Abstract
Peltigera canina, a cyanolichen containing Nostoc as cyanobiont, produces and secretes arginase to a medium containing arginine. Secreted arginase acts as a lectin by binding to the surface of Nostoc cells through a specific receptor which develops urease activity. The enzyme urease has been located in the cell wall of recently isolated cyanobionts. Cytochemical detection of urease is achieved by producing a black, electron-dense precipitate of cobalt sulfide proceeding from CO2 evolved from urea hydrolysis in the presence of cobalt chloride. This urease has been pre-purified by affinity chromatography on a bead of active agarose to which arginase was attached. Urease was eluted from the beads by 50 mM α-D-galactose. The experimentally probed fact that a fungal lectin developing subsidiary arginase activity acts as a recognition factor of compatible algal cells in chlorolichens can now been expanded to cyanolichens.
Key words: arginase, lectin, Peltigera canina, recognition, urease
Introduction
Lichens are symbiotic associations between a fungus and a cyanobacterium (cyanolichens) or a green alga (chlorolichens), joined to form a new biological entity different from its individual components. Both bionts appear in nature among a mixture of millions of non-symbiotic microorganisms, and biochemical mechanisms for compatible combination are required.1 Thus, specificity is required for the lichen association. Specificity can be defined in this context as the preferential, but not exclusive, association of a biont with another.2 Recognition of compatible algal cell is carried out by specific lectins produced and secreted by the potential mycobiont.3 Lichen phenolics are not involved in the recognition process,4 in contrast to that found for other plant symbioses, such as mycorrhizal or Rhizobium legume associations.5
Some lectins from chlorolichens have been characterized as glycosylated arginases which bind to an algal cell wall receptor identified as an α-1,4-polygalactosylated urease.6 The binding is improved by Ca+2 and Mn+2 in a similar way to that described for legume lectins.7 Two forms of glycosylated arginases seem to be involved in the cell contact between chlorobiont and mycobiont, one of them, located in the cell wall of fungal hyphae and involved in cell contact between phyco- and mycobiont, whereas a second, secreted arginase produces recruitment of compatible algal cells near the surface of fungal hyphae.7 When glycosylated urease is lacking from the algal cell wall, fungal arginase is internalised, increasing the levels of algal putrescine, which promotes chloroplast disorganization, activation of glucanases and breakdown of the cell wall with loss of the protoplast.8 The evolution of symbiotic relationships implies then the synchronization of cell division and lectin receptor production,9 probably as a consequence of the perception of some environmental factors, such as short photoperiodic timing and low temperature values.
In addition, algal cell walls contain ligands other than polygalactosylated proteins, since concanavalin A, a legume lectin that specifically binds to mannose-containing ligand, is able to recognize potential chloro- and cyanobionts.10,11 Its will be of great interest to assess the roles of the non-galactose binding lectins in the process of discrimination between compatible and incompatible partners.
Concerning to cyanobiont recognition, Rikkinen12 described a model for signaling between symbionts in cyanolichens. Two types of signaling elicitors are known: general elicitors without major differences in sensitivity among responding organisms and specific elicitors that function is specific ways. General elicitors are usually substances associated to primary metabolism and include glucans, chitin oligomers, glycopeptides, cell wall fragments and many phenolics. Specific elicitors may include proteins, peptides, syrincolides and phenolics compounds. However, as described above, there is no any experimental evidence about the involvement of phenolics in lichen symbionts recognition and, in addition, no specific phenolics are produced by cyanolichens. Cell recognition of trigger molecules involves trans-membrane proteins, a part of which may function as a protein kinase, and transmitting signals to cellular mechanisms resulting in interbiotic responses. Unfortunately, no experimental probes about this mechanism have been as yet published.
Recognition of compatible cyanobiont can also be effected by lectins. In ferns and bryophytes, the plant produces lectins wich recognize sugar residues on the cell surface of a pre-symbiotic Nostoc.13 Kardish et al.14 studied the binding of a lectin isolated from the mycobiont of Nephroma laevigatum to Nostoc cells from different origins and concluded that proteins are involved in the control and regulatory processes of the symbiotic balance in the lichen thallus. Lectins have also been isolated from the mycobionts of some Peltigera species.15,16 In P. aphthosa, a lectin recognizes compatible Nostoc cells at the initial stage of the cephalodium formation and this process is highly specific.17 The specificity for cyanobionts has been confirmed in a more recent inoculation study attempting introduce foreign Nostoc strains into cephalodia of P. aphthosa.18
In this study, we attempt to probe that the role of a lectin-like arginase in the first step of the recognition of compatible bionts is also functional in a cyanolichen, such as Peltigera canina.
