Abstract
This paper reports a new phenomenon connected with the influence of green light (GL) on biological systems. Our experiments have revealed an antioxidant effect of GL on cells subjected to lethal doses of UV at the cellular level and a protective effect of GL on DNA denatured by UV, coupled with a structural modification of DNA macromolecules under GL irradiation, at the molecular level. Mouse melanocyte cultures are subjected to UV irradiations with L50 fluxes of 16.0 J m − 2 s − 1. GL is obtained from a strontium aluminate pigment, which emits GL under UV activation. Cells grown in GL, prior to UV irradiation, present a clear surprising protective effect with surviving values close to the controls. A GL antioxidant effect is suggested to be mediated through GL influence on cellular water cluster dynamics. To test this hypothesis, reactive oxygen species (ROS) are determined in cell cultures. The results revealed a decrease of cellular ROS generation in the UV-irradiated samples protected by a previous 24 h of GL irradiation. At the DNA level, the same type of GL protection against UV damage is recorded by gel electrophoresis and by UV spectroscopy of the irradiated DNA molecules. Two physical methods, impedance spectroscopy and chronoamperometry, have revealed at the level of GL-irradiated DNA molecules spectral modifications that correlate with the UV spectroscopy results. The interaction between the chargeless photons and the field of water molecules from the cellular compartments is discussed in relation with the new field of macroscopic quantum coherence phenomena.
Keywords: Green light antioxidant effect, Water clusters, UV cellular effects, UV DNA irradiation effects, Green light protective effects
Introduction
This paper reports a new phenomenon connected with the influence of green light (GL) on biological systems. The experiments presented here show:
An antioxidant effect of GL on cells subjected to lethal doses of UV at the cellular level.
A protective effect of GL against DNA denaturation by UV and a structural modification of DNA under GL irradiation at the molecular level.
These results, obtained through the influence of an electromagnetic (EM) field from the visible domain which is not absorbed, in the conventional meaning, and accordingly does not induce electronic transitions, suggested their possible connection with the new area of macroscopic quantum coherence (MQC).
The possibility to extend the quantum domain outside the atomic realm has frequently haunted physicists. Wigner [1], in a famous paper, excluded this conjecture as contradicting the very foundations of quantum mechanics. Nevertheless, the question still lingered. The problem concerns the nature and the effects of quantum systems that may retain their manifest quantum features at the macroscopic level. Within classic formalisms, the main required characteristic would refer to the wave function’s oscillatory nature, which should reveal quantum coherence, i.e., oscillations at different places beat time with one another. In our context, we refer to MQC in the meaning introduced by Penrose [2]. He considers quantum coherence the circumstances when large number of particles can collectively cooperate as a single quantum state which remains essentially disentangled with its environment. This MQC is found in Bose–Einstein condensation (which occurs in laser action) where the entire system behaves as a whole. The discussion on the occurrence of MQC phenomena resumed in the 1970s with the noted paper of Fröhlich [3] on long-range coherence in enzymes kinetics. Fröhlich suggested that since the energy of cellular metabolic drive is large enough and the dielectric properties of biological materials unusually extreme, large-scale quantum coherence, similar to the Bose–Einstein condensation, may occur. Indeed, 1011-Hz oscillations, as predicted by Fröhlich’s model, have been observed within cells [4].
A radically new idea emerged within the Society of Quantum Biology (San Francisco) concerning the possibility of generating molecular activations, different from the canonical electronic excitation level. A mathematical model developed by Rosen [5, 6] served as the blueprint for these theoretical considerations. He presented an elegant formal proof that a chemical reaction of the [ΣAi→P] type may be interpreted as a microphysical system in which appropriate “perturbations” may induce shifts in the eigenvalues of a particular observable set. Using these conjectures, Comorosan [7–9] designed an experimental setup in which the “perturbation” of the chemical target observables would not induce structural modifications of the molecule (the classic chemical domain) but would generate changes in its energetic profile (the quantum domain). In this context, “perturbations” by irradiation with light which is not absorbed (hence, no classic electronic transitions) appeared as an ideal technique. He published the first experimental evidence, revealing kinetic changes in enzyme activity, through low-level visible irradiations of their substrates. He also observed that within the visible domain, green photons (λ = 530–550 nm) display the highest quantum yield as compared with blue or yellow ones. A rather dynamic field of research was active up to the 1990s; see Comorosan [10, 11] for a review. The technological level of the epoch precluded the extension of investigations toward a possible connection with MQC, that these phenomena were suggesting. Starting in 2000, the situation changed, and a wealth of new experimental results appeared. Leggett [12] observed MQC in small superconducting quantum interference devices. The modern field of quantum computing provided data on biological quantum coherence and molecular superposition states [13].
