from the ratio of the rates of exchange catalysis measured in the RNA fragments and for the isolated nucleoside representing the base in the open pair situation. The apparent lifetime of the open pair, τopen, is equal to the product 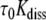 . The ammonia contribution to imino proton exchange in the r(CACCUGG CGACAGGUG) hairpin displayed in Fig. 6 is typical of the exchange measurements performed in the presence or in the absence of Mg2 + . The linear dependence of
. The ammonia contribution to imino proton exchange in the r(CACCUGG CGACAGGUG) hairpin displayed in Fig. 6 is typical of the exchange measurements performed in the presence or in the absence of Mg2 + . The linear dependence of 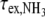 vs. the inverse of the ammonia concentration is consistent with an exchange process occurring from a single open state. Without Mg2 + , the base-pair lifetime of r(G553·C) obtained by extrapolation of the NH3 exchange contribution to infinite ammonia concentration is 22 ms at 15°C. Upon addition of 1 molar equivalent of Mg2 + , the r(G553·C) lifetime increases to 41 ms.
vs. the inverse of the ammonia concentration is consistent with an exchange process occurring from a single open state. Without Mg2 + , the base-pair lifetime of r(G553·C) obtained by extrapolation of the NH3 exchange contribution to infinite ammonia concentration is 22 ms at 15°C. Upon addition of 1 molar equivalent of Mg2 + , the r(G553·C) lifetime increases to 41 ms.
Fig. 6.
Ammonia contribution to the exchange time of the r(G553·C) imino proton of the 16-mer hairpin with and without Mg2 + . The slopes of the plots indicate a slight decrease of base-pair stability, whereas extrapolations to infinite ammonia concentration show an increase of the base-pair lifetimes upon addition of Mg2 +
r(G·C) base-pair kinetics with and without Mg2 +
Several differences emerge from comparison of base-pair kinetics and stability in the RNA fragments with and without Mg2 + (Table 1).
-
Kinetics of the r(G·C) base-pairs without Mg2 +
The r(G·C) base-pair lifetimes fall in the same range of values as those of RNA duplexes previously studied [10]. The base-pairs r(G546·C) and r(G572·C) are compared to the r(G·C) base-pairs in the second position of the previously studied duplexes [10], in order to get a first understanding of the stabilization provided by a loop with regard to a terminal r(G·C) base-pair influenced by fraying effects. Additionally, the inner-stem base-pair r(G553·C) is compared to r(G·C) base-pairs in the third position of the previously studied duplexes [10], which represent typical inner-stem base-pairs. This approach offers a set of reference values to serve as benchmarks for undertaking the more subtle comparison when Mg2 + is brought into play later on. More precisely, r(G553·C) behaves like an RNA duplex inner r(G·C) pair but r(G546·C) and r(G572·C) show a second position base-pair character. The similarity between the double-stranded stems of the two hairpins and short RNA duplexes is largely explained by their common right-handed helices composed of Watson–Crick base-pairs with an A-form geometry. The 16-mer r(G546·C) apparent dissociation constants are much greater than those of control RNA duplexes, whereas the 11-mer r(G572·C) apparent dissociation is just slightly greater. The lifetimes of the open r(G546·C), which are about ten times longer than those internal to RNA duplexes, indicate an increased end-fraying propagation.
-
Kinetics of the r(G·C) base-pairs with Mg2 +
The 11-mer r(G572·C) base-pair lifetime is comparable to that obtained without Mg2 + . In contrast, the 16-mer r(G553·C) and r(G546·C) base-pair lifetimes are longer upon addition of 3 molar equivalents of Mg2 + , respectively increasing from 22 to 43 ms and from 4.8 to 11 ms. An Mg2 + preferential effect for the 16-mer hairpin may explain the variations of r(G553·C) and r(G546·C) opening–closing kinetics which strongly suggest a structural effect of this cation. This observation supports the hypothesis of Mg2 + /GCGA tetraloop binding, which may provide a nucleation site to ensure proper folding of the larger RNA [15, 18, 19]. The 16-mer r(G546·C) apparent dissociation constant is much lower in the presence of Mg2 + and the r(G553·C) apparent dissociation constant remains unaltered, suggesting that Mg2 + serves to enhance stability of the 16-mer [16]. It is also unlikely that the changes observed upon addition of 3 molar equivalents of Mg2 + could be due to intermolecular effects, because no indication for such interactions could be found on the 2D NMR spectra. They more probably might be attributed to further structural changes. Their characterization and the distinction between global/local structural effects induced by the addition of Mg2 + constitute an important problem for the future and may be solved by determining the 16-mer hairpin structures without and with Mg2 + . Furthermore, Mg2 + magnetic relaxation dispersion measurements combined with residual dipolar coupling determinations in oriented media may help in completing the dynamical aspects provided by the base-pair kinetics results. The 11-mer r(G572·C) apparent dissociation constant is slightly higher. The lifetimes of the open r(G572·C) argue for it being comparable to those in the absence of Mg2 + .