Results
Arginase secretion, labeling and binding to cyanobiont cells.
Samples of lichen thallus of 1.0 g dry weight were floated on 20 mL 40 mM arginine in 0.1 M Tris-HCl buffer, pH 9.15, for 8 h at 25°C maintained in the dark. Protein secretion was estimated as 0.99 mg mL−1 developing an arginase activity of 0.13 units.
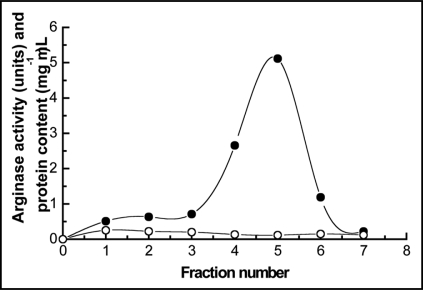
Secreted arginase was pre-purified by affinity chromatography on a bed of activated agarose containing bound urease purified from E. prunastri, as described in methods. Elution was carried out by using 100 mM α-D-galactose and a main peak of arginase activity was recovered at 15 mL filtrate (Fig. 1).
Figure 1.
Elution profile of secreted arginase chromatographied onto a bead of activated agarose containing bound urease purified from Evernia prunastri. Elution was carried out by using 100 mM α-D-galactose in 0.1 M phosphate buffer, pH 7.4. (○) protein content; (●) arginase activity.
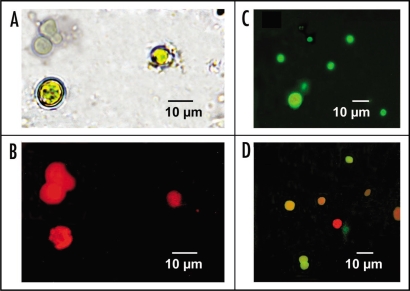
Purified arginase was labelled with FITC and freshly isolated cyanobionts from P. canina were incubated with FITC-arginase in the dark, at 30°C, for 2 h. Isolated and untreated algal cells (Fig. 2A) showed strong, red autofluorescescence of photo-excited chlorophylls in absence of the added fluorescent arginase (Fig. 2B). When cyanobionts were incubated with FITC-arginase, algal cells showed intense emission of green fluorescence (Fig. 2C). Desorption of the lectin from corresponding cell walls, achieved by incubation of FITC-lectin-labelled cells with 50 mM α-D-galactose for 2 h, resulted in almost total recovery (92.2%) of superficially adhered fluorescence in the supernatant.
Figure 2.
(A) Recently isolated Nostoc cells from Peltigera canina thallus without any treatment, observed at light microscope. (B) The same untreated Nostoc cells observed under fluorescence microscope. (C) Isolated Nostoc cells incubated for 2 h in the dark with 20 g of FITC-arginase secreted from P. canina thalli and observed under fluorescence microscope. (D) Isolated Nostoc cells incubated for 2 h in the dark with 20 g of FITC-arginase secreted from P. canina thalli, for 1 h on 100 mM α-D-galactose in 0.1 M phosphate buffer, pH 7.4, and observed under fluorescence microscope.
The enzymatic activity of cell wall urease.
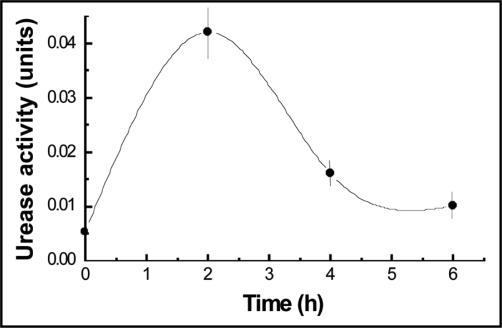
Urease activity has been assayed by using cells walls prepared from isolated cyanobionts cultured from 0, 2, 4 and 6 h on urea. As shown in the Figure 3, maximum of activity was achieved at 2 h incubation to decrease and stabilize later.
Figure 3.