The new available powerful monochromatic irradiation sources (light-emitting diodes, LEDs), as well as the new theoretical grounds for macroscopic quantum behavior, renewed the interest in the investigation of visible light effects. This is the scientific context in which we have designed our experimental models in order to connect biology to the domain of modern physics domain.
Materials and methods
The cellular system
A line of standardized mouse melanocyte cultures (ECACC-85011438) is used. Cells are cultured in the PromoCell growth medium kit for melanocytes (Heidelberg, Germany) as a monolayer in Petri dishes (diameter 90 mm), with medium changed twice a week and passages realized once a week in a humidity atmosphere containing 5% CO2 in air. Cultures at fifth passage are used. Adherent cells in the logarithmic phase were detached by 3 ml of 0.05% (w/v) trypsin and 0.02% (w/v) EDTA. Cellular density is determined by counting in a hemocytometric chamber.
UV irradiations
A Vilbert Lourmat UV lamp, model VL-204G, 16 W, emitting 90% of the energy at λ = 254 nm is used. A special setup with time monitored by an electronic shutter is arranged in order to deliver irradiation fluxes between 10 and 16 J m − 2 s − 1, with fixed doses obtained through different exposure times for each particular setup.
GL irradiations
In the cellular experiment, the GL irradiation is accompanied by a simultaneous UV irradiation. The GL emission is generated by a chemical fluorescence mechanism. A strontium aluminate pigment (that emits GL under UV irradiation) coated on a specially designed UV transparent disk is used. The Petri dishes with the cell cultures are placed on top of it. The pigment is activated by a 10-min UV irradiation and reactivated every 6 h. After 24 h of GL treatment, the cells were irradiated with the L50 UV dose. At that point, a standard procedure is used to measure the cell viability. The means, standard deviation, and deviation from median have been computed by analysis of variance software to determine the variance between groups. For the GL irradiations of solutions, powerful LEDs of 100 lumens mounted on copper-ventilated radiators are used. In these setups, designed for each particular type of experiment, a monochromatic light of λ = 527 nm is obtained with intensities of 3 × 105 − 4 × 105 lx, determined with a digital Luxmeter LX-1102, Lutron, with 4 × 10- to 4 × 105-lx range.
Cellular viability determination
The effect of UV irradiation and the potential GL effect on cellular viability were assessed by the standard neutral red colorimetric method and by fluorescence microscopy. The cellular sample is marked with a 1.7% solution of neutral red prepared in Hank’s medium and incubated for 3 h at 37°C. The excess color is eliminated from the medium with 1% formaldehyde and the cells are rinsed twice with phosphate-buffered saline. Finally, the color is extracted from the live cells by 1% acetic acid and the absorption measured at 540 nm.
For the fluorescence microscopy, we have used a two-color fluorescence viability assay [14] with SYTO 63 (Molecular Probes, Invitrogen, Karlsruhe, Germany), a low-affinity nucleic acid stain that diffuses through the membranes of living/dead cells, and propidium iodide (PI; Sigma-Aldrich, Deisenhofen, Germany), a DNA-selective dye, membrane-impermeable, but penetrating dead cells’ membranes. The fluorescent stain is performed by adding to the culture medium 0.2 μg/ml PI and 1.25 μM SYTO. Cells are incubated for 30 min at 37°C and analyzed immediately without further washing steps by confocal microscopy on a LCS microscope (Leica Lasertechnik) interfaced with lasers emitting at 488, 543 and 633 nm. The detection channels are 570 nm for SYTO and 590 nm for PI. Confocal images at 600× magnification are evaluated by a direct counting of the live and dead cells. Two sets of experiments were performed, each one on five individual cell cultures, for control and the irradiated samples. The same two sets of experiments were designed for the investigation of the prospective GL effect. The Petri dishes are left under GL light, from the activated strontium aluminate pigment, for 24 h before the UV exposure. The UV irradiations were delivered for 3 min in air, perpendicular to the plane of the Petri dish, at a 2-cm distance above the plate surface.
