Abstract
The last decade has brought major advances in our knowledge of the structures and mechanisms of MAO A and MAO B, which are pharmacological targets for specific inhibitors. In both enzymes, crystallographic and biochemical data show their respective C-terminal transmembrane helices anchor the enzymes to the outer mitochondrial membrane. Pulsed EPR data show both enzymes are dimeric in their membrane-bound forms with agreement between distances measured in their crystalline forms. Distances measure between active-site directed spin labels in membrane preparations show excellent agreement with those estimated from crystallographic data. Our knowledge of requirements for development of specific reversible MAO B inhibitors is in a fairly mature status. Less is known regarding the structural requirements for highly specific reversible MAO A inhibitors. In spite of their 70% sequence identity and similarities of Cα-folds, the two enzymes exhibit significant functional and structural differences that can be exploited in the ultimate goal of the development of highly specific inhibitors. This review summarizes the current structural and mechanistic information available that can be utilized in the development of future highly specific neuroprotectants and cardioprotectants.
The flavoenzymes monoamine oxidase A (MAO A) and monoamine oxidase B (MAO B)1 have been extensively studied with approximately 20,000 published papers listed in data bases such as PubMed. This long-term interest stems from their roles in the oxidative catabolism of important amine neurotransmitters including serotonin, dopamine, and epinephrine. Since the original discovery by Zeller (1) that hydrazines are MAO inhibitors that can lead to mood elevation, many pharmaceutical and academic laboratories have developed MAO inhibitors with potential clinical use; these include seven different MAO-inhibitors approved by the Food and Drug Administration. The concept that humans and other mammals contain two distinct MAOs was a controversial topic until the demonstration (2) that MAO A and MAO B are encoded by separate genes that correspond to different amino acid sequences with ~70% identity. All mammals contain both MAO A and MAO B, which are localized in the outer membrane of mitochondria in most tissues. It is currently believed that the respective genes encoding MAO A and MAO B evolved by a duplication event from a single ancestral gene. Teleost (fish) and lower invertebrates are found to contain only a single MAO gene. Limited studies of zebrafish MAO suggest the single MAO exhibits properties of both MAO A and of MAO B although little is known regarding its specific enzymological and structural properties (3).
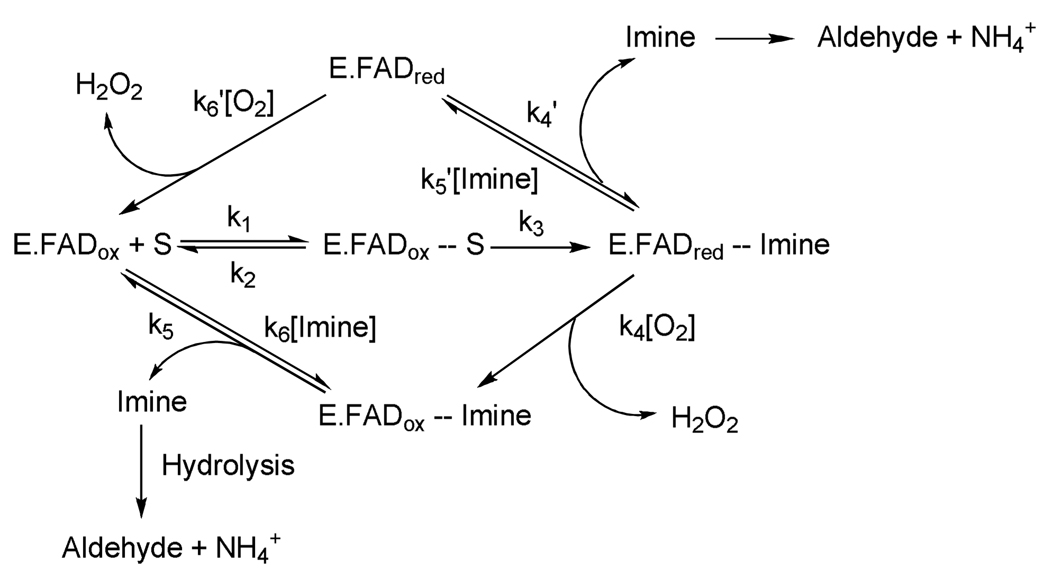
MAO A and MAO B catalyze the oxidative deamination of amine neurotransmitters using O2 as electron acceptor. Table 1 shows the relative rates of biological amine oxidation by MAO A and by MAO B. There are two general catalytic reaction pathways as shown in Scheme 1. For most substrates both MAO A and MAO B follow the lower loop of the pathway shown in Scheme 1 in which oxygen reacts with the enzyme-product complex before product has dissociated. There is general consensus that the deprotonated (rather than the protonated) amine moiety on the substrate binds to the active site on the enzymes and is oxidized to the protonated imine (4) (found with both MAO B and MAO A) with the covalent 8α-S-cysteinyl FAD cofactor being reduced to its hydroquinone form. To complete the catalytic cycle, the reduced FAD cofactor reacts with O2 to generate oxidized flavin and H2O2. The dissociated protonated imine is released from the enzyme and undergoes a non-catalyzed hydrolysis to form NH4+ and the corresponding aldehyde. The products of the MAO-catalyzed reactions (especially the H2O2) have sufficient deleterious reactivities to account for associated health-related problems. Therefore, a considerable effort has gone into the design and development of specific MAO inhibitors that could function in a protective role. Since the original observation that MAO inhibition results in mood elevation, six (and a recent 7th) FDA approved MAO inhibitors have been available for clinical use. The use of these compounds as pharmacological agents has declined because of the side effects, which require careful dietary restrictions to reduce the intake of biogenic amines in foods that could serve as false neurotransmitters. This review summarizes available knowledge on the molecular and catalytic properties of MAO A and MAO B relevant to current and future development of protective agents that could be effective in both neuro- and cardio-therapies.
Table 1.
Turnover numbers for the oxidation of naturally-occurring amine neurotransmitters by purified human MAO A and MAO B.a
| Amine Substrate | MAO A | MAO B | ||
|---|---|---|---|---|
| kcat (min−1) | Km (µM) | kcat (min−1) | Km (µM) | |
| Serotonin | 198 (183) | 295 ± 46 | 61 (33) | 2,270 ± 310 |
| Dopamine | 75 (71) | 240 ± 43 | 277 (65) | 128 ± 16 |
| Histamine | Ki = 2.1 mM b | 3.5 (2.0)c | ~ 4,000 | |
| N-Methylhistamine | Ki > 15 mM b | 35 (17) | 166 ± 8.1 | |
kcat values at saturating concentrations of organic substrate and of O2. Values in parenthesis are for saturating amine levels at air saturation. All data are determined following the rate of O2 uptake polarographically.
Histamines are poorly oxidized by MAO A and the Ki values listed are competitive inhibition constants with kynuramine as substrate. A histamine kcat value of 0.6 min−1 is found and N-Me-histamine is oxidized with a kcat value of ~0.1 min−1. Both values were determined at saturating amine concentrations at air saturation.
Values determined at 5 mM substrate concentrations.
Scheme 1.
General Reaction Scheme for MAO Catalysis.
Medical and Pharmacological Importance
MAO B
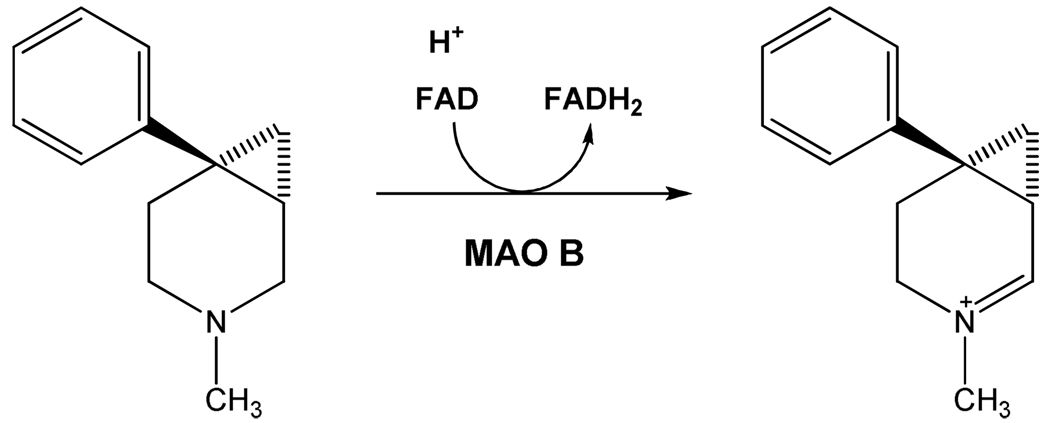
The observation by Fowler’s laboratory (5) that MAO B levels in the human increase ~4–5 fold on aging had been also observed earlier in rats and presents a rationale for the involvement of MAO B in age-related neurological disorders such as Parkinson’s Disease (6). Increased MAO B levels would be expected to diminish dopamine levels and to increase levels of the catalytic reaction products, dopanal and H2O2. Dopanal has been implicated in α-synuclein aggregation involved in the etiology of Parkinson’s Disease (7). Increased levels of H2O2 in the cell promote apoptotic signaling events resulting in the decreased levels of dopamine-producing cells and the development of Parkinson’s Disease (6). A classic example of the MAO B-catalyzed bioactivation of a compound that results in the genesis of Parkinson-like syndrome is the oxidation of MPTP to MPP+ (8) which functions as a mitochondrial toxin and results in the destruction of glial cells in the substantia nigra. Inhibition of MAO B results in a protective effect from this cell-destructive bio-activation.
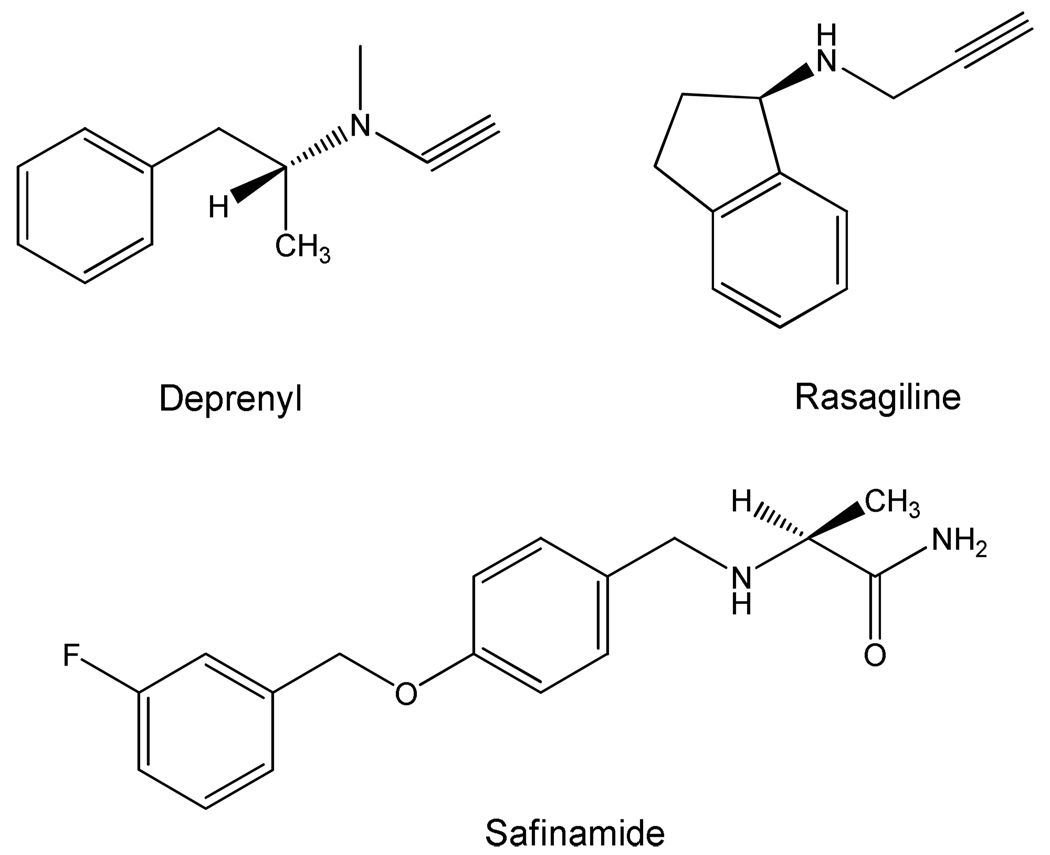
Agents that specifically target the inhibition of MAO B are likely to serve as neuroprotectants. Indeed, treatment of pre-Parkinson’s patients with deprenyl (and, more recently, with rasagiline) (see Scheme 2 for structures) have been shown to be effective in reducing the development (but not prevention) of the disease (9). Both of the above-mentioned inhibitors are acetylenic compounds that form covalent adducts with the N(5) of the covalent FAD of MAO B, and are pharmacologically inert as MAO A inhibitors. Recent results with safinamide (see Scheme 2 for structure), a very specific MAO B non-covalent inhibitor, suggest it may function effectively as a neuroprotectant.
