Abstract
Two RecA homologs, Rad51 and Dmc1, assemble as cytologically visible complexes (foci) at the same sites on meiotic chromosomes. Time course analysis confirms that co-foci appear and disappear as the single predominant form. A large fraction of co-foci are eliminated in a red1 mutant, which is expected as a characteristic of the interhomolog-specific recombination pathway. Previous studies suggested that normal Dmc1 loading depends on Rad51. We show here that a mutation in TID1/RDH54, encoding a RAD54 homolog, reduces Rad51-Dmc1 colocalization relative to WT. A rad54 mutation, in contrast, has relatively little effect on RecA homolog foci except when strains also contain a tid1/rdh54 mutation. The role of Tid1/Rdh54 in coordinating RecA homolog assembly may be very direct, because Tid1/Rdh54 is known to physically bind both Dmc1 and Rad51. Also, Dmc1 foci appear early in a tid1/rdh54 mutant. Thus, Tid1 may normally act with Rad51 to promote ordered RecA homolog assembly by blocking Dmc1 until Rad51 is present. Finally, whereas double-staining foci predominate in WT nuclei, a subset of nuclei with expanded chromatin exhibit individual Rad51 and Dmc1 foci side-by-side, suggesting that a Rad51 homo-oligomer and a Dmc1 homo-oligomer assemble next to one another at the site of a single double-strand break (DSB) recombination intermediate.
During meiosis, homologous chromosomes recombine to produce the reciprocal crossovers needed for accurate reductional segregation during the first meiotic division (MI). In the budding yeast Saccharomyces cerevisiae, most recombination is initiated by double-strand breaks (DSBs; reviewed in refs. 1 and 2). The ends at DSB sites are processed by nucleolytic activity that forms 3′ single-stranded DNA (ssDNA) (reviewed in refs. 1, 3, and 4). The ssDNAs are then converted to homologous joint molecules (JMs), a recombination intermediate with two Holliday junctions between homologous duplex DNAs (2, 5). The conversion of DSBs to JMs requires a homology search, followed by strand invasion and exchange. JMs are eventually resolved into mature recombination products.
Rad51 and Dmc1 are yeast homologs of RecA, the major bacterial strand exchange protein (reviewed in refs. 6 and 7). Rad51 and Dmc1 proteins play critical roles in the conversion of DSBs to JMs in meiosis. Both Rad51 and Dmc1 can promote the formation of recombinants in the absence of the other, indicating that they have overlapping functions (8–11). On the other hand, the finding that single mutants have qualitatively different effects on recombination intermediates indicates that the two play distinct roles in the formation of recombinants (10, 11). Rad51 forms right-handed helical filaments on ssDNA that carry out strand exchange reactions in vitro. The Rad51-mediated strand exchange reaction is stimulated by protein cofactors RPA, Rad52, Rad55–57, and Rad54 (12–16). Whereas there is less biochemical data on Dmc1 function, human Dmc1 has been shown to promote formation of homologous joint molecules and to form toroidal oligomers on DNA (17, 18).
Rad51 physically interacts with other recombination proteins including Rad54 (6). Tid1/Rdh54 is a structural and functional relative of Rad54 (19–21). Hereafter, we will refer to Tid1/Rdh54 as Tid1 for simplicity. Both Rad54 and Tid1 belong to the Swi2/Snf2 family of helicase-related proteins (22). Both proteins have DNA-unwinding activity that can promote Rad51-dependent JM formation in vitro (refs. 15 and 23; P. Sung, personal communication). Tid1 interacts directly with both Dmc1 and Rad51, with the Dmc1–Tid1 interaction being stronger than the Rad51–Tid1 interaction (19). Tid1's function appears to be partially specialized to promote recombination between homologous chromatids rather than between sister chromatids (19–21, 24, 25). The same type of specialization has been observed in meiosis for Dmc1, as compared with Rad51 (10, 11).
Immunostaining of spread of meiotic yeast nuclei has revealed that both Rad51 and Dmc1 proteins form subnuclear assemblies called “foci” (26). Several observations support the view that these foci correspond to sites of functioning recombination complexes (10, 26, 27). Rad51 and Dmc1 foci often colocalize, suggesting they function together in the same recombination event (19, 26). RecA homolog foci in lily and mouse mark sites of “zygotene nodules” or “early nodules” (28, 29), which are proteinaceous structures detected by ultrastructural analysis (30, 31).
