Abstract
O-succinylbenzoyl-CoA (OSB-CoA) synthetase (EC 6.2.1.26) catalyzes the ATP-dependent condensation of o-succinylbenzoate (OSB) and CoA to form OSB-CoA, the fourth step of the menaquinone biosynthetic pathway in Bacillus anthracis. Gene knockout studies have highlighted this enzyme as a potential target for the discovery of new antibiotics. Here we report the first studies on the kinetic mechanism of B. anthracis OSB-CoA synthetase, classifying it as an ordered Bi Uni Uni Bi ping-pong mechanism. Through a series of pre-steady-state and steady-state kinetic studies in conjunction with direct-binding studies, it is demonstrated that CoA, the last substrate to bind, strongly activates the first half-reaction after the first round of turnover. The activation of the first-half reaction is most likely achieved by CoA stabilizing conformations of the enzyme in the ‘F’ form, which slowly isomerize back to the E form. Thus, the kinetic mechanism of OSB-CoA synthetase may be more accurately described as an ordered Bi Uni Uni Bi Iso ping-pong mechanism. The substrate specificity of OSB-CoA synthetase was probed using a series of OSB analogs with alterations in the carboxylate groups. OSB-CoA shows a strong preference for OSB over all of the analogs tested as none were active except 4-(2-trifluoromethylphenyl)-4-oxobutyric acid which exhibited a 100-fold decrease in kcat/Km. Based on an understanding of OSB-CoA synthetase’s kinetic mechanism and substrate specificity, a reaction intermediate analog of OSB-AMP, 5’-O-(N-(2-trifluoromethylphenyl)-4-oxobutyl) adenosine sulfonamide (TFMP-butyl-AMS), was designed and synthesized. This inhibitor was found to be an uncompetitive inhibitor to CoA and a mixed-type inhibitor to ATP and OSB with low micromolar inhibition constants. Collectively, these results should serve as an important forerunner to more detailed and extensive inhibitor design studies aimed at developing lead compounds against the OSB-CoA synthetase class of enzymes.
The bacterial menaquinone biosynthetic enzymes are potentially attractive targets for antibiotics discovery because they are essential for bacterial growth and are absent from humans. Menaquinone is an essential lipophilic molecule that shuttles electrons between dehydrogenases and cytochromes in the bacterial electron transport chain (1). The Bacillus family of bacteria cannot acquire menaquinone from the environment and therefore require the menaquinone biosynthetic pathway for the production of this critical cofactor (1, 2). Studies on Bacillus subtilis have demonstrated that deletion of any of its menaquinone biosynthetic genes compromises its growth in LB medium (3). Although knockout studies on the menaquinone biosynthetic genes from Bacillus anthracis have not yet been reported, by analogy it is possible that these genes are also essential for growth of B. anthracis since it is a close relative of B. subtilis (4). Finally, since the entire bacterial menaquinone biosynthetic pathway is absent from the human genome as humans utilize ubiquinone for the electron transport chain in the mitochondria, the enzymes in the menaquinone pathway are unique and therefore attractive targets for the discovery of new antibiotics against B. anthracis and other bacteria.
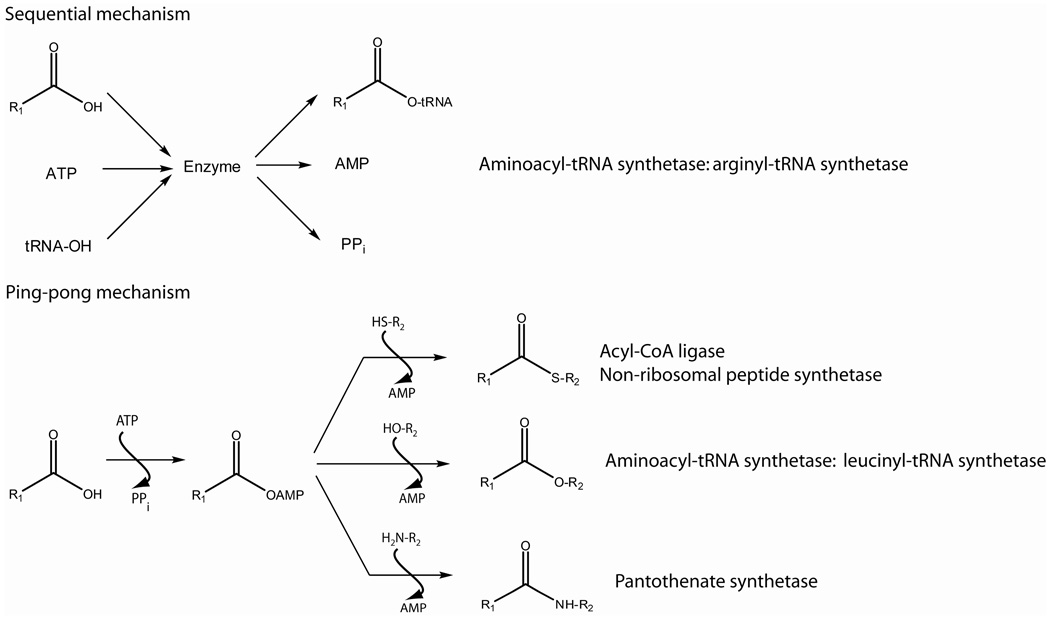
Among all the B. anthracis menaquinone biosynthetic enzymes, o-succinylbenzoyl-coenzyme A (OSB-CoA) synthetase was chosen as a potential drug target of our studies based on the availability of various assays to detect the enzymatic reaction products (5, 6), and the fact that the substrate OSB and various analogs can be readily obtained by chemical synthesis (7). OSB-CoA synthetase (EC 6.2.1.26) catalyzes the fourth reaction in the menaquinone biosynthetic pathway that converts the substrates OSB, ATP, and CoA to the products AMP, PPi, and OSB-CoA (8). Mg2+ is also required for the enzymatic activity (9, 10). Enzymes using ter-reactant mechanisms usually catalyze either a ter-ter sequential reaction (11) or a Bi Uni Uni Bi ping-pong reaction (12–15) (Scheme 1). Escherichia coli arginyl-tRNA synthetase utilizes a ter-ter sequential mechanism for its reaction whereby all three substrates (ATP, arginine, and tRNA) must bind to the enzyme before any of the three products (PPi, AMP, and arinyl-tRNA) is released into the bulk solution (11). In contrast, other enzymes follow a Bi Uni Uni Bi ping-pong reaction with the formation of an acyl-AMP intermediate (12–15). As illustrated in Scheme 1, the reaction proceeds in two steps. In the first step, a carboxylic acid substrate reacts with ATP to form an acyl-AMP intermediate and release the first product, PPi. In the second step, the acyl-AMP intermediate reacts with the third substrate to form the last two products. In this case, the third substrate is not required for the first step of the reaction. As expected, the formation of an acyl-AMP intermediate has been experimentally confirmed in the enzymes that utilize ping-pong reaction mechanisms but not in the enzymes utilizing sequential mechanisms (14–17). Indeed, analogs that mimic the reaction intermediate have been found to be potent inhibitors of the enzymes that follow a ping-pong mechanism (18, 19).
Scheme 1.
Analogous to the reaction mechanism mentioned above, OSB-CoA synthetase may catalyze either a ping-pong or a sequential reaction. To determine which kinetic mechanism is utilized by B. anthracis OSB-CoA synthetase, the steady-state kinetics of the reaction were studied by initial velocity, product inhibition and direct binding studies. The pre-steady-state kinetics of the first half-reaction and the overall reaction was also studied, and the substrate specificity of the enzyme was probed using a series of OSB analogs. Based on these experimental results, a potent reaction intermediate analog was designed and synthesized, which is the first potent inhibitor of an OSB-CoA synthetase from any organism.
MATERIALS AND METHODS
Materials
ATP, AMP, CoA, benzoyl-CoA, yeast inorganic pyrophosphatase (IPP), and bacterial purine nucleoside phosphorylase (PNP) were purchased from Sigma (St. Louis, MO). The compound 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG) was purchased from Berry & Associates (Dexter, MI). Restriction enzymes NdeI and BamHI were purchased from Fermentas (Hanover, MD). T4 DNA ligase, and Pfx DNA polymerase were purchased from Invitrogen Corporation (Carlsbad, CA). Plasmid vector pET15b and Escherichia coli BL21 (DE3) strain were purchased from Novagen (Madison, WI). All chromatographic instruments and columns were purchased from Amersham Biosciences (Piscataway, NJ).
