Abstract
Cellular carbon (C) and nitrogen (N) metabolism must be tightly coordinated to sustain optimal growth and development for plants and other cellular organisms. Furthermore, C/N balance is also critical for the ecosystem response to elevated atmospheric CO2. Despite numerous physiological and molecular studies in C/N balance or ratio response, very few genes have been shown to play important roles in C/N balance signaling. During recent five years, exciting progress was made through genetic and genomic studies. Several DNA microarray studies have shown that more than half of the transcriptome is regulated by C, N and the C-N combination. Three genetic studies involving distinct bioassays have demonstrated that a putative nitrate transporter (NTR2.1), a putative glutamate receptor (GLR1.1) and a putative methyltransferase (OSU1) have important functions in the C/N balance response. OSU1 is identical to QUA2/TSD2 which has been implicated to act in cell wall biogenesis, indicating a link between cell wall property and the C/N balance signaling. Given that many investigations are only focused on C alone or N alone, the C/N balance bioassays and gene expression patterns are discussed to assist phenotypic characterization of C/N balance signaling. Further, re-examination of those previously reported sugar or nitrogen responsive genes in C/N balance response may be necessary to dissect the C/N signaling pathways. In addition, key components involved in C-N interactions in bacterial, yeast and animal systems and whether they are functionally conserved in plants are discussed. These rapid advances have provided the first important step towards the construction of the complex yet elegant C/N balance signaling networks in plants.
Key words: carbon, carbon dioxide, sugar, nitrogen, amino acid, C/N balance, C:N ratio, OSU1, Arabidopsis, cell wall, hormone
Introduction
Plants are non-motile organisms and therefore through evolution they have developed the complex sensing and signaling mechanisms to robustly monitor and appropriately respond to the dynamic changes of their surrounding environments. Among many environmental factors, carbon (C) and nitrogen (N) are crucial for plants to perform the routine and fundamental cellular activities. C compounds include various carbohydrates in particular sucrose (Suc) and glucose (Glc), and these photosynthetic products provide both the energy and the C-skeletons for ammonium assimilation during amino acid biosynthesis. N nutrients include inorganic compounds (nitrate and ammonium) and the organic compounds (i.e., all amino acids) which are synthesized by incorporating ammonium into the C-skeletons. Amino acids and the resulting proteins are the key building blocks of the cell. Both C and N nutrients are essential for various cellular functions, and therefore adequate supply of these two nutrients are critical for plant growth, development and response to a wide array of stresses and ultimately for the completion of life cycle and the production of harvestable organs.
Monitoring the Dynamic C/N Balance Status is Critical for Plant Growth and Development and Ecosystem Stabilization
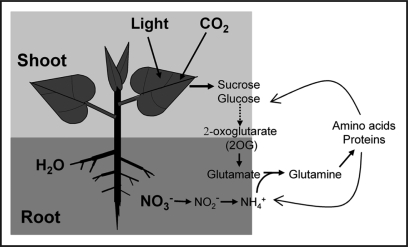
It has been well recognized that cellular C and N metabolism must be tightly coordinated. This coordination occurs at different levels (Fig. 1). From the biochemical point of view, CO2 is assimilated through photosynthesis, and the resulting Suc and Glc are converted through glycolysis and the tricarboxylic acid cycle to 2-oxoglutarate (2OG) or α-ketoglutarate, while nitrate (NO3−) is reduced by nitrate reductase to nitrite (NO2−) and further by nitrite reductase to ammonium (NH4+). 2OG serves as a C skeleton for the synthesis of glutamate (Glu) by incorporating photorespiratory NH4+. NH4+ from the primary N assimilation is then incorporated to Glu, resulting in the production of glutamine (Gln). Glu and Gln donate NH4+ used for the synthesis of all other amino acids, including aspartate (Asp) or asparagine (Asn), which serves as either an active NH4+ donor (Asp) or a transport/storage compound (Asn). Proteins, in particular enzymes, are essential for almost all cellular activities, including various steps of metabolic reactions involved in C and N metabolism. Therefore, maintaining an appropriate balance or ratio of C and N nutrients is critical from the metabolic point of view.
Figure 1.
A simplified whole plant view of tightly coordinated C and N metabolism. C assimilation and N uptake occur in the leaf and the root systems, respectively. 2-oxoglutarate (2OG), an important intermediate product of C metabolism, serves as the C-skeleton for the synthesis of glutamate (which uses photorespiratory ammonium; not drawn here). Ammonium (NH4+) resulted from primary N assimilation from nitrate (NO3−) is then incorporated to glutamate, and glutamine is synthesized. Other amino acids are then synthesized by using NH4+ donated from glutamate and glutamine, and therefore proteins can be synthesized. Proteins are essential for almost all of cellular activities, including C and N metabolism.
Another level of tight coordination involves the sensing and signaling of C/N balance through the long distance. NO3− and/or NH4+ are usually taken up by the root system and transported to the leaf, while C assimilation and metabolism primarily occur in the leaf (Fig. 1). Physiological and biochemical studies have concluded that when plants are deficient in N, the photosynthetic output was negatively affected, which can then be recovered if N is provided back to the growth media or the soil.1,2 Similarly, increasing C supply will promote N uptake and assimilation. Furthermore, genetic evidence have been obtained to demonstrate the importance of C on N metabolism or vice versa.3–5 Therefore, plants must develop a mechanism to sense the status of N in the root system and the surrounding soil environment, and coordinate with the sensory machinery in the leaf where photosynthetic output will be determined.