Normal appearance of brightly staining Dmc1 foci in yeast depends on RAD51; the Dmc1 foci that form in a rad51 mutant went undetected in an initial study but were later detected and found to stain faintly compared with those in WT (10, 26). These results suggest that Rad51 promotes normal assembly of Dmc1 and also show that at least some Dmc1 assembly can occur in the absence of Rad51. Rad51 foci form normally in dmc1 mutants (26). Whereas evidence for structural and functional interactions indicates that the two yeast RecA homologs can, and often do, contribute to the same recombination event, it remains unknown how assembly of two RecA homologs on meiotic chromosomes is coordinated.
In this paper, we present evidence that Tid1 promotes Rad51-Dmc1 colocalization. Tid1 and Rad54 also promote timely disappearance of Rad51 and Dmc1 foci. In addition, we show that Red1, a major meiosis-specific chromosome component (11, 32–34), is also required for the normal codistribution of the two proteins. Together with the results of previous studies, these findings suggest that Tid1 acts to coordinate assembly of strand exchange proteins during both Red1-dependent interhomolog recombination and during Red1-independent recombination.
Materials and Methods
Strains.
All strains described here are derivatives of SK1. The strains used contain the same markers as NKY1551 (MATa/α ho∷LYS2/′′, ura3/′′, leu2∷hisG/′′, lys2/′′, his4X-LEU2- BamHI-URA3/his4B-LEU2, arg4-nsp/arg4-bgl); MSY241 (rad54∷hisG-URA3-hisG/′′), MSY134 (tid1∷LEU2/′′), MSY147 (tid1∷LEU2/′′, rad54∷hisG-URA3-hisG/′′), MSY443 (tid1∷LEU2/′′, red1∷LEU2/′′), MSY439 (red1∷LEU2/′′), NKY2515 (zip1∷LEU2/′′). Also used were NKY1314 (MATa/α ho∷LYS2/′′, ura3/′′, leu2∷hisG/′′, lys2/′′, his4X-LEU2/his4B-LEU2) and an isogenic derivative, DKB968, containing red1∷LEU2/′′. Double prime means “homozygous.”
Anti-Serum.
Guinea pig anti-Rad51 anti-serum was raised against yeast Rad51 protein purified from Escherichia coli. The anti-serum was prepared by SP Co. Ltd., Kobe, Japan. The rabbit anti-Dmc1 and anti-Rad51 were previously described (26). Both Rad51 and Dmc1 antibodies were purified as described (35). Crossreactivity of each primary antibody was tested by using rad51 and dmc1 deletion mutant strains, and none was detected. Antibodies were used at a concentration of 2 and 5 μg/ml for anti-Rad51 and anti-Dmc1, respectively.
Cytology.
Induction of synchronous meiosis was carried out as described previously (10, 27). Meiotic cells were spherolasted and surface-spread on glass slides in the presence of detergent (Lipsol) and fixative (4% paraformaldehyde; ref. 36). After drying, spread nuclei were immunostained as described (26). The slides were incubated with both guinea pig anti-Rad51 and rabbit anti-Dmc1 antibodies simultaneously overnight at 4°C, followed by incubation with secondary antibodies for 2 h at 4°C. Epifluorescence microscopy was carried out by using a Zeiss Axiovert 135 M or a Zeiss Photomicroscope III. Images were captured with a cooled charge-coupled device digital camera.
Scoring of Immunostained Nuclei.
Scoring of foci and determination of colocalization frequency was as described previously (26, 27). Pairs of foci in which >50% of the two signals overlapped were scored as double-staining foci. The predicted frequency of colocalization resulting from random distribution of foci was also determined by generation of random focal staining patterns by using dot-stat software (27, 37).
Results
Codistribution of Rad51 and Dmc1 During Wild-Type Meiosis.
The two RecA homologs, Rad51 and Dmc1, were reported previously to colocalize on meiotic chromosomes in wild-type (WT) yeast nuclei (19, 26). To study structures containing both RecA homologs in more detail, we carried out time course analysis of synchronized meiotic cultures. Chromosome spreads from various times in meiosis were prepared and double immunostained with anti-Rad51 and anti-Dmc1 antibodies (Fig. 1). Three types of subnuclear staining foci can be detected by this method: Rad51 foci (R foci), Dmc1 foci (D foci), and foci that stain for both of these proteins (“co-foci” or RD foci).