Scheme 2.
Structures of MAO B-Specific Inhibitors.
MAO A
MAO A functions specifically in the oxidative metabolism of serotonin although it also oxidizes dopamine effectively. Complete elimination of MAO A activity by genetic deletion in mice (10) (and one case in humans (11), results in the expression of aggressive behavior (12). Recent studies on MAO A knockout mice demonstrated a marked effect of higher serotonin levels on cardiac remodeling (13). MAO A levels have also been found to dramatically increase (~9 fold) in the heart of aged rats, thereby suggesting the probability of age-dependent increases in MAO A levels in humans (14). The consequence of this large increase of MAO A in the heart is suggested to involve increased apoptosis and necrosis of cardiac cells due to increased levels of reactive oxygen species from the H2O2 produced. As suggested from the work of Parini and co-workers (15), the identification of MAO A involvement in cardiac cellular degeneration thus presents a potential drug target for the development of cardio-protective agents for an aging population. Initial attempts have been published (16); however, this area of MAO research is still in its infancy.
Molecular Structures of Human MAO B and MAO A
Expression and Purification
Both MAO A and MAO B are known to be membrane-associated enzymes that are located specifically to the outer mitochondrial membrane. The purification of these enzymes, in the past, represented a formidable challenge since most mammalian tissues contained both enzymes that are not readily separable from one another due to their molecular similarities. Prior to 1999, human MAO A was either purified from human placental tissue (required 6–7 placentas for one enzyme purification in a 35% yield) (17) or purified from a Saccharomyces cerevisiae expression system (18) which required the growth of 20 liters of culture to provide sufficient cells for isolation of a reasonable quantity (50–100 mg) of enzyme. During this same time period, the most convenient source of MAO B was bovine liver mitochondria (19).
The development of expression systems for human MAO B and MAO A provided a stable source of either enzyme and greatly facilitated both structural and mechanistic work. Procedures and conditions for the expression of human MAO B (20) and human MAO A (21) in the Pichia pastoris yeast system have been published. This system allows the purification of ~200 mg of pure enzyme from 2 liters of fermentation medium and also allows incorporation of site-directed mutations on either enzyme as another tool for structure and mechanistic probes. Expressed full length human MAO A or MAO B is situated in the outer mitochondrial membrane of the yeast cell and constitutes ~50% of the total protein content of mitochondrial outer membrane preparations. The availability of large quantities of homogeneous protein led to the determination of their structural properties of both enzymes by X-ray crystallography.
Structures of Human MAO A and MAO B
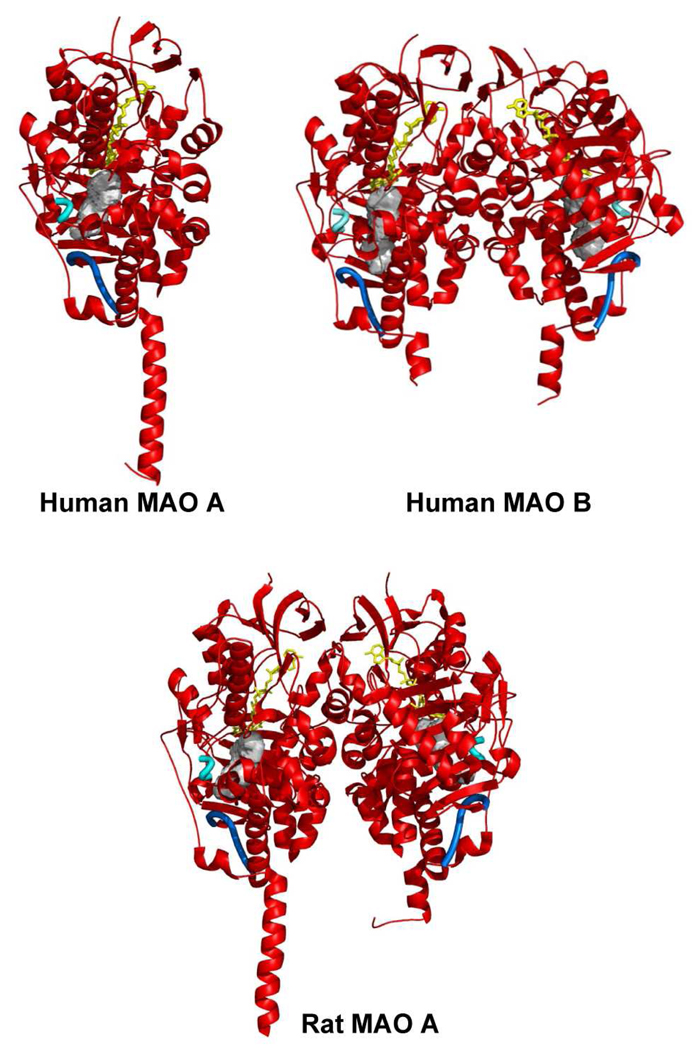
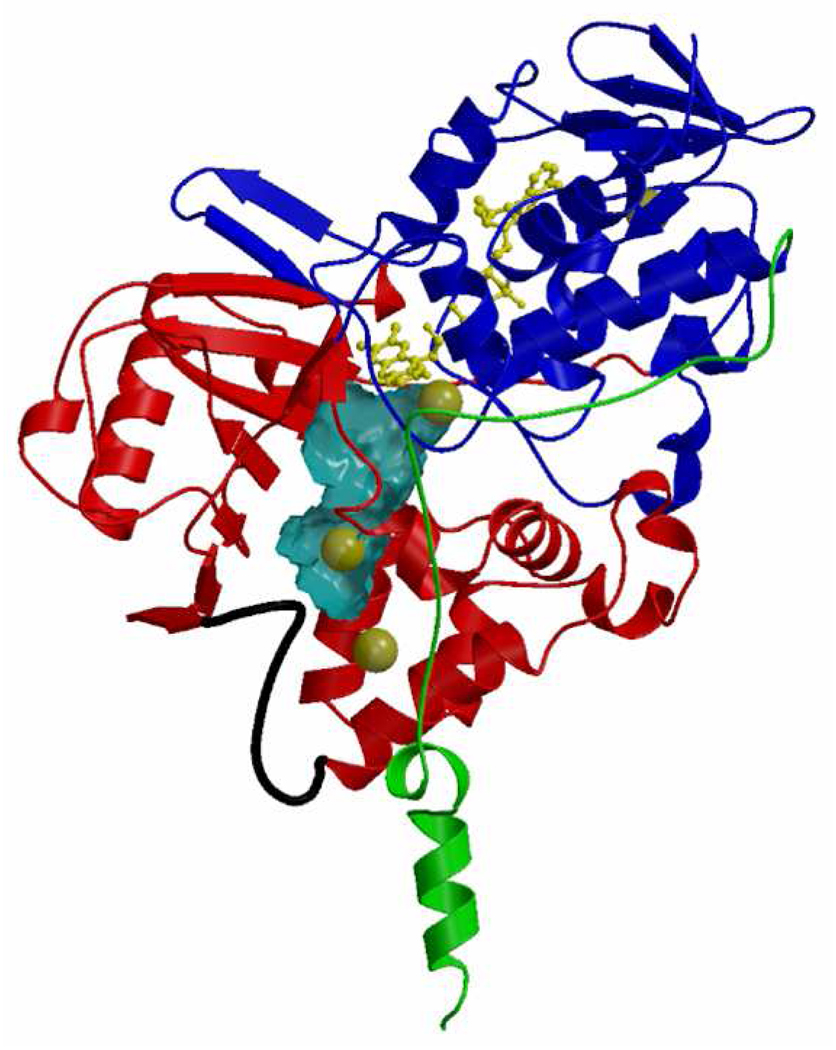
With the availability of reagent quantities of purified enzymes, the 3-dimensional structures of human MAO B (22), human MAO A (23, 24), and rat MAO A (26) have been determined and are shown as ribbon diagrams in Figure 1. The X-ray structure of human MAO B has been determined in complex with a range of inhibitors (see below) to a resolution of 1.65 Å for the best structure. Human and rat MAO A X-ray structures have been solved to 2.2 and 3.3 Å resolution, respectively. All of the enzymes have structures with similar folds and with a high degree of identities in their respective Cα coordinates. These structures show that the membrane binding motifs of the enzymes are located in the C-terminal 35–40 residues. In human MAO B this trans-membrane region folds into an α-helix protruding perpendicularly from the main globular body of the protein, although the last 20 residues of this motif are too disordered to provide definitive electron density (22). The C-terminal part of human MAO A has a similar topology to rat MAO A with the electron density sufficiently defined to provide a complete view of the trans-membrane α-helix (24). These structural data support previous biochemical data (29) that identified the C-terminal helix as the mode of MAO A or MAO B binding to the outer mitochondrial membrane. It is of interest that chimeric enzymes constructed using fragments of MAO A and MAO B show that “swaps” of the respective C-terminal helices between MAO B to MAO A result in inactive enzyme (30, 31) suggesting that there are differences in the specific membrane-binding architectures between the two isozymes.
Figure 1.
Ribbon diagram showing the three-dimensional structures of human MAO A (24), human MAO B (25) and rat MAO A (26). All structures are oriented with the C-terminal trans-membrane helices pointing downwards. The FAD cofactor is in yellow ball-and-stick representation. Active site cavity in each enzyme molecule is drawn as a gray surface. The cavity-shaping loop (residues 210–216 in human and rat MAO A and the corresponding residues 201–207 in human MAO B) is highlighted in cyan. The loop lining the entrance cavity space in human MAO B (residues 99–110) (27) is featured in blue. The corresponding residues in rat and human MAO A adopt the same conformation. The cavities were calculated with VOIDOO (28) and the structural representations produced with Pymol (www.pymol.org).
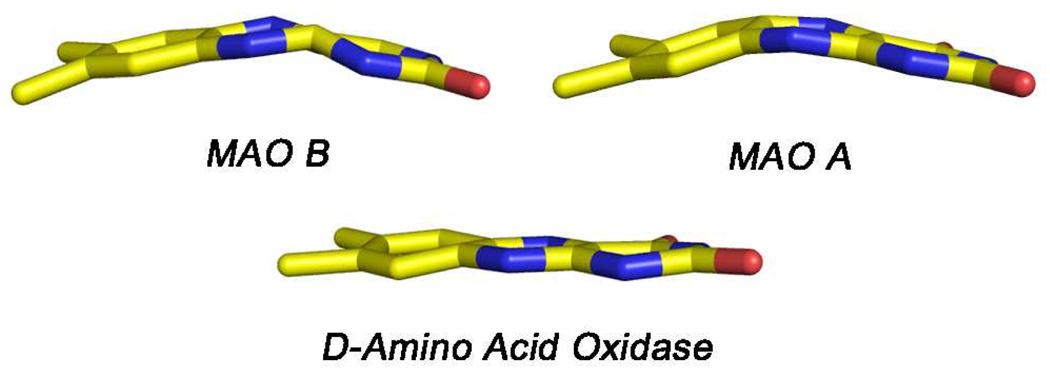
The protein structures around the active site covalent FAD coenzymes are quite similar among the three known MAOs. The position of the FAD cofactor with respect to the overall structure is highly conserved, and the substrate-binding sites consist of elongated cavities (Figure 1), whose features will be thoroughly discussed in the next section of this review. In all three enzymes, the flavin rings exist in “bent” rather than the more common planar configurations about the N(5)-N(10) axis (Figure 2) demonstrating strain at the coenzyme binding sites that may have catalytic relevance (see below). In all cases, the flavin side chains are in extended conformations, as is commonly observed in the structures of other flavoenzymes containing FAD as cofactor (32). Another example of strain about the FAD site is the finding in human MAO B (27) and in human MAO A (24) that the amide linkage between Cys397 (Cys406 in MAO A) (the site for the covalent flavin 8α thioether linkage) and Tyr398 (Tyr407 in MAO A) is in a cis conformation.