Chemical synthesis
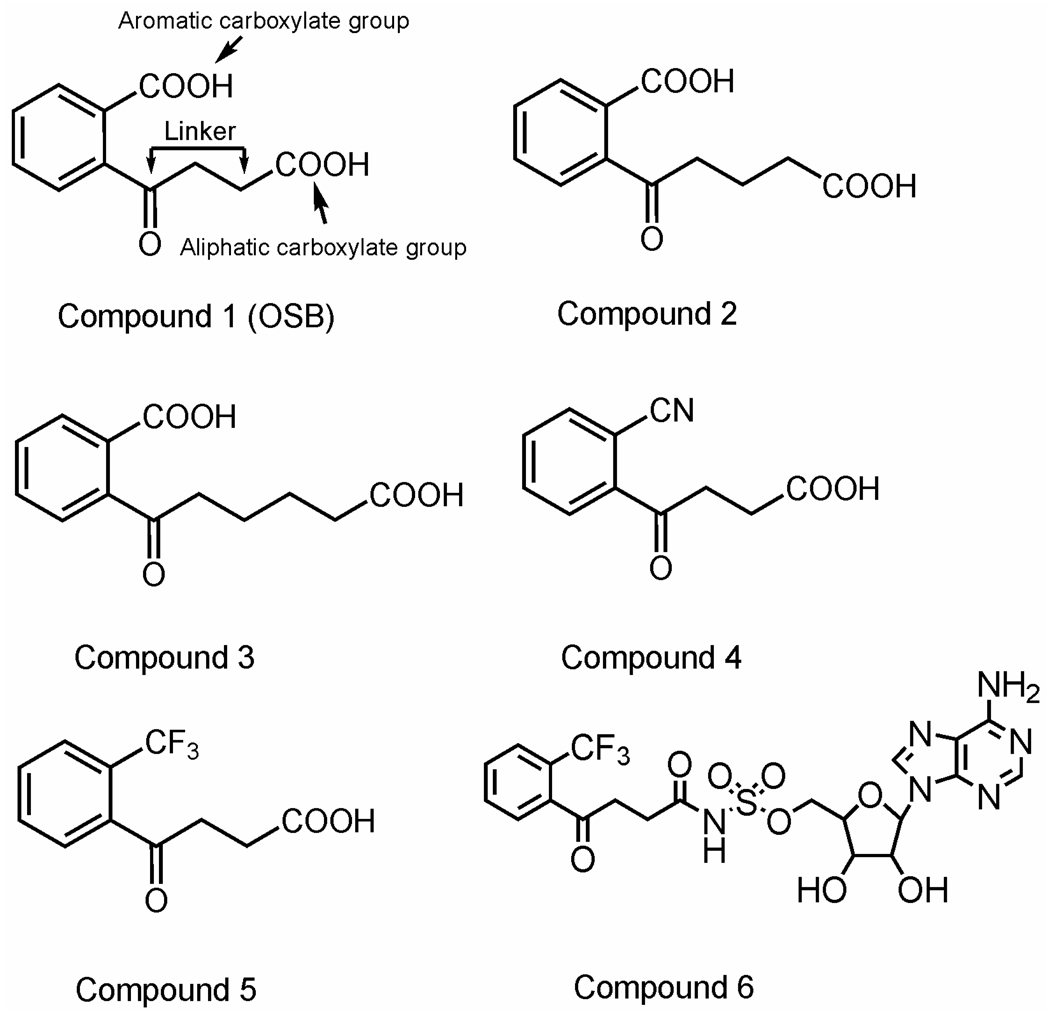
OSB, 1, was synthesized as previously described (7). Four OSB analogs, 2–5, and 6, an OSB-AMP analog, 5’-O-(N-(2-trifluoromethylphenyl)-4-oxobutyl) adenosine sulfonamide (TFMP-butyl-AMS), were synthesized and the procedures are described in Supporting Information. The structures for these compounds are given in Figure 1.
Figure 1. Structures of OSB, OSB analogs and an OSB-AMP analog.
Compound 6 (TFMP-butyl-AMS) is an OSB-AMP analog that was designed and synthesized by covalently linking compound 5 and a sulfonamide adenosine. Synthetic protocols for these compounds are given in Supporting Information.
Cloning, expression, and purification of 1,4-dihydroxy-2-naphthoate-CoA (DHNA-CoA) synthetase (MenB) from E. coli
The full details for the expression and purification are given in Supporting Information.
Cloning, expression, and purification of OSB-CoA synthetase from B. anthracis
The B. anthracis OSB-CoA synthetase gene (menE) sequence was obtained from Genbank by accession number BA5108. The menE gene was obtained by PCR amplification from genomic DNA isolated from the B. anthracis Sterne strain which was a generous gift from Dr. Alexander A. Neyfakh (University of Illinois at Chicago) using Pfx DNA polymerase. Two primers (5’-GACACGACATATGGAGACGATGCCAAATTGGTTA-3’; 5’-CGGGATCCTTACATCTCCTCCACTAATTGTCTTAACTCTCG-3’) containing NdeI and BamHI sites (underscored in the primer sequences) were used for the PCR reactions. The PCR products were cloned into the pET15b plasmid using the above two restriction sites which placed the menE gene in frame with an N-terminal (His)6-tag sequence. The entire menE gene was sequenced at the DNA sequencing facility at Research Resource Center (RRC) of University of Illinois at Chicago. The mutation-free construct was transformed into E. coli strain BL21 (DE3) for expression. Cells were grown at 37°C in 4 L LB medium until the OD600 reached 0.6, when IPTG was added to the cell culture at a final concentration of 0.1 mM for induction of protein expression. After two hours of induction, cells were collected by centrifugation at 3,300g for 10 minutes at 4°C. For purification, 12 grams of cell pellet were resuspended in 30 mL of Buffer A (50 mM Tris-HCl, 10 mM imidazole, 0.5 M NaCl, pH 7.5) and lysed by sonicating using a GEX-600 sonics ultrasonic processor (Sonics and Materials, Inc., Newtown, CT) with a 0.5’’ probe. The sonication lasted for 6 minutes with a repeating pulse of 6.6 seconds on and 9.9 seconds off at 65% amplitude. The cell lysate was centrifuged at 39,000g for 40 minutes at 4°C, after which the supernatant was collected and filtered through a 0.22 µm filter (Millipore, Carrigtwohill, Co.Cork, Ireland). The clear filtrate (∼30 mL) was loaded onto a 5 mL HiTrap affinity column (1.6 × 2.5 cm) (Amersham Biosciences, Piscataway, NJ) charged with cobalt and equilibrated with Buffer A. The column was washed with 100 mL Buffer A to remove any weakly bound proteins. OSB-CoA synthetase was then eluted using a linear gradient of 0–100% Buffer B (50 mM Tris-HCl, 1 M imidazole, 0.5 M NaCl, pH 7.5) for 120 mL. Fractions containing OSB-CoA synthetase were pooled, and 3.8 M ammonium sulfate was added to a final concentration of 1 M. The resulting solution was loaded on a HP Phenyl Sepharose column (1.6 × 20) (Amersham Biosciences, Piscataway, NJ) equilibrated with Buffer C (50 mM Tris-HCl, 1 M ammonium sulfate, 0.2 M NaCl, pH 7.5). The column was washed with 200 mL Buffer C to remove weakly bound proteins. OSB-CoA synthetase was eluted using a linear gradient of 0–100% Buffer D (50 mM Tris-HCl, 0.1 M NaCl, pH 7.5). Fractions were analyzed by SDS-PAGE and those containing pure OSB-CoA synthetase were pooled. Prior to storage at −80°C, the buffer was exchanged with 50 mM Tris-HCl (pH 7.5), and the protein was concentrated to about 100 µM using a protein concentrator (Millipore, Carrigtwohill, Co. Cork, Ireland). Protein concentrations were determined using a BIO-RAD protein assay kit (BIO-RAD Laboratories Inc., Hercules, CA) with bovine serum albumin (BSA) as a standard.
OSB-CoA synthetase activity assay
Two assays were utilized for the measurement of the OSB-CA synthetase activity. In the first assay (see Figure S1a in the Supporting Information), the release of PPi was coupled to the reactions of IPP and PNP as previously described (5). The increase in absorbance of 2-amino-6-mercapto-7-methylpurine at 360 nm was monitored on a SpectraMax 384 Plus plate reader (Molecular Devices, Sunnyvale, CA). A molar extinction coefficient of 11,000 M−1cm−1 was used to calculate the amount of product formation. The assays were performed in 96-well plate format with an assay volume of 200 µL. Each assay contained 50 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, 0.5 U PNP, 0.075 U IPP, 300 µM MESG, pH 7.5, and different concentrations of OSB, ATP, and CoA. The above reaction mixture was pre-incubated for 10 minutes, and the reaction was initiated with a final concentration of 50 nM of OSB-CoA synthetase. The reaction rates were obtained from the initial, linear region of the reaction progress curves. A unit of enzyme activity is defined as one µmole of PPi formed per minute with the specific activity defined as one unit per mg of enzyme. All assays were performed at 21°C. The assay conditions described above were used throughout this report, unless specified otherwise.