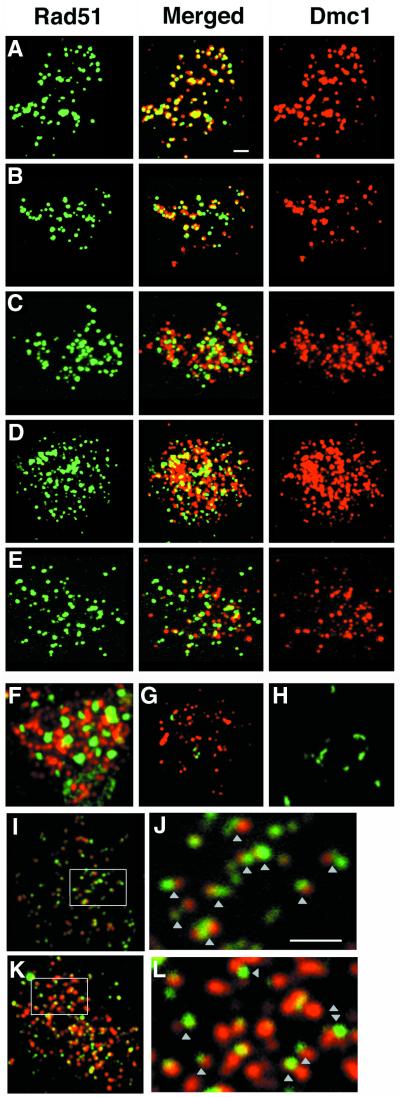
Figure 1.
Colocalization of Rad51 and Dmc1 foci. Spreads of meiotic nuclei were prepared and indirectly immunostained with antibodies against Rad51 (green; Left) and Dmc1 (red; Right). Single-staining images are pseudocolored from the gray scale images. Merged images are a two-channel combination of original image pairs (Middle). Representative nuclei are shown. (A) WT, 3 h; (B) red1, 3 h; (C) tid1, 3 h; (D) rad54 tid1, 3 h; (E) red1 tid1, 3 h; (F) rad54 tid1, 12-h nucleus showing Rad51 aggregates; (G) WT, 4.5-h nucleus with mostly D foci; (H) rad54, 4.5-h nucleus with mostly R foci; (I and J) WT, 4.5-h nucleus showing side-by-side R and D foci; (K and L) rad54 tid1, 7.5-h nucleus showing side-by-side R and D foci. J and L are photographic enlargements of the areas indicated in I and K, respectively. (White scale bars = 2 μm long.)
First, samples taken at regular intervals during meiosis were spread, and the number of foci of the three classes was determined in a large sample (n = 100) of randomly selected nuclei. This analysis shows that 40% to 82% of foci observed are RD foci. These foci appear and disappear in a single peak with maximal abundance at t = 3 h. An amount equal to 7.8% and 7.9% of foci observed were R and D foci, respectively. The two types of single-staining foci were initially present in similar numbers, but, at late times, D foci outnumbered R foci.
In a second analysis, samples taken at regular intervals during synchronous meiosis were scored in a binary way as either “focus positive” or “focus negative,” according to whether they contained a minimum number of foci of a given type, Rad51 or Dmc1, irrespective of colocalization. These two categories are referred to as R+ and D+, respectively. This analysis was done because most focus-positive nuclei contain more than 15 foci (M.S., unpublished data), and, thus, transitions into and out of the foci-positive stage are fairly abrupt in most cases. When the frequencies of the two classes are plotted as a function of time in meiosis, the appearance and disappearance of R+ or D+ nuclei reflects the overall timing with which the two proteins assemble on and disassemble from the chromosomes. The area under each curve reveals the average life span of the stage. In addition, integration of this curve provides a cumulative curve that reveals when cells “enter” the stage, which they then “exit” one life span later (see legend of Fig. 2 for details). This analysis reveals that, during wild-type meiosis, R+ and D+ nuclei appear with very similar kinetics, with 50% of cells having entered the corresponding stage at t = 2.5 h. The average life spans were also similar, 1.6 and 2.1 h, respectively. Whereas the difference in life span might imply late-stage conversion of RD foci to D foci, a more likely possibility is that foci containing only Dmc1 are defective in disassembly (see below).
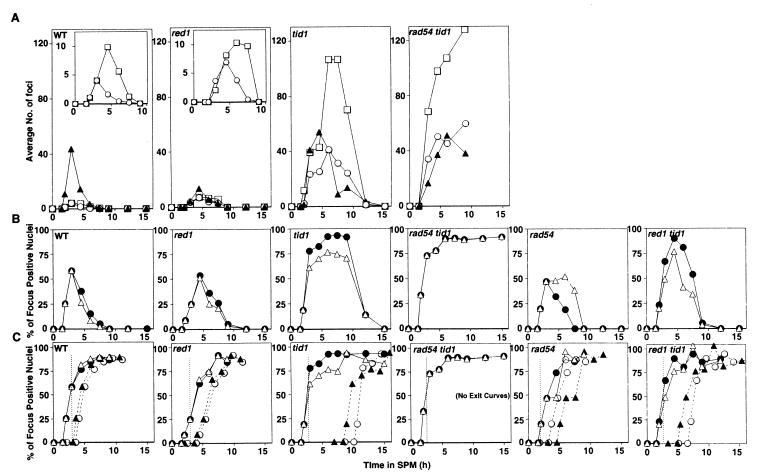
Figure 2.