Figure 2.
Relative conformations of the isoalloxazine rings of the 8α-S-cysteinylFAD coenzymes in human MAO B and MAO A (top). For comparison, the ring conformation of the FAD cofactor of Rhodotorula gracilus D-amino acid oxidase is also shown (below). These structures are taken from coordinates deposited in the PDB with codes: 1OJA, 2Z5X, and 1C0K, respectively.
The largest difference in structures among the three known structures of MAO are the respective natures of their oligomeric states (Figure 1) where human MAO B and rat MAO A are dimers while human MAO A is a monomer. A survey of the literature shows membrane proteins are generally oligomeric in the membrane, with dimerization being most common (33). Theoretical modeling studies and species-dependent genetic analyses have led Andres et al. (34) to propose that the monomeric structure of human MAO A could be due to a human exclusive Glu151Lys mutation at a site located near the dimer interface. All other species of MAO A and MAO B have a conserved Glu at this position. The agreement of the experimental observation of a monomeric human MAO A structure with the prediction by Andres et al. (34) resulted in questions regarding the functional significance of this structural difference and whether the monomeric form of human MAO A existed in the membrane or whether it was an artifact of protein solubilization and purification from its membrane environment.
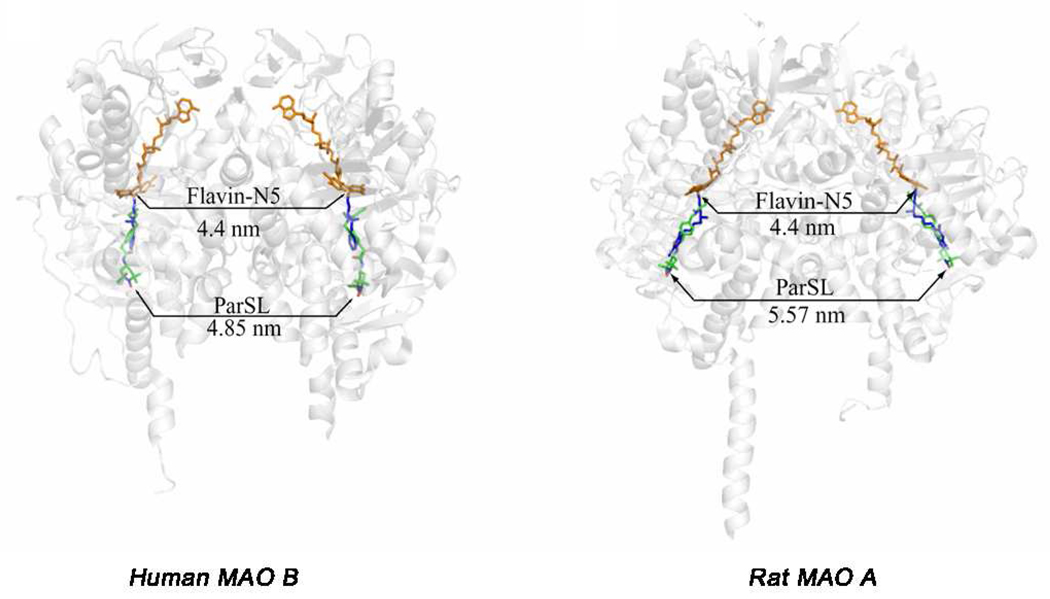
To address these questions, pulsed EPR (DEER) experiments were performed using a nitroxide spin labeled pargyline analogue that was covalently attached to the FAD coenzyme in both MAO A and MAO B. This technique can measure distances between two paramagnetic species separated by distances up to 60 Å and is applicable for detergent-solubilized as well as membrane preparations (35). This approach provides additional distance data to address the question of whether the enzyme structure determined by X-ray crystallography is the same as that in a membrane environment. Recently published pulsed EPR data show that all rat and human MAO A and MAO B enzymes are dimeric in their membrane-bound forms and that dimer formation is independent of whether Glu or Lys occurs at position 151 (or its analogous site) (36). In the detergent-solubilized, purified preparations, both human and rat MAO B are found to remain 100% dimeric, whereas human and rat MAO A exist only fractionally (50%) in their dimeric forms. Dissociation of oligomeric structures of membrane proteins in detergent micelles has precedent in the literature as reported for the bovine mitochondrial ADP/ATP carrier (37) and is found to also be the case for MAO A but not for MAO B. The human MAO A monomer species is more likely to crystallize than its dimeric form, while the dimer structure of rat MAO A is more readily crystallized. The similarities of DEER measured distances (Figure 3) with those measured from crystallographic data of human MAO B and rat MAO A provide definitive evidence for the biological significance of their respective crystal structures.
Figure 3.
Distances calculated from X-ray structures and determined from analysis of DEER dipolar couplings between the flavin cofactors (orange) and between the pargyline analogue spin labels (green). The positions of the pargyline spin labels in each active site are based on super-positions with the known structures of pargyline covalent adducts (blue).
Structures of Active Site Cavities in MAO B and in MAO A and Influence of Inhibitor Binding on Those Structures
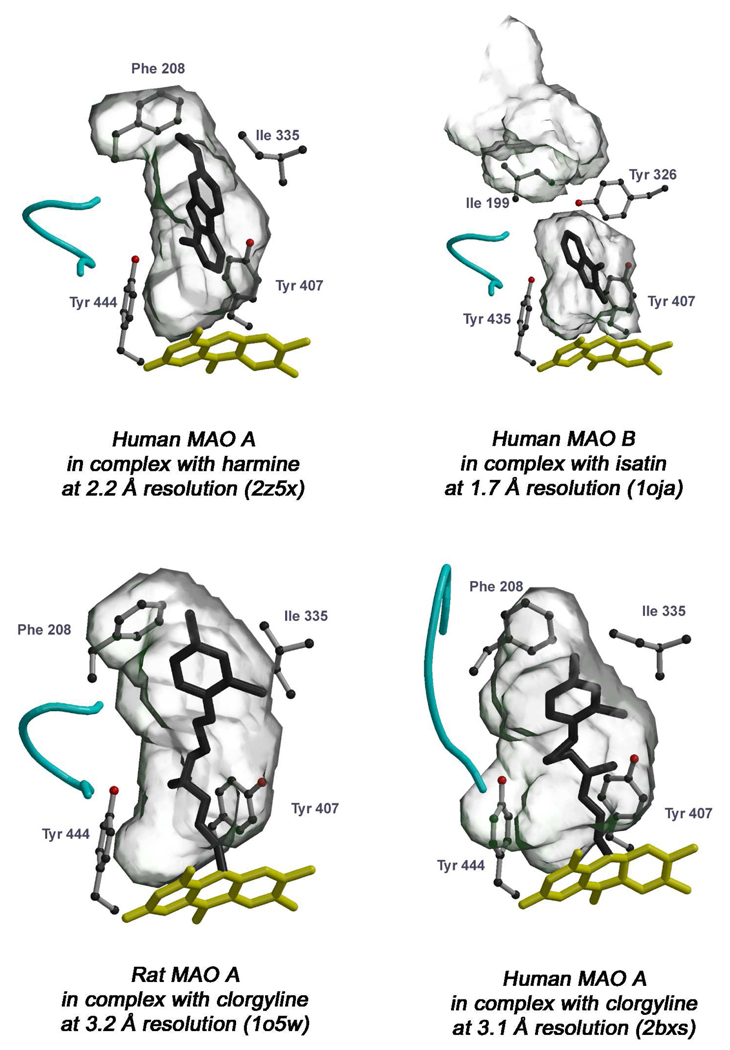
The structural elucidation of human and rat MAOs revealed that substrate binding and oxidation occurs in elongated cavities extending from the flavin site at the core of the enzyme to the surface of the protein on the opposite side of the FAD adenosine ring (Figure 1 and Figure 4). Although in all three enzymes the cavities are generally hydrophobic, details of the active site architectures demonstrate differences in their respective structural properties that account for their distinct substrate and inhibitor specificities. The recently reported 2.2 Å crystal structure of human MAO A in complex with harmine (24) allows a more detailed comparison of the active sites between the two human enzymes (Figure 4, top panel). The volume of the human MAO A cavity is ~400 Å3, whereas that of the combined two cavities of human MAO B is ~700 Å3. The MAO B cavity is bipartite and is comprised of two separate spaces, the substrate cavity (~400 Å3) and the entrance cavity (~300 Å3) with the latter facing the solvent after movement of loop 99–110 (in blue in Figure 1). Whether the active site of MAO B is a large single cavity or a bipartite cavity is an example of catalytic site plasticity determined by the conformation of Ile199. The side chain of this “gating” residue may, depending on the nature of the bound ligand, adopt two different conformations (“closed” and “open”) (Figure 5) (27). The corresponding residue in human and in rat MAO A is Phe208, but it does not function as a gating residue. Mutagenesis experiments were performed on human MAO B Ile199Phe since the well-studied bovine enzyme contains a Phe at 199 position rather than Ile (38). Structural and inhibitor binding experiments on this mutant form of human MAO B demonstrated that the bulky Phe side chain impedes such conformational flexibility, reduces the space of the entrance cavity and interferes with the binding of MAO B-specific inhibitors (38). These findings provide a structural rationale for differential binding affinities reported in the literature on comparison of bovine and human MAO B preparations (39) and presents a warning on pitfalls that can be encountered on the use of differing MAO sources for development of inhibitors of human MAO (40).
Figure 4.
Active site cavity and inhibitor binding in MAO A and MAO B. The protein molecule is rotated approximately 180° with respect to the orientation of Figure 1 (i.e. with the C-terminal α-helix pointing upwards). In the top panel, among the known structures of human MAO B in complex with various inhibitors (Figure 5), that with isatin (27) has been chosen to be compared with binding of harmine to human MAO A because they are both non-covalent inhibitors of similar size and binding affinity (MAO B-isatin Ki = 3 µM, MAO A-harmine Ki = 0.6 µM). In the bottom panel the structures of human and rat MAO A in complex with clorgyline are compared. The flavin and inhibitor molecules are in yellow and black ball-and-stick representation, respectively. The Tyr side chains beside the flavin (Tyr407 and Tyr444 in MAO A, Tyr398 and Tyr435 in MAO B) form the conserved “aromatic sandwich” (41). Among the other active site residues, Phe208 (Ile199 in MAO B) and Ile335 (Tyr326 in MAO B) are drawn to highlight their role in determining substrate/inhibitor specificity between MAO A and MAO B (see text). The PDB codes for each structure are in parentheses. The cavity-shaping loop (residue 210–216 in MAO A) is colored in cyan as in Figure 1. The cavities were calculated with VOIDOO (28) and the pictures were produced with Bobscript (42) and Raster3D (43).