Time course of urease extracted from isolated cells walls of P. canina cyanobionts obtained from thalli floated 50 mM phosphate buffer, pH 6.9, containing 40 mM urea, at 30°C in the dark. Values are the mean of three replicates. Vertical bars give standard error where larger than the symbols.
Cytochemical detection of urease.
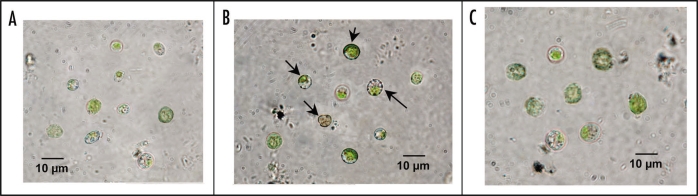
Urease activity was cytochemically located by using recently isolated cyanobionts from P. canina thalli floated for 2 h in the dark, at 30°C, on 40 mM urea (Fig. 4A). Then, cells were incubated with 10 mM cobalt choride during 30 min. Carbon dioxide produced during urease-catalyzed reaction formed insoluble cobalt carbonate when it was produced in the presence of cobalt choride. Then, cobalt sulphide was revealed as a black, electron-dense precipitate after addition of ammonium sulphide (Fig. 4B). However, cells without detectable urease activity, used as a control without a previous incubation on urea, did not show any deposit on their cell walls (Fig. 4C).
Figure 4.
Cytochemical detection of cell wall urease. (A) Nostoc cyanobionts recently isolated from P. canina thallus. (B) Cyanobionts incubated for 2 h on 40 mM urea and after this, on 10 mM CoCl2 and then revealed for CoCO3 with (NH4)2SO4. Black deposits (arrows) are mainly located at the cell wall. (C) Cyanobionts without urease induced in their cell walls treated for cytochemical localization of the enzyme.
Isolation of secreted urease by affinity chromatography.
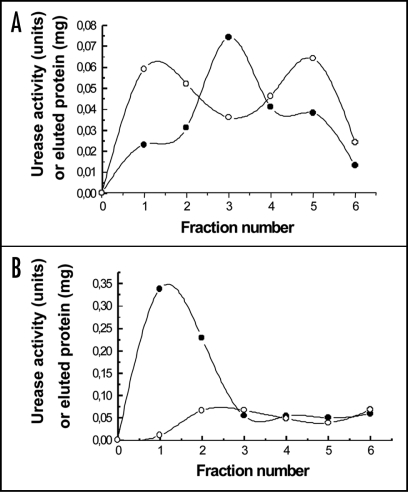
Cell wall ligand was isolated, by using protein extracted from these cell walls, by affinity choromatography using cyanogen bromide-activated agarose bearing arginase secreted from P. canina thalli. Protein was eluted with 50 mM α-D-galactose showing the first peak of eluate high urease activity (Fig. 5B). However, protein eluted with 50 mM phosphate buffer, pH 6.9, showed very low urease activity values in the totality of eluate fractions, as shown in Figure 5A, in which the range of urease activity was 1/10 with respect to that shown in Figure 5B.
Figure 5.
Elution profile of cell wall urease chromatographed onto a bead of activated agarose containing bound arginase from P. canina. Elution was carried out with 50 mM phosphate buffer, pH 6.9 (A) and 50 mM α-D-galactose in the same buffer (B). (○) protein content; (●) urease activity.