(A) Time course analysis of Rad51 and Dmc1 foci. One hundred nuclei were randomly selected. The number of foci in each of the three possible focus classes, RD foci (closed triangles); R foci (open circles), and D foci (open squares), were counted in each nucleus examined. The average numbers of the foci at each time point are shown. (B and C) Time course analysis of Rad51 and Dmc1 focus-positive nuclei. The percentages of nuclei containing more than five foci were derived from the images of more than 100 random nuclei from each time point. The threshold of five is set to avoid including nuclei that display a small amount of background staining in the focus-positive class (ref. 26; and M.S., unpublished data). Noncumulative curves (B) are converted into cumulative curves (C) by using the method of Padmore et al. (44). The duration of a stage is given by the area under the corresponding noncumulative curve, divided by the total fraction of nuclei progressing through meiosis (44), assumed to be 90% at all time points. The average life span of Rad51-positive nuclei was 1.77 ± 0.24 h (n = 7). Cumulative curves describe what fraction of cells have either entered or completed a given event as a function of time. The curve is identical to the noncumulative curve plotted up to the first nonzero time point and one life span thereafter. Then, for any time point (t) after one life span has elapsed, the cumulative curve value at t is equal to the noncumulative curve value at t plus the cumulative curve value at t minus one life span. The exit curve for any event is given by plotting entry curve at a rightward displacement along the x axis by one life span. From the left: WT, red1, tid1, rad54 tid1, rad54, and red1 tid1 mutants. In B, percentages of R+ and D+ nuclei are indicated by open triangles and closed circles, respectively. In C, the fraction of nuclei that had entered R+ and D+ stages are indicated by open triangles and closed circles, respectively. The fraction that had exited R+ and D+ stages are indicated as hatched lines with closed triangles and open circles, respectively. The vertical line in each panel highlights the 3-h time point. Construction of cumulative curves assumes that the life span of a particular stage is constant as time progresses. The bi-phasic shape of the R+ curve in the tid1 mutant suggests that the life span of this stage is not uniform in this mutant.
Analysis of Codistribution During Meiosis in Mutant Strains.
Mutant strains have been analyzed by the approaches described above for WT. Analysis of focus-positive nuclei indicated that nuclei examined at t = 3 h have recently acquired foci; 50% or less of all cells had acquired Rad51- and Dmc1-containing foci by this time point (Fig. 2). Thus, differences in the frequency of colocalization are expected to reflect primarily differences in the outcome of focus assembly rather than disassembly. Because quantification of individual focus types throughout a meiotic time course is very laborious, we began by analyzing the distribution of different focus types at this time (Table 1). As a control, we examined a mutant lacking Zip1 protein, a component of the synaptonemal complex (SC). Because the SC normally appears after focus formation, concomitant with disassembly of RecA homologs from recombination complexes, a zip1 mutant might be expected to have no effect. Accordingly, wild-type and zip1 mutant nuclei exhibit the same numbers and distributions of focus types, with 77% of foci detected being RD foci (compare lines 1 and 7). In contrast, analysis of mutants lacking Red1, Tid1, and Rad54 all exhibit important differences in both the absolute and relative numbers of all three types of foci. In all of these mutants, the fraction of foci containing both Rad51 and Dmc1 (percentage of RD foci) is reduced. Colocalization is only slightly reduced in rad54 (61%); but in all of the other cases, the fraction of RD is dramatically reduced, to ≈30% in red1 and tid1 single mutants and to ≈15% in red1 tid1 and rad54 tid1 double mutants.
Table 1.
Double staining analysis of Rad51 and Dmc1 in various mutants
| Strain | Number of foci per
nucleus at 3 h*
|
% of RD foci† | Time of entry, h‡
|
Life span, h§
|
||||
|---|---|---|---|---|---|---|---|---|
| RD foci | R foci | D foci | R+ stage | D+ stage | R+ stage | D+ stage | ||
| WT | 60 ± 18 | 6.7 ± 2.9 | 11 ± 2.8 | 77 (3.8) | 2.5 | 2.5 | 1.6 | 2.1 |
| red1 | 13 ± 4.1 | 16 ± 3.4 | 11 ± 4.8 | 32 (2.9) | 3.5 | 3.5 | 2.1 | 2.5 |
| tid1 | 43 ± 22 | 44 ± 15 | 59 ± 21 | 30 (11) | 2.6 | 2.5 | 6.9 | 8.5 |
| rad54 | 43 ± 16 | 14 ± 2.4 | 13 ± 5.9 | 61 (3.7) | 3.0 | 3.0 | 3.1 | 1.7 |
| rad54 tid1 | 22 ± 9.9 | 47 ± 17 | 93 ± 31 | 13 (12) | 2.3 | 2.3 | NA | NA |
| red1 tid1 | 14 ± 4.1 | 35 ± 12 | 47 ± 19 | 15 (13) | 3.0 | 2.6 | 3.4 | 5.1 |
| zip1 | 64 ± 3.2 | 5.8 ± 1 | 13 ± 5.8 | 77 (ND) | ND | ND | ND | ND |
ND and NA, not determined, and not available, respectively.