C/N balance in plants also affects the ecosystem response to CO2. Short-term laboratory experiments have shown that N limitation in the growth medium inhibits the shoot growth-promoting effect of high CO2.6 Consistently, a recent meta analysis of published physio-ecological studies have indicated that plants grown under elevated CO2 have lower leaf tissue concentrations of N than that grown under ambient CO2, which may negatively impact photosynthesis, plant community dynamics and the performance of herbivores or soil microorganisms.7–9 Indeed, a recent study which investigated many types of plant species for the long-term (six years) effects of CO2 revealed that N limitation is a major constraint for the sustainability of plant ecosystem response to elevated CO2.7 Elevated atmospheric CO2 has been both a significant challenge to the society and potentially an exciting opportunity to increase plant biomass for the purpose of C sequestration, food security and bioenergy production. Therefore, increasing the N input so as to maintain an optimal C/N balance in plants seems necessary to sustain the CO2 utilization efficiency at the ecosystem level.
Despite the central role of maintaining an appropriate C/N balance or ratio in plants, the C/N balance sensing and signaling mechanisms remain largely unknown. With the use of molecular biology tools during 1990s, some genes, such as ASN1 which encodes an isoform of Asn synthetase, are shown to be activated by N and suppressed by C.10 There are two excellent reviews on the C/N ratio or interactive response published eight years ago,1,2 and some up-to-date comprehensive reviews on C or N sensing or signaling.11,12 Therefore, this mini-review only focuses on the most recent findings from genetic and genomic studies that address the C/N balance or C:N ratio response. For convenience of comparing various reports, in this mini-review the C/N balance condition is represented by milimolar concentrations of Suc (or Glc) and of total N. For example, 30C/15N represents the combination of 30 mM Suc (or Glc in some instances) and 15 mM total N (NO3− and/or NH4+).
DNA Microarray Studies Reveal Complex Regulatory Patterns of Gene Expression in Response to Various C/N Conditions
Given the importance of C/N balance in the control of metabolism and cellular response to external nutrient status, one would expect that many genes involved in C and/or N metabolism and the associated sensing/signaling pathways are affected by different status of C and/or N nutrients. Gene expression analyses using traditional molecular biology tools have shown that several genes, such as ASN1 and GS2 (Gln synthetase), are reciprocally regulated by C (or light) and N.10,13 Indeed, during the recent five year period, DNA microarray-based transcriptomic studies involving N response have suggested that many genes involved in C metabolism are affected by N.14–16 Direct testing of the C/N interactive effects on genome-wide expression studies were performed in two groups, and their results have supported that more than half of the Arabidopsis genes are either up or downregulated by C alone, N alone, or the C-N combinations.17–19 However, these studies used different C/N regimes in their treatments and different tissue samples for RNA extraction and chip hybridization. In the first experiment performed by the Jang group,17 the C- and N-starved whole seedlings were treated by 60 mM N (40 mM NO3− and 20 mM NH4+) and/or 3% Glc (167C) for 3 hours in the dark before being harvested for analysis. In this experiment, it was reported that in the absence of Glc, N (0C/60N) only affected expression of 235 genes, compared to the 0C/0N control, while Glc alone (167C/0N) regulated the expression of almost 1,000 genes. More interestingly, N was found to modulate the expression of 8% of the Glc-regulated genes, illustrating the interactive effect of C and N in the control of gene expression. Among a total of nine clusters of C and N regulatory patterns illustrated in the report, surprisingly only a very small cluster of seven genes exhibited an interesting C-N interactive pattern: upregulated by 167C/0N, downregulated by 0C/60N, while 167C/60N brought the expression level close to that in the 0C/0N control. Although not shown in these clusters, it seems that some genes might exhibit an opposite pattern, such as SGA21, which was downregulated by 167C/0N (4.6 folds) and slightly upregulated by 0C/60N (1.3 folds), but similar under 167C/60N (down 1.4-fold) compared to 0C/0N.
The studies (in particular the second study) performed by the Gloria group are the most comprehensive so far as they tested a wide spectrum of C/N ratios and analyzed the data extensively using various bioinformatic or systems biology tools.18,19 It is noteworthy that in these studies different experimental setups were used compared to the aforementioned study.17 Palenchar et al.18 first reported the use of the C (Suc) and N-treated whole seedlings which were grown in hydroponics, and recently Gutierrez et al.19 only used the roots of the treated mature plants for RNA extraction and microarray analysis. In the first study,18 four C/N conditions, 0C/0N, 30C/0N, 0C/6N (NO3− and NH4+ in the ratio of 2:1), and 30C/6N, were used to treat the young seedlings which had been dark-adapted for two days. GeneChip analysis revealed a total of 3,652 detected genes which were then clustered into 60 different InterAct classes. Not surprisingly, close to 70% of the genes showed various responses to C and/or N, and among these differentially affected genes, 47% of them are regulated by the C and N interaction (30C/6N). This result strongly suggests that C-N interaction is an important response mechanism for plants. They also identified a very small number (nine) of genes that were only activated or repressed by the combination of C and N, which alone did not seem to have any effect. Like the study done by the Jang group,17 Palenchar et al.18 found that many genes (1,310) were affected by C only, but very few (4 only) genes were shown to be regulated by N alone, supporting that C has a profound effect in gene expression. However, N affected the expression of many C-activated or -repressed genes. Another promising finding is the identification of some novel, putative C-responsive cis elements, CN-responsive elements and N-dependent enhancers of C regulation (NDEs). Experimental verification of these regulatory elements will help to establish how C and/or N control gene expression.