Discussion
Usually, plant lectins are involved in cell recognition, but specific enzyme activities have also been found associated to some lectins.19 In lichens, it could be expected that the recognition mechanism in chlorolichens7 could be expanded to cyanolichens on the basis of structural and functional similarities between lectins from cyano- and chlorolichens. In fact, a cyanolichen such as Peltigera canina produces and secretes arginase that, labelled with FITC, is able to bind to the cell wall of cyanobionts, apparently using a polygalactosylated urease as specific cell wall ligand, as it has previously been shown in chlorolichens.20 Desorption of arginase from the cyanobacterial cell wall is achieved by 50 mM α-D-galactose, indicating that the lectin uses the same polygalactosilated ligand that specifically uses in chlorolichens. Obviously, no desorption is achieved by using other hexoses, such as glucose, mannose, fructose, fucose and rhamnose, or even some aminosugars, such as glucosamine, N-acetylglucosamine and N-acetyl galactosamine. This is not surprising because Marx and Peveling11 found that cyanobionts isolated from different cyanolichen species were able to bind some lectins, such as ricin A, this being specific of α- and β-D-galactose. Thus, the occurrence of a polygalactosylated receptor to glycosylated arginases seems to affect to both chloro- and cyanobionts. In addition, other receptors can be developed by both photobionts, since concanavalin A, a lectin from Canavalia ensiformis specific from α-D-glucose and α-D-mannose, also binds to several cyanobionts11 and chlorobionts.10
Several attempts to locate urease in lichens symbionts have been achieved in the past.21–23 Cytochemical reaction described by Gomori (1952)24 has been used in the present work, using cyanobionts recently isolated from P. cannina. Images obtained in this way show that black deposits of cobalt carbonate, revealing urease activity, are mainly located at the cell wall of cyanobionts, although the occurrence of some polydisperse, small granules inside the cells indicates intracellular urease activity. This distribution of urease can be seen as surprising. However, it must be considered that a lichen thallus is composed by about 90% of a fungus (mycobiont) and 10% of algal cells in terms of total volume.25 Thus, the bulk of exogenous urea probably is captured by the fungal component from the intercellular spaces whereas the main opportunity for algal cells is the superficial hydrolysis of this urea in contact with the external surface of the cell wall urease. This is in agreement with the conservation of enzyme activity by urease immobilization in inert matrices, such as cellulose,26 alginate27 or gelatin,28 matrices that conserve a hydrophilic environment sufficient to carry out catalysis depending on the immobilized enzyme.
The nature of the ligand, defined as glycosylated urease, has been confirmed by affinity chromatography using a stationary phase constituted by arginase immobilized on agarose, resulting in the enrichment of urease activity. Since desorption of urease has been achieved by using a one-step elution, a unique peak of desorbed protein would be expected.
Urease is currently induced by urea in laboratory conditions using intact lichen thalli.29 Algal incompatibility has been previously defined as the internalization of lichen lectins into incompatible chlorobionts cells without specific ligand(s) in their cell walls.30 Internalization induces a supplement of intracellular putrescine synthesized by the action of fungal arginase, which increases the pool of endogenous putrescine synthesized by algal cells by action of their own arginine decarboxylase. These facts are supported by previous studies on enzyme localization.29 Very high levels of putrescine then lead to chloroplast disorganization, activation of algal glucanases, breakdown of the cell wall, loss of the chloroplast and finally, cell death.8,30 However, retention of fungal arginases outside algal cells, on the external surface of the cell wall, blocks the overproduction of putrescine and minimizes ultra-structural changes of compatible algal cells. Thus, compatibility depends on the retention of lectin on the external surface outside the algal cell, and this correlates to the occurrence of the a specific ligand in the cell wall of he algal cells rather than a specific feature based on the phylogenetic position of the potential chlorobiont.7
The genetic background for photobiont selection requires further studies to assess taxonomic relationships to selectivity patterns. Only in a few cases have a chlorobiont binding protein been found to mediate photobiont choice.7,31 Two principal strategies may be possible drivers for the evolution of symbiont selectivity patterns. Lower selectivity for symbiont lineages may increase the probability of finding an appropriate partner in varying habitants, whereas higher degree of specificity may resulting an optimization of the symbiotic interaction.32 In any way, the ability of fungal lectins developing arginase activity seems to be functional also in cyanobiont containing thalli. The first hypothesis would be supported by our, and some of previous, results. The universality of a unique receptor, a polygalactosylated urease, with different degrees of affinity for the ligand, high affinity for homologous and low affinity for heterologous arginases,6,7 as well as the existence of other receptors for different glycoproteins10,11 indicate that the increase of the probability to find the appropriate partner is the preferred strategy for the initiation of lichen association. This probably implies that the process described herein constitutes a first step in the global recognition mechanism in which potential cyanobionts (or chlorobionts) produces the lectin ligand as a defence response to the enzymatic attack of a fungus. After the development of this resistance reaction, other mechanisms of cellular communication must be developed in order to achieve the complete compatibility program, as suggested by Rikkinen.12
Material and Methods
Plant material.
Peltigera canina (Ach.) Schrad., is a cyanolichen fron Peltigerales, containing Nostoc as cyanobyont. Thalli were colleted at El Escorial (Madrid, Spain), and used throughout this work. Thalli were dried in air and stored in the dark, at room temperature, no longer than two weeks.