In the second assay, (see Figure S1b in Supporting Information), OSB-CoA synthetase activity was measured by coupling the formation of the product, OSB-CoA, to the reaction catalyzed by DHNA-CoA synthetase (20). In this reaction, OSB-CoA is converted into DHNA-CoA which has a molar extinction coefficient of 4,000 M−1 cm−1 at 392 nm (20). For the 96-well, plate-based assay, a 200 µL standard assay mixture contained 50 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, 88 µM E. coli DHNA-CoA synthetase, pH 7.5, with varying concentrations of OSB, ATP and CoA. The reaction was initiated by the addition of OSB-CoA synthetase to a final concentration of 1 µM. The reaction rates were determined from the initial linear region of the reaction progress curve. One unit of activity is defined one µmole of DHNA-CoA formed per minute.
Single substrate kinetics of B. anthracis OSB-CoA synthetase
The Km and kcat values for each of the three substrates (OSB, ATP, and CoA) were determined by single substrate kinetics. The first set of enzyme reaction rate measurements were made by varying OSB concentration (1–256 µM) and fixing ATP (512 µM) and CoA (1024 µM) concentrations. For the second set of initial velocity measurements, the ATP concentration was varied (2–512 µM) at fixed OSB (256 µM) and CoA (1024 µM). For the third set of initial velocity measurements, the CoA concentration was varied (8–1024 µM) at fixed OSB (256 µM) and ATP (512 µM). Finally, a kinetic study for the OSB analog (compound 5 in Figure 1) was performed by varying the concentration of 5 from 12 to 3000 µM and fixing ATP (512 µM) and CoA (1024 µM) concentrations. The data were fit to Equation 1:
| (1) |
where v is the reaction rate (μmole/min/mg), Vmax is the maximum reaction rate (μmole/min/mg), [S] is the varying substrate or substrate analog concentration (μM), and Km is the Michaelis-Menten constant. The data were fit to equation 1 using the Enzyme Kinetics module in SigmaPlot 2000 (SYSTAT Software, Inc., Point Richmond, CA).
Initial velocity studies
Bisubstrate kinetic studies were performed for the initial velocity study. The reaction rates were studied by varying the concentrations of two substrates in the presence of a fixed, variable concentration of the third substrate. The experimental data were fit to equation 2–equation 5 using nonlinear regression analysis. The goodness-of-fit was evaluated on the basis of Akaike Information Criterion corrected for small sample size (AICc) values and the standard errors of the parameter estimates (21). AICc is used for comparing non-nested enzyme kinetic equations. AICc improves the performance of AIC when the sample size is small compared to the number of equation parameters. The equation with the lower or more negative AICc value is considered to be the equation that best fits the dataset. A difference of ≥ 7 in the AICc values is considered statistically significant in distinguishing between models (21). If two the equations have nearly the same AICc values or the differences between their AICc values are less than 7, AICc values alone cannot be used to distinguish between models. In such cases, the equation with the better estimate of the fitted parameters and with random distribution of residuals in residual plots can be classified as the better fit and to infer the more appropriate kinetic model. The kinetic mechanism and relevant kinetic parameters were derived from a best-fit to one of the following equations:
| (2) |
| (3) |
| (4) |
| (5) |
In each of the equations, v is the reaction rate (μmole/min/mg), Vmax is the maximum velocity of the reaction (μmole/min/mg), [A] is the substrate A concentration (μM), [B] is the substrate B concentration (μM), Ka and Kb are dissociation constants of substrate A and B (μM), KmA and KmB are Michaelis-Menten constants for substrate A and B (μM), respectively. The data were processed using the Enzyme Kinetics module in SigmaPlot 2000 (SYSTAT Software, Point Richmond, CA).
Direct binding measurements for ATP, OSB, and CoA to B. anthracis OSB-CoA synthetase
The dissociation constants (Kd) of ATP, OSB, and CoA from OSB-CoA synthetase were determined by the centrifugal ultrafiltration method (22, 23). Prior to the binding studies, the purified enzyme was dialyzed (40,000-fold) in a buffer containing 50 mM Tris-HCl and 20 mM NaCl (pH7.5) to remove any trace amount of endogenous ligands associated with the enzyme during expression and purification. The binding studies were performed using the same buffer conditions at 21 °C. For the ATP to OSB-CoA synthetase binding study, the enzyme (70 µM) was incubated with increasing concentrations of ATP (0–800 µM) in the presence and absence of 2 mM MgCl2. For the OSB to OSB-CoA synthetase binding study, the enzyme (70 µM) was incubated with increasing concentrations of OSB (0–800 µM) and 2 mM MgCl2. For the CoA to enzyme binding study, the enzyme (200 µM) was incubated with different concentrations of CoA (0–1280 µM) and 2 mM MgCl2 in the presence and absence of 2 mM ATP. After 20 minutes of binding and equilibration, the free substrates were separated from the bound substrates by passing ∼60 µL of the 500 µL binding mixture through a Microcon YM-10 centrifugal filter device (Millipore Corporation, Bedford, MA). The free ATP concentration was determined directly by measuring the absorbance of the filtrate at 260 nm and then extrapolating the concentration values from an ATP concentration versus absorbance standard curve. The concentration of free OSB in the filtrate was measured enzymatically by the phosphate detection assay with the presence of saturating concentration of ATP (512 µM) and CoA (1024 µM). The increase in absorbance at 360 nm was converted into the concentration of OSB in the filtrate using a molar extinction coefficient of 11,000 M−1cm−1. The free CoA concentration was determined by reacting CoA directly with 5, 5'-dithiobis-2-nitrobenzoic acid (DTNB). Briefly, equal volumes of filtrate and DTNB stock solution (4 mM) were mixed and the absorbance was read at 412 nm. A standard curve of free CoA concentration versus absorbance at 412 nm was constructed and used to obtain the CoA concentration in the filtrates. The fraction of binding v was calculated based on equation 6 and the data were fit to equation 7 for Kd determination (23):
| (6) |
| (7) |
where [L]t is the total substrate concentration in the binding assay (μM), [L]f is the free substrate concentration in the filtrate (μM), [E]t is the total OSB-CoA synthetase concentration (μM), v is the fraction of binding, and Kd is the dissociation constant of the substrate to the enzyme (μM).
Product inhibition study
The product inhibition studies were performed with AMP, benzoyl-CoA, and TFMP-butyl-AMS. Benzoyl-CoA was used as a mimic to the product OSB-CoA since the latter compound is not stable (10). The effects of these inhibitors on reaction rates were studied by varying the concentration of one product inhibitor and one substrate while keeping the other two substrates at constant subsaturating concentrations. The experimental data were fit to equation 8–equation 11 by nonlinear regression analysis. The goodness-of-fit was evaluated on the basis of standard errors of the parameter estimates and AICc values. The kinetic mechanism and relevant kinetic parameters were derived from the best-fit to one of the following equations: specifically, equation 8 for competitive inhibition, equation 9 for uncompetitive inhibition, equation 10 for mixed-type inhibition, and equation 11 for noncompetitive inhibition.
| (8) |
| (9) |
| (10) |
| (11) |
In each of the equations, v is the reaction rate (μmole/min/mg), Vmax is the maximum velocity of the reaction (μmole/min/mg), [S] is the substrate S concentration (μM), [I] is the inhibitor I concentration (μM), Km is the Michaelis-Menten constants for substrate S, Ki is the dissociation constant of the inhibitor I to free enzyme E (mM), and αKi is the dissociation constant for inhibitor I to ES complex (mM). The data were fit to the above equations using the Enzyme Kinetics module in SigmaPlot 2000 (SYSTAT Software, Point Richmond, CA).
Pre-steady-state kinetics of enzyme with ATP and OSB
The phosphate detection assay was used to study the pre-steady-state reaction kinetics of OSB-CoA synthetase with the substrates ATP and OSB. The assays were performed in a 100 µL quartz cuvette with 1 cm pathlength. The assay contained 50 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, 0.5 U PNP, 0.075 U IPP, 300 µM MESG, pH 7.5, 256 µM OSB, and 512 µM ATP. The reactions were initiated with various concentrations of the enzyme (2.2 µM, 4.4 µM, and 8.8 µM), and the absorbance change at 360 nm was monitored for 2.5 minutes on a Cary 50 Bio UV-Visible Spectrophotometer (Varian, Inc., Palo Alto, CA). The enzyme concentrations were kept high in this experiment so that the product, PPi, from a single enzyme turnover could be detected and quantitated. The amount of PPi formed (μM) over time (minutes) was calculated using a molar extinction coefficient of 11,000 M−1cm−1. The time course data were fit to equation 12, which describes a single exponential followed by a linear steady-state component (24):
| (12) |
where y is the concentration of PPi released (μM), t is time (minutes), A is the observed burst amplitude (μM), k1 is the observed exponential burst rate constant (min−1), and k2 is the observed linear steady-state rate (μM min−1).