In the second study19 which did not involve dark adaptation, the experimental space increased dramatically to include various C/N ratios at different levels of C (0, 30, 60 and 90) or N (0, 5, 10 and 15 NO3−). Approximately 1/3 of the detected genes (around 5,300) were affected by C alone, N alone and CN combinations. Furthermore, much more genes were regulated by N alone (15%) and C alone (about 50%). These suggest that the root system has a slightly different gene expression profile than the whole seedlings, if NO3− does not show a drastic difference than the combination of NO3− and NH4+. One significant finding from this report is the construction of the C, N or C-N regulatory networks using all of approximately 5,300 genes as queries in the network assembly. Although it remains unknown whether using specific cluster(s) of genes will result in similar or distinct gene-gene interaction networks, the assembled network based on all of these C, N responsive or C-N interactive genes indicates that the major subnetwork is primary and secondary metabolism. This provides the convincing genomic evidence that C/N balance (or imbalance) profoundly affects many aspects of cellular metabolism. Another intriguing finding is the possible involvement of auxin in C/N balance response. There are several auxin-related genes regulated by C alone or N alone and at least two genes are affected by C-N interaction. Although auxin has been shown to be important for signaling of C and N nutrients respectively,11,20 it remains to be demonstrated whether these effects are due to C/N balance.
It should be noted that it is difficult to compare these three sets of DNA microarray data, but two common features emerge. The first one is the lack of a pattern in which genes might respond similarly to the same ratio of C/N. While it is impossible to assess the C/N ratios for the two early studies because 0C and 0N were used,17,18 the most recent study did not seem to identify the genes that were responsive to the C/N ratios regardless of the absolute C or N levels.19 However, two clusters of genes exhibiting the expression patterns that may reflect a C/N balance regulatory response.19 One cluster consists of 71 genes that are repressed by C alone and N alone but not by the combinations of C and N, and conversely the other cluster includes 33 genes which are activated by C and N, respectively, but not by the C and N combinations. This is consistent with a preliminary DNA microarray study using 0C/0N, 90C (Suc)/0N, or 0C/60N-treated Arabidopsis wild-type seedlings (Zheng and Yang, unpublished data), where some genes were found to be upregulated by both of the imbalanced C:N ratios, i.e., 90C/0N and 0C/60N, while 90C/60N brought the expression level close to that under 0C/0N. Therefore, it is likely that distinct experimental conditions would lead to contrasting conclusions, which is reasonable given the ability and necessity for non-motile plants to precisely monitor and accurately respond to the dynamic changes in environmental conditions. The second feature is that the genes involved in metabolism, protein synthesis and degradation, RNA metabolism, signal transduction and hormone (such as auxin, ethylene and abscisic acid) pathways may play an important role in the C/N balance or ratio response.17–19 Although gene network assembly and regulatory element analysis provide insightful clues to the functions of these genes, it remains a daunting task to determine which genes play a critical role in the C/N balance response network. Furthermore, those key regulatory genes might not be affected at the transcriptional level and therefore can only be identified through different approaches.
Genetic Analyses Identify Three Novel Components in C/N Balance Response Using Three Distinct Bioassays
While many genes affected by various C/N treatments await functional dissection, the following three exciting breakthroughs have been made using either the classical mutant screens or the reverse genetic approach. Interestingly, in these studies, three different bioassays were used to demonstrate the functions of a putative nitrate transporter, a putative Glu receptor and a putative methyltransferase in the C/N balance response.
LIN1/NRT2.1.
The Malamy group first demonstrated that Arabidopsis lateral root initiation was sensitive to high C/low N.21 In wild-type plants, the lateral root densities were similar under low C/low N (0.5% Suc/0.02 mM NH4+ NO3−, equivalent to 15C/0.04N) and high C/high N (131C/60N) conditions, but under the high C/low N (131C/0.04N) condition, lateral root number and density decreased dramatically (about 10–20-fold decrease). Using this innovative bioassay, they isolated a recessive mutant designated lin1 (lateral root initiation1) that was insensitive to high C/low N, suggesting that the LIN1 gene product likely acts as a positive regulator of high C/low N response. Subsequently, work from the same group22 confirmed that lin1 affected the high C/low N response in lateral root initiation under a different C/N regime, i.e., 219C/0.2N. These results thus clearly demonstrate that the lin1 lateral root initiation phenotype is dependent on the high C:N ratios. Molecular cloning revealed that LIN1 encodes a putative high affinity NO3− transporter designated NRT2.1.22 Surprisingly, the presumed transporter function of NRT2.1 did not seem to be the primary mechanism for the C/N imbalance response phenotype observed in lin1/nrt2.1–4 mutants. Instead, they proposed that NRT2.1 has an additional function in signaling or even acting as a sensor of high C/low N, and that this function is more likely responsible for the C/N balance response in lateral root formation. This ground-breaking work provided the first evidence that C/N balance response is complex and yet genetically tractable. As C/N imbalance can go to the other extreme, i.e., low C/high N, it will be interesting to determine whether lateral root initiation is similarly repressed by low C/high N and, if so, whether LIN1/NRT2.1 is involved in low C/high N imbalance signaling or specifically in high C/low N response.