Isolation of lichen cyanobionts.
Cyanobionts were isolated from thalli of P. canina according to a protocol modified from Wastlhuber and Loos.34 Samples of dry thalli (1.0 g) were rinsed distilled water to remove superficial contaminations. Samples were then macerated in a mortar with 15 mL of an extraction buffer (50 mM HEPES-NaOH buffer, pH 7.0, containing 0.25 M sorbitol with the addition of 1% polyvinylpirrolidone 40,000 and 0.25% bovine serum albumin). To sediment larger fragments, the homogenate was brought to 700 xg, and then the centrifuge was turned off. Grinding in new buffer and centrifugation were repeated four times. All supernatants were pooled and centrifuged (1,200 xg, 10 min). The pellet obtained contained only few traces of fungal hyphae.
Arginase secretion.
Samples of lichen thallus of 1.0 g dry weight were floated on 20 mL 40 mM arginine in 0.1 M Tris-HCl buffer, pH 9.15, for 8 h at 25°C maintained in the dark. Media were filtered through Whatman paper 3 MM and dialyzed against distilled water for 24 h at 4°C in the dark.35 Protein estimated according Lowry et al.36 Peltigera arginase was pre-purified by affinity chromatography using cyanogen bromide-activated agarose bearing urease secreted from Evernia prunastri thalli, according to Legaz et al.7 About 2.0 g of agarose were fully hydrated with MilliQ water and mixed with 20 mL of a solution of purified urease containing 0.5 mg protein mL−1 in 0.1 M phosphate buffer (pH 7.4) for 16 h at 4°C. Glycine (0.6 g) was added to the mixture and stored at room temperature for 8 h.37 Beads of activated agarose containing bound urease were packed in columns (4.0 cm × 1.0 cm i.d.) and washed with 300 mL 0.1 M phosphate buffer (pH 7.4) at a flux rate of 1.0 mL min−1. Samples of 2.0 mL of media after incubation of P. canina thalli on 40 mM arginine were loaded onto the bead and kept for 2 h at room temperature. Protein was eluted with 100 mM α-D-galactose in buffer, collecting fractions of 3.0 mL volume and monitored by absorbance at 280 nm. Eluted protein from agarose beads was assayed for arginase activity.
Assay of arginase activity.
Both dialyzed media and cell-free extracts were assayed for arginase activity according to Legaz and Vicente,38 including crystalline urease in the reaction mixtures. These contained 10 µmol Tris-HCl buffer (pH 9.1), 7.5 µmol maleic acid, 5.0 µmol Mn2+ (as manganese sulfate), 0.4 µmol arginine, 8.1 mg crystalline urease, and 10.0 µg purified protein in a final volume of 3.0 mL. The reaction was carried out at 37°C for 30 min and stopped by addition of 0.5 mL of a saturated potassium carbonate solution. Ammonia produced was measured by the microdiffusion method.39 A unit of specific activity was defined as 1.0 µmol ammonia produced per mg protein per min.
Labeling of lichen arginases with fluorescein isothiocynate and binding to cyanobiont cells.
Purified arginase was labelled with fluorescein isothiocyanate (FITC) according Molina et al.20 A buffer solution of arginase, containing 10 µg protein mL−1, was mixed with 100 µg FITC and maintained in the dark, for 1 h at 30°C with continuous shaking. This mixture was dialyzed with distilled water to remove free fluorophore.
Cyanobionts cells isolated from samples of 1.0 g of P. canina thalli were incubated with fluorescent, secreted arginase (10 µg protein mL−1) in a final volume of 2.0 mL for 2 h at 30°C in the dark and continuous shaking. Cells were then colleted by centrifugation at 10,000 xg for 5 min at 2°C, resuspended in 1 mL extraction buffer (50 mM HEPES-NaOH buffer, pH 7.0, containing 0.25 M sorbitol with the addition of 1% polyvinylpirrolidone 40,000 and 0.25% bovine serum albumin).
Samples of collected cells were immobilized on a slide with 30 µL poly-L-lysine (1.0 µM in MiliQ water, Milipore)-coated slide 12 h, embedded with Mowiol-DABCO (Calbiochem, La Jolla, CA, USA) to avoid fluorescence decay, and examined in Zeiss Axioplan 2 fluorescence microscope (Jena, Germany) equipped with CCD. Images were captured with Viewfinder Lite program (Zeiss).