Free radical determination
In this study, we have used dichlorofluorescin diacetate (DCFDA, Sigma-Aldrich, D.6883 min.97%) as a fluorescence probe. Since DCFDA enters the cells rapidly, in order to avoid cytotoxicity, low concentrations are loaded. We used the fluorescence technique as described in Li [15]. Cultured cells are detached with trypsin/EDTA and resuspended in HEPES-buffered saline solution, 25 mmol/l, pH 7.04. Cells are loaded with 5 μmol DCFDA for 45 min at 37°C and samples of 103 cells distributed for spectrofluorescence determination. Excitation intensities are measured at 500-nm wavelength and the fluorescence emission at 520-nm wavelength in a photon-counting spectrofluorimeter (PC1, ISS Inc., Champaign, IL, USA). In two sets of experiments (for control and irradiated samples), each one performed on five individual cell cultures, we have loaded the cells with a fluorescent probe (DCFDA) and measured subsequently the respective fluorescence emission. In a similar setup (experiment performed on five individual cell cultures), we have loaded the cells grown previously under a 24-h GL from the activated strontium aluminate pigment before the UV exposure.
DNA samples
Two types of DNA samples are used in this study: commercial DNA (Sigma-Aldrich) in a 0.9% NaCl stock solution, 1 mg/ml, prepared under refrigeration for 24 h with gentle stirring, and DNA extracted from human whole blood. The extraction and purification of the blood DNA samples are realized according to the standard Qiagen technique [16] by sample vortexing and spin column centrifugation up to 14 × 103 rpm. Nucleic acid yield is determined by absorbance at 260 nm and purity by the A260/A280 ratio. Samples with the ratio of 1.7–1.9 are stored in Qiagen buffer AE at − 20°C.
Gel electrophoresis
An agarose reagent sufficient to resolve 50- to 2,000-bp fragments of DNA is used. The gel is prepared in 0.5X TBE buffer with ethidium bromide. Five microliters of DNA at 1 mg/ml in 0.9% NaCl is mixed with 1 μl Blue Orange and run at 10 V cm − 1 gel length. Samples are photographed over an UV transilluminator and stored on a computer.
The irradiation setup is organized with four probes: (1) control, (2) DNA denatured by UV, (3) DNA denatured by UV and simultaneously protected by GL irradiation, and (4) DNA irradiated by GL.
For the denatured probe, five micro spots each of 20 μl nucleic acid are placed on a thin glass plate and irradiated between 40 and 50 min in air, perpendicular, at 2-cm distance, with the UV lamp 10.00 J m − 2 s − 1 placed above the plate. The spots are recuperated and 1 μl nucleic acid is mixed with 5 μl color reagent for electrophoretic migration.
For the simultaneous irradiations, the UV light is collimated at the LED level to give a spot of ~3 cm and the irradiation performed as described above. The simultaneous GL irradiation is performed from below, through the thin glass plate, at an intensity of 2 × 105 lx.
UV spectroscopy
UV spectroscopy is recorded on a UV–vis instrument, GBC-Cintra 10B (Australia) with 200- to 1,100-nm range in 3 ml quartz cells. A stock DNA solution, 1 mg/ml, is used. In a small Petri dish (25-mm diameter), a volume of 3 ml 1/20 dilution of DNA is UV-irradiated at 1 cm in air (10 J m − 2 s − 1, 15 min.), perpendicular, from above. For the GL protection, the simultaneous irradiation is performed from below, through the glass Petri dish, with an intensity of 4 × 105 lx. Volumes of 2.5 ml are recuperated and used in the spectrophotometric determinations.
Impedance spectroscopy
A Zahner IM6 Khronach (Germany) electrochemical unit with <0.0025% frequency stability is used, with a specially designed miniaturized cell (50 μl) and two platinum bands (1-mm width, 0.1-mm thickness). A 1% DNA solution in 0.9% NaCl is used for GL experiments. A specially designed miniaturized irradiation cell is elaborated from a Teflon 30/30-cm cylinder with a mini hole of 4-mm height/5-mm width in the middle of it. A volume of 70 μl is GL-irradiated for 30 min. Forty-five microliters is recuperated and introduced into the electrochemical cell for impedance determinations. The GL beam is collimated, from above in air, on the target at an intensity of 3 × 105 lx.
Chronoamperometry
Chronoamperometry is recorded with a high-level performance electrochemical combined (±0.2% accuracy) VoltaLab 40 analytical radiometer with platinum plate electrode, saturated calomel electrode, and a cell with external thermostatic jacket. A 0.9% NaCl solution is used as electrolyte. A rigorously measured volume of 3.0 ml is placed on two identical electrochemical cells (control and irradiated). Irradiations are performed through a hole, in a Faraday cage, with the irradiation device fixed outside, on top of it. Two LEDs of 100 lumens each on a ventilated copper radiator, emitting at λ = 527 nm with 3 × 105 lx, are used. The variation of current density in time at a fixed potential is determined. Variations of temperature (<0.3°C) and evaporation of solutions (<0.015 g) are rigorously controlled and included in the results.