GLR1.1.
The second component involved in high C/low N response is the putative glutamate receptor 1.1 (GLR1.1) in Arabidopsis.23 The function of GLR1.1 in high C/low N response is suggested by the seed germination phenotype of the antisense AtGLR1.1 plants.23 Seeds from these antisense transgenic lines germinated and seedlings developed similarly as wild-type on the N-free MS media without supplemental Suc (0C/0N). However, only about 13% of the seeds of the antisense lines germinated when 25 mM or 87 mM (3%) of Suc was provided to the medium (25C/0N or 87C/0N), while germination percentage of the wild-type seeds did not seem to be affected. Interestingly, this inhibitory effect in the antisense lines could be reversed by adding 15 mM N (10 mM of NO3− and 5 mM of NH4+). The seed germination suppression under 25C/0N (or 87C/0N), but not under 0C/0N, 25C/15N or 87C/15N, strongly suggests that GLR1.1 may function to negatively modulate the high C/low N response in seed germination. Surprisingly, 25 mM Glc failed to exert the same effect as 25 mM Suc. Likewise, only NO3− but not NH4+ could reverse the high C/low N effect. These indicate that the C/N imbalance response phenotype might be due to the Suc- or NO3−-specific signaling effects rather than the general C or N nutritional status. Further support for the role of GLR1.1 as an important regulator of C and N metabolism is that the activities or transcript levels for the genes encoding several enzymes involved in C or N metabolism are suppressed in the mature antisense plants. It will be interesting to determine whether this gene expression effect is related to various C/N conditions. In addition, as in the case of LIN1/NRT1;2, it is not clear whether GLR1.1 is also involved in another aspect of C/N imbalance response, i.e., low C//high N. Results from a recent report indicate that low C/high N imbalance does not greatly affect seed germination kinetics.24 Therefore, the seed germination bioassay might not be suitable for testing whether GLR1.1 is also involved in low C/high N response. As abscisic acid is an important hormone involved in seed germination control and sugar signaling, it was further shown that GLR1.1 acts to affect ABA biosynthesis.23,25 However, how C/N balance affects GLR1.1 function remains to be investigated, although Glu is believed to be the major ligand for this putative Glu receptor.
OSU1/QUA2/TSD2.
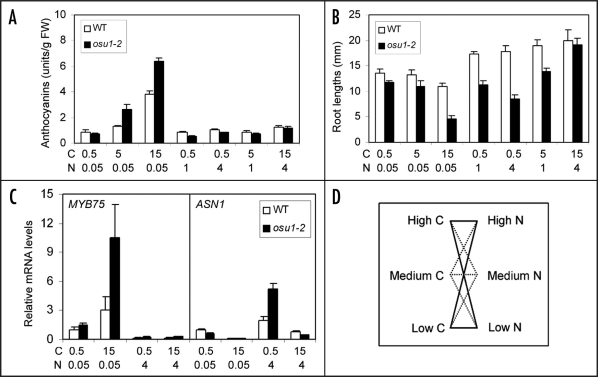
In contrast to the aforementioned two players, LIN1/NRT1;2 and GLR1.1, my group recently reported that a putative methyltransferase called OVERSENSITIVE TO SUGAR1 (OSU1) is a critical component involved in the negative modulation of both high C/low N and low C/high N imbalance responses.24 The osu1 mutants were initially isolated based on their sugar oversensitive phenotypes in shoot anthocyanin accumulation and primary root growth suppression. These mutants were hypersensitive to various metabolisable sugars such as Suc, Glc and maltose, but not to unmetabolisable or slowly metabolisable sugars including mannitol and a Glc analog, 3-O-methylglucose, suggesting that the sugar response is not due to the osmotic stress but instead the C nutrient status. By modifying the anthocyanin accumulation and primary root growth assays reported earlier,26 Gao et al.24 tested a wide spectrum of balanced and imbalanced C/N ratios (C ranging from 0.5 to 30, and N from 0.05 to 30). These results showed that osu1 mutants were indeed more sensitive to both increasing C/N ratios and decreasing C/N ratios in most of the cases (Fig. 2A and B). However, under the balanced C/N ratios, i.e., low C/low N, medium C/medium N, and high C/high N, the differences in cotyledon anthocyanin contents and root lengths were very similar between osu1 and wild-type. Furthermore, compared to wild-type, the osu1 mutants displayed similar or identical germination profiles under various C/N conditions and responded similarly to high C/low P (phosphate) or high C/low S (sulphate) conditions. Taken together, these results unequivocally demonstrate that OSU1 encodes a critical, negative modulator in the C/N imbalance response.
Figure 2.