Cells treated as described above were later incubated for 2 h at 30°C with 1 mL of 50 mM α-D-galactose in buffer extraction for desorbing labelled-fluorescein arginase. Cells were observed using Zeiss Axioplan 2 fluorescence microscope (Jena, Germany) equipped with CCD. Images were captured with Viewfinder Lite program (Zeiss). Fluorescence in the supernatant was measured by using a spectrofluorimeter Kontron SF25. Samples were excited at 468 nm and the fluorescence emission was measured at 512 nm.
Cytochemical detection of urease.
Isolated cyanobionts were collected by centrifugation, washed and incubated with 50 mM phosphate buffer, pH 6.9, containing 40 mM urea for 2 h at 30°C. Then, cells were incubated in 2.0 mL 10 mM Cl2CO during 30 min at 30°C. After this, cyanobionts were centrifuged and washed, incubated for 30 s at room temperature with (NH4)2S, newly washed with 50 mM phosphate buffer, pH 6.9 and observed under light microscope.24
Isolation of algal cell walls and extraction of protein from cyanobiont cell walls.
Recently isolated cyanobionts were collected by centrifugation, washed and incubated with 50 mM phosphate buffer, pH 6.9, containing 40 mM urea for 0, 2, 4 and 6 h at 30°C. Then, algal cells were mechanically disrupted with alumina. Mixtures were subjected to ultrasonic disruption (10 × 5 s for a total of 2 min at 20 Kcycles s−1) on ice. Pellets were resuspended in 5 mL of distilled water with 0.05% Tween 20 (v/v) during 12 h at 4°C. After this, homogenates were centrifuged at 3,200 xg for 10 min at 2°C. Pellets were resuspended in 5.0 mL of aqueous 2% NaOH (w/v). Mixtures were subjected to ultrasonic disruption (10 × 5 s for a total of 2 min at 20 Kcycles s−1) on ice and immediately centrifuged at 3,200 xg form 10 min at 2°C. Supernatants were dialyzed against distilled water for 12 h at 4°C.40
Isolation of cell wall urease by affinity chromatography.
Cell wall ligands were isolated by affinity chromatography using cyanogen bromide-activated agarose bearing arginase secreted from P. canina thalli, according to Legaz et al.7 About 2.0 g of agarose were fully hydrated with MiliQ water, mixed with 20 mL of a solution of purified arginase containing 0.5 mg protein in 50 mM phosphate buffer, pH 6.9 for 16 h at 4°C. Glycine (0.6 g) was added to the mixture and stored at room temperature for 8 h.37 Beads of activated agarose containing bound arginase were packed in columns (3.0 cm × 1.5 cm i.d.) and washed with 300 mL 50 mM phosphate buffer, pH 6.9 t a flux rate of 1.0 mL min−1. Samples of 2.0 mL of media after incubation of P. canina thalli on 40 Mm urea were loaded onto the bead and kept for 2 h at room temperature. Protein was eluted with 50 mM phosphate buffer, pH 6.9 and 50 mM α-D-galactose in buffer, and monitored buy absorbance at 280 nm.
Urease activity was assayed in reaction mixtures containing 0.5 mL urea 40 mM, 2.2 mL phosphate buffer, pH 6.9 and 20 µg de protein, measured according to Lowry et al.36 in a final volume of 5 mL. The reaction was carried out 37°C for 30 min and stopped by addition of 0.5 mL of saturated potassium carbonate solution. Ammonia produced was measured by microdiffusion method.39 A unit of specific activity was defined as 1.0 µmol ammonia produced per mg protein per min.
Acknowledgements
This work was supported by a Grant from the Dirección General de Investigación Científica y Tecnológica (Ministerio de Ciencia y Tecnología, Spain) BFI2006-14263.