Pre-steady-state kinetics of enzyme with ATP, OSB and CoA
The phosphate detection assay was used to study the pre-steady-state reaction kinetics of OSB-CoA synthetase with the substrates ATP, OSB and CoA. The assays were performed in a 100 µL quartz cuvette with 1.0 cm pathlength. The first assay contained 50 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, 2.5 U PNP, 1 U IPP, 300 µM MESG, pH 7.5, 256 µM OSB, and 512 µM ATP and 1024 µM CoA. The reactions were initiated with 1 µM OSB-CoA synthetase, and the absorbance change at 360 nm was monitored for 0.4 minute on a Cary 50 Bio UV-Visible Spectrophotometer (Varian, Inc., Palo Alto, CA). The amount of excess coupling enzyme necessary in this assay was determined to ensure that the rate determining step of the coupled assay was the OSB-CoA synthetase catalyzed reaction. The amount of PPi formed (μM) over time (minutes) was calculated using a molar extinction coefficient of 11,000 M−1cm−1. In the second assay, the OSB-CoA synthetase (1 µM) was preincubated with 50 mM Tris-HCl, 20 mM NaCl, 2 mM MgCl2, 2.5 U PNP, 1 U IPP, 300 µM MESG, pH 7.5, 256 µM OSB, and 512 µM ATP for 1 minute so that the steady-state reaction was achieved. An amount of 1024 µM CoA (final concentration) was then added into the reaction and the absorbance at 360 nm was monitored for 0.4 minutes.
RESULTS
Single substrate kinetics of B. anthracis OSB-CoA synthetase
The release of PPi from the OSB-CoA synthetase reaction was coupled to the reactions of IPP and PNP, resulting in an increase of absorbance at 360 nm. Using this pyrophosphate detection assay, the purified enzyme showed maximum activity between pH 7.25 to 7.5 (Supporting Information Figure S2). As a result, all the experimental studies were performed at pH 7.5. The divalent metal Mg2+ was found to be required for enzyme activity and showed maximum activities between 1 to 2 mM Mg2+ (Supporting Information Table S1). A concentration of 2 mM total Mg2+ was used therefore throughout all the studies. To determine the initial estimates of the values for the kinetic parameters of OSB, ATP, and CoA, we performed a single substrate kinetic study by varying one substrate concentration and fixing the other two substrate concentrations. The single substrate kinetic parameters are listed in Table 1. CoA showed the highest Km values among the three substrates, while ATP and OSB showed similar Km values. The enzyme has kcat values in the range of ∼155 min−1. Using the OSB-CoA detection assay, the purified enzyme showed a kcat value of 134.0 ± 12.5 min−1 for the OSB-CoA release rate. All of the kinetic parameters are comparable to E. coli OSB-CoA synthetase (9).
Table 1.
Kinetic parameters and patterns for B. ANTHRACIS OSB-CoA synthetase from the single substrate and bisubstrate initial velocity studiesa
| Fixed concentration (μM) | Apparent Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) or kinetic patterns | ||||
|---|---|---|---|---|---|---|---|
| OSB | ATP | CoA | OSB | ATP | CoA | ||
| - | 512 | 1024 | 21.9 ± 2.0 | - | - | 155 ± 3.6 | 7.1 |
| 256 | - | 1024 | - | 26.8 ± 2.7 | - | 153 ± 3.2 | 5.7 |
| 256 | 512 | - | - | - | 304 ± 22.1 | 156 ± 4.5 | 0.5 |
| 8 | - | - | - | 19.3 ± 1.3 | 231 ± 18.4 | 80.7 ± 2.7 | ping-pong |
| 64 | - | - | - | 10.6 ± 0.7 | 279 ± 19.3 | 108 ± 3.3 | ping-pong |
| - | 8 | - | 12.8 ± 0.8 | - | 194 ± 14.9 | 63.6 ± 2.1 | ping-pong |
| - | 128 | - | 5.8 ± 0.5 | - | 269 ± 24.9 | 66.1 ± 4.5 | ping-pong |
| - | - | 256 | 1.5 ± 0.2 | 3.2 ± 0.5 | - | 52.7 ± 0.8 | sequential |
| - | - | 1024 | 5.0 ± 0.9 | 8.7 ± 0.9 | - | 80.0 ± 1.9 | sequential |
Data presented as mean ± standard error in the fit parameters
OSB substrate analogs
To probe the contribution of the functional groups of OSB to substrate reactivity, we focused on the modification of the two carboxylate groups of OSB: the aromatic carboxylate group and the aliphatic carboxylate group. Compounds 2 and 3 have longer linkers for the aliphatic carboxylate group than that of OSB (Figure 1). Compounds 4 and 5 have the same aliphatic carboxylate group as OSB, but they have a CN or CF3 group that replaces the aromatic carboxylate group of OSB (Figure 1). Compounds 2–5 were first tested for their ability to serve as substrates for OSB-CoA synthetase. At a concentration of 1 mM for each of the OSB analogs, OSB-CoA synthetase showed 5.8% residual activity only towards compound 5 (4-(2-trifluoromethylphenyl)-4-oxobutyric acid) and little to no measurable activity with compounds 2–4‥ OSB-CoA synthetase has a Km value of 166 ± 18 µM and a kcat value of 12.5 ± 3.6 min−1 for compound 5. Compared to OSB, OSB-CoA has a significant decrease (100-fold) in rate constant for capture, kcap or kcat/Km, for compound 5 in addition to a reduced (10-fold) rate constant, krel or kcat, for its release (25, 26).
Initial velocity study
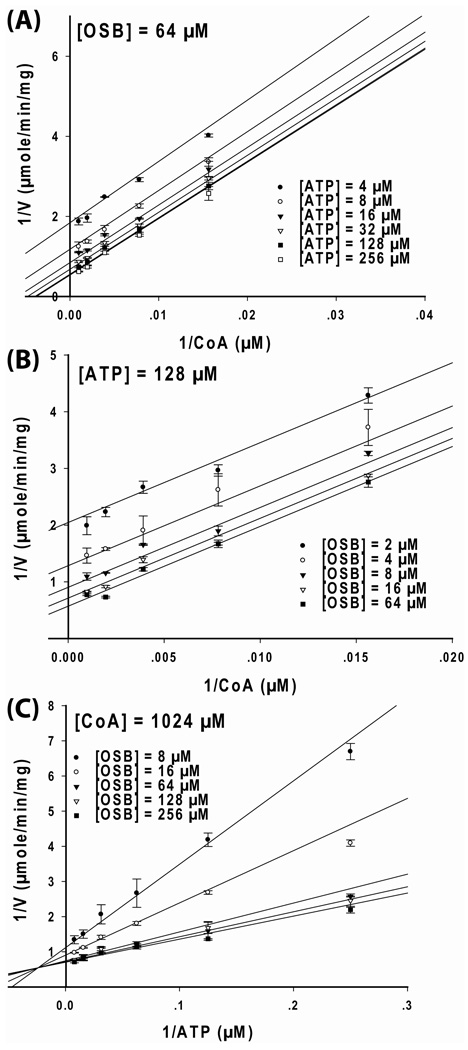
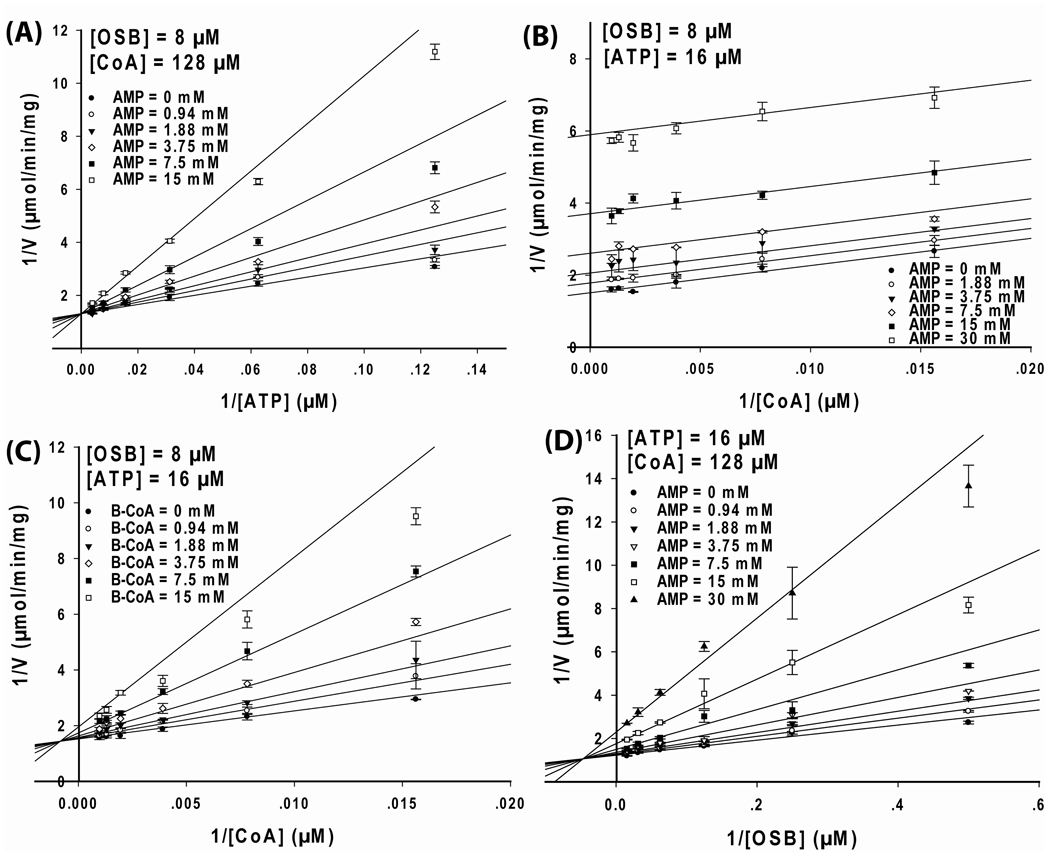
To study the order of substrate addition for OSB-CoA synthetase, bisubstrate kinetic studies were performed. The three possible combinations of the variable substrate pairs were first studied at a fixed, unsaturating level of the third substrate. Nonlinear regression was performed on the experimental data using equation 2–equation 5, and the resulting goodness-of-fit values for each equation are listed in Table S2 in Supporting Information. When the OSB concentration was fixed at a constant, near saturating level (64 µM) and the CoA concentrations were varied at several fixed concentrations of ATP, the data were best fit to equation 2, and the Lineweaver-Burk plot of the data produced a series of parallel lines (Figure 2A). A similar result and plot were also obtained when the ATP concentration was fixed at a constant, near saturating level (128 µM), and CoA concentrations were varied at several, fixed concentrations of OSB (Figure 2B). The observation of parallel lines in those plots indicated a ping-pong kinetic mechanism where a chemistry step and transformation of the enzyme into a new enzyme forms occurs between the addition of the first two substrates (OSB and ATP) and the addition of CoA. Based on our observation that the first-half reaction is catalyzed by OSB-CoA synthetase in the absence of CoA (see below), these steps could be the transformation of ATP and OSB in to OSB-AMP and PPi, and the release of PPi or PPi and AMP from a ternary enzyme complex. The resulting plots (Figure 2A and 2B) also indicated that CoA was the final substrate of addition in the kinetic scheme.