For each strain, at least 100 unselected 3-h meiotic nuclei were counted. Numbers are averages of three to six independent experiments. Experimental errors are also indicated as ±.
The predicted frequency of fortuitous superposition, as estimated by the computer program dot-stat (37), is given in parenthesis.
Time at which 50% of nuclei had entered the focus-positive stage, as determined from the cumulative curves in Fig. 2C.
Life spans of focus-positive stages as determined from the noncumulative curve data in Fig. 2B.
Differences in the percentage of RD foci imply that the behavior of the two proteins is less tightly coupled in the mutant cases than in WT. We interpret these reductions to represent a reduced probability that both proteins will assemble onto the same site. The alternative possibility is that co-assembly occurs at the same frequency in the mutant, but RD foci have shorter life spans compared with R and D foci. The following considerations lead us to disfavor this alternative (as outlined in more detail below). First, the life spans of the R+ and D+ are the same or greater in the mutants than in WT. Second, there is no indication that R+ and D+ nuclei occur in sequential waves, as would be required for a reduction in the co-residency time. Third, in the case of tid1, Dmc1 foci form before Rad51 foci rather than simultaneously as they do in WT, indicating a loss of dependency of Dmc1 assembly on Rad51. Thus, red1 and tid1 appear to reduce the frequency of colocalized foci.
Time Course Analysis of Mutant Strains.
For RED1, analysis of the timing of entry into the focus-positive stage showed that the red1 mutation delays entry into R+ and D+ by about 1.0 h (Fig. 2C). However, the life spans of both focus-positive stages were similar to those in WT. Nuclei entered R+ and D+ simultaneously as in WT. Time course analysis of individual foci revealed that the peak in RD foci was reduced about 4-fold in relation to WT (from 43 to 13 in the experiment shown; Fig. 2A). The effect of red1 on R and D foci was modest by comparison to the effect on RD foci. The peaks of R and D foci were slightly elevated, but the shapes of the curves were similar to WT, and the order of the peaks was the same (Fig. 2A Insets). The data indicate that Red1 is specifically required for the appearance of normal numbers of RD foci.
For RAD54 and TID1, cultures of rad54 mutant cells showed a delay in entry into both R+ and D+ relative to WT. Whereas the degree of the delay varied somewhat from one experiment to the next, a delay of about 0.5 h in the time of entry into both R+ and D+ was seen in three of four experiments (including the experiment shown in Fig. 2). These results indicate that RAD54 accelerates assembly of the RecA homologs.
In marked contrast to the role of RAD54, time course analysis of mutants suggests that TID1 delays the time of entry into D+. This conclusion is supported by comparison of tid1, red1 tid1, and rad54 tid1 mutant strains with the three corresponding TID1 strains. In all three cases, the strain containing the tid1 mutation enters the D+ phase earlier than its counterpart. This difference was particularly obvious when the red1 and rad54 cumulative curves were compared with the red1 tid1 and rad54 tid1 curves, respectively; entry into D+ was about 0.7–0.9 h earlier in the double mutants. Comparison of WT and tid1 curves also suggests that entry into D+ is earlier in the tid1 mutant than WT. Whereas we favor this interpretation, we note that the R+ cumulative curve constructed from tid1 data has a reproducibly bi-phasic shape. This bi-phasic nature of the curve raises the possibility that the life span of the R+ stage is not uniform in the culture, with nuclei entering R+ early exiting that stage more rapidly than cells entering R+ late. Such heterogeneity in the life span of the R+ stage is expected to shift the upper portion of the R+ curve downward, exaggerating the difference between the D+ and R+ curves (see Fig. 2 legend for details). Regardless of this complication, there is more independent evidence that tid1 forms D foci earlier than WT; about 20% of focus-positive tid1 nuclei contain D foci but not R foci at 3 h (Fig. 2B). Given that the average life span of the R+ stage in the tid1 is 6.9 h, the 3-h R− D+ subpopulation is very unlikely to represent nuclei that have exited the R+ stage and likely to represent nuclei that have acquired D foci before acquiring R foci. In contrast to tid1, R− D+ nuclei were not detected during the assembly phase in WT (Fig. 2B). Thus, TID1 appears to slow entry into D+ not just in red1 and rad54 mutant cells, but in WT cells as well.
