Abstract
The proper regulation of enlargement and patterning of plant lateral organs is essential for plant functionality. In an earlier work, we characterized the role of a microRNA (miRNA)-transcription factor regulatory module, miRNA164-CUC2, in the enlargement and patterning of multiple lateral organs in Arabidopsis. This regulatory module genetically interacts with another transcription factor, CRC, in fruit development patterning. Here, we characterize the genetic interaction of this module with a homeodomain transcription factor, BREVIPEDICELLUS (BP), that has been shown to play roles in leaf development patterning.
Key words: CUC2, miRNA164, BREVIPEDICELLUS, KNAT, lateral organ patterning, microRNAs, Arabidopsis
Plant development is an intricate process. Multiple lateral organs are generated that carry out specialized functions. These lateral organs must be properly patterned and correctly sized relative to the entire plant to fulfill their functions. Intensive work over the last several decades has made significant progress in uncovering players that function in these processes. However, our understanding of the regulatory mechanisms that direct these complex developmental processes are far from complete.
In a screen for lateral organ development mutants, we uncovered a central regulatory module that functions as a positive regulator of lateral organ enlargement and patterning.1 This regulatory module consists of a NAC-domain transcription factor, CUC2, and its known targeting miRNA, miRNA164. The module was isolated as a single point mutation in the miRNA target site of CUC2 from an Arabidopsis mutant screen. As predicted, this mutation results in a dominant allele of CUC2, and thus was named cuc2-1D. The cuc2-1D allele enabled us to uncover and characterize a previously unknown role this module plays in regulating the size of plant lateral organs. A disruption of miRNA targeting of CUC2, due to the mutant miRNA target site, results in overaccumulation of CUC2 transcript and enlarged vegetative and reproductive lateral organs. This increased size is due to a disruption of a cell proliferation checkpoint; cellular proliferation continues beyond its usual developmental timeframe in expanding lateral organs, resulting in larger organs.
In addition to the role of miRNA164 and CUC2 as a regulatory module in lateral organ enlargement, this module also regulates aspects of both vegetative and floral lateral organ patterning. Silique patterning is disrupted, resulting in reduced seedset. Leaf patterning is also disrupted; the leaves display a lobed leaf margin. Work using a triple loss-of-function mutant line of all three miRNA164 loci and transgenic cuc2 alleles with mutant miRNA target sites have also shown similar patterning phenotypes.2,3 Recent work has also uncovered a role for NAC-domain transcription factors in the regulation of leaflet formation in species having compound leaves.4,5 In our earlier work, we characterized a genetic interaction with another transcription factor, CRC in flower developmental patterning. Here we report on the genetic interaction with another transcription shown to play a role in leaf patterning.
Several of the class 1 KNOTTED-like members from Arabidopsis thaliana (KNAT) homeodomain proteins have been shown to play roles in shoot apical meristem maintenance and organ patterning and development. BP (KNAT1) has been shown to play a role in multiple developmental processes. BP was first described as a genetic marker in Arabidopsis,6 and was later cloned as KNAT1.7 Loss of function alleles of bp show several flowering phenotypes including downward pointing siliques.7 In addition, overexpression of BP in Arabidopsis results in deeply lobed leaves, similar to an overexpression phenotype observed in maize.8 Other KNAT members SHOOT-MERISTEMLESS (STM) and KNAT6 play important roles in plant development and genetically interact with CUC genes.9–11 Furthermore, during flowering, BP plays a role in restricting KNAT6 and KNAT2 expression to properly guide floral development.12 The transcription factor, ASYMMETRIC LEAVES1 and gradients in the hormone auxin repress BP and thus promote leaf fate.13 In the gain of function mutant jagged lateral organs-D, BP and STM were upregulated and bulk auxin transport reduced resulting in strongly lobed leaves.14 BP is also regulated by the BEL1-LIKE HOMEODOMAIN (BLH) members SAWTOOTH1 (BLH2/SAW1) and SAWTOOTH2 (BLH4/SAW2).15 In loss of function saw1 saw2 double mutants, BP is ectopically expressed and the leaves display a serrated margin. Thus, BP clearly plays a role in the development of lobed leaves and genetically interacts with multiple other transcription factors. Additionally, since CUC genes have been shown to interact with other KNAT members, we investigated the genetic interactions between the miRNA164-CUC2 regulatory module and BP during leaf development.
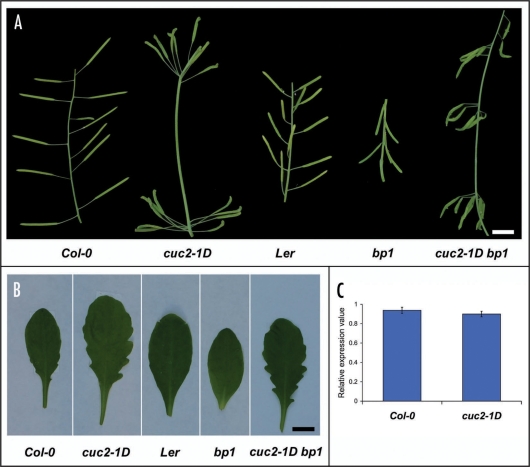
To investigate the genetic interactions between the miRNA164-CUC2 regulatory module and BP, we characterized the segregating F2 population from a cross using cuc2-1D and bp1. bp1 is a loss of function mutant in a Landsberg erecta (Ler) background.6,7 However, er loss of function mutants where observed to suppress both the leaf enlargement and leaf lobation phenotype in cuc2-1D.1 Therefore, we selected for plants in which ER was present at least as a heterozygote. In this population, we isolated plants which were homozygous cuc2-1D and bp1. In the flowering stalks, these double mutant plants displayed an additive phenotype; the silique patterning was disrupted and the arrangement along the inflorescence stalk was clustered as in cuc2-1D (Fig. 1A). The siliques were also downward orientated as in bp1. The leaves displayed the typical lobed margin found in cuc2-1D single mutants (Fig. 1B). These data suggest miRNA164-CUC2 and BP either act independently of each other or miRNA164-CUC2 acts downstream of BP.