Figure 2. Initial velocity study of B. anthracis OSB-CoA synthetase catalyzed reaction.
The kinetic data were displayed as Lineweaver-Burk plots of the reaction rate vs substrate concentrations. For each plot, one substrate was kept at constant level while varying the other two substrate concentrations. (A) OSB was kept constant at 64 µM while varying CoA and ATP concentrations (•=4, ○=8, ▼=16, □=32, ■=128, □=256 µM). (B) ATP was kept constant at 128 µM while varying CoA and OSB concentrations (•=2, ○=4, ▼=8, □=16, ■=64 µM). (C) CoA was kept constant at 1024 µM while varying ATP and OSB concentrations (•=8, ○=16, ▼=64, □=128, ■=256 µM). Each data point in the figures represents the mean of a duplicate test. The upper and lower bars represent the duplicate measurements. The kinetic parameters and patterns are summarized in Table 1.
When CoA was fixed at a constant, near saturating level (1024 µM) and ATP concentrations were varied at several, fixed OSB concentrations, the data were best fit to equation 4 and equation 5 (equation 4 and 5 are the same equations but with a different definition of the parameters in the denominator). A Lineweaver-Burk plot of the data produced a series of converging lines (Figure 2C), indicating a sequential mechanism in which there is no product released between the addition of OSB and ATP. Although differentiation between a steady-state ordered (equation 4) and a rapid-equilibrium random (equation 5) mechanism for the OSB and ATP substrate pair remained unclear based solely upon these experiments, these data did rule out a ter-ter sequential and a hexa-uni ping-pong mechanism and were suggestive of either a Bi Uni Uni Bi or Bi Bi Uni Uni ping-pong reaction. In both mechanisms, OSB and ATP bind to the free enzyme in a sequential manner, which is then followed by the release of either PPi, or PPi and AMP, before CoA combines with the enzyme.
To support these mechanisms, the kinetic experiments were repeated using an 8-fold and 16-fold decrease (subsaturating) in the concentrations of OSB and ATP. Plots similar to those in Figures 2A and 2B were constructed and parallel lines were also obtained (Figure S3A and S3B). These results further supported the ping-pong mechanism for the OSB/CoA substrate pair and the ATP/CoA substrate pair. Next, the kinetic studies presented in Figure 2C were repeated with a 4-fold decrease in CoA concentration. These data are plotted in Figure S3C and the same pattern of intersecting lines was obtained. Together, the results presented in Figure 2 support a sequential mechanism for the OSB-ATP substrate pair. The initial velocity data were fit to equation 2 (ping-pong mechanism) for the OSB/CoA substrate and the ATP/CoA substrate pairs. For the OSB/ATP substrate pair, the data were fit to equation 4 (steady-state ordered mechanism, see below). The kinetic parameters were derived and are summarized in Table 1.
Binding of ATP, OSB, and CoA to OSB-CoA synthetase
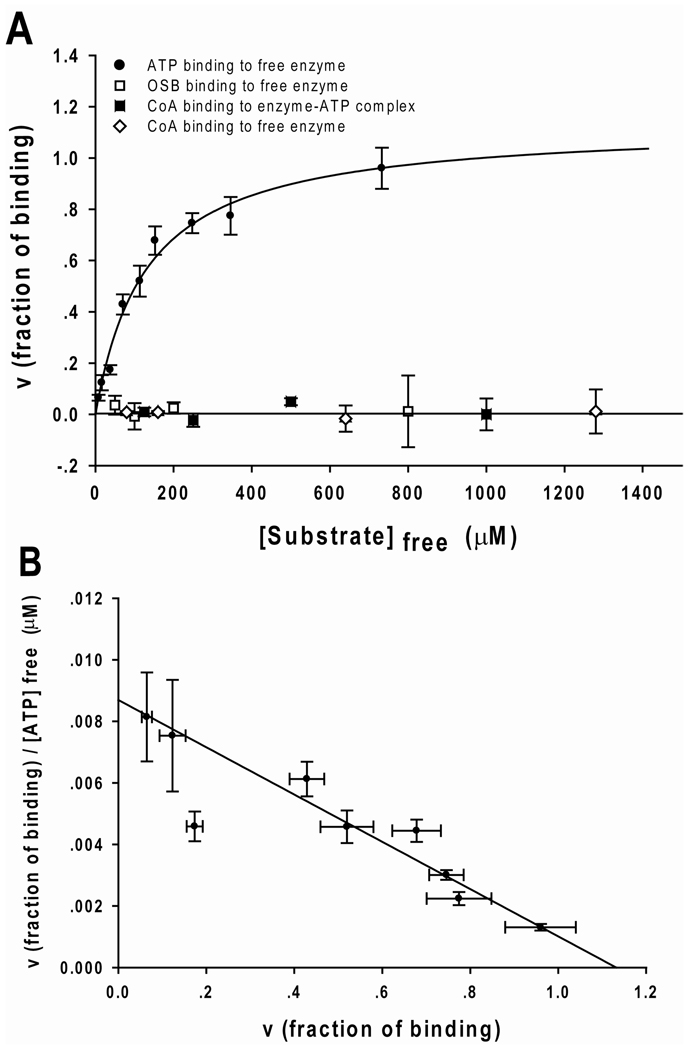
In order to differentiate between the steady-state ordered and rapid-equilibrium random mechanisms for the OSB and ATP substrate pair, the binding of each substrate to the free enzyme was independently studied by an equilibrium ultrafiltration method. No significant binding of ATP to the free enzyme in the absence of Mg2+ was observed (data not shown). However, in the presence of 2 mM Mg2+, a plot of free ATP-Mg2+ versus the fractional saturation produced a binding curve with a Kd value of 131 ± 22 µM for ATP-Mg2+ (Figure 3A). A Scatchard plot of the binding data indicates a binding ratio of ATP-Mg2+ to the free enzyme of 1:1 (Figure 3B).
Figure 3. Direct binding measurements of the interaction of ATP, OSB and CoA with B. anthracis OSB-CoA synthetase.
(A) The binding to ATP (•), OSB (□) and CoA (◇) to free enzyme and the binding of CoA (■) to enzyme-ATP complex were measured using the ultrafiltration method described in Materials and Methods. The resulting binding curve for ATP was obtained by nonlinear regression analysis of the raw data fitting to equation 7. The Kd for ATP to free enzyme was determined to be 131 ± 23 µM. No significant binding of OSB or CoA to the free enzyme or to the enzyme-ATP complex was observed. Each data point represents the average of 4 replicates and the error bars represent the standard deviation of the data at each concentration. (B) Scatchard plot transformation of the ATP-binding data shown in A. The plot intersects the x-axis between 1.0 and 1.2, indicating a binding stoichiomtery of ATP to free enzyme of 1:1.
For the substrate OSB, no significant binding to the free enzyme was observed (Figure 3A) which could result from a high Kd for OSB and therefore a lack of a defined binding site for OSB to the free enzyme. Binding of OSB to the enzyme-Mg2+ATP complex could not be studied since significant turnover during the incubation period made it difficult to measure the free OSB concentration in the filtrate. Based upon these direct binding data, an ordered binding mechanism of ATP followed by OSB to the enzyme is suggested. These results rule out a random-sequential mechanism and support an ordered-sequential mechanism for the first half-reaction. Finally, no significant binding of CoA to the free enzyme or to the enzyme-Mg2+ATP complex was observed (Figure 3A) indicating that CoA must combine with a form of the enzyme generated after the binding of Mg2+-ATP. From this point on, we will refer to Mg2+-ATP as ATP.