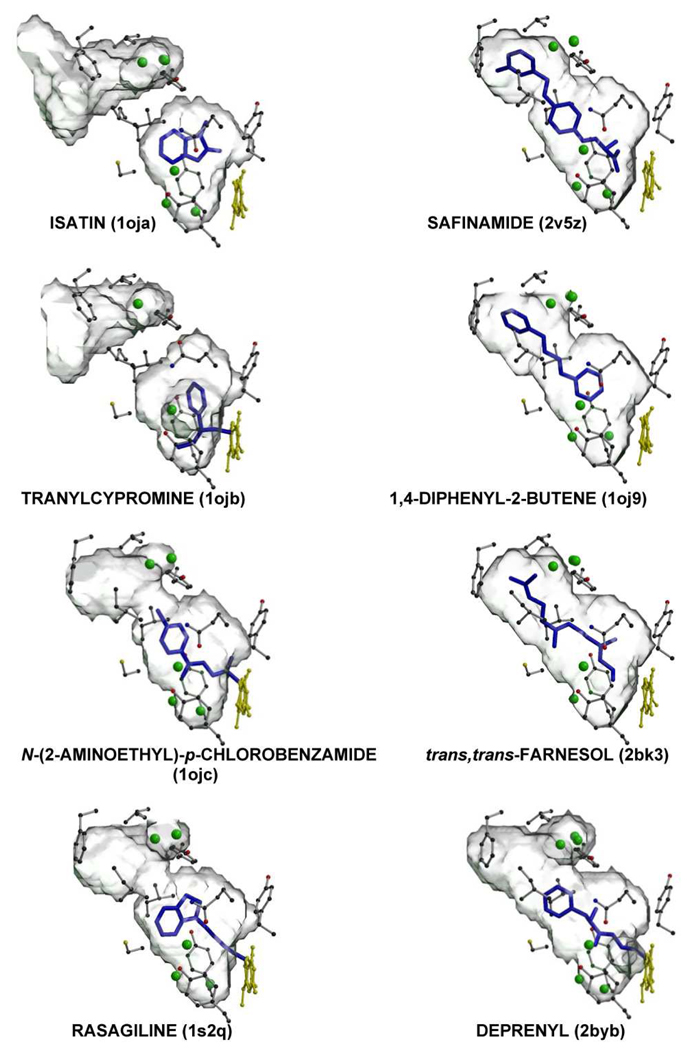
Figure 5.
Comparison of active site cavity structures of human MAO B in different enzyme-inhibitor complexes. The PDB codes for each structure are in parentheses. The protein molecule is rotated approximately 90° around an axis perpendicular to the plane of picture with respect to Figure 4. The residues lining the cavity are in gray ball-and-stick with carbon, oxygen, nitrogen and sulfur atoms in black, red, blue and yellow, respectively. Water molecules included in the cavity are represented as green spheres. The flavin is in yellow and the inhibitor molecule is in blue.
Another difference between MAO A and MAO B structures is also involved with the active site cavity structures. The Tyr326 side chain in MAO B, although not directly involved in the partition of the two cavities, does produce a restriction that is less pronounced in human MAO A where Ile335 occupies that position. Conserved active site residues in both enzymes include the Tyr pair of the “aromatic sandwich” (Figure 4) at the re face of the flavin coenzyme isoalloxazine ring and a Lys residue hydrogen bonded via a water molecule to the N(5) position of the flavin (Lys305 in MAO A and Lys296 in MAO B). Other non-conserved residues in the active sites include Asn181 and Ile180 in MAO A (Cys172 and Leu171 in MAO B, respectively), which, however, do not significantly affect the shapes of the cavities. Therefore, the MAO A Phe208-Ile335 and the MAO B Ile199-Tyr326 pairs appear to be major determinants in dictating the differential substrate and inhibitor specificities of the two enzymes.
An intriguing issue in MAO enzymology is to understand where and how the substrates (or inhibitors) are admitted to the active sites. In human MAO B the cavity is extended and substrate binding is likely to occur in proximity to the outer mitochondrial membrane surface region with the entrance loop (residues 99–110) involved in the access (colored in blue in Figure 1). In both human and rat MAO A the active site cavity is more compact. However, Son et al. (24) have recently demonstrated by site-directed mutagenesis that the conserved Gly110 is important for ensuring flexibility of the entrance loop required to provide access for the substrate. Therefore, the structural data suggest that MAO A and MAO B ligands follow similar pathways in binding. The cavity-shaping loop (residues 210–216 in MAO A, cyan in Figure 4 and Figure 5) adopts a helical conformation that is conserved in all structures except for human MAO A in complex with clorgyline (Figure 4). This is unlikely to be due to the inhibitor because rat MAO A complexed with clorgyline retains the folded conformation of loop 210–216, although the inhibitor adopts slightly different conformation. However, this observation suggests that this loop may be more susceptible to structural flexibility in MAO A than in MAO B.
The high-resolution crystal structures of human MAO B in complex with different inhibitors of clinical interest allow a more in-depth analysis of the active site cavity, which is important because of the pharmacological relevance of these enzymes. The elongated cavity of human MAO B is mostly hydrophobic, with a small hydrophilic area in front of the re face of the flavin cofactor (27) that is occupied by highly conserved water molecules (Figure 5). Human MAO B can bind compounds of different size, and a number of them react with the flavin cofactor to form a covalent adduct (Figure 5), an example of the classical definition of mechanism-based inhibitor. In general, all inhibitors contain an aromatic moiety (as expected considering the aromatic nature of the neurotransmitter substrates), but the cavity showed a significant affinity also for aliphatic ligands such as trans,trans-farnesol. Recent data show inhibition of MAO B by agents found in common laboratory plasticware; these include the slip agent, oleamide, and the biocide, di(2-hydroxyethyl)methyldodecylammonium ion (44). This inhibitory versatility is related to the plasticity of the human MAO B active site cavity. As described above, depending on the conformation of the gating residue Ile199, the cavity can host either small inhibitors, such as isatin and tranylcypromine, or cavity-filling ligands such as safinamide, trans,trans-farnesol and 1,4-diphenyl-2-butene (a component of polystyrene plasticware) (45). In the former case the active site is restricted to a small cavity separated from an entrance cavity space that opens to the exterior like a funnel, whereas in the latter the cavity shapes has a compact ellipsoidal entity (Figure 5). Binding of inhibitors such as rasagiline (46), N-(2-aminoethyl)-p-chlorobenzamide (27), and deprenyl (23) induce a mid-span type of cavity because they are large enough to push the gating residue Ile199 into the open conformation. With these inhibitors, the shape of the resulting cavity has a more bipartite character than that induced by the cavity-filling inhibitors. It is important to note that in all these MAO B-inhibitor complexes, the positions of the active site residues, apart from the gating residue Ile199, are highly conserved, and this cavity plasticity is probably determined by subtle conformational changes. From a pharmacological viewpoint, this inhibitory versatility has important implications. Small compounds such as isatin and tranylcypromine, which can be hosted in a limited pocket, show similar binding affinities for both MAO A and MAO B, whereas cavity-filling ligands are highly specific MAO B inhibitors. Less self-explanatory are the inhibitory properties of rasagiline, which is highly specific for MAO B although it occupies only one half of the entire cavity (Figure 5). Even more surprising is the evidence that a rasagiline analog methylated on the amino moiety of the propargyline chain connecting the inhibitor to the flavin loses this characteristic specificity for MAO B (47). Moreover, the covalent nature of some MAO inhibitors raises additional questions. What is the mechanism for covalent adduct formation of the propargyl moiety of compounds such as rasagiline or deprenyl with flavin? Unpublished work in this laboratory shows the reaction occurs without consumption of O2, suggesting no catalytic turnover in the inhibition reaction.
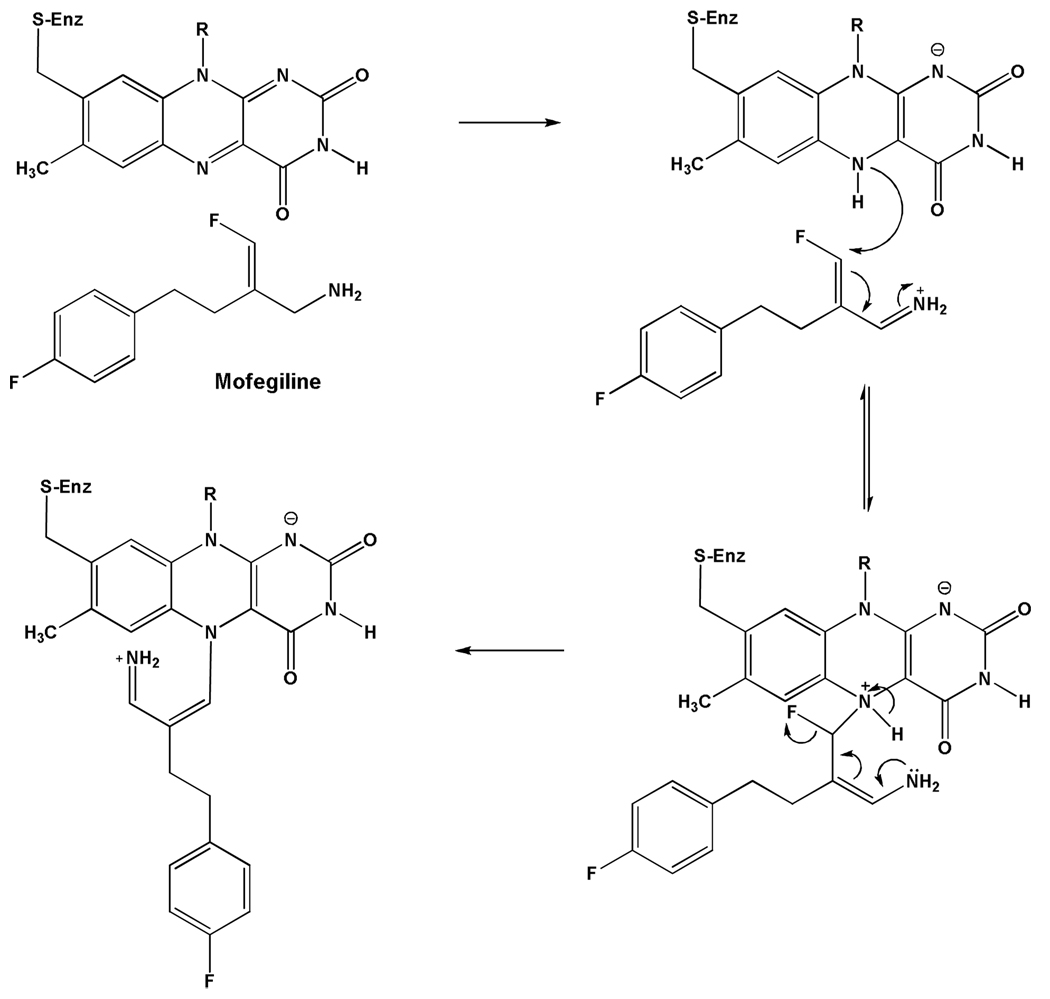
Recent studies on the differential inhibition of MAO B and MAO A with the vinyl fluoro amine Mofegiline (48) show covalent adduct formation with the flavin in MAO B but only non-covalent competitive inhibition with MAO A. This inhibitor is one of the most effective MAO B active site inhibitors the authors have encountered. The proposed mechanism of inhibition is shown in Scheme 3 where the adduct formed is at flavin N(5) with the vinyl side chain of the inhibitor with accompanying loss of F−. This adduct exhibits unique spectral properties in that the absorption bands of the adduct resemble those of the oxidized flavin (48) rather than the “bleached” form generally found with flavins alkylated at the N(5) position (49). The proposed reason this compound does not react irreversibly with MAO A is that this enzyme is unable to oxidize arylalkylamine analogues with an extended side chain (50). α-C-H Bond cleavage of the amine side chain of Mofegiline is required for formation of reduced flavin, which then reacts nucleophilically with the vinyl fluoride to form the covalent adduct.
Scheme 3.
Proposed Mechanism for Mofegiline Inhibition of Human MAO B
To date, essentially all irreversible MAO inhibitors form N(5) flavin adducts. The only exception is the flavin C(4a) adduct formed on inhibition and ring opening of tranylcypromine with MAO B (27). Previous model system studies of phenylethylhydrazine inhibition of MAO A or MAO B suggested the alkylflavin to be a C(4a) adduct; however, recent structural and mechanistic studies show MAO inhibition to involve covalent addition to the flavin N(5) position (49). Structural data on tranylcypromine inhibition of other flavin-dependent amine oxidases such as Lysine-Specific histone Demethylase 1 (51) show the ring-opened cyclopropyl moiety to be linked to the flavin at both the N(5) and C(4a) positions. Therefore, the possibility remains that the adduct observed in tranylcypromine-inhibited MAO B may have originated from the initial formation of an N(5) adduct with subsequent migration occurring during the time period required for crystallization of the enzyme. Previous model system studies with alkyl adduct of flavins that were produced via photoreaction with phenylacetates show thermal isomerization between N(5) and C(4a) to occur (52), so the possible isomerization occurring in the amine oxidases remains a concern in addressing mechanistic studies.