C/N balance bioassays and gene expression patterns. (A) An example of osu1–2 hypersensitivity to high C/low N in anthocyanin accumulation. (B) An example of osu1–2 hypersensitivity of root growth inhibition in response to high C/low N and low C/high N. (C) Example of gene expression patterns in response to various C/N ratios. Relative mRNA levels of MYB75 and ASN1 genes, using ACTIN2 as an internal control, are shown. WT, wild-type. C (Suc) and N (total N) are in minimolar concentrations. Note that (A–C) were redrawn using the data published elsewhere.24 For more details, please see ref. 24. (D) A proposed mathematical model for C/N balance bioassay and gene expression analysis. Solid lines indicate the four C/N conditions which are recommended the minimum for showing the C/N balance response phenotype, while the dotted lines include other C/N set-ups which will convincingly demonstrate the C/N balance phenotype at various C or N levels or C/N ratios.
Molecular cloning showed that OSU1 is allelic to QUA2/TSD2,27,28 which encodes a putative methyltransferase but its substrate information is still lacking. In the Arabidopsis genome, there are a total of 29 members of this family of putative methyltransferase, most of which are functionally uncharacterized. Although how OSU1 functions in the C/N balance response remains to be investigated, it is shown to be expressed preferentially in the root system and in the vascular tissues of roots, stems and leaves, which is consistent with a function in nutrient response. However, OSU1 is less likely regulated by the C/N balance conditions at the transcriptional level.24 Instead, OSU1 is more likely regulated at the enzyme activity level, as implicated from the nature of the osu1–2 allele which caused a missense mutation in a conserved seven amino acid motif but did not suppress its transcript level. Gene expression analyses show that there likely exist two operating pathways modulated by OSU1 (Fig. 2C). These pathways are the low C/high N response pathway affecting ASN1 expression and the high C/low N pathway mediated by MYB75/PAP1 transcription factor.
A Proposed Mathematical Model for C/N Balance Assay
The fact that NRT2.1, GLR1.1 and OSU1, the three genes involved in C/N balance response, were identified using three distinct bioassays21,23,24 suggests that robust perception and signaling of C/N balance are critical for regulating many aspects of metabolism, growth and development. It is anticipated that more of this type of C/N balance bioassay will be uncovered in the future, given its central importance in various cellular activities and physiological functions. However, identifying the major components and pathways for the C/N balance response remains one of the most important tasks. Therefore, it is necessary to develop a relatively universal or complete experimental setup for both bioassays and gene expression analyses. There are at least three important aspects that need to be considered. First, a C/N balance bioassay should be focused on the default or major C/N balance response to simplify the mutant screen or phenotypic characterization. These could be shoot anthocyanin accumulation, root growth or lateral root initiation, or even shoot:root biomass ratio. For example, it will be interesting to test whether GLR1.1 transgenic lines and NRT2.1 mutants also alter the response to shoot anthocyanin accumulation or root growth. If so, this simple bioassay might be used in genetic screens to uncover more of the key players in C/N balance signaling.
Second, a complete or thorough C/N balance design will help demonstrate unequivocally whether the genes of interest are actually involved in the response to C/N balance or just C alone or N alone. A simplified mathematical model is proposed (Fig. 2D). Ideally, if one gene is involved in C/N balance response, one would expect that it must alter the response to different C/N ratios under various C or N levels, but the mutants or transgenic lines may behave similarly under the balanced C/N conditions, such as low C/low N, medium C/medium N and high C/high N. A dose curve response phenotype can then be characterized, as reported for the osu1 mutants,24 but a minimum of four C/N conditions (low C/low N, low C/high N, high C/low N and high C/high N) should be used to characterize the C/N balance response phenotypes. However, it should be noted that it might be difficult or in some instances even impossible to obtain the same phenotypic output under the balanced C/N conditions or the same C/N ratios where C and N have different absolute levels (for example, low C/low N vs. high C/high N). This is because biological responses are quite complex and in many cases they are also affected by increasing doses of C and/or N even though the C/N ratios are kept constant. In this case, the comparison of the mutants or transgenic lines with wild-type will be necessary because if they behave similarly as wild-type under the balanced C/N conditions, the conclusion that they do not affect the response to the balanced C/N conditions can be drawn, as in the case of osu1. Regarding the gene expression outputs, besides the models presented by the DNA microarray studies,18,19 two other patterns need to be seriously considered. The first one is shown in Figure 2C, where high C/low N have an opposite effect as low C/high N, but the transcript levels should be the same or similar under low C/low N and high C/high N. The second pattern, which shows the upregulation (or downregulation) by both of the imbalanced C:N ratios (i.e., high C/low N and low C/high N) but similar expression levels under high C/high N and low C/low N, will suggest the existence of a common C/N imbalance response pathway rather than the interaction of two imbalance pathways (e.g., high C/low N and low C/high N pathways).
Third, to demonstrate that the genes of interest are involved in sensing or signaling the internal C/N ratios, it is important to test whether the mutants or transgenic lines also respond similarly to organic N (such as major amino acids) as inorganic N. In addition, metabolomic study,29 coupled with transcriptomic analysis, will be more informative regarding how the genes are involved in regulating or modulating the C and N metabolism in response to various C/N ratios.