Results
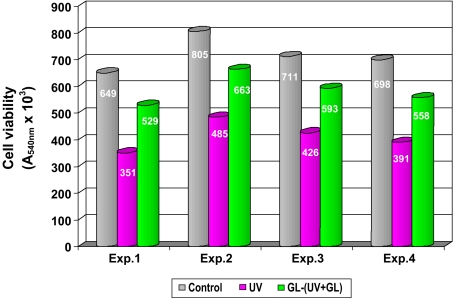
The cellular system The main result revealed in the experiments on cell viability is represented by a clear protective effect of GL against cellular damage induced by UV irradiation (Fig. 1).
Fig. 1.
Protective green light effect on cell viability under L50 UV irradiation. Experimental setup: 16.0 J m − 2 s − 1 UV irradiation, 3-min exposure at 2-cm distance, in air, perpendicular, from above. Green light λ = 546 nm applied for 24 h before UV irradiation and simultaneously with the UV irradiation. Each experiment performed with five separate standardized cellular samples; viability determined by neutral red colorimetric method. Mean values (A540 nm × 103 ± SD): control, 716 ± 65; UV-irradiated, 413 ± 57 GL; and UV irradiations, 586 ± 58, p < 0.05 between groups
As clearly seen from the figure, in the UV irradiation experiment, the cell viability decreased to a mean value of 0.413 AU with the difference between mean values in control and in UV irradiated samples of 0.303 AU, whereas in the GL experiment, viability decreased to a mean value of 0.586 AU with a mean difference versus control of 0.130 AU.
The results obtained with the two-color fluorescence viability assay are presented in Table 1. These results correlate well with the protective effect obtained with the neutral red viability assay. As seen from the table, the ratio of vital cells/total cells drops from the control value of 0.92 to 0.46 in the UV-irradiated experiment, significantly lower than the 0.80 value for the GL-protected experiment.
Table 1.
GL-protective effect on UV-irradiated cells
| Dose | Vital cells/total cells | |
|---|---|---|
| Control | 0 | 0.92 |
| UV | 16.0 J m − 2 s − 1 | 0.46 |
| GL+UV | 24 h GL+16.0 J m − 2 s − 1 | 0.80 |
Double-fluorescence viability assay
Results are computed from the ratios between SYTO cells/total cells and PI cells/total cells. In each experiment, numbers are computed from the average of five different investigated samples. UV exposure for 3 min at 2-cm distance, perpendicular, from above (n = 5, p < 0.002)
Since the UV irradiation induces well-known oxidizing phenomena and since the green photons protect the biological structures against them, a green light antioxidant effect is suggested. To test this hypothesis, we designed a fluorescence experiment to check reactive oxygen species (ROS) in cells.
Free radicals The results are presented in Table 2. As seen from the table, the recorded ROS generation in the UV-irradiated cultures ranges between 310 and 350 RFU × 103 cells, whereas this value drops to 108–138 RFU × 103 cells for the GL-protected cultures. These data support our suggestion that green light induces antioxidant effects.
Table 2.
Fluorescence yields, determined by the DCFA-probe, in culture cells UV-irradiated and in cultures protected by GL
| Relative fluorescence units (RFU × 103cells) | ||
|---|---|---|
| UV (± SD) | GL+UV (± SD) | |
| Exp.1 | 350 ± 0.04 | 138 ± 0.05 |
| Exp.2 | 310 ± 0.03 | 108 ± 0.06 |
Results represent averages computed from yields of reactive species generated in two sets of experiments, each one of five different culture samples UV-irradiated, and yields of reactive species generated in two sets of experiments, each one of five different culture samples UV-irradiated and previously protected by 24 h GL illumination. UV flux, 16.0 J m − 2 s − 1, 3-min exposure at 2-cm distance, perpendicular, from above
DNA gel electrophoresis This technique revealed a surprising experimental result. As seen from Fig. 2, the DNA molecules are split, as expected, by the UV irradiation in small fragments that leave a shadow in the electrophoretic path for the 40-min UV irradiation [3] and disappear completely from the field for the 50-min UV irradiation [4]. As seen from the figure, in both cases of simultaneous irradiations, for 40-min UV+GL [2] and 50-min UV+GL [5], the DNA spots present the same electrophoretic migration and compactness as the control [1]. An interesting result is recorded on the GL irradiation of DNA macromolecules. In the electrophoretic field, the DNA spot [6] appears with a higher brightness (indicating a possible stronger coupling with the color reagent) and compactness, which may be correlated with the effect revealed on the nucleic acids’ UV and impedance spectra, suggesting modifications of the medium wrapped around the nucleic acid macromolecules.