Product inhibition studies
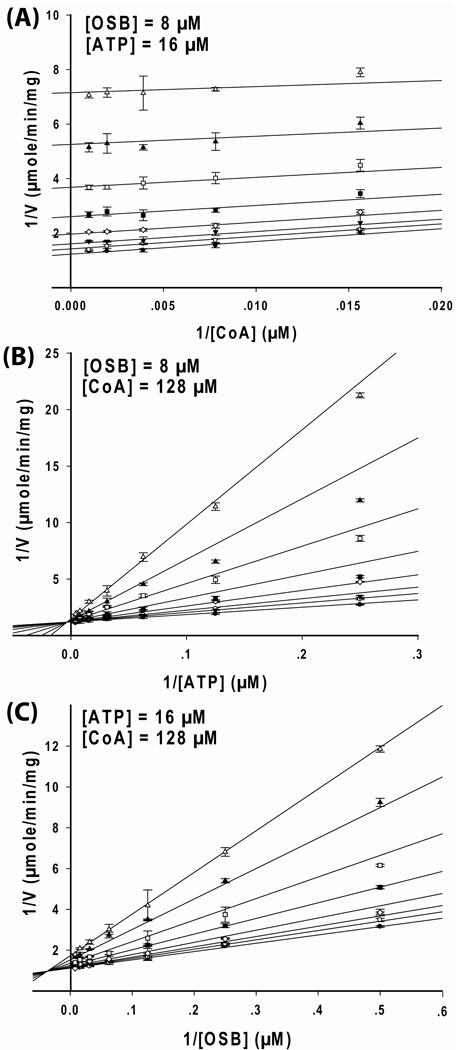
In order to differentiate between the Bi Bi Uni Uni ping-pong and the Bi Uni Uni Bi ping-pong mechanisms and to study the order of product release, product inhibition studies were performed. Nonlinear regression was performed on the experimental data using equation 8–equation 11 and the resulting goodness-of-fit values are listed in Table S3 in Supporting Information. When the ATP concentrations were varied at fixed concentrations of AMP with both OSB and CoA concentrations fixed at subsaturating levels, the data fit best to equation 8. A Lineweaver-Burk plot of the data produced a series of lines that converge on the y-axis (Figure 4A), indicating AMP is a competitive inhibitor to ATP. This result was consistent with a Bi Uni Uni Bi ping-pong mechanism and ruled out the Bi Bi Uni Uni ping-pong mechanism. As a result, OSB-CoA and AMP are the last two products to be released (the Q and R products). There were two possible mechanisms for the release of products, random or ordered. When CoA concentrations were varied at fixed concentrations of AMP with both OSB and ATP kept at subsaturating levels, the data were best fit to equation 9 and the Lineweaver-Burk plot produced a series of parallel lines (Figure 4B), indicating AMP is an uncompetitive inhibitor to CoA.
Figure 4. Product inhibition study of the B. anthracis OSB-CoA synthetase catalyzed reaction.
The kinetic data were displayed as Lineweaver-Burk plots of the reaction rate vs substrate concentrations at different concentrations of the product inhibitor. For each plot, two substrates were kept at constant level while varying the third substrate and the product inhibitor concentrations. (A) AMP is a competitive inhibitor to ATP. OSB and CoA were kept constant at 8 µM and 128 µM, respectively while varying ATP and AMP concentrations (•=0, ○=0.94, ▼=1.88, ◇=3.75, ■=7.5, □=15.0 mM). The inset is a zoom-out of the region where the lines cross at the y-axis. (B) AMP is an uncompetitive inhibitor to CoA. OSB and ATP were kept constant at 8 µM and 16 µM, respectively while varying CoA and AMP concentrations (•=0, ○=1.88, ▼=3.75, ◇=7.5, ■=15.0, □=30.0 mM). (C) Benzoyl-CoA (B-CoA) is a mixed-type inhibitor to CoA. OSB and ATP were kept constant at 8 µM and 16 µM, respectively while varying CoA and benzoyl-CoA concentrations (•=0, ○=0.94, ▼=1.88, ◇=3.75, ■=7.5, □=15.0 mM). (D) AMP is a mixed-type inhibitor to OSB. CoA and ATP were kept constant at 128 µM and 16 µM, respectively while varying OSB and AMP concentrations (•=0, ○=0.94, ▼=1.88, ◇=3.75, ■=7.5, □=15.0, ▲ =30 mM). Each data point in the figures represents the mean of a duplicate test. The upper and lower bars represent the duplicate measurements. The kinetic parameters and patterns are summarized in Table 2.
It has been reported that OSB-CoA is an unstable compound which decomposes spontaneously in solution into a spirodilactone (10). To circumvent this problem, we used an OSB-CoA analog, benzoyl-CoA, as an alternative for the product inhibition studies. When CoA concentrations were varied at fixed concentrations of benzoyl-CoA with both ATP and OSB fixed at subsaturating levels, the data were best fit to equation 10. A Lineweaver-Burk plot of the data produced a series of lines that converged to the left of y-axis (Figure 4C), indicating mixed-type inhibition. These results are consistent with an ordered mechanism whereby OSB-CoA (the Q product) is released after the addition of CoA and before the release of AMP (the R product). Finally, when OSB concentrations were varied at fixed concentrations of AMP with both ATP and CoA fixed at subsaturating levels, the data were best fit to equation 10 and a Lineweaver-Burk plot of the data produced a series of lines that converged to the left of y-axis (Figure 4D), further supporting the ordered release of OSB-CoA and AMP. The kinetic parameters and product inhibition patterns are summarized in Table 2. AMP and benzoyl-CoA have similar Ki values in the millimolar range, compared to the micromolar range of the Km values for all three substrates. Thus, the product inhibitors AMP and benzoyl-CoA bind to the enzyme much weaker than the substrates.
Table 2.
Inhibition kinetic parameters and patterns for the inhibitors of B. anthracis OSB-CoA synthetasea
| Varied substrate | Inhibitors | Fixed substrates (μM) |
Km (μM) | Ki (mM) | αKi (mM) | Inhibition pattern | ||
|---|---|---|---|---|---|---|---|---|
| OSB | ATP | CoA | ||||||
| ATP | AMP | 8 | - | 128 | 13.1 ± 1.0 | 3.6 ± 0.4 | - | competitive |
| CoA | AMP | 8 | 16 | - | 49.9 ± 4.9 | - | 10.3 ± 0.6 | uncompetitive |
| OSB | AMP | - | 16 | 128 | 2.8 ± 0.2 | 4.6 ± 0.6 | 34.5 ± 4.5 | mixed |
| CoA | Benzoyl-CoA | 8 | 16 | - | 66.2 ± 6.3 | 3.0 ± 0.5 | 15.3 ± 2.6 | mixed |
| CoA | TFMP-butyl-AMS | 8 | 16 | - | 39.3 ± 1.8 | - | 0.0089 ± 0.0005 | uncompetitive |
| ATP | TFMP-butyl-AMS | 8 | - | 128 | 5.4 ± 0.5 | 0.0052 ± 0.0008 | 0.108 ± 0.017 | mixed |
| OSB | TFMP-butyl-AMS | - | 16 | 128 | 3.6 ± 0.2 | 0.0056 ± 0.0008 | 0.025 ± 0.004 | mixed |
Data presented as mean ± standard error in the fit parameters
Pre-steady-state kinetics of B. anthracis OSB-CoA synthetase reacting with ATP and OSB in the first half-reaction
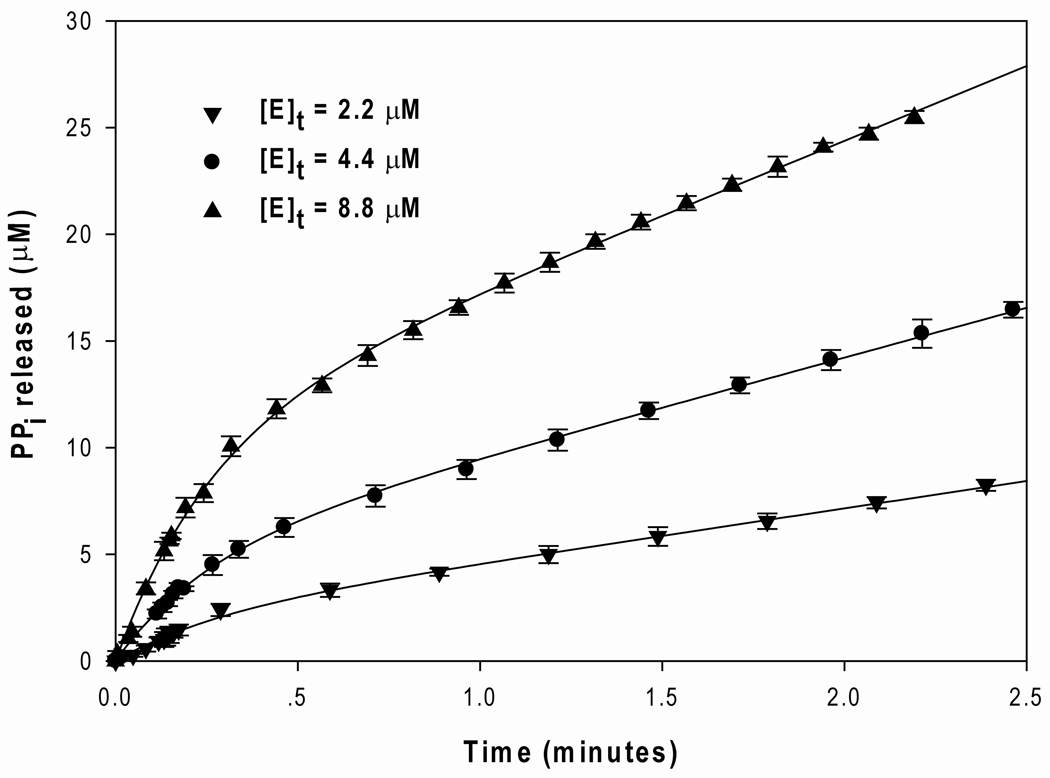
The production and release of PPi was observed for the OSB-CoA synthetase half-reaction with ATP and OSB as substrates in the absence of CoA. To measure the rate of PPi production in the absence of CoA, the phosphate detection assay was utilized to monitor the increase in concentration of PPi over time and at several high concentrations of the enzyme ([E]t = 2.2 µM, 4.4 µM, and 8.8 µM) in the presence of saturating amounts of ATP (512 µM) and OSB (256 µM). The results of these studies are presented in Figure 5. The time courses for PPi production were fit to equation 12 and the values of the pre-steady-state kinetic parameters are summarized in Table 3. A set of distinct, biphasic curves was obtained whereby a fast burst phase is followed by a slower and linear steady-state phase. The magnitude of the observed burst amplitudes for the curves in Figure 5, which are proportional to the concentration of PPi produced, closely approached the concentrations of the enzyme utilized in the experiments (Table 3). A flat line was observed for the E+ATP experiment, suggesting that no PPi release occurred without the addition of OSB. These results indicate that the burst phase corresponds to a single-turnover event of the first half-reaction. The observed burst rate constant (k1) obtained from those curves was approximately 3 times greater than the observed steady-state rate constant (k2/[E]t). The rate constants of the burst stage (∼4 min−1) and the following steady-state linear stage (∼1 min−1) determined in the absence of CoA were approximately 37 and 142 times lower than the steady-state rate constant determined in the presence of CoA (∼155 min−1). These results suggest that CoA influences the rate constants associated with the first half-reaction.