A Surprising Link between Cell Wall Biogenesis and C/N Balance Response
OSU1 turns out allelic to QUA2 and TSD2, which were identified independently through distinct mutant screens as being important for cell adhesion and other developmental functions.27,28 In addition, QUA2/TSD2 has been shown to localize to the Golgi apparatus,27,28 and QUA2 is co-expressed with many genes involved in pectin biosynthesis.28 The most interesting finding is the 50% decrease of homogalacturonan in qua2, compared to wild-type,28 although it remains to be determined whether qua2 also affects other components of cell wall. These results indicate that OSU1/QUA2/TSD2 may act as a putative pectin methyltransferase in cell wall biogenesis, as proposed.28 However, the effort to investigate its enzymatic property has been hampered by the difficulty of expressing fusion proteins in prokaryotic systems.24,28 Therefore, its proposed role as a pectin methylatransferase remains to be vigorously tested.
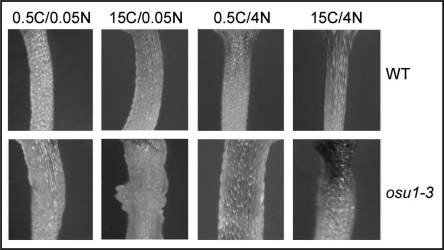
However, the fact that the mutations in the same gene result in both cell wall defect and hypersensitivity to imbalanced C/N ratios raises an intriguing possibility that cell wall biogenesis is linked to the C/N balance response. This potential link is also implicated by a recent report which shows a sugar hypersensitive mutant (hsr8) is allelic to a hemicellulose mutant mur4.30 Indeed, two other hemicellulose mutants, mur1 and mur3, were also involved in part of sugar response (hypocotyl growth in the dark). Although it remains to be tested whether this sugar response is a bono fide C/N imbalance response and whether the mutations in the genes involved in the biosynthesis of other cell wall components (such as cellulose) also affect C/N balance response, these results together with osu1/qua2/tsd2 do suggest that there could be a link between cell wall biogenesis and C/N balance response. There are at least three possibilities to explain the functional link. First, the cell wall defect could be part of the imbalanced C/N responses, as DNA microarray studies show that the N deficiency stress affected expression of some cell wall-related genes, such as expansins.14 Second, the altered C/N imbalance responses might be one of the consequences of modified cell wall biosynthesis or structure. It is possible that alterations in the biosynthesis of these complex carbohydrates might affect the overall C metabolism and thus impact the C/N responses. Third, the direct link between these two processes might not exist but they could affect each other rather indirectly. A preliminary experiment seems to exclude the first possibility as the balanced C/N conditions did not rescue the osu1 phenotype of hypocotyl swelling (Fig. 3). Indeed, the hypocotyl swelling phenotype was quite similar under these four C/N conditions, although 15C/0.05N seemed to exert a slightly stronger effect in ous1. In this case, although the second possibility cannot be excluded, the third possibility might very well be the underlying mechanism for the distinct roles of OSU1/QUA2/TSD2 in cell wall biogenesis and C/N balance response. If this is the case, OSU1 might have different substrates or targets which in turn regulate pectin biosynthesis and C/N balance responses, respectively. Clearly, it is crucial to understand the biochemical functions of the OSU1/QUA2/TSD2-encoded putative methyltransferase.
Figure 3.
Hypocotyl phenotypes of osu1–3 under various C/N conditions. Shown are the representative hypocotyls (right below the hypocotyl-cotyledon junction) from the seven-day-old wild-type (WT) and osu1–3 seedlings grown under four different C/N conditions.
C/N Interactions in Bacterial, Yeast and Animal Systems
As C/N balance perception and signaling are critical for optimal metabolism, growth and development for all cellular organisms, one would expect that studies in other systems such as bacteria, yeast and animals should provide some insights into the plant-based research. In bacteria, PII protein has been demonstrated to act as a central processor for integrating N and C metabolism.31 However, in yeast or animal systems, very little is known regarding the C and N signaling cross-talk or pathway interactions, although it has been well recognized that Ras/Rho GTPases, Snf1/AMP-activated kinases (AMPK) or the “hexosamine signaling pathway” and other components have various roles in the sensing/signaling of glucose or amino acid status.32–34 Nonetheless, it is encouraging to know that the C and N nutrient coincidence detection system does exist, such as the one reported recently.35 Some examples of the C/N cross-talk include the convergence of Snf1-glucose and TOR-nitrogen signaling pathways onto the Gln3 transcription factor,36 and the TOR pathway-mediated carbon catabolite repression of amino acid permeases.37 However, the regulatory mechanism for sensing the balance between C and N status and subsequently transmitting the signal remains to be investigated in yeast and animal systems.