Abbreviations
- FITC
fluorescein isothiocyanate
- Tris-HCl
tris (hydroxymethyl) aminomethane hydrochloride
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/9164
References
- 1.Galun M, Kardish N. Lectins as determinants of symbiotic specificity in lichens. Cryptog Bot. 1995;5:144–148. [Google Scholar]
- 2.Bubrick P. Effects of symbiosis on the photobionts. In: Galun, editor. Handbook of Lichenology. Vol. 2. Boca Raton FL: CRC Press; 1988. pp. 130–133. [Google Scholar]
- 3.Lockhart CM, Rowell P, Stewart WDP. Phytohaemagglutinins from the nitrogen-fixing lichens Peltigera canina and P. polydactyla. FEMS Microbiol Lett. 1978;3:127–130. [Google Scholar]
- 4.Millanes A, Vicente C. Photoprotective strategies in lichens: an experimental approach using Evernia prunastri. J Hattori Bot Lab. 2003;94:293–302. [Google Scholar]
- 5.Doyle JJ, Luckow MA. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003;131:900–910. doi: 10.1104/pp.102.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacristán M, Millanes AM, Vicente C, Legaz ME. Synchronic production of fungal lectin, phycobiont lectin receptors and algal division in Evernia ptunastri. J Hattori Bot Lab. 2006;100:739–751. [Google Scholar]
- 7.Legaz ME, Fontaniella B, Millanes AM, Vicente C. Secreted arginases from phylogenetically far-related lichen act as cross-recognition factor for two different algal cells. Eur J Cell Biol. 2004;83:1–12. doi: 10.1078/0171-9335-00384. [DOI] [PubMed] [Google Scholar]
- 8.Molina MC, Stoker-Wörgötter E, Turk R, Bakon C, Vicente C. Secreted, glycosylated arginase from Xanthoria parietina thallus induces loss of cytoplasmic material from Xanthoria photobionts. Cell Adh Commun. 1998b;6:481–490. doi: 10.3109/15419069809010796. [DOI] [PubMed] [Google Scholar]
- 9.Czeczuga B, Krukowska KR. Effect of habitat condition on phycobionts and the content of photosynthesizing pigments in five lichen species. J Hattori Bot Lab. 2001;90:293–305. [Google Scholar]
- 10.Fontaniella B, Millanes AM, Vicente C, Legaz ME. Concanavalin A binds to a mannosecontaining ligand in the cell wall of some lichen phycobionts. Plant Physiol Biochem. 2004;42:773–779. doi: 10.1016/j.plaphy.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Marx M, Peveling E. Surface receptors in lichen symbionts visualized by fluorescence microscopy after use of lichens. Protoplasma. 1983;114:52–61. [Google Scholar]
- 12.Rikkinen J. Cyanolichens: an evolutionary overview. In: Rai AN, Bergman B, Rasmussen, editors. Cyanobacteria in Symbiosis. Amsterdam: Kluwe Academic Publ; 2002. pp. 31–72. [Google Scholar]
- 13.Bergman B, Rai AN, Johanson C, Söderbäck E. Cyanobacterial-plant symbioses. Symbiosis. 1993;14:61–81. [Google Scholar]
- 14.Kardish N, Silberstein L, Flemminger G, Galum M. Lectin from the lichen Nephroma laevigatum. Localization and function. Symbiosis. 1991;11:47–62. [Google Scholar]
- 15.Lehr H, Fleminger G, Galum M. Lectin from the lichen Peltigera menbranacea Characterization and function. Symbiosis. 1995;18:1–13. [Google Scholar]
- 16.Rai AN, Söderbäck E, Bergman B. Cyanobacterium-plant symbioses. New Phytol. 2000;147:449–481. doi: 10.1046/j.1469-8137.2000.00720.x. [DOI] [PubMed] [Google Scholar]
- 17.Lehr H, Galum M, Ott S, Jahns HM, Fleminger G. Cephalodia of the lichen Peltigera aphthosa. Specific recognition of the compatible photobiont. Symbiosis. 2000;29:357–365. [Google Scholar]
- 18.Paulsrud P, Rikkinen J, Lindblad P. Field experiments on cyanobacterial specificity in Peltigera aphthosa. New Phytol. 2001;152:117–123. doi: 10.1046/j.0028-646x.2001.00234.x. [DOI] [PubMed] [Google Scholar]
- 19.Shannon LM, Hnakins CN. Enzymatic properties of phytohemagglutinins. In: Loewus FA, Ryan CA, editors. The Phytochemistry of Cell Surface Interactions. New York and London: Plenum Press; 1981. pp. 93–114. [Google Scholar]