The effect of a tid1 mutation on entry into R+ was more restricted than its effect on entry into D+; tid1 allowed early formation of faintly staining R foci in a rad54 mutant background (data not shown) but had little effect in a WT or red1 mutant strain background.
Mutations in tid1 and rad54 had dramatic effects on the timing of RecA homolog disassembly. Based on the area under noncumulative curves, the average life spans of R+ and D+ are 4.3- and 4.0-fold longer in tid1 than in WT, respectively. The time between exit from R+ and D+ was increased in tid1 (Fig. 2C). The data indicate that TID1 plays a role in disassembly of both Rad51 and Dmc1 foci and that the effect on disassembly of Dmc1 is more pronounced than the effect on Rad51. The single focus analysis revealed a strong peak of D foci in tid1 shortly before cells exit the R+ stage (Fig. 2A). This peak may reflect the fact that the foci containing only Dmc1 may be particularly defective in disassembly. It is also possible that the peak of D foci reflects an intermediate stage in disassembly of RD foci when detectable amounts of Rad51 have been lost, but detectable amounts of Dmc1 remain.
The life span of R+ in rad54 was increased 2-fold to about 3 h whereas the life span of D+ was decreased slightly. Exit from D+ proceeded exit from R+, a reversal of the normal order (Fig. 1H). These results imply that RAD54 is more important for the normal timing of Rad51 disassembly whereas TID1 is more important for Dmc1 disassembly.
Disassembly of foci appeared to be completely blocked in the rad54 tid1 double mutant, indicating that TID1 and RAD54 share a function needed for RecA homolog disassembly. Whereas Rad51 foci did not disappear at late times, their staining intensity increased until approaching that of WT; then, starting at 6 h, much larger Rad51-containing structures began to appear and these structures became increasingly predominant as time progressed (Fig. 1F). It seemed that, rather than disappearing, R foci aggregated into large structures. This formation of large agglegates was not true for the Dmc1 staining pattern, which remained punctate.
Combination of a red1 mutant and a tid1 mutant yielded an intermediate phenotype with respect to the average life span of R+ and D+. These results indicate that TID1 is required for timely exit from the R+ and D+ stage for RED1-independent as well as RED1-dependent foci, although disassembly of RED1-independent foci is somewhat less dependent on TID1 than is disassembly of RED1-dependent structures.
Side-by-Side Appearance of Rad51 and Dmc1 in WT and rad54 tid1.
In some of our experiments, a small fraction of spread WT nuclei were observed in which foci of Rad51 and Dmc1, rather than being precisely colocalized, lay side-by-side in closely spaced pairs (Fig. 1 I and J). The frequency of such R–D pairs suggests that they could be closely related to the structures scored as co-foci (RD foci) in the majority of spread nuclei. The spread chromatin (as detected by staining with the DNA-specific dye 4,6-diamidino-2-phenylindole; DAPI) occupied a larger than average area in nuclei containing R–D pairs, suggesting that such pairs might be visible at a brief stage during which chromatin is decondensed. It is also possible that these structures are actually the same as those seen as co-foci, with the difference reflecting subtle differences in spreading conditions across the surface of the slide. We also note that rad54 tid1 double mutant nuclei selected on the basis of having relatively few D foci were often found to contain multiple side-by-side R–D pairs (Fig. 1 K and L).
Discussion
Colocalization of Rad51 and Dmc1 in WT.
The results here confirm earlier work that showed a high degree of temporal and spatial coordination of the two RecA homologs Rad51 and Dmc1 (26). Foci containing both Rad51 and Dmc1 are a predominant structure involved in meiotic recombination. Importantly, the data show that other proteins coordinate assembly of Rad51 and Dmc1. The average frequency of RD foci observed here was 77%. This value lies between previously reported values of 92% (26) and 40% (19). The differences in the reported values are likely to result from selection of a subset of nuclei with bright overall staining in the first case and use of a different strain background combined with a more restrictive definition of RD foci in the second case.
Red1 Is Required for a PreDSB Process That Promotes Coordinated Assembly of the Two RecA Homologs.