On the other hand, attempts to investigate whether the plant orthologs of PII and TOR have conserved functions in C/N ratio sensing or signaling have only implicated that plants might evolve a distinct system to monitor and respond to the dynamic C/N conditions. For example, GLB1, the only Arabidopsis homolog of PII, was first implicated in the C/N ratio sensing, based on an ectopic expression study,38 but it was later shown that the T-DNA knockout mutants of GLB1 only exhibited weak phenotypes.39 These indicate that the plant PII protein may play a subtle role in the regulation of some steps of primary C and N metabolism in plants, such as Arg biosynthesis.39,40 Similarly, the role, if any, of the Arabidopsis TOR homolog in vegetative development does not seem to be critical,41 and the function of its regulatory proteins, RAPTOR1A and RAPTOR1B, in nutrient signaling has not been reported.42,43 However, it should be noted that the limited number of studies described above cannot exclude the alternative possibility that the evolutionally conserved components shared by bacteria, yeast, plants and animals do exist but they remain to be identified. For example, the role of methylation regulation in human nutrient response and obesity has been demonstrated by the recent studies involving the FTO (fat mass and obesity associated)-encoded demethylase.44
Future Perspective
Despite exciting progress towards dissecting the C/N balance response regulatory network, little is known regarding what signal is actually sensed. Several outstanding questions remain. Is there a C/N balance signal, or the C/N balance response is simply the outcome of the cross-talk of the C pathways and the N pathways? In either case, what are the sensor(s)? If it is due to the cross-talk of C and N pathways, do the two operating pathways, high C/low N and low C/high N, exist and if so, how do they interact with each other? In the case of OSU1, one explanation for osu1 to retain the normal response under the balanced C/N conditions is that OSU1 may be involved in the two distinct response pathways (high C/low N and low C/high N) or a common C/N imbalance response pathway, while the pathway for the balanced C/N response is distinct and operates independently of OSU1. This is because of the observations that anthocyanin accumulation and root growth are two separable responses and that expression of MYB75/MYB90 and ASN1 are differentially modulated by OSU1 under the imbalanced C/N24 (also see Fig. 2C). Therefore, OSU1 likely acts as a negative modulator either in distinct pathways leading to high C/low N or low C/high N responses or in a single pathway regulating various transcription factors in response to these distinct C/N imbalance conditions.
Clearly, in order to define which signal is sensed or which pathway operates in the complex C/N balance response, it is imperative to identify more players, which would allow us to place them in the context of the C/N balance response network. The established distinct bioassays and the proposed experimental design will help to reveal more of key players in the C/N balance response, which may act dependently or independently of OSU1, NRT2.1 and GLR1.1. It also should be noted that re-examination of the previously reported C or N response mutants is also important as it will help to determine whether the genes represented by the mutants or transgenic plants are involved in signaling to the C/N balance or specifically to C alone or N alone. Of particular interest are those mutants or transgenic plants showing reasonably strong phenotypes and representing regulatory genes such as hexokinases and their associated components,45–47 heterotrimeric G-proteins and their associated components,48 monomeric signaling GTPase ROP2,49 and ANR1 transcription factor.50 Some of these proteins have been extensively studied, with some signaling components identified, and therefore it will quickly put forth a C/N signaling pathway if they are shown to be also involved in the C/N balance. In addition, hormones have been shown to affect various nutrient responses including C and N,11,20 and therefore it is also necessary to assess which hormone(s) and which of their signaling pathways are involved in regulating or modulating C/N balance response. Through a combination of genetic, genomic and other approaches, it is perceivable that important breakthroughs towards establishing the complex yet elegant regulatory network for plant C/N balance response will be made in the next five to ten years.
Acknowledgements
Work in my laboratory was supported by a grant from the National Institutes of Health (NIH-SCORE, grant number S06GM008225, Project 12 to Z.L.Z.). I am grateful to Dr. Zeyu Xin, a former lab member, for providing Figure 3.
Abbreviations
- C
carbon
- N
nitrogen
- Suc
sucrose
- Glc
glucose
- ASN1
Asn synthetase isoform 1
- OSU1
OVERSENSITIVE TO SUGAR1
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8540
References
- 1.Coruzzi G, Bush DR. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 2001;125:61–64. doi: 10.1104/pp.125.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coruzzi GM, Zhou L. Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Curr Opin Plant Biol. 2001;4:247–253. doi: 10.1016/s1369-5266(00)00168-0. [DOI] [PubMed] [Google Scholar]
- 3.Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, et al. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006;142:1574–1588. doi: 10.1104/pp.106.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigelt K, Kuster H, Rutten T, Fait A, Fernie AR, Miersch O, et al. ADP-glucose pyrophosphorylase-deficient pea embryos reveal specific transcriptional and metabolic changes of carbon-nitrogen metabolism and stress responses. Plant Physiol. 2009;149:395–411. doi: 10.1104/pp.108.129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schofield RA, Bi YM, Kant S, Rothstein SJ. Overexpression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant Cell Environ. 2009;32:271–285. doi: 10.1111/j.1365-3040.2008.01919.x. [DOI] [PubMed] [Google Scholar]
- 6.Sun J, Gibson KM, Kiirats O, Okita TW, Edwards GE. Interactions of nitrate and CO2 enrichment on growth, carbohydrates and rubisco in Arabidopsis starch mutants. Significance of starch and hexose. Plant Physiol. 2002;130:1573–1583. doi: 10.1104/pp.010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich PB, Hobbie SE, Lee T, Ellsworth DS, West JB, Tilman D, et al. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature. 2006;440:922–925. doi: 10.1038/nature04486. [DOI] [PubMed] [Google Scholar]