- 20.Molina MC, Muñiz E, Vicente C. Enzymatic activities of algal-bilding protein and its algal cell wall receptor in the lichen Xanthoria parietina. An approach to the parasitic basis of mutualism. Plant Physiolo Biochem. 1993;31:131–142. [Google Scholar]
- 21.Legaz ME, Vicente C. Location of several enzymes of L-arginine catabolism in Evernia prunastri thallus. Z Naturforsch. 1981;36:692–693. [Google Scholar]
- 22.Pérez-Urria E, Vicente C. Purification and some properties of the secreted urease from Evernia prunastri. J Plant Physiol. 1989;133:692–695. [Google Scholar]
- 23.Millanes AM, Fontaniella B, García ML, Solas MT, Vicente C, Legaz ME. Cytochemical location of urease in the cell wall of two different lichen phycobionts. Tissue and Cell. 2004;36:373–377. doi: 10.1016/j.tice.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Gomori G. Microscopic Histochemistry. Chicago: The University of Chicago Press; 1952. p. 243. [Google Scholar]
- 25.Collins CA, Farrar JF. Structural resistances to mass transfer in the lichen Xanthoria parietina. New Phytologist. 1978;81:71–83. [Google Scholar]
- 26.Wang J, Song XJ, Ren ZL, Yuan B, Lei ZL. Studies on immobilization of urease on cellulose acetate membrane. Chem J Chin Univ. 1998;19:1104–1106. [Google Scholar]
- 27.Das N, Kayasth AM, Malhotra OP. Immobilization of urease from pigeon pea (Cajanus cajan L.) in polyacrylamide gels and calcium alginate beads. Biotechniol Appl Biochem. 1998;28:251. doi: 10.1111/j.1470-8744.1998.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 28.El-Shora HM. Properties an immobilization of urease from leaves of Chenopodium album. Bot Bull Acad Sin. 2001;42:251–258. [Google Scholar]
- 29.Vicente C, Legaz ME. Lichen enzymology. In: Galun M, editor. Handbook of Lichenology. Vol. 1. Raton, FL: CR Press Boca; 1988. pp. 239–284. [Google Scholar]
- 30.Molina MC, Bajon C, Sauvanet A, Robert D, Vicente C. Detection of polysaccharides and ultrastructural modification of the photobiont cell wall produced by two arginase isolectins from Xanthoria parietina. J Plant Res. 1998a;11:191–197. [Google Scholar]
- 31.Doyle JJ. Phylogenetic perspectives on nodulation: Evolving views of plants and symbiotic bacteria. Trends Plant Sci. 1998;3:473–478. [Google Scholar]
- 32.Grube M, Hawksworth DL. Trouble with lichen: the re-evaluation and re-interpretation of thallus form and fruit body types in the molecular era. Mycol Res. doi: 10.1016/j.mycres.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Sacristán M, Millanes AM, Legaz ME, Vicente C. A lichen lectin specifically binds to the α-1,4-polygalactoside moiety of urease located in the cell wall of homologous algae. Plant Signaling & Behavior. 2006;1:23–27. doi: 10.4161/psb.1.1.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wastlhuber R, Loos E. Differences between cultured and freshly isolated cyanobiont from Peltigera is there symbiosis-specific regulation of glucose carrier. Lichenologist. 1996;28:67–68. [Google Scholar]
- 35.Planelles V, Legaz ME. Purification and some proprieties of the secreted arginase of the lichen Evernia prunastri and its regulation by usnic acid. Plant Sci. 1987;51:9–16. [Google Scholar]
- 36.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 37.Walters RR. Ligands forinmobilization. In: Dean PGD, Johnson WS, Middle FA, editors. Affinity Chromatography, a Practical Approach. Oxford: IRL Press; 1985. pp. 114–166. [Google Scholar]
- 38.Legaz ME, Vicente C. Two forms of arginase in Evernia prunastri. Biochem Biophys Res Commun. 1982;104:1441–1446. doi: 10.1016/0006-291x(82)91411-5. [DOI] [PubMed] [Google Scholar]
- 39.Conway EJ. Microdiffusion Analysis and Volumetric Error. London: Crosby Loockwood; 1962. [Google Scholar]
- 40.Legaz ME, Vicente C. Regulation of urease activity of Cladina dendroides and its photobiont by lichen pheno. Plant Sci. 1989;63:15–24. [Google Scholar]