Fig. 2.
Gel electrophoresis of DNA samples, UV-irradiated and GL-protected. 1 Control; 2 UV 40 min simultaneously with GL 40 min; 3 UV 40 min; 4 UV 50 min; 5 UV 50 min simultaneously with GL 50 min; 6 GL 50 min
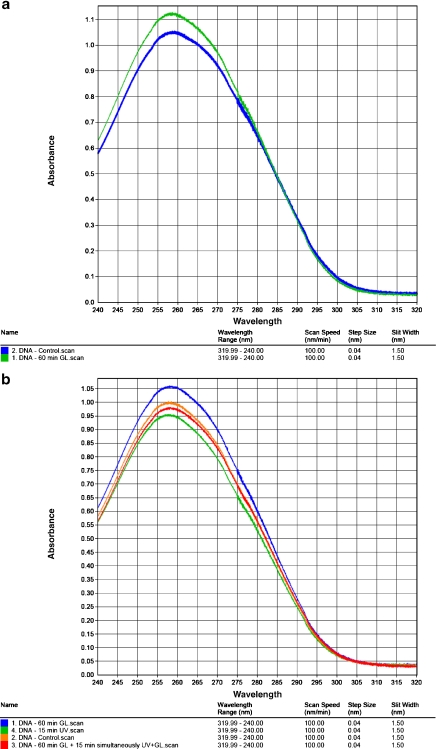
UV spectroscopy As seen from Fig. 3a, the UV spectrum of the DNA macromolecules reveals a clear modification under a 60-min GL irradiation with an increase of absorbance at λ = 258.36 nm. As seen from Fig. 3b, the 15-min UV irradiation of DNA solution induces a clear-cut modification of the spectrum, with a significant decrease of absorbance at λ = 258.36 nm. When the DNA solution is previously protected by a 60-min GL irradiation and subsequently simultaneously irradiated 15 min by UV+GL, the absorbance decrease is diminished. The protective effects of GL against the UV denaturation of DNA macromolecules correlate well with the same type of effects, as manifested in cellular systems.
Fig. 3.
a The effect of GL on the DNA-UV spectrum. For GL irradiation, the DNA probe is irradiated for 60 min in air, from above, with GL at an intensity of 4 × 105 lx on 3 ml DNA solution in a small Petri dish (diameter 30 mm). b UV spectra of UV-irradiated DNA samples and of DNA samples which have been UV-irradiated and simultaneously protected by GL. The DNA probe is UV-irradiated for 15 min in air, perpendicularly, from above, with a flux of 10 J m − 2 s − 1 in a volume of 3 ml on small Petri dishes. The GL protective irradiation is performed from below, through the glass Petri dish, with an intensity of 4 × 105 lx
Impedance spectroscopy This physical technique is significant for nucleic acid investigation. The DNA macromolecule is an electrical conductor, hence appropriate for electrochemical measurements, and impedance spectroscopy links three important parameters: resistance, inductance, and capacitance.
We report here the investigations of the changes in electrical impedance that occur when DNA solutions are exposed to green light irradiation. The impedance can be expressed as a complex number, 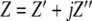 , where
, where 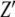 and
and 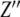 are the real and imaginary components of the impedance, respectively.
are the real and imaginary components of the impedance, respectively.
The changes in impedance values are difficult to detect at low frequencies, but become apparent at frequencies higher than approximately 1 kHz.
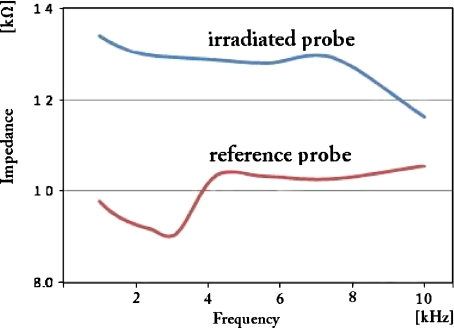
Figure 4 represents the electrochemical response for the control and the irradiated solutions measured at open circuit potential with a 50-mV amplitude sinusoidal modulation.
Fig. 4.