- 8.Strengbom J, Reich PB. Elevated [CO2] and increased N supply reduce leaf disease and related photosynthetic impacts on Solidago rigida. Oecologia. 2006;149:519–525. doi: 10.1007/s00442-006-0458-4. [DOI] [PubMed] [Google Scholar]
- 9.Taub DR, Wang X. Why are nitrogen concentrations in plant tissues lower under elevated CO2? A critical examination of the hypotheses. J Integr Plant Biol. 2008;50:1365–1374. doi: 10.1111/j.1744-7909.2008.00754.x. [DOI] [PubMed] [Google Scholar]
- 10.Lam HM, Peng SS, Coruzzi GM. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol. 1994;106:1347–1357. doi: 10.1104/pp.106.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramon MRF, Sheen J. Sugar sensing and signaling. In: Somerville CRME, editor. The Arabidopsis Book. Rockville, MD: American Spciety of Plant Biologists; 2008. www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal EA, Gutierrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol. 2008;11:521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira IC, Coruzzi GM. Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 1999;121:301–310. doi: 10.1104/pp.121.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron and sulfate metabolism. Plant Physiol. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price J, Laxmi A, St Martin SK, Jang JC. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell. 2004;16:2128–2150. doi: 10.1105/tpc.104.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol. 2004;5:91. doi: 10.1186/gb-2004-5-11-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubio V, Bustos R, Irigoyen ML, Cardona-Lopez X, Rojas-Triana M, Paz-Ares J. Plant hormones and nutrient signaling. Plant Mol Biol. 2009;69:361–373. doi: 10.1007/s11103-008-9380-y. [DOI] [PubMed] [Google Scholar]
- 21.Malamy JE, Ryan KS. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 2001;127:899–909. [PMC free article] [PubMed] [Google Scholar]
- 22.Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA. 2005;102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang J, Turano FJ. The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:6872–6877. doi: 10.1073/pnas.1030961100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao P, Xin Z, Zheng ZL. The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS ONE. 2008;3:1387. doi: 10.1371/journal.pone.0001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang J, Mehta S, Turano FJ. The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 2004;45:1380–1389. doi: 10.1093/pcp/pch159. [DOI] [PubMed] [Google Scholar]
- 26.Martin T, Oswald O, Graham IA. Arabidopsis seedling growth, storage lipid mobilization and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 2002;128:472–481. doi: 10.1104/pp.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krupkova E, Immerzeel P, Pauly M, Schmulling T. The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 2007;50:735–750. doi: 10.1111/j.1365-313X.2007.03123.x. [DOI] [PubMed] [Google Scholar]
- 28.Mouille G, Ralet MC, Cavelier C, Eland C, Effroy D, Hematy K, et al. Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 2007;50:605–614. doi: 10.1111/j.1365-313X.2007.03086.x. [DOI] [PubMed] [Google Scholar]
- 29.Stitt M, Fernie AR. From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr Opin Biotechnol. 2003;14:136–144. doi: 10.1016/s0958-1669(03)00023-5. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, et al. Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell. 2007;19:2500–2515. doi: 10.1105/tpc.106.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Commichau FM, Forchhammer K, Stulke J. Regulatory links between carbon and nitrogen metabolism. Curr Opin Microbiol. 2006;9:167–172. doi: 10.1016/j.mib.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Schneper L, Duvel K, Broach JR. Sense and sensibility: nutritional response and signal integration in yeast. Curr Opin Microbiol. 2004;7:624–630. doi: 10.1016/j.mib.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Avruch J, Lin Y, Long X, Murthy S, Ortiz-Vega S. Recent advances in the regulation of the TOR pathway by insulin and nutrients. Curr Opin Clin Nutr Metab Care. 2005;8:67–72. doi: 10.1097/00075197-200501000-00010. [DOI] [PubMed] [Google Scholar]
- 34.Marshall S. Role of insulin, adipocyte hormones and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity and cancer. Sci STKE. 2006;2006:7. doi: 10.1126/stke.3462006re7. [DOI] [PubMed] [Google Scholar]
- 35.Xue C, Bahn YS, Cox GM, Heitman J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol Biol Cell. 2006;17:667–679. doi: 10.1091/mbc.E05-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertram PG, Choi JH, Carvalho J, Chan TF, Ai W, Zheng XF. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol Cell Biol. 2002;22:1246–1252. doi: 10.1128/MCB.22.4.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peter GJ, During L, Ahmed A. Carbon catabolite repression regulates amino acid permeases in Saccharomyces cerevisiae via the TOR signaling pathway. J Biol Chem. 2006;281:5546–5552. doi: 10.1074/jbc.M513842200. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh MH, Lam HM, van de Loo FJ, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrario-Mery S, Bouvet M, Leleu O, Savino G, Hodges M, Meyer C. Physiological characterisation of Arabidopsis mutants affected in the expression of the putative regulatory protein PII. Planta. 2005;223:28–39. doi: 10.1007/s00425-005-0063-5. [DOI] [PubMed] [Google Scholar]
- 40.Ferrario-Mery S, Besin E, Pichon O, Meyer C, Hodges M. The regulatory PII protein controls arginine biosynthesis in Arabidopsis. FEBS Lett. 2006;580:2015–2020. doi: 10.1016/j.febslet.2006.02.075. [DOI] [PubMed] [Google Scholar]
- 41.Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson GH, Veit B, Hanson MR. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 2005;3:12. doi: 10.1186/1741-7007-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang JC, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 47.Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 48.Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, et al. The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell. 2006;18:1226–1238. doi: 10.1105/tpc.105.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang G, Gao P, Zhang H, Huang S, Zheng ZL. A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE. 2007;2:1074. doi: 10.1371/journal.pone.0001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]