Differences in Active Site Cavity Accessibilities between Human MAO B and MAO A and Comparison with Rat Enzymes
In spite of their high sequence identities (>90%) and the general assumption that rat MAO A and MAO B are reasonable substitutes for the human enzymes in drug development studies, there have been a number of reports in the literature that the rat enzymes exhibit different affinities (40) relative to the human enzymes. Confirmation of these differences has been shown by experiments designed to monitor similarities and differences between rat and human MAO A and MAO B. Using a spin labeled pargyline analogue (53), the accessibility of the bound nitroxide in human MAO A is found to be considerably higher (~10-fold) than that in human MAO B. Rat MAO A and MAO B exhibit similar accessibilities to a neutral paramagnetic probe at a level intermediate between the two human enzymes (53). These data support the observation of differing affinities for selected inhibitors published in the literature and also demonstrate structural and/or dynamic differences between rat and human enzymes either in their soluble, purified forms or in their membrane bound forms. Therefore, a measure of caution is urged by investigators in the field in attempting to extrapolate detailed molecular data to the human from data collected in rats.
Mechanistic Studies on MAO Catalysis
Analysis of the Different Proposed Mechanisms of C-H Bond Cleavage
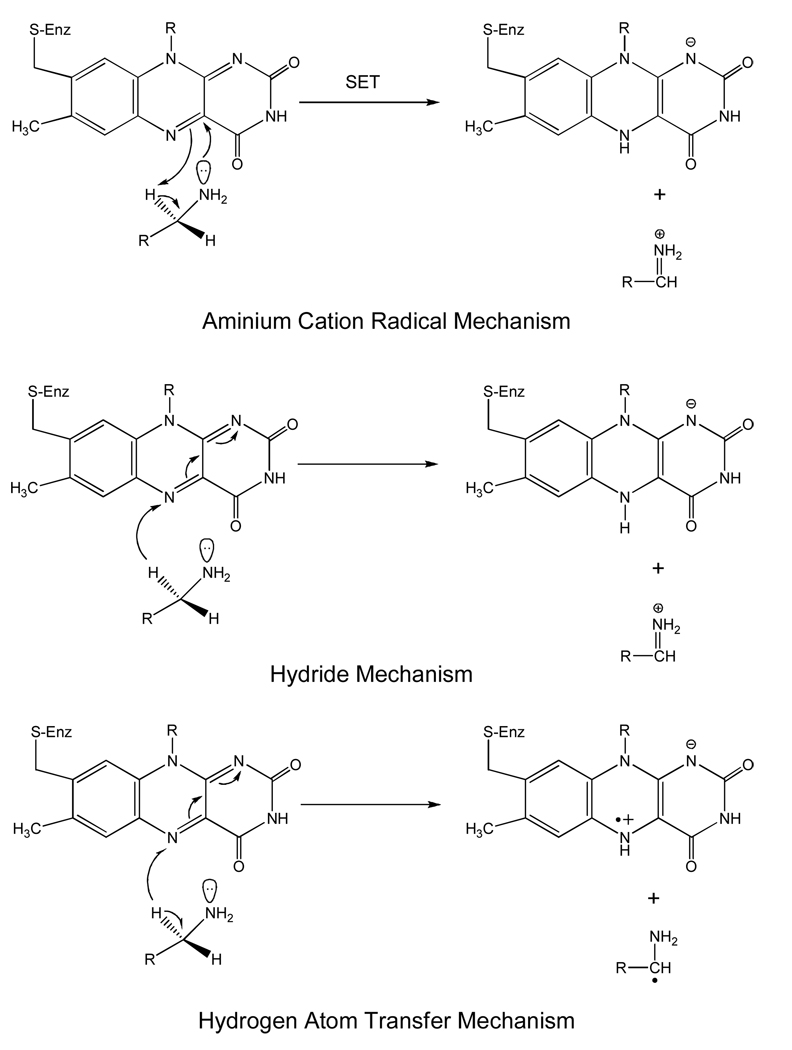
C-H bond cleavage reactions occur by three possible mechanisms: the heterolytic hydride transfer, the heterolytic H+ abstraction (includes the proposed ammonium cation radical as well as the polar nucleophilic mechanism), and a homolytic H· abstraction (see Scheme 4). Of these possibilities, the two heterolytic reactions that have been proposed have resulted in a good deal of controversy as to whether MAO catalysis follows a hydride mechanism as suggested for the amino acid oxidase and the alcohol oxidase classes of flavoenzymes. Hydrogen atom abstractions are known to occur involving the flavin triplet state, however, no convincing evidence is known for ground state flavins in either model systems or flavoenzymes. The stereochemistry of the hydrogen transfer step is well documented to be the pro-R H of the substrate for both MAO A and MAO B (54, 55). This property differs from the Cu-quinoprotein amine oxidases where both R and S stereochemistry of hydrogen transfer is found (56). As is the case with the Cu-quinoprotein amine oxidases, MAO oxidation of benzylamines (57, 58) and phenethylamines (50) exhibit large deuterium kinetic isotope effects showing, in most instances, that C-H bond cleavage is rate limiting in catalysis. Previous studies on bovine MAO B have demonstrated H tunneling to contribute in the H transfer step (59). The evidence supporting a hydride transfer from substrate to flavin in other flavoprotein oxidases is suggested from 15N kinetic isotope effect experiments (60, 61). Other kinetic and structural data on the amino acid oxidases also support this mechanism (62). A strong argument against the hydride mechanism for MAO catalysis is the demonstration that either MAO A or MAO B readily oxidizes arylalkylhydrazines to form diazenes (49). It is difficult to envision the oxidation of a hydrazine to a diazene by a hydride ion abstraction from a nitrogen atom. In fact, the amine oxidases appear to be unique among flavoenzymes in their ability to catalyze hydrazine oxidation reactions. The polar nucleophilic mechanism discussed below would be compatible with the oxidation of a hydrazine moiety.
Scheme 4.
General Mechanisms for C-H Bond Cleavage Reactions in Flavoenzymes.
Similarities in high field EPR and ENDOR spectral properties of the anionic FAD semiquinones of both human MAO A and yeast D-amino acid oxidase (63) indicate similar distributions of spin densities among their flavoenzyme semiquinones. The significance of these studies is to definitively rule out any flavin radical-tyrosyl radical equilibria in MAO A as suggested in a previous study (64) and thought to have implications in radical mechanism. Recent work on a blue light sensor flavoprotein demonstrate that a tyrosyl radical adjacent to a neutral flavin radical (65) exhibits considerable spin coupling that is readily observed in X-band EPR spectra. Such spin coupling is not observed in MAO A and this constitutes additional evidence that no tyrosyl radical is apparent in MAO A or in MAO B. EPR and ENDOR studies of model flavin semiquinones and flavoenzyme semiquinones have long been known from the pioneering work of Ehrenberg and colleagues (66), and the spin distribution of the radical about the isoalloxazine ring in a number of flavoenzymes with different functions is found to be quite similar. Therefore, the spectral properties of flavoenzyme semiquinones do not provide definitive evidence to dictate whether they follow similar catalytic mechanisms.
Of the H+ abstraction mechanistic pathways proposed, perhaps the aminium cation radical mechanism or SET mechanism proposed by Silverman and co-workers (67) has received the most attention. This mechanism is based on the premise that the pKa of α-C-H proteons of an amine require a very strong base for abstraction that would not likely be fulfilled by a basic amino acid side chain. In order to lower the pKa of these protons for H+ abstraction by a basic amino acid residue, the amine is first oxidized to the amine cation radical form, which results in acidification of the α-C-H group, and thus allows H+ abstraction. To date, experiments designed to test this mechanism have failed to provide supporting evidence that this mechanism is operational in MAO A or in MAO B. No flavin radical intermediates are detected spectrally in stopped-flow experiments (50, 57, 58), no influence of magnetic field on the rate of enzyme reduction is observed (68), and the oxidation-reduction potential of the FAD cofactor is too low (~+ 40 mV) (69) to be an effective oxidant of the deprotonated amine (Em = + 1.5 V) (70). In addition to these mechanistic probes, the structures of MAO A and of MAO B show no obvious basic amino acid residues that could serve as an active site bases in catalysis. The main supporting evidence for the hemolytic one-electron mechanism is the observation that MAO A or MAO B is inactivated by cyclopropylamine analogues with subsequent ring opening, a reaction characteristic of radical reactions. Recent studies of the MAO B-catalyzed oxidation of “Rimoldi’s Amine”, which consists of a cyclopropyl group fused to an N-methyl-pyridinium ring show the cyclic imine formation to occur without opening of the cyclopropyl ring (Scheme 5) (71) would be expected if the reaction were to involve radical intermediates. In the aggregate, these studies do not support the concept that the mechanism of amine oxidation in MAO is initiated by a hemolytic single electron transfer step.
Scheme 5.
Reaction of Rimoldi’s Amine with MAO B.
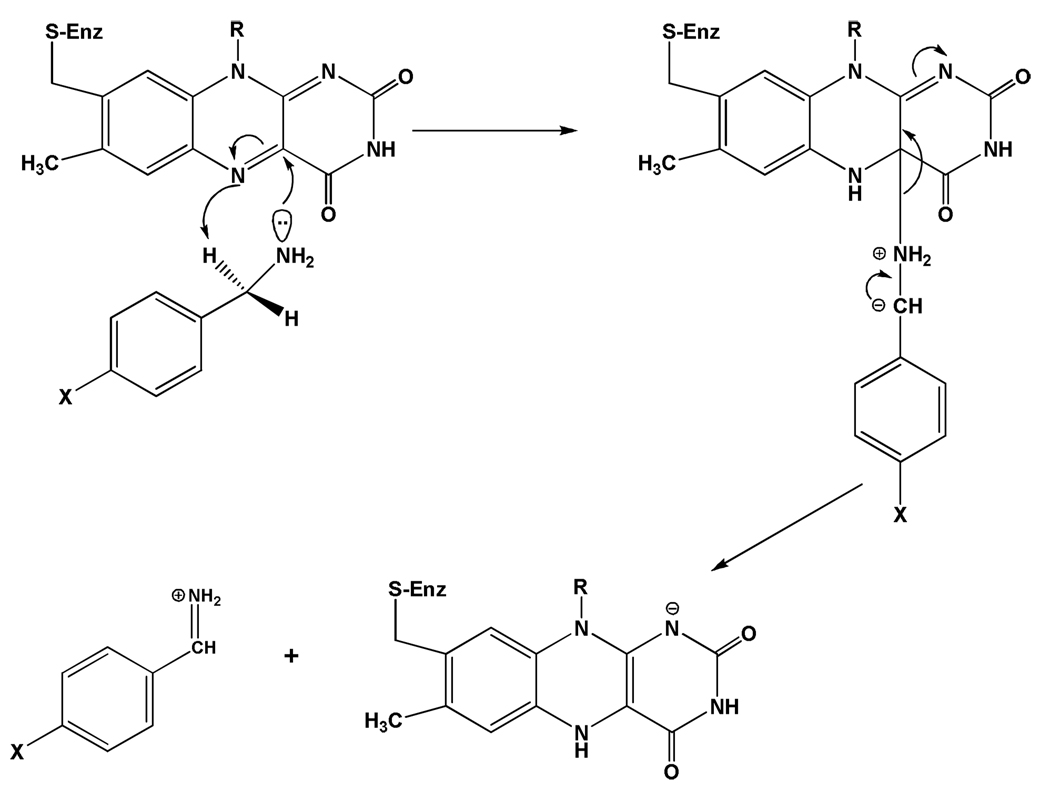
Current mechanistic data do support the polar nucleophilic mechanism proposed by Miller and Edmondson (58) for human MAO A catalysis, which is based on the observation of a ρ value of ~ +2 from analysis of Hammett Plots of steady-state and rapid-reaction kinetic data on the oxidation of a series of para-substituted benzylamine analogues. This behavior is also exhibited by rat MAO A (72) as well as mutants of both human and rat MAO A and appears to be the most definitive evidence for the abstraction of the pro-R α-C-H as a H+ as the mode of C-H bond cleavage. Previous data on the Cu-quinoprotein plasma amine oxidase with a series of para-substituted benzylamine analogues show a ρ value of ~ +1.4 for C-H bond cleavage (73). As most investigators accept a proton abstraction mechanism from the Schiff’s base intermediate in the quinoprotein-catalyzed amine oxidation, one can suggest a similar type of H+ abstraction mechanism for the flavin-dependent amine oxidation. In order to accomplish this difficult proton abstraction (pKa for a benzyl hydrogen is ~25 (74), no basic amino acid residues are in the catalytic site and therefore, the flavin must be activated to become a strong base. This could be accomplished by C(4a) addition of the amine followed by or concerted with abstraction of the α-C-H by the resulting basic N(5), which is estimated to exhibit a pKa ~25 from NMR studies of reduced flavin models (75). The reaction mechanism for this proposed reaction is shown in Scheme 6.