Impedance spectrum of a GL-irradiated DNA probe. The DNA probe, 70 μl of 1% DNA in 0.9% NaCl is GL-irradiated for 30 min in a miniaturized irradiation cell with an intensity of 3 × 105 lx. Volumes of 45 μl are used in the impedance measurements
The irradiation-induced changes in electrical properties are clearly observed over the frequency range of 1–10 kHz, showing an increase of the impedance for the irradiated sample. Since the irradiation is performed in a specially designed miniaturized cell, to accommodate very small nucleic acid volumes different from the classic electrochemical ones, the observed changes may be attributed both to modifications in the bulk solution and in the topology of DNA macromolecules, which may in turn induce modifications of the molecular electrochemical properties.
Chronoamperometry
As seen from Table 3, the GL irradiation of the NaCl solution clearly reveals a decrease of the current density from the 31.02 ± 4.12 μA cm − 2 control value to 18.65 ± 5.01 μA cm − 2 for the GL-irradiated sample. This may be a significant result, suggesting water cluster formation around the Na + and Cl − ions. These relatively large water structures would then induce a slower ionic mobility, as revealed by the decrease in current density.
Table 3.
Effect of a 60-min GL irradiation on current density of a 0.9% NaCl solution
| Current density (μA cm − 2) | |
|---|---|
| Control sample | 60 min GL-irradiated sample |
| 31.02 ± 4.12 | 18.65 ± 5.01 |
The values represent results of chronoamperometry at a fixed potential of 300 mV vs. saturated calomel electrode. Mean ± SD of ten individual determinations
Discussion
The basic problem raised by our experimental results represents their innovative character. The main cellular structures, proteins, nucleic acids, lipids, and components of cell membranes do not absorb in the green domain. There are studies concerning biological effects induced on animal systems by visible light. They are mostly the result of light absorption by cellular chromophores (hence, photochemical reactions), with point-like consequences [17], or linked with a subsequent biological functional alteration [18, 19].
Since our observed phenomena are connected with results obtained with a radiation which is not conventionally absorbed and which induces a whole set of biophysical effects that do not have a direct interpretation under current scientific knowledge, a new mechanism has to be elaborated. Such a mechanism should elucidate the nature of the interactions involved in our experiments and the integration of the results into existing scientific models. Our experimental setup was designed to investigate the effects of visible light. The EM spectrum has established effects in the absorption domain (IR and UV), inducing electronic transitions, and in the high-energy ionizing domain (X-rays, γ-rays...), inducing molecular ionization. Our experimental model based on GL irradiations is not part of this canon, and accordingly, it represents a new field of investigation.
The elaboration of a suggested mechanism is based on a main hypothesis: we assume that the green photons may react—via a field-mediated interaction—with the water aggregates, the ubiquitous ingredient of biological compartments, and modify their structures. We are in the significant domain of weak interactions. We advance here an analytical analysis of the results. Water is a polar molecule, forming layers of clusters by hydrogen bridges (binding energy ~20 KJ/mol) and van der Waals forces. The topology of the unit represents an energetic cloud with a shape determined by the angle of 104.45° realized between oxygen and the two hydrogen atoms, extending in space with a van der Waals radius of 1.2 Å in the H–direction and a van der Waals radius of 1.4 Å in the O–direction. The directional properties of H– bonds are well known. When the vector along the covalent O–H bond points directly at the acceptor oxygen, a strong bond is formed. When the vector points away from the oxygen atom, weaker bonds are generated. The interaction of a low-intensity EM field—which is not absorbed and does not induce electronic transitions—may in turn induce displacement of charges at the target’s level, thus modifying the topology of the energetic profile. This is a known fact of classical physics. The modification of molecular geometry will then change the water dipole moment and bring about fluctuations in the water aggregate dynamics. The recent methods of solution dynamics and quantum mechanics have generated complex water clusters (available at http://www.lsbu.ac.uk/water/clusters.html) with a 14-water molecule as a tetrahedral unit and with icosahedral assemblies of 280 molecules, inducing long-range effects up to 10 Å from the center [20]. It is suggested that hydrogen bonding configurations may then produce fluctuations in the cluster’s network between an expanded (ES) and a collapsed (CS) form [21]. In this theoretical context, we advance a conjecture: we assume that GL may change the 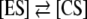 equilibrium, toward the ES configuration, with larger cavities that may accommodate the UV-induced hydroxyl radical (H:O·) and fix it in the network, resulting a free radical scavenger GL effect. Let us now discuss a direct physical interpretation of this conjecture derived from the chronoamperometry experiment. The GL irradiation of the NaCl solution pumps into the system the green photon energy that may extend the 104.45° angle of the water molecule during the interaction time with the hydrogen bridges. Such an extension would then induce structures with long-range alignment and the ability to maintain long coherence processes. This experiment connects, in our opinion, our results with MQC.
equilibrium, toward the ES configuration, with larger cavities that may accommodate the UV-induced hydroxyl radical (H:O·) and fix it in the network, resulting a free radical scavenger GL effect. Let us now discuss a direct physical interpretation of this conjecture derived from the chronoamperometry experiment. The GL irradiation of the NaCl solution pumps into the system the green photon energy that may extend the 104.45° angle of the water molecule during the interaction time with the hydrogen bridges. Such an extension would then induce structures with long-range alignment and the ability to maintain long coherence processes. This experiment connects, in our opinion, our results with MQC.