The prominent defect in RecA homolog assembly in red1 is the 4-fold reduction in the peak of RD foci. This reduction in RD foci can be explained based on previous observations. First, mutations in RED1 reduce DSBs 3-fold or more (11, 25, 38, 39). Second, analysis of JMs in red1 indicated that the reduction in DSBs resulted in a corresponding reduction in JMs between homologous chromatids whereas JMs between sister chromatids were unaffected (11). Third, mutant analysis showed that both Rad51 and Dmc1 make separate contributions to the high ratio of interhomolog to intersister JMs characteristic of WT cells (11). Based on these observations, Schwacha and Kleckner proposed that RED1 is required before or during the DSB step on a “highly differentiated, interhomolog-only” pathway that requires participation of both RecA homologs. The present results are consistent with their proposal if RED1-dependent RD foci are intermediates on the interhomolog-only pathway. Mutation of RED1 blocks a subset of recombination events that would normally involve assembly of RD foci, whereas the remainder of events, which involve the appearance of all three focus types, occur with relatively little perturbation.
Rad54 Contributes to Assembly of Rad51.
The appearance of Rad51 foci is delayed in a rad54 mutant, indicating that it may have a modest role in assembly. Kanaar and colleagues have reported a much more dramatic role for Rad54 in assembly of Rad51 in mouse embryonic stem cells treated with ionizing radiation (40). Appearance of Dmc1 foci is also delayed in rad54, probably as an indirect result of the defect in Rad51 assembly.
Regulation of Dmc1 Assembly by Tid1.
In WT, assembly of Dmc1 is promoted by Rad51 and occurs most often at the sites of Rad51 assembly. Mutation of tid1 increases the tendency of Dmc1 to assemble at sites that do not contain Rad51 assemblies. The high degree of coordination of Rad51/Dmc1 assembly is likely to involve specific protein–protein contacts. Direct interaction between Dmc1 and Rad51 has been detected for the human homologs (17, 29), but not in yeast (19). However, two-hybrid data have provided evidence that Tid1 can interact directly with both Rad51 and Dmc1 (19). Thus, Tid1 may serve as an intermolecular bridge between structures formed by the two RecA homologs, thereby directing Dmc1 to sites of Rad51 assembly or by promoting simultaneous assembly of both proteins. The rad54 tid1 double mutant shows a lower colocalization frequency than the tid1 single mutant, indicating that Rad54 is likely to be able to substitute for Tid1 in colocalization of RecA homologs to a certain degree.
Dmc1 foci form earlier in tid1 mutants than in TID1+ (WT) cells. These results suggest that, in addition to a positive role in directing Dmc1 to the sites of Rad51, TID1 may also act as a negative regulator by preventing assembly of Dmc1 at sites devoid of Rad51. There is precedent for factors that confer specificity by reducing nonspecific interactions. For example, the transcription factors σ in bacteria and TFIIF in eukaryotes employ direct contacts with RNA polymerase to lower the affinity of the polymerase for random DNA sequences (41, 42). In addition to this negative role in determining specificity, σ factor also plays a positive role by binding promoter sequences. Perhaps association of Tid1 with Dmc1 lowers the affinity of Dmc1 for recombination sites devoid of Rad51.
Tid1 and Rad54 Share a Function Needed for Disassembly of Rad51 and Dmc1.
In addition to promoting timely assembly of Rad51 foci, Rad54 promotes timely disassembly. Tid1 has an even more pronounced role in disassembly; a tid1 mutant retains both Rad54 and Dmc1 foci for long periods. RAD54 and TID1 can substitute for one another in promoting disappearance of foci, just as they have been shown to substitute for one another in converting meiotic DSBs to recombinants (21). The defects in the timing of focus disappearance observed in rad54 and tid1 mutants follow the general rule that disappearance of RecA homolog foci during yeast meiosis occurs shortly after conversion of DSBs to homologous JMs (26, 27). It is possible that the disassembly defect and the defect in converting DSBs to JMs are both indirect consequences of failing to properly assemble the RecA homologs into a structurally normal complex. Alternatively, Rad54 and Tid1 may have two distinct roles, one that is nonessential for assembly of foci (but nonetheless coordinates such assembly) and a second role, which has been demonstrated biochemically (15, 23, 40), that is essential for the ability of the RecA homologs to invade duplex DNA and form stable joints.
In addition to revealing a function shared by Rad54 and Tid1, the life spans of the focus-positive stages in the single mutants provide evidence for preferred functional interaction between Tid1 and Dmc1 and between Rad51 and Rad54. A rad54 mutation increases R+ life span more than D+ life span whereas the opposite is true of a tid1 mutation.
How Are Rad51 and Dmc1 Arranged in RD Foci?