Scheme 6.
Proposed Polar Nucleophilic Mechanism for the Reductive Half Reaction of MAO Catalysis.
Since deprotonated amines do not appear to exhibit the nucleophilicity to readily add to the flavin C(4a) position in model systems and no direct evidence for a stable amine-flavin adduct has been found, one must examine factors in the enzyme that would facilitate this type of reaction. Structural data show the isoalloxazine ring of the flavin to exist in a conformation ~30° bent from planarity in the oxidized form of the enzyme (Figure 2). This bending has been shown by NMR studies of other flavoenzymes to result in the C(4a) position to be more electrophilic and the N(5) nitrogen to be more nucleophilic (76). In addition, the presence of two tyrosyl rings approximately perpendicular to the re face of the flavin demonstrates the amine moiety of the substrate must pass between them to approach the flavin. Results from our laboratory show that mutagenesis of Tyr435 in MAO B and the corresponding Tyr444 in MAO A results in a functional enzyme of lowered activity that has been correlated with dipole-dipole interactions of the phenolic ring and the deprotonated amine (77). This suggests that the combined dipole moments of the two tyrosyl residues would result in the distribution of the lone pair of electrons on the amine nitrogen to become more elongated, which would increase the effective nucleophilicity of the amine nitrogen. Both of the above factors discussed would fit the proposed mechanism in Scheme 6.
In contrast to MAO A, analysis of the MAO B-catalyzed oxidation of a series of para-substituted benzylamine analogues show the reaction rate to depend mainly on steric parameters with no detectable influence of substituent electronic effects (57). This behavior is suggested, by structural data, to reflect the steric constraints of the benzyl ring of the substrate in MAO B, which prevents the full expression of electronic parameters on the benzyl C-H group (58). In contrast, the MAO A structure shows a more open and less restricted active site, which allows the substrate more steric freedom and therefore overlap of the π-orbitals for the full expression of para-substituent electronic effects. In principle, alterations to the MAO B active site could decrease this proposed steric constraint thereby allowing the expression of electronic effects on C-H bond cleavage. To date, such an approach has not been done.
The detailed mechanism of flavin-dependent amine oxidation remains a subject for additional studies. Not only is it important for the understanding of MAO A and MAO B catalysis, but is biologically important for our understanding of other amine oxidases such as LSD-1, the flavoenzyme involved in the epigenetic regulation of gene experession through histone demethylation (51).
Reaction of MAO with O2: the Oxidative Half Reaction
The second half reaction of MAO A or MAO B catalysis is the oxidative half reaction where O2 is reduced to H2O2 on reaction with the flavin hydroquinone. As discussed in the introductory section of this review, the formation of H2O2 by MAO constitutes a potential source of toxic reactive oxygen species leading to apoptopic cellular responses. A key difference in the two forms is that the Km for O2 with MAO B is ~240 µM (57) while the corresponding value for MAO A is ~12 µM (78). Thus, under normal cellular conditions, MAO A is operating at saturating concentrations of O2 while MAO B is probably less than half saturated. The molecular basis for this difference is still unknown. Unpublished structural data show Xe (a proposed probe of O2 binding sites in enzymes) to bind to MAO B both in the substrate cavity and in the entrance cavity (Figure 7). These data suggest the path for O2 to approach the reduced flavin is the entrance and substrate cavities in MAO B.
Stopped-flow experiments with bovine MAO B show that the reduced enzyme reacts with O2 in a second-order manner with a rate constant of 3.6×105 M−1 min−1 that is 10-fold slower than that where the imine product is still bound (2×106 M−1 min−1) (79). This oxygen rate acceleration in MAO by product is presumably due to the stabilizing effect of the protonated imine on the formation of superoxide anion (see below) in the first step of the oxygen reaction, analogous to the role of an active site histidine residue in glucose oxidase (80) (Scheme 7). Ramsay (78) has found that the MAO A rate of reaction with O2 is accelerated by the presence of substrate to achieve a rate similar to that observed with MAO B. The reaction of reduced flavin coenzymes with O2 is still an active area of investigation, especially among the flavoenzyme oxidase class where the reaction product is H2O2. From a structural view, both MAO A and MAO B contain lysine residues hydrogen bonded via an intervening H2O molecule to the N(5) position of their respective flavin coenzymes. This feature is also observed in the structures of a number of flavoenzyme oxidases. The mechanistic significance of this linkage has been suggested to be involved in the O2 reaction (81).
Scheme 7.
Proposed Mechanisms for O2 Reaction in the Oxidative Half Reaction of MAO Catalysis.
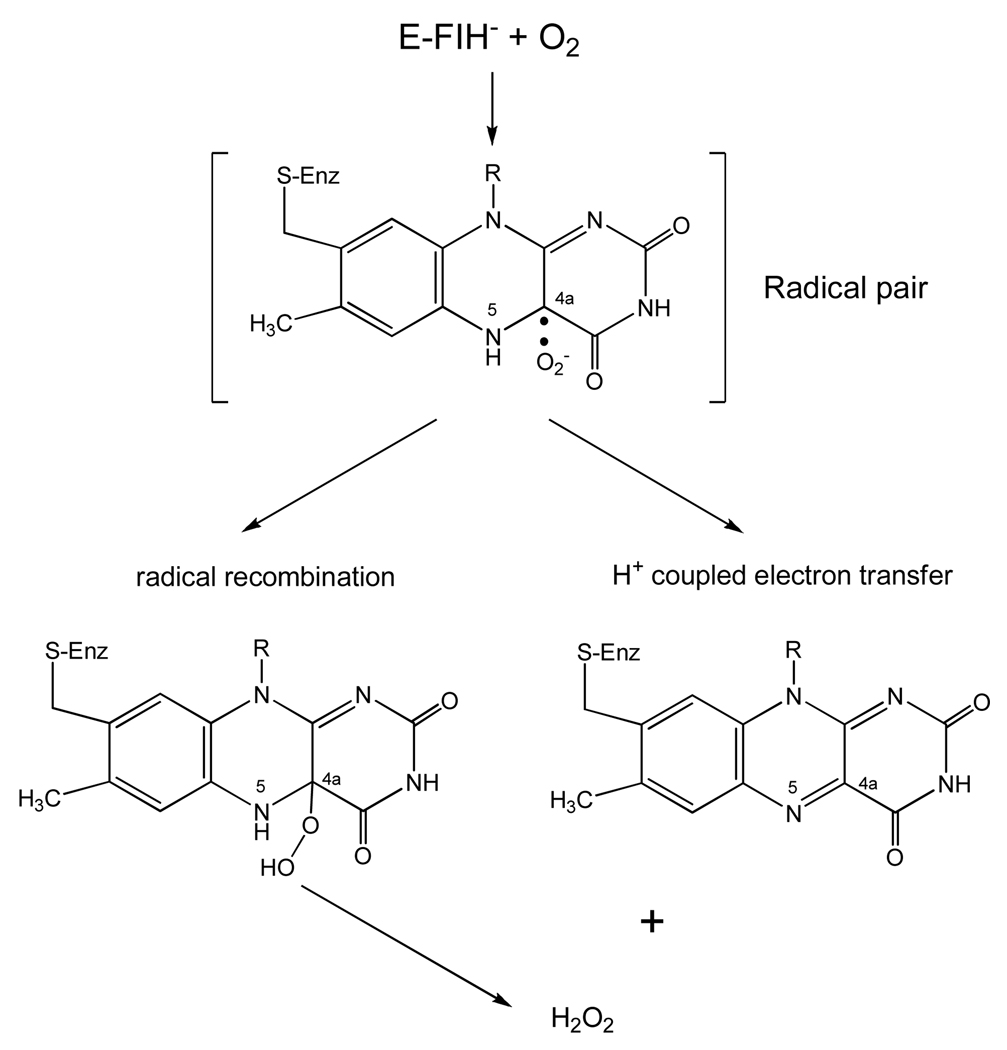
It is now generally accepted that the reaction of triplet O2 with singlet ground state reduced flavin must involve a rate limiting single electron transfer to form flavin semiquinone (the neutral form) and superoxide anion (82). This mechanism circumvents classical symmetry rules in such chemical reactions and has been investigated in detail with glucose oxidase (80). The reaction can then proceed via two different mechanisms (Scheme 7). One is the recombination of the radical pair to form the C(4a) flavin peroxide which would subsequently release H2O2 and the oxidized coenzyme. This intermediate is formed in the flavoenzyme hydroxylase family (83) and has recently been observed in flavoenzyme oxidases (84, 85). The other possibility is the reaction of superoxide with the flavin neutral radical to form H2O2 and oxidized flavin in a proton-coupled electron transfer step without an intermediate covalent linkage. Theoretical calculations for such a mechanism have been published (81) and recent experimental work on monomeric sarcosine oxidase supports these calculations. For example, mutagenesis of the H2O-bonded lysine residue to the flavin results in a dramatic reduction of the rate for O2 reoxidation of the reduced flavin (86). These results suggest that the reactions of MAO with O2 will be vigorously investigated and will be important for fully explaining how these membrane-bound enzymes function in their oxidative half-reactions.
Figure 6.
Location of three bound Xe atoms in the entrance cavity and substrate cavity of human MAO B (only one monomeric unit is shown). Xenon is denoted as dark yellow spheres. FAD is in yellow. The substrate binding domain is in red, FAD binding domain in blue, and the membrane binding domain in green. The substrate and entrance cavities of MAO B are in cyan. Unpublished data from this laboratory.
Acknowledgements
The authors wish to thank former colleagues including Drs. P. Newton-Vinson, J.R. Miller, R. Nandigama, L. De Colibus and M. Li for their contributions to the project over the years. Ms. Milagros Aldeco has provided essential technical support for a major number of studies cited in this review. Finally, the authors wish to acknowledge the contributions of the numerous collaborators who are cited in the references.
Footnotes
Support for much of the work cited in this review is from the National Institutes of Health (GM-29433) and is gratefully acknowledged. A.M. acknowledges grant support from Associazione Italiana per la Ricerca sul Cancro and the Cariplo foundation.
Abbreviations: MAO A, monoamine oxidase A; MAO B, monoamine oxidase B; DEER, double electron-electron resonance; ENDOR, electron-nuclear double resonance; EPR, electron paramagnetic resonance; NMR, nuclear magnetic resonance; LSD-1, lysine-specific histone demethylase-1; SET, single electron transfer.