UV effects on nucleic acids are well studied (see the monographs of Halliwell and Gutteridge [22] and Sieh [23]). The biochemistry of UV-mutagenic damages has been investigated in detail. Cyclobutane–pyrimidine dimers (CPDs) 6-4 photoproducts (6-4 PPs) and their Dewar valence isomers are the main cytotoxic-induced lesions (see the review of Sinha and Donat [24]). It is clear, however, that there is no chemical mechanism for the green photons to participate directly and in a point-like manner in these reactions. The high-energy short-wavelength photons absorbed by chromophoric molecules lead to the formation of singlet oxygen and free radicals known to destroy membranes and cellular structures. It is also clear that a direct green photon/UV-photon interaction is not, at least for the moment, a physical alternative. Some of the biochemical details may be relevant in this context. CPDs and 6-4 PPs induce a bend or kink of 7–9° and 44°, respectively, in the DNA helix and distort it [25]. Sequences that facilitate bending and unwinding are favorable sites for damage inducement, and CPDs form at higher yields at the flexible ends of poly (dA)-(dT) tracts [26]. Another direct effect of DNA damage on the TBP-binding protein induces the selective formation of 6-4 PPs at sites where DNA is bent [27]. These types of biochemical reactions may account for the UV effect recorded by us on the DNA-UV spectrum, suggesting drastic changes in the topology of the chemical groups that absorb in λ = 260 nm and correlate with the fragmentation of nucleic acids chains revealed in our electrophoretic determination. Nevertheless, the GL protective effect may not be linked by a direct physical interaction with this type of biochemistry. The general nature of the protection, precluding a cascade of various damaging chemical reactions, strongly suggests a common actor, the cellular (molecular) water, acting through the cluster dynamics as described in our model.
As a final point, we elaborate the connections with the new emerging field of MQC. Let us first observe that changes in the foundation of quantum field theory were long anticipated. In the 1970s, Primas [28] remarked that biological systems represent a mixture of classical and quantum characteristics and provided examples in which the Hamiltonian—the classic observable of molecular theory—is not necessarily itself an observable.
The macroscopic coherence (in the quantum meaning) considered as well at the theoretical level as at the experimental one is now an actively investigated problem with already spectacular results; see the Zeilinger experiment [29] on superposition of states. Our experimental results may be of interest for this domain, at the cellular and at the molecular level.
In the cellular systems, the GL irradiations induce a general type of energetic modifications which act in a cooperative mode to block a cascade of dangerous UV effects. The modern techniques of quantum mechanical simulations have revealed an active dynamics of ions in water solutions. Our GL irradiations induce in the experiments with NaCl solutions large areas of clusters, with extended spatial stability and long coherence times. This experiment may be linked with the work performed on lattices of neuronal microtubules [30] where quantum coherence phenomena may account for nervous activity, with water molecules in clustered form as an active ingredient.