Figure 5. Pre-steady-state kinetics of B. anthracis OSB-CoA synthetase.
The time course of PPi release was plotted for the pre-steady-state of the first half-reaction (E+ATP+OSB). The reactions were initiated with a final concentration of 2.2 µM (▼), 4.4 µM (•), and 8.8 µM (▲) of the enzyme. For clarity, the continuous lines of the experimental data were represented by discrete data points. The data from the E+ATP experiment (control) was subtracted from those from the E+ATP+OSB experiment. The data point and error bar represent the average and standard deviation of a triplicate measurement. The curves display biphasic stages: a faster burst stage followed by a slower linear stage. The data were fit to equation 12, and the values for the kinetic parameters (burst amplitude, burst rate constant, and linear rate constant) are summarized in Table 3.
Table 3.
Kinetic parameters obtained from the pre-steady-state studies of B. anthracis OSB-CoA synthetase.
| Parameters | Enzyme concentrations [E]t (μM)a |
Mean ± S. D.b | ||
|---|---|---|---|---|
| 2.2 | 4.4 | 8.8 | ||
| Burst amplitude A (μM) | 2.0 ± 0.1 | 4.8 ± 0.1 | 10.3 ± 0.2 | - |
| Burst rate constants k1 (min−1) | 3.6 ± 0.5 | 3.9 ± 0.3 | 3.9 ± 0.2 | 3.9 ± 0.6 |
| Linear rate constants k2/[E]t (min−1) | 1.2 ± 0.05 | 1.0 ± 0.05 | 1.1 ± 0.2 | 1.1 ± 0.2 |
Data presented as mean ± standard error.
S. D. stands for standard deviation. The Mean and S. D. of three numbers a ± σa, b ± σb, and c ± σc were calculated as (a + b + c)/3 and square root of (σa2 + σb2 + σc2), respectively.
Inhibition kinetics of TFMP-butyl-AMS
Collectively, the kinetic studies support a Bi Uni Uni Bi ping-pong reaction mechanism for B. anthracis OSB-CoA synthetase with the formation of an OSB-AMP reaction intermediate. We therefore hypothesized that compounds that mimic the OSB-AMP intermediate may exhibit potent inhibitory effects on the enzyme reaction. To test this hypothesis, a TFMP-butyl-AMS inhibitor (Compound 6 in Figure 1) was synthesized by covalently linking compound 5 (Figure 1) and a sulfonamide adenosine to mimic the OSB-AMP. The inhibition of OSB-CoA synthetase by TFMP-butyl-AMS against each of the three substrates was then probed to determine inhibition mechanisms. Nonlinear regression was performed on the inhibition kinetic data using equation 8–equation 11 and the resulting goodness-of-fit values for each equation are summarized in Table S3 of Supporting Information.
When CoA concentrations were varied at fixed concentrations of the inhibitor with ATP and OSB concentrations fixed at subsaturating levels, the data were best fit to equation 9. The Lineweaver-Burk plot of the data produced a series of parallel lines (Figure 6A), indicating that TFMP-butyl-AMS is an uncompetitive inhibitor to CoA. When ATP concentrations were varied at fixed concentrations of the inhibitor with both CoA and OSB concentrations held at subsaturating levels, the data were best fit to equation 10 (Figure 6B) indicating that TFMP-butyl-AMS is a mixed-type inhibitor respect to ATP. When OSB concentrations were varied at fixed concentrations of the inhibitor with both ATP and CoA fixed at subsaturating levels, the data were best fit to equation 10 indicating that mixed-type inhibition with respect to OSB is also observed (Figure 6C). These kinetic patterns of inhibition are in agreement with an ordered Bi Uni Uni Bi ping-pong mechanism. The resulting kinetic constants for this study are summarized in Table 2. The inhibitor has αKi and Ki values in the lower micromolar range compared to AMP and benoyl-CoA, which only have millimolar inhibition values.
Figure 6. Inhibition study of TFMP-butyl-AMS on B. anthracis OSB-CoA synthetase activity.
The kinetic data were displayed as Lineweaver-Burk plots of the reaction rate vs substrate concentrations at different inhibitor concentrations. For each plot, two substrates were kept at constant level while varying the third substrate and the inhibitor concentrations. (A) Uncompetitive inhibition to CoA. OSB and ATP were kept constant at 8 µM and 16 µM, respectively, while varying CoA and inhibitor concentrations (•=0, ○=1.56, ▼=3.13, ◇=6.25, ■=12.5, □=25, ▲ =50, Δ=100 µM). (B) Mixed-type inhibition to ATP. OSB and CoA were kept constant at 8 µM and 128 µM, respectively, while varying ATP and inhibitor concentrations (•=0, ○=1.56, ▼=3.13, ◇=6.25, ■=12.5, □=25, ▲ =50, Δ =100 µM). (C) Mixed-type inhibition to OSB. ATP and CoA were kept constant at 16 µM and 128 µM, respectively, while varying OSB and inhibitor concentrations (•=0, ○=0.78, ▼=1.56, ◇=3.13, ■=6.25, □=12.5, ▲ =25, Δ=50 µM). Each data point in the figures represents the mean of a duplicate test. The upper and lower bars represent the duplicate measurements. The kinetic parameters and patterns are summarized in Table 2.
DISCUSSION
Studies on OSB-CoA synthetases from E. coli and Mycobacterium phli suggest that both the aromatic carboxylate group and the aliphatic carboxylate group, especially the linker length, are important for OSB substrate reactivity (9, 27). Our data indicate that the linker length of the aliphatic carboxylate group is critical for OSB substrate recognition by B. anthracis OSB-CoA synthetase. OSB-CoA synthetase was also found to exhibit moderate selectivity for the aromatic carboxylate group of OSB: a replacement of the aromatic carboxylate with a CF3 group results in a viable but less efficient substrate.