References
- 1.Zeller EA, Bardsky J. In vivo inhibition of liver and brain monoamine oxidase by 1-isonicotinyl-2-isopropyl hydrazine. Proc. Soc. Exp. Biol. Med. 1952;81:459–461. doi: 10.3181/00379727-81-19910. [DOI] [PubMed] [Google Scholar]
- 2.Bach AWJ, Lan NC, Johnson DL, Abell CW, Bembenek ME, Kwan SW, Seeburg PH, Shih JC. cDNA Cloning of human liver monoamine oxidases A and B: Molecular basis of differences in enzymatic properties. Proc. Natl. Acad. Sci. USA. 1988;85:4934–4938. doi: 10.1073/pnas.85.13.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setini A, Pierucci F, Senatori O, Nicotra A. Molecular characterization of monoamine oxidase in zebrafish (Danio rerio) Comp. Biochem & Physiol. B. 2005;140:153–161. doi: 10.1016/j.cbpc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Edmondson DE, Bhattacharyya AK, Walker MC. Spectral and kinetic studies of imine product formation in the oxidation of p-(N,N-dimethylamino)-benzylamine analogues by monoamine oxidase B. Biochemistry. 1993;32:5196–5202. doi: 10.1021/bi00070a031. [DOI] [PubMed] [Google Scholar]
- 5.Fowler JS, Logan J, Volkow ND, Wang GJ, MacGregor RR, Ding YS. Monoamine oxidase: radiotracer development and human studies. Methods. 2002;27:263–277. doi: 10.1016/s1046-2023(02)00083-x. [DOI] [PubMed] [Google Scholar]
- 6.Mallajosyula JK, Kaur D, Chinta SJ, Rajagopalan S, Rane A, Nicholls DG, Di Monte DA, Macarthur H, Andersen JK. MAO B elevation in mouse brain astrocytes results in Parkinson’s pathology PloS One. 2008;3:e1616. doi: 10.1371/journal.pone.0001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke WJ, Kumar VB, Pandey N, Panneton WM, Gan Q, Franko MW, O’Dell M, Li SW, Pan Y, Chung HD, Galvin JE. Aggregation of α-synuclein by DOPAL, the monoamine oxidase metabolite of dopamine. Acta Neuropathol. 2008;115:193–203. doi: 10.1007/s00401-007-0303-9. [DOI] [PubMed] [Google Scholar]
- 8.Jenner P, Marsden CD. MPTP-induced Parkinsonism as an experimental model of Parkinson’s Disease. In: Jankovic J, Tolosa E, editors. Parkinson’s Disease and Movement Disorders. Baltimore-Munich: Urban & Schwarzenberg; 1988. pp. 37–48. [Google Scholar]
- 9.Chen JJ, Swope DM. Pharmacotherapy for Parkinson's disease. Pharmacotherapy. 2007;27:161S–173S. doi: 10.1592/phco.27.12part2.161S. [DOI] [PubMed] [Google Scholar]
- 10.Cases O, Seif I, Grimsby J, Gasper P, Chen K, Muller U, Aguet M, Babinet C, Shih JC, De Maeyer E. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAO A. Science. 1995;268:1763–1766. doi: 10.1126/science.7792602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunner HG, Nelen MR, van Zandvoort NG, Abeling NGGM, van Gennip AH, Wolters EC, Kuiper MA, Rogers HH, van Oost BA. X-Linked borderline mental retardation with prominent behavioral disturbance: phenotype, genetic localization, and evidence for disturbed monoamine metabolism. Am. J. Hum. Genet. 1993;52:1032–1039. [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott R, Tingley D, Cowden J, Frazetto G, Johnson DDP. Monoamine oxidase A gene (MAO A) predicts behavioral aggression following provocation. Proc. Natl. Acad. Sci. USA. 2009;106:2118–2123. doi: 10.1073/pnas.0808376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lairez O, Calise D, Bianchi P, Ordener C, Spreux-Varoquaux O, Guilbeau-Frugier C, Escourrou G, Seif I, Roncalli J, Pizzinat N, Galinier M, Parini A, Mialet-Perez J. Genetic deletion of MAO-A promotes serotonin-dependent ventricular hypertrophy by pressure overload. J. Mol. Cell Cardiol. 2009 Jan 21; doi: 10.1016/j.yjmcc.2008.12.017. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Maurel A, Hernandez C, Kunduzova O, Bompart G, Cambon C, Parini A, Francés B. Age-dependent increase in hydrogen peroxide production by cardiac monoamine oxidase A in rats. Am. J. Physol. Heart Circ. Physiol. 2003;284:H1460–H1467. doi: 10.1152/ajpheart.00700.2002. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi P, Kunduzova O, Masini E, Cambon C, Bani D, Raimondi L, Seguelas MH, Nistri S, Colucci W, Leducq N, Parini A. Oxidative stress by monoamine oxidase mediates receptor-independent cardiomyocyte apoptosis by serotonin and postischemic myocardial injury. Circulation. 2005;112:3297–3305. doi: 10.1161/CIRCULATIONAHA.104.528133. [DOI] [PubMed] [Google Scholar]
- 16.Gentili F, Pizzinat N, Ordener C, Marchal-Victorion S, Maurel A, Hofmann R, Renard P, DeLagrange P, Pigini M, Parini A, Giannella M. 3-[5-(4,5-dihydro-1H-imidazol-2-yl)-furan-2-yl]phenylamine (amifuraline), a promising reversible and selective peripheral MAO-A inhibitor. J. Med. Chem. 2006;49:5578–5586. doi: 10.1021/jm060605r. [DOI] [PubMed] [Google Scholar]
- 17.Weyler W, Salach JI. Purification and properties of mitochondrial monoamine oxidase type A from human placenta. J. Biol. Chem. 1985;260:13199–13207. [PubMed] [Google Scholar]
- 18.Weyler W, Titlow CC, Salach JI. Catalytically active monoamine oxidase type A from human liver expressed in Saccharomyces cerevisiae contains covalent FAD. Biochem. Biophys. Res. Commun. 1990;173:1205–1211. doi: 10.1016/s0006-291x(05)80914-3. [DOI] [PubMed] [Google Scholar]
- 19.Salach JI. Monoamine oxidase from beef liver mitochondria: simplified isolation procedure, properties, and determination of its cysteinyl flavin content. Arch. Biochem. Biophys. 1979;192:128–137. doi: 10.1016/0003-9861(79)90078-x. [DOI] [PubMed] [Google Scholar]
- 20.Newton-Vinson P, Hubálek F, Edmondson DE. High-level expression of human liver monoamine oxidase B in Pichia pastoris. Prot. Exp. and Purif. 2000;20:334–345. doi: 10.1006/prep.2000.1309. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Hubálek F, Newton-Vinson P, Edmondson DE. High-level expression of human liver monoamine oxidase A in Pichia pastoris: comparison with the enzyme expressed in Saccharomyes cerevisae. Prot. Exp. and Purif. 2002;24:152–162. doi: 10.1006/prep.2001.1546. [DOI] [PubMed] [Google Scholar]
- 22.Binda C, Newton-Vinson P, Hubálek F, Edmondson DE, Mattevi A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol. 2002;9:22–26. doi: 10.1038/nsb732. [DOI] [PubMed] [Google Scholar]
- 23.De Colibus L, Li M, Binda C, Lustig A, Edmondson DE, Mattevi A. Three- dimensional structure of human monoamine oxidase A (MAO A): relation to the structures of rat MAO A and human MAO B. Proc. Natl. Acad. Sci. USA. 2005;102:12684–12689. doi: 10.1073/pnas.0505975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Son SY, Ma J, Kondou Y, Yoshimura M, Yamashita E, Tsukihara T. Structure of human monoamine oxidase A at 2.2-Å resolution: the control of opening the entry for substrates/inhibitors. Proc. Natl. Acad. Sci. USA. 2008;105:5739–5744. doi: 10.1073/pnas.0710626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Binda C, Wang J, Pisani L, Caccia C, Carotti A, Salvati P, Edmondson DE, Mattevi A. Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: Safinamide and coumarin analogs. J. Med. Chem. 2007;50:5848–5852. doi: 10.1021/jm070677y. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Yoshimura M, Yamashita E, Nakagawa A, Ito A, Tsukihara T. Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J. Mol. Biol. 2004;338:103–114. doi: 10.1016/j.jmb.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Binda C, Li M, Hubálek F, Restelli N, Edmondson DE, Mattevi A. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high resolution crystal structures. Proc. Natl. Acad. Sci. USA. 2003;100:9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. 1994;D50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 29.Mitoma J, Ito A. Mitochondrial targeting signal of rat liver monoamine oxidase B is located at its carboxy terminus. J. Biochem. (Tokyo, Japan) 1992;111:20–24. doi: 10.1093/oxfordjournals.jbchem.a123712. [DOI] [PubMed] [Google Scholar]
- 30.Chen K, Wu HF, Shih JC. Influence of C terminus on monoamine oxidase A and B catalytic activity. J. Neurochem. 1996;66:797–803. doi: 10.1046/j.1471-4159.1996.66020797.x. [DOI] [PubMed] [Google Scholar]
- 31.Gottowik J, Malherbe P, Lang G, Da Prada M, Cesura AM. Structure/function relationships of mitochondrial monoamine oxidase A and B chimeric forms. Eur. J. Biochem. 1995;230:934–942. doi: 10.1111/j.1432-1033.1995.tb20639.x. [DOI] [PubMed] [Google Scholar]
- 32.De Colibus L, Mattevi A. New frontiers in structural flavoenzymology. Curr. Opin. Struct. Biol. 2006;16:722–728. doi: 10.1016/j.sbi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Ann. Rev. Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 34.Andres AM, Soldevila M, Navarro A, Kidd KK, Olivia B, Bertranpetit J. Positive selection in MAO A gene is human exclusive: determination of the putative amino acid change selective in the human lineage. Hum. Genet. 2004;115:377–386. doi: 10.1007/s00439-004-1179-6. [DOI] [PubMed] [Google Scholar]
- 35.Fajer PG, Brown L, Song L. Practical Pulsed Dipolar ESR (DEER) In: Hemminga MA, Berliner LJ, editors. ESR spectroscopy in membrane biophysics. New York: Springer; 2007. pp. 95–128. [Google Scholar]
- 36.Upadhyay AK, Borbat PP, Wang J, Freed JH, Edmondson DE. Determination of the oligomeric states of human and rat monoamine oxidase in the outer mitochondrial membrane and octyl-β-D-glucopyranoside micelles using pulsed dipolar electron spin resonance spectroscopy. Biochemistry. 2008;47:1554–1566. doi: 10.1021/bi7021377. [DOI] [PubMed] [Google Scholar]
- 37.Kunji ERS, Harding M. Projection structure of atractyloside-inhibited mitochondrial ADP/ATP carrier of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:36985–36988. doi: 10.1074/jbc.C300304200. [DOI] [PubMed] [Google Scholar]
- 38.Hubálek F, Binda C, Khalil A, Li M, Mattevi A, Castagnoli N, Edmondson DE. Demonstration of isoleucine 199 as a structural determinant for the selective inhibition of human monoamine oxidase B by specific reversible inhibitors. J. Biol. Chem. 2005;280:15761–15766. doi: 10.1074/jbc.M500949200. [DOI] [PubMed] [Google Scholar]
- 39.Krueger MJ, Mazouz F, Ramsay RR, Milcent R, Singer TP. Dramatic species differences in the susceptibility of monoamine oxidase B to a group of powerful inhibitors. Biochem. Biophys. Res. Commun. 1995;206:556–562. doi: 10.1006/bbrc.1995.1079. [DOI] [PubMed] [Google Scholar]
- 40.Navaroli L, Daina A, Favre E, Bravo J, Carotti A, Leonetti F, Catto M, Carrupt PA, Reist M. Impact of species-dependent differences on screening, design, and development of MAO B inhibitors. J. Med. Chem. 2006;49:6264–6272. doi: 10.1021/jm060441e. [DOI] [PubMed] [Google Scholar]
- 41.Binda C, Mattevi A, Edmondson DE. Structure-function relationships in flavoenzyme-dependent amine oxidations: a comparison of polyamine oxidase and monoamine oxidase. J. Biol. Chem. 2002;277:23973–23976. doi: 10.1074/jbc.R200005200. [DOI] [PubMed] [Google Scholar]