Three types of observation lead us to favor the view that Rad51 and Dmc1 each form homo-oligomers at recombination sites rather than a uniform hetero-oligomer composed of subunits of both proteins. (i) A tid1 mutation has a more severe effect on Dmc1 foci than on Rad51 foci, whereas the opposite is true of rad54. These observations add to others indicating that the two RecA homologs can readily assemble and disassemble multimeric forms independently of one another (10, 27). (ii) We detected a subset of nuclei that contain multiple side-by-side R and D foci rather than co-foci. (iii) Whereas homotypic interactions of Rad51 and Dmc1 are readily detected by two-hybrid methods, heterotypic interactions between yeast Rad51 and Dmc1 are too weak to be detected with the same constructs (19, 43). We propose that the side-by-side arrangement of Rad51 and Dmc1 homo-oligomers is a key feature of meiotic recombination and that Tid1 promotes the assembly of this structure. One attractive possibility is that Dmc1 assembles on one DNA end at the site of a DSB, whereas Rad51 assembles on the other end.
Acknowledgments
We thank Ms. Itagaki and Ms. Taniguchi for their technical assistance. We particularly thank the editor for many helpful suggestions, especially her suggestion to apply cumulative curve analysis to the data. We also thank members of the Bishop laboratory for critical reading of the manuscript. This work was supported by a grant from the Ministry of Education, Science, and Culture of Japan to A.S. and by National Institutes of Health Grant GM50936 (to D.K.B.). M.S. was supported by a Human Frontier Postdoctoral Research Fellowship.
Abbreviations
- DSB
double-strand break
- JM
joint molecule
- R foci
Rad51-only foci
- D foci
Dmc1-only foci
- RD foci
Rad51-Dmc1 double-staining foci
- R+ stage
Rad51-positive stage
- D+ stage
Dmc1-positive stage
- WT
wild type
References
- 1.Lichten M, Goldman A S. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 2.Schwacha A, Kleckner N. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 3.Kleckner N. Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roeder G S. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 5.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 6.Shinohara A, Ogawa T. Mutat Res. 1999;435:13–21. doi: 10.1016/s0921-8777(99)00033-6. [DOI] [PubMed] [Google Scholar]
- 7.Kowalczykowski S C, Eggleston A K. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 8.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 10.Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop D K. Genes Cells. 1997;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- 11.Schwacha A, Kleckner N. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 12.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 13.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 14.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 15.Petukhova G, Stratton S, Sung P. Nature (London) 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 16.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Nature (London) 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 17.Masson J Y, Davies A A, Hajibagheri N, Van Dyck E, Benson F E, Stasiak A Z, Stasiak A, West S C. Embo J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Golub E I, Gupta R, Radding C M. Proc Natl Acad Sci USA. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dresser M E, Ewing D J, Conrad M N, Dominguez A M, Barstead R, Jiang H, Kodadek T. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein H L. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinohara M, Shita-Yamaguchi E, Buerstedde J M, Shinagawa H, Ogawa H, Shinohara A. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen J A, Sweder K S, Hanawalt P C. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 24.Arbel A, Zenvirth D, Simchen G. Embo J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop D K, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 26.Bishop D K. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 27.Gasior S L, Wong A K, Kora Y, Shinohara A, Bishop D K. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson L K, Offenberg H H, Verkuijlen W M H C, Heyting C. Proc Natl Acad Sci USA. 1997;94:6868–6873. doi: 10.1073/pnas.94.13.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarsounas M, Morita T, Pearlman R E, Moens P B. J Cell Biol. 1999;147:207–220. doi: 10.1083/jcb.147.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Wettstein D, Rasmussen S W, Holm P B. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 31.Carpenter A T. Bioessays. 1987;6:232–236. doi: 10.1002/bies.950060510. [DOI] [PubMed] [Google Scholar]
- 32.Rockmill B, Roeder G S. Proc Natl Acad Sci USA. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockmill B, Roeder G S. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith A V, Roeder G S. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 36.Klein F, Laroche T, Cardenas M E, Hofmann J F, Schweizer D, Gasser S M. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S M. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao-Draayer Y, Galbraith A M, Pittman D L, Cool M, Malone R E. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L, Weiner B M, Kleckner N. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 40.Tan T L, Essers J, Citterio E, Swagemakers S M, de Wit J, Benson F E, Hoeijmakers J H, Kanaar R. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 41.Conaway J W, Conaway R C. Science. 1990;248:1550–1553. doi: 10.1126/science.2193400. [DOI] [PubMed] [Google Scholar]
- 42.deHaseth P L, Lohman T M, Burgess R R, Record M T., Jr Biochemistry. 1978;17:1612–1622. doi: 10.1021/bi00602a006. [DOI] [PubMed] [Google Scholar]
- 43.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 44.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]