Figure 1.
Genetic interactions between miRNA164-CUC2 and BP. (A) cuc2-1D displays a clustered silique spacing on the flowering stalk relative to the background Col-0. bp1 displays downward orientated siliques on the flowering stalk relative to the background Ler. In the double mutant, cuc2-1D bp1, the plants display an additive phenotype with clustered and downward orientated siliques. (B) cuc2-1D displays a lobed leaf phenotype relative to Col-0, while bp1 has a smooth leaf margin. In the double mutant, cuc2-1D bp1, the lobed leaf phenotype is retained. (C) qPCR was used to track BP transcript levels in cuc2-1D plants. No significant differences were seen in cuc2-1D relative to the background Col-0. The average and SE of three biological replicates are shown. qPCR was completed as in (Larue et al. 2009) except that the following primers were used for BP: forward 5′-CTA ACA AGG CCC ATT CAG GA-3′ and reverse 5′-TGT CAC TCT TCC CAT CAG GA-3′. Scale bars in (A and B): 1 cm.
It is possible that cuc2-1D increases the expression or results in misexpression of BP, resulting in a lobed leaf phenotype. However, in the bp1 loss of function line, other unknown factors may still be modulated by cuc2-1D and these could be sufficient for the lobed leaf phenotype. Therefore as a further check, we also determined if cuc2-1D modulates the expression of BP using quantitative reverse transcription polymerase chain reaction (qPCR). However, there was no significant change in BP expression levels of cuc2-1D relative to Col-0 plants (Fig. 1C). Additionally, RNA in situ analysis did not detect any misexpression of BP in developing leaves of cuc2-1D (data not shown). This suggests the miRNA164-CUC2 regulatory module does not directly modulate BP expression to affect the lobed leaf phenotype.
In conclusion, we propose two possible genetic interactions between miRNA164-CUC2 and BP. The first is these two important developmental regulators act independently. This would be unique from that shown with some of the genetic interactions with CUC2 and other KNAT members in Arabidopsis.9–11 In this case, cuc2-1D must act through alternative unknown players to direct leaf lobation. The second possible interaction is that miRNA164-CUC2 acts downstream of BP. Thus, cuc2-1D bypasses the need for BP in the leaf lobation phenotype. If this were true, we would expect a modulation of the downward orientated silique phenotype found in bp1. Although we observed no change of this phenotype in the double mutant, we can not preclude that the silique orientation phenotype and the leaf phenotype are independent with respect to miRNA164-CUC2. Thus, much additional work remains to fully characterize the role of miRNA164-CUC2 in leaf development. Additional mutant screening in a cuc2-1D background will be helpful in allowing us to begin to uncover additional players in these important developmental processes. Future work to dissect the complex regulatory pathways that direct plant developmental patterning and enlargement will likely yield more critical pieces in this puzzle.
Acknowledgements
We would like to acknowledge the Walker Laboratory members for their helpful discussions. C.T.L. was supported by a MU-Monsanto Graduate Research Fellowship. This research was supported by grants from DOE (DE-FG02-05ER15652) and the University of Missouri Food for the 21st Century Program.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/9037
References
- 1.Larue CT, Wen J, Walker JC. A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis. Plant J. 2009;58:450–463. doi: 10.1111/j.1365-313X.2009.03796.x. [DOI] [PubMed] [Google Scholar]
- 2.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development. 2007;134:1051–1060. doi: 10.1242/dev.02817. [DOI] [PubMed] [Google Scholar]
- 4.Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, et al. A conserved molecular framework for compound leaf development. Science. 2008;322:1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- 5.Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, et al. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development. 2009;136:823–832. doi: 10.1242/dev.031625. [DOI] [PubMed] [Google Scholar]
- 6.Koornneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. Linkage map of Arabidopsis thaliana. J Hered. 1983;74:265–272. [Google Scholar]
- 7.Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, et al. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. PNAS. 2002;99:4730–4735. doi: 10.1073/pnas.072626099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long J, Moan E, Medford J, Barton M. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 10.Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell. 2006;18:1900–1907. doi: 10.1105/tpc.106.041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- 12.Ragni L, Belles-Boix E, Gunl M, Pautot V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell. 2008;20:888–900. doi: 10.1105/tpc.108.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay A, Barkoulas M, Tsiantis M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development. 2006;133:3955–3961. doi: 10.1242/dev.02545. [DOI] [PubMed] [Google Scholar]
- 14.Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell. 2007;19:1795–1808. doi: 10.1105/tpc.106.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell. 2007;19:2719–2735. doi: 10.1105/tpc.106.048769. [DOI] [PMC free article] [PubMed] [Google Scholar]