References
- 1.Wigner, E.: The Probability of a Self-Reproducing Unit. Symmetries & Reflections. Indiana University Press, Bloomington (1967)
- 2.Penrose, R.: The Road to Reality, Ch. 23, The Entangled Quantum World. Alfred Knopf, New York (2005)
- 3.Fröhlich, H.: Long range coherence in the activity of enzymes. Nature 228, 228–234 (1970) [DOI] [PubMed]
- 4.Grundler, W., Keilman, F.: Sharp resonances in yeast growth. Phys. Rev. Lett. 51, 1214–1216 (1983) [DOI]
- 5.Rosen, R.: A relational theory of biological systems. Bull. Math. Biophys. 21, 109–128 (1959) [DOI]
- 6.Rosen, R.: A note on the quantum-theoretic basis of primary genetic activity. Bull. Math. Biophys. 25, 183–187 (1963) [DOI] [PubMed]
- 7.Comorosan, S.: The biochemical flip-flop. Nature 227, 64–65 (1970) [DOI] [PubMed]
- 8.Comorosan, S., Vieru, S., Sadru, D.: Evidence for a new biological effect of low level irradiations. Int. J. Radiat. Biol. 17, 105–115 (1970) [DOI] [PubMed]
- 9.Comorosan, S., Vieru, S., Murgoci, P.: The effect of electromagnetic field on enzymic substrates. Biochim. Biophys. Acta 268, 620–621 (1972) [DOI] [PubMed]
- 10.Comorosan, S.: Biological observables. In: Rosen, R. (ed.) Progress in Theoretical Biology, pp. 161–203. Academic, New York (1976)
- 11.Comorosan, S.: A novel interaction of light with matter. J. Biol. Phys. 17, 151–164 (1990) [DOI]
- 12.Leggett, A.J.: Testing the limits of quantum mechanics. J. Phys. Condens. Matter 14R, 415–451 (2002) [DOI]
- 13.Mavromatos, N.E., Powell, A.K.: On the Possibility of Quantum Coherence in Biological Systems. Decoherence & Entropy in Complex Systems. Springer, Berlin (2000)
- 14.van der Kuip, H., Mürdter, T.M., Sonnenberg, M., McClellan, M., Gutzeit, S., Gerteis, A., Simon, W., Fritz, P., Aulitzky, W.E.: Short term culture of breast cancer tissues. BMC Cancer 6, 86–98 (2006) [DOI] [PMC free article] [PubMed]
- 15.Li, J.M.: Essential role of the NADPH oxidase subunit 47phox in endothelial cell superoxide production. Circ. Res. 90, 143–150 (2002) [DOI] [PubMed]
- 16.Sicinschi, L.A., Correa, P., Bravo, L.E., Schneider, B.G.: Detection and typing of Helicobacter pylori cagA/vacA genes by radioactive, one-step polymerase chain reaction. J. Microbiol. Methods 52(2), 197–207 (2003) [DOI] [PubMed]
- 17.Douglas, A.D., Kraves, S., Deisseroth, K., Schier, A.F., Engert, F: Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr. Biol. 18, 1133–1137 (2008) [DOI] [PMC free article] [PubMed]
- 18.Alilain, W.J., Li, X., Horn, K.P., Dhingra, R., Dick, T.E., Herlitze, S., Silver, J.: Light-induced rescue of breathing after spinal cord injury. J. Neurosci. 28(46), 11862–11870 (2008) [DOI] [PMC free article] [PubMed]
- 19.Huber, D., Petreanu, L., Ghitani, N., Ranade, S., Hromádka, T., Mainen, Z., Svoboda, K.: Sparse optical microstimulation in barrel cortex drives learned behavior in freely moving mice. Nature 451, 61–64 (2007) [DOI] [PMC free article] [PubMed]
- 20.Tanaka, H.: Simple physical model of liquid water. J. Chem. Phys. 112, 799–809 (2000) [DOI]
- 21.Stratt, R.M., Maroncelli, M.: Nonreactive dynamics in solution. J. Phys. Chem. 100, 12981–12996 (1996) [DOI]
- 22.Halliwell, B., Gutteridge, J.M.C.: Free Radicals in Biology and Medicine, 2nd edn. Clarendon, Oxford (1988)
- 23.Sieh, H.: Oxidative Stress, Oxidants and Antioxidants. Academic, New York (1991)
- 24.Sinha, R.P., Donat, P.H.: UV-induced damage and repair. Photochem. Photobiol. Sci. 1, 225–235 (2002) [DOI] [PubMed]
- 25.Wang, C.I., Taylor, J.S.: Site-specific effect of thymine dimer formation on dAn.dTn tract bending and its biological implications. Proc. Natl. Acad. Sci. U. S. A. 88, 9072–9076 (1991) [DOI] [PMC free article] [PubMed]
- 26.Becker, M.M., Wang, Z.: Origin of ultraviolet damage in DNA. J. Mol. Biol. 210, 429–438 (1989) [DOI] [PubMed]
- 27.Aboussekhra, A., Thoma, F.: TATA-binding protein promotes the selective formation of UV-induced (6-4)-photoproducts. EMBO J. 18, 433–443 (1999) [DOI] [PMC free article] [PubMed]
- 28.Primas, H.: Probleme der Interpretation der Quantenmechanik grosser Systeme. E.T.H. Zürich (1970)
- 29.Zeilinger, A., Gähler, R., Shull, C.G., Treimer, W., Mampe, W.: Single & double slit diffraction of neutrons. Rev. Mod. Phys. 60, 1067–1073 (1988) [DOI]
- 30.Hameroff, S.R., Watt, R.C.: Information in processing in microtubules. J. Theor. Biol. 98, 546–561 (1982) [DOI] [PubMed]