Table 1.
Lifetimes, apparent dissociation constants, and apparent open pair lifetimes in r(G·C) base-pairs of the 16-mer and the 11-mer hairpins at 15°C
| [Mg2 + ] |
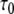 (ms) (ms) |
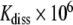 |
τopen (ns) | ||
|---|---|---|---|---|---|
| 16-mer simplex | G546·C | 0 eq | 4.8 (3.4, < < 1)a | 21 (2.2, 20)a | 101 (7.5, < < 20)a |
| 1 eq | 6.5 | 7.6 | 49 | ||
| 3 eq | 11 | 15 | 160 | ||
| G553·C | 0 eq | 22 (33, 7)b | 1.6 (0.28, 0.57)b | 35 (9.2, 4)b | |
| 1 eq | 41 | 4.0 | 160 | ||
| 3 eq | 43 | 2.2 | 95 | ||
| 11-mer simplex | G572·C | 0 eq | 11 (3.4, < < 1)a | 4.9 (2.2, 20)a | 54 (7.5, < < 20)a |
| 1 eq | 13 | 9.5 | 120 | ||
| 3 eq | 11 | 11 | 120 |
The uncertainties on base-pair lifetimes are estimated as ±30%. The uncertainties on dissociation constants are about ±20%
aAs a comparison, the parameters measured for base-pair (G2· C) respectively in [r(CGCGAUCGCG)]2 and [d(CGCGAUCGCG)]2 are given inside parentheses
bThe parameters measured for base-pair (C3· G) respectively in [r(CGCGAUCGCG)]2 and [d(CGCGAU CGCG)]2 are given inside parentheses
Conclusion
Divalent Mg2 + cations are efficient and specific in promoting RNA folding and stability. Their distribution around an RNA molecule may be represented as continuous and dynamic. However, the observation of different values of r(G553·C) and r(G546·C) base-pair lifetimes with and without Mg2 + shows that this alteration in exchange kinetics is related to the local presence or absence of Mg2 + . Moreover, the chemical shift of 16-mer r(G547) showed a variation of around 0.3 ppm after incremental addition of molar equivalents of Mg2 + [8]. Crystallographic studies [20] have already led to an understanding of the stereochemistry of metal ion binding to bases, to nucleosides and to nucleotides, recognizing that the phosphate oxygen, the sugar 2′ hydroxyl, or the base keto oxygen and ring nitrogens are good ligands for alkaline earth cations, since these atoms carry lone electron pairs. Mg2 + seems to bind or at least to have a preferential effect on a specific region of the 16-mer hairpin, involving r(G553·C) and r(G546·C), and may be classified as a specific ion for the 16-mer hairpin. The strength of the Mg2 + -induced force appears strong enough to slow down the neighboring base-pair kinetics against random motion. Our NMR approach is local, although the possibility cannot be totally excluded that the observed Mg2 + -induced changes in exchange rate constants and dissociation constants may originate from long-range effects of Mg2 + binding/affecting other sites. On the other hand, the addition of Mg2 + was not accompanied by chemical shift changes of the 11-mer imino protons (less than 0.01 ppm) and the constant value of 11-mer r(G572·C) base-pair lifetime indicates that Mg2 + seems to move diffusively without altering any specific sites in the 11-mer hairpin. Interest in the role of Mg2 + in promoting IRES function has been rekindled recently, since it may bring light to bear on the active structure of the picornavirus genome. Recent experiments have shown that RNA folding mechanisms depend critically on the initial conditions [21]. We are now therefore characterizing the other 16-mer and 11-mer base-pairs which seem to open independently of each other, both with and without Mg2 + , a result which would appear to be in agreement with general views on RNA behavior. Their exchange kinetics at different temperatures is still under analysis but seem to strengthen the interpretation of the preferential and specific effect of Mg2 + and will be presented elsewhere.
Acknowledgements
Julien A. Dupont is a studentship recipient of the F.R.I.A. (Fonds pour la formation à la Recherche dans l’Industrie et l’Agriculture). This work was supported by the Belgian National Foundation for the Scientific Research (F.N.R.S.) and the Catholic University of Louvain (Fonds Spéciaux de Recherche).