- 42.Esnouf RM. Further additions to molScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. 1999;D55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 43.Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Meth. Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 44.McDonald GR, Hudson AL, Dunn SMJ, You HT, Baker GB, Whittal RM, Martin JW, Jha A, Edmondson DE, Holt A. Bioactive contaminants leach from disposable laboratory plasticware. Science. 2008;322:917. doi: 10.1126/science.1162395. [DOI] [PubMed] [Google Scholar]
- 45.Hubálek F, Binda C, Li M, Mattevi A, Edmondson DE. Polystyrene microbridges used in sitting drop crystallization release 1,4-diphenyl-2-butene, a novel inhibitor of human MAO B. Acta Crystallogr. 2003;D59:1874–1876. doi: 10.1107/s0907444903016883. [DOI] [PubMed] [Google Scholar]
- 46.Hubálek F, Binda C, Li M, Herzig Y, Sterling J, Youdim MB, Mattevi A, Edmondson DE. Inactivation of purified human recombinant monoamine oxidases A and B by rasagiline and its analogues. J. Med. Chem. 2004;47:1760–1766. doi: 10.1021/jm0310885. [DOI] [PubMed] [Google Scholar]
- 47.Binda C, Hubálek F, Li M, Herzig Y, Sterling J, Edmondson DE, Mattevi A. Binding of rasagiline-related inhibitors to human monoamine oxidases: a kinetic and crystallographic analysis. J. Med. Chem. 2005;29:8148–8154. doi: 10.1021/jm0506266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milczek EM, Bonivento D, Binda C, Mattevi A, McDonald IA, Edmondson DE. Structural and mechanistic studies of mofegiline inhibition of recombinant human monoamine oxidase B. J. Med. Chem. 2008;51:8019–8026. doi: 10.1021/jm8011867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binda C, Wang J, Li M, Hubálek F, Mattevi A, Edmondson DE. Structural and mechanistic studies of arylalkylhydrazine inhibition of human monoamine oxidases A and B. Biochemistry. 2008;47:5616–5625. doi: 10.1021/bi8002814. [DOI] [PubMed] [Google Scholar]
- 50.Nandigama RK, Edmondson DE. Structure-activity relations in the oxidation of phenethylamine analogues by recombinant human liver monoamine oxidase A. Biochemistry. 2000;39:15258–15265. doi: 10.1021/bi001957h. [DOI] [PubMed] [Google Scholar]
- 51.Yang MJ, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu HT. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 52.Walker WH, Hemmerich P, Massey V. Reductive photoalkylation of flavin nucleus and flavin catalyzed photodecarboxylation of phenylacetate. Helv. Chim. Acta. 1967;50:2269–2279. doi: 10.1002/hlca.19670500812. [DOI] [PubMed] [Google Scholar]
- 53.Upadhyay AK, Wang J, Edmondson DE. Comparison of the structural properties of the active site cavities of human and rat monoamine oxidases A and B in their soluble and membrane-bound forms. Biochemistry. 2008;47:526–536. doi: 10.1021/bi7019707. [DOI] [PubMed] [Google Scholar]
- 54.Yu PH, Bailey BA, Durden DA, Boulton AA. Stereospecific deuterium substitution at the α-carbon position of dopamine and its effect on oxidative deamination catalyzed by MAO A and MAO B from different tissues. Biochem. Pharm. 1986;35:1027–1036. doi: 10.1016/0006-2952(86)90094-8. [DOI] [PubMed] [Google Scholar]
- 55.Li M. Ph. D. Dissertation. Emory University; 2006. Comparative mechanistic and structural approaches to investigate the membrane-bound enzymes monoamine oxidase A and monoamine oxidase B. [Google Scholar]
- 56.Farnum MF, Klinman JP. Stereochemical probes of bovine plasma amine oxidase: evidence for mirror-image processing and a syn abstraction of hydrogens from C-1 and C-2 of dopamine. Biochemistry. 1986;25:6028–6036. doi: 10.1021/bi00368a029. [DOI] [PubMed] [Google Scholar]
- 57.Walker MC, Edmondson DE. Structure-activity relationships in the oxidation of benzylamine analogues by bovine liver monoamine oxidase B. Biochemistry. 1994;33:7088–7098. doi: 10.1021/bi00189a011. [DOI] [PubMed] [Google Scholar]
- 58.Miller JR, Edmondson DE. Structure-activity relationships in the oxidation of para-substituted benzylamine analogues by recombinant human liver monoamine oxidase A. Biochemistry. 1999;38:13670–13683. doi: 10.1021/bi990920y. [DOI] [PubMed] [Google Scholar]
- 59.Jonsson T, Edmondson DE, Klinman JP. Hydrogen tunneling in the flavoenzyme monoamine oxidase B. Biochemistry. 1994;33:14871–14878. doi: 10.1021/bi00253a026. [DOI] [PubMed] [Google Scholar]
- 60.Kurtz KA, Rishavy MA, Cleland WW, Fitzpatrick PF. Nitrogen isotope effects as probes of the mechanism of D-amino acid oxidase. J. Am. Chem. Soc. 2000;122:12896–12897. [Google Scholar]
- 61.Ralph EC, Hirschi JS, Anderson MA, Cleland WW, Singleton DA, Fitzpatrick PF. Insights into the mechanism of flavoprotein-catalyzed amine oxidation from nitrogen isotope effects on the reaction of N-methyltryptophan oxidase. Biochemistry. 2007;46:7655–7664. doi: 10.1021/bi700482h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris CM, Pollegioni L, Ghisla S. pH and kinetic isotope effects in D-amino acid oxidase catalysis: evidence for a concerted mechanism in substrate dehydrogenation via hydride transfer. Eur. J. Biochem. 2001;268:5504–5520. doi: 10.1046/j.1432-1033.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- 63.Kay CWM, El Mkami H, Molla G, Pollegioni L, Ramsay RR. Characterization of the covalently bound anionic flavin radical in monoamine oxidase A by electron paramagnetic resonance. J. Am. Chem. Soc. 2007;129:16091–16097. doi: 10.1021/ja076090q. [DOI] [PubMed] [Google Scholar]
- 64.Rigby SEJ, Hynson RM, Ramsay RR, Munro AW, Scrutton N. A stable tyrosyl radical in monoamine oxidase A. J. Biol. Chem. 2005;280:4627–4631. doi: 10.1074/jbc.M410596200. [DOI] [PubMed] [Google Scholar]
- 65.Nagai H, Fukushima Y, Okajima K, Ikeuchi M, Mino H. Formation of interacting spins on flavosemiquinone and tyrosyl radical in photoreaction of a blue light sensor BLUF protein TePixD. Biochemistry. 2008;47:12574–12582. doi: 10.1021/bi8010187. [DOI] [PubMed] [Google Scholar]
- 66.Ehrenberg A, Eriksson LEG, Hyde JS. Electron-nuclear double resonance from radicals of flavins and flavoproteins. In: Kamin H, editor. Flavins and Flavoproteins. Baltimore: Univ. Park Press; 1971. pp. 141–152. [Google Scholar]
- 67.Lu X, Rodriguez M, Ji H, Silverman R, Vintem APB, Ramsay RR. Chapman S, Perham R, Scrutton N, editors. Irreversible inactivation of mitochondrial monoamine oxidases. Flavins and Flavoproteins. 2002:817–830. [Google Scholar]
- 68.Miller JR, Edmondson DE, Grissom CB. Mechanistic probes of monoamine oxidase B catalysis: rapid-scan stopped-flow and magnetic field independence of the reductive half reaction. J. Am. Chem. Soc. 1995;117:7830–7831. [Google Scholar]
- 69.Newton-Vinson P, Edmondson DE. High-level expression, structural, kinetic, and redox characterization of recombinant human liver monoamine oxidase B. In: Ghisla S, Kroneck P, Macheroux P, Sund H, editors. Flavins and Flavoproteins. Berlin: Agency for Scientific Publications; 1999. pp. 431–438. [Google Scholar]
- 70.Hull LA, Davis GT, Rosenblatt DH, Mann CK. Oxidation of amines: chemical and electrochemical correlations. J. Phys. Chem. 1969;73:1389–1393. [Google Scholar]
- 71.Bissel P, Khalil A, Rimoldi JM, Igarashi K, Edmondson DE, Miller A, Castagnoli N. Stereochemical studies on the novel monoamine oxidase B substrates (1R,6S)- and (1S,6R)-3-methyl-6-phenyl-3-aza-bicyclo[4.1.0]heptane. Bioorg. Med. Chem. 2008;16:3557–3564. doi: 10.1016/j.bmc.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 72.Wang J, Edmondson DE. Do monomeric vs dimeric forms of MAO A make a difference? Comparison of the catalytic properties of rat and human MAO A’s. J. Neur. Trans. 2007;114:721–724. doi: 10.1007/s00702-007-0678-8. [DOI] [PubMed] [Google Scholar]
- 73.Hartmann C, Klinman JP. Structure-function studies of substrate oxidation by bovine serum amine oxidase: relationship to cofactor structure and function. Biochemistry. 1991;30:4605–4611. doi: 10.1021/bi00232a035. [DOI] [PubMed] [Google Scholar]
- 74.Bordwell FG, Cheng JP, Satish AV, Twyman CL. Acidities and homolytic bond dissociation energies (BDE’s) of benzyl-type C-H binds in sterically congested substrates. J. Org. Chem. 1992;57:6542–6546. [Google Scholar]
- 75.Macheroux P, Ghisla S, Sanner C, Rüterjans H, Müller F. Reduced flavin: NMR investigations of N(5)-H exchange mechanism, estimation of ionization constants and assessment of properties as biological catalyst. BMC Biochemistry. 2005;6:26. doi: 10.1186/1471-2091-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eisenreich W, Kemter K, Bacher A, Mulrooney SB, Williams CH, Müller F. 13C-, 15N-, and 31P-NMR studies of oxidized and reduced low mass thioredoxin reductase and some mutant proteins. Eur. J. Biochem. 2004;271:1437–1452. doi: 10.1111/j.1432-1033.2004.04043.x. [DOI] [PubMed] [Google Scholar]
- 77.Li M, Binda C, Mattevi A, Edmondson DE. Functional role of the “aromatic cage” in human monoamine oxidase B: structures and catalytic properties of Tyr435 mutant proteins. Biochemistry. 2006;45:4775–4784. doi: 10.1021/bi051847g. [DOI] [PubMed] [Google Scholar]
- 78.Ramsay RR. Kinetic mechanism of monoamine oxidase A. Biochemistry. 1991;30:4624–4629. doi: 10.1021/bi00232a038. [DOI] [PubMed] [Google Scholar]
- 79.Husain M, Edmondson DE, Singer TP. Kinetic studies on the catalytic mechanism of liver monoamine oxidase. Biochemistry. 1982;21:595–600. doi: 10.1021/bi00532a028. [DOI] [PubMed] [Google Scholar]
- 80.Roth JP, Klinman JP. Catalysis of electron transfer during activation of O2 by the flavoprotein glucose oxidase. Proc. Natl. Acad. Sci. USA. 2003;100:62–67. doi: 10.1073/pnas.252644599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prabhakar R, Li M, Musaev DG, Morokuma K, Edmondson DE. A density functional theory (DFT) study of the spin forbidden dioxygen activation in monoamine oxidase B (MAO B) In: Nishino T, Riura R, Tanokura M, Fukui K, editors. Flavins and Flavoproteins. Tokyo: ArchiTect, Inc.,; 2005. pp. 127–131. [Google Scholar]
- 82.Bruice TC. Oxygen-flavin chemistry. Isr. J. Chem. 1984;24:54–61. [Google Scholar]
- 83.Massey V. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 84.Sucharitakul J, Prongjit M, Haltrich D, Chaiyen P. Detection of a C(4a)-hydroperoxyflavin intermediate in the reaction of a flavoprotein oxidase. Biochemistry. 2008;47:8485–8490. doi: 10.1021/bi801039d. [DOI] [PubMed] [Google Scholar]
- 85.Orville AM, Lountos GT, Finnegan S, Gadda G, Prabhakar R. Crystallographic, spectroscopic and computational analysis of a flavin C(4a)-oxygen adduct in choline oxidase. Biochemistry. 2009;48:720–728. doi: 10.1021/bi801918u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao GH, Bruckner RC, Jorns MS. Identification of the oxygen activation site in monomeric sarcosine oxidase: Role of Lys265 in catalysis. Biochemistry. 2008;47:9124–9135. doi: 10.1021/bi8008642. [DOI] [PMC free article] [PubMed] [Google Scholar]