References
- 1.Van Der Velden, A., Kaminski, A., Jackson, R., Belsham, G.: Defective point mutants of the encephalomyocarditis virus internal ribosome entry site can be complemented in trans. Virology 214, 82–90 (1995). doi:10.1006/viro.1995.9952 [DOI] [PubMed]
- 2.Hoffman, M.A., Palmenberg, A.C.: Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J. Virol. 69, 4399–4406 (1995) [DOI] [PMC free article] [PubMed]
- 3.Witherell, G.W., Schultz-Witherell, C.S., Wimmer, E.: Cis-acting elements of the encephalomyocarditis virus internal ribosomal entry site. Virology 214, 660–663 (1995). doi:10.1006/viro.1995.0081 [DOI] [PubMed]
- 4.Roberts, L.O., Belsham, G.: Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the co-expression of fragments of the IRES. Virology 227, 53–62 (1997). doi:10.1006/viro.1996.8312 [DOI] [PubMed]
- 5.Kolupaeva, V., Pestova, T., Hellen, C., Shatsky, I.: Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273, 18599–18604 (1998). doi:10.1074/jbc.273.29.18599 [DOI] [PubMed]
- 6.Robertson, M., Seamons, R., Belsham, G.: A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5, 1167–1179 (1999). doi:10.1017/S1355838299990301 [DOI] [PMC free article] [PubMed]
- 7.Fernandez-Miragall, O., Ramos, R., Ramajo, J., Martinez-Salas, E.: Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 12, 223–234 (2006). doi:10.1261/rna.2153206 [DOI] [PMC free article] [PubMed]
- 8.Phelan, M., Banks, R.J., Conn, G., Ramesh, V.: NMR studies of the structure and Mg2 + binding properties of a conserved RNA motif of EMCV picornavirus IRES element. Nucleic Acids Res. 32, 4715–4724 (2004). doi:10.1093/nar/gkh805 [DOI] [PMC free article] [PubMed]
- 9.Snoussi, K., Leroy, J.-L.: Alteration of AT base-pair opening kinetics by the ammonium cation in DNA A-tracts. Biochemistry 41, 12467–12474 (2002). doi:10.1021/bi020184p [DOI] [PubMed]
- 10.Snoussi, K., Leroy, J.-L.: Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry 40, 8898–8904 (2001). doi:10.1021/bi010385d [DOI] [PubMed]
- 11.Leroy, J.-L., Charretier, E., Kochoyan, M., Guéron, M.: Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry 27, 8894–8898 (1988). doi:10.1021/bi00425a004 [DOI] [PubMed]
- 12.Kochoyan, M., Leroy, J.-L., Guéron, M.: Processes of base-pair opening and proton exchange in Z-DNA. Biochemistry 29, 4799–4805 (1990). doi:10.1021/bi00472a008 [DOI] [PubMed]
- 13.Guéron, M., Leroy, J.-L.: Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 261, 383–413 (1995). doi:10.1016/S0076-6879(95)61018-9 [DOI] [PubMed]
- 14.Cantor, C.R., Warshaw, M.M., Shapiro, H.: Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers 9, 1059–1077 (1970). doi:10.1002/bip.1970.360090909 [DOI] [PubMed]
- 15.Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H.M., Pardi, A.: A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264, 968–980 (1996). doi:10.1006/jmbi.1996.0690 [DOI] [PubMed]
- 16.Searle, M.S., William, D.H.: On the stability of nucleic acid structures in solution: enthalpy–entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 21, 2051–2056 (1993). doi:10.1093/nar/21.9.2051 [DOI] [PMC free article] [PubMed]
- 17.Hermann, T., Westhof, E.: Exploration of metal binding sites in RNA folds by Brownian-dynamics simulations. Structure 6, 1303–1314 (1998). doi:10.1016/S0969-2126(98)00130-0 [DOI] [PubMed]
- 18.Uhlenbeck, O.C.: Nucleic-acid structure tetraloops and RNA folding. Nature 20, 613–614 (1990). doi:10.1038/346613a0 [DOI] [PubMed]
- 19.Antao, V.P., Tinoco, I., Jr.: Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 20, 819–824 (1992). doi:10.1093/nar/20.4.819 [DOI] [PMC free article] [PubMed]
- 20.Saenger, W.: Principles of Nucleic Acid Structure. Springer, New York (1984)
- 21.Russell, R., Zhuang, X., Babcock, H.P., Millett, I.S., Doniach, S., Chu, S., Herschlag, D.: Channels in the folding landscape. Proc. Natl. Acad. Sci. U. S. A. 99, 155–160 (2002). doi:10.1073/pnas.221593598 [DOI] [PMC free article] [PubMed]