The initial velocity, product inhibition and direct binding studies for B. anthracis OSB-CoA synthetase are consistent with an ordered Bi Uni Uni Bi ping-pong mechanism with the ordered addition of the first two substrates, ATP followed by OSB, and the ordered release of the last two products, OSB-CoA followed by AMP. Based on this kinetic mechanism, the reaction contains two half-reactions where the enzyme first reacts with ATP and OSB to form PPi and OSB-AMP, and then OSB-AMP reacts with CoA to form OSB-CoA and AMP. Therefore, OSB-CoA synthetase can be classified as a member of the acyl-AMP forming family of enzymes. Although we do not have direct experimental evidence for the formation of an OSB-AMP intermediate during the reaction, indirect evidence supports the formation of the OSB-AMP intermediate. First, the Bi Uni Uni Bi ping-pong mechanism suggests the formation of an OSB-AMP intermediate. Second, the pre-steady-state kinetics of the first half-reaction suggests the presence of a rate-limiting step which is likely the slow release of a tightly bound OSB-AMP intermediate from the active site. Finally, the formation of an acyl-AMP intermediate has been characterized in several members of the acyl-AMP forming family of enzymes including acetyl-CoA synthetase (28), malonyl-CoA synthetase (14) and 4-chlorobenzoate-CoA synthetase (29).
It has been observed that acyl-AMP intermediates are capable of binding to their target enzymes between 2 to 3 orders of magnitude tighter than their carboxylate and ATP substrates (30, 31). Various non-hydrolyzable acyl-AMP analogs have been synthesized for different enzymes of this family. For instance, salicyl-AMS was found to be potential inhibitors to Mycobacterium tuberculosis enzyme MbtA and Yersinia pestis enzyme YbtE, both of which belong to the acyl-AMP-forming family of enzymes involved in the bacterial siderophore biosynthesis (19). We therefore designed and synthesized a chemically stable OSB-AMP analog, the TFMP-butyl-AMS (compound 6 in Figure 1), by covalently linking compound 5 (Figure 1) with a sulfonamide adenosine moiety.
The resulting analog is predicted to be able to occupy the ATP site via its “AMS” portion and to the OSB site via its “TFMP-butyl” portion. Since the enzyme follows an ordered mechanism for ATP and OSB binding, it is expected that TFMP-butyl-AMS can bind to the free enzyme as a competitive inhibitor to ATP and a mixed-type inhibitor to OSB. However, TFMP-butyl-AMS showed mixed-type inhibition to both ATP and OSB (Figure 6B and 6C) suggesting that it can bind to the ATP and OSB sites of the free enzyme as well as to the OSB site in the enzyme-ATP complex using its “TFMP-butyl” portion in an ordered mechanism. Our observation that the affinity of TFMP-butyl-AMS to enzyme-ATP complex (αKi = 108 ± 17.0 µM) is comparable to the affinity of compound 5 (the “TFMP-butyl” portion of the inhibitor) to the enzyme-ATP complex (Km = 168 ± 18.0 µM) further supports this model. In addition, our observation that the TFMP-butyl-AMS is an uncompetitive inhibitor to CoA (Figure 6A) is consistent with this binding model. As expected, the affinity of TFMP-butyl-AMS to the enzyme-ATP complex (αKi = 108.0 ± 17.0 µM) is much lower than to the free enzyme (Ki = 5.2 ± 0.8 µM). To the best of our knowledge, this is the first reaction intermediate analog against OSB-CoA synthetase from any organism with low, micromolar inhibition constants. Unfortunately, the TFMP-butyl-AMS inhibitor was found to have limited whole cell activity against B. anthracis ∆ANR strain presumably due to poor penetration through the cell wall (data not shown).
Several members of the acyl-CoA synthetase subfamily of enzymes utilize Bi Uni Uni Bi ping-pong mechanisms (13, 14, 32, 33). Interestingly, there are significant differences among those enzymes in terms of the order of addition of the first two substrates and the order of release of the last two products. OSB-CoA synthetase from B. anthracis shares a very similar kinetic mechanism with Bradyrhizobium japonicum malonyl-CoA synthetase (14) and bovine liver medium-chain fatty acid-CoA synthetase (32). Their reaction mechanisms all proceed with an ordered addition of the first two substrates and an ordered release of the last two products. The kinetic mechanism of pig liver bile acid-CoA synthetase, however, contains the random release of the last two products (33) while yeast acetyl-CoA synthetase follows random addition of the first two substrates and random release of the last two products (13).
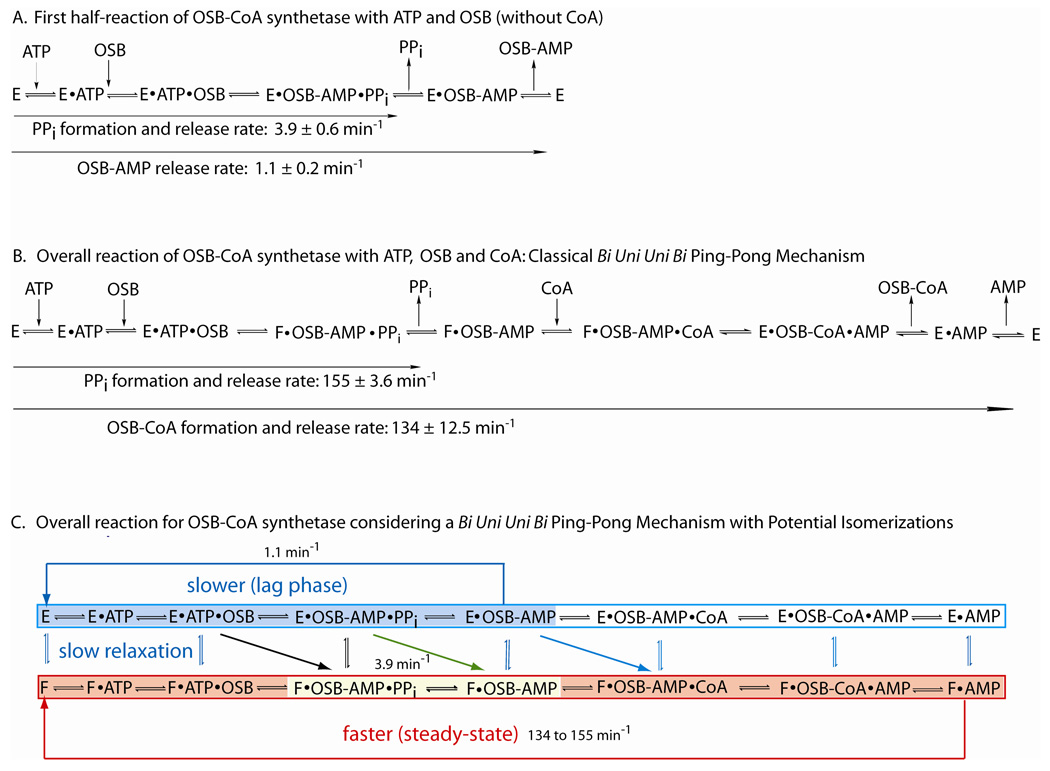
We have found that CoA is not absolutely required for the production and release of PPi and OSB-AMP in the first half-reaction. The results of the pre-steady-state kinetic studies indicate that in the absence of CoA, OSB-CoA synthetase reacts with ATP and OSB in a biphasic manner characterized by a burst phase with a rate constant about three times greater than the following linear, steady-state phase (Figure 5 and Table 3). The burst kinetics suggests that a rapid step in catalysis is followed by a slower step. Our interpretation of these data is that in the first phase of the half-reaction, ATP and OSB combine with the enzyme and react rapidly to form OSB-AMP and PPi. Since the assay we routinely utilize detects the formation of PPi, the more rapid initial step would represent the formation and release of PPi. In the second and slower linear phase of the first half-reaction, OSB-AMP is released slowly from the enzyme-OSB-AMP complex so as to regenerate the free enzyme for a second round of turnover. The difference in rate constants for these two steps in the absence of CoA is approximately 3 to 4 fold. Thus, for the first half reaction in the absence of CoA, the slow release of OSB-AMP (1 min−1) from the active site most likely represents the rate-limiting step compared to the production and release of PPi in the first turnover of (∼ 4 min−1) (Scheme 2A).
Scheme 2.
When the OSB-CoA synthetase reaction is performed in the presence of CoA, the turnover number, kcat, as determined via the phosphate detection assay, is approximately 155 min−1 which is approximately 147-fold faster than the steady-state rate for the first half-reaction (1.1 min−1) in the absence of CoA (Scheme 2A and B). This value is nearly identical to the kcat, value of 134 min−1 for the overall reaction as determined by the direct detection of the final product OSB-CoA (Scheme 2B). The increase in rate of the first half-reaction by the presence of CoA would suggest that CoA must somehow bind to the enzyme during the first half-reaction or immediately at the time of OSB-AMP and PPi formation and/or release. From a simplistic view of kinetic schemes, this would at first seem to conflict with a Bi Uni Uni Bi ping-pong mechanism, i.e. the third substrate always binds to the enzyme after the first half-reaction. In the case of OSB-CoA synthetase, this would require that OSB-AMP and PPi be formed and PPi be released before the binding of CoA. Our data strongly support the Bi Uni Uni Bi ping-pong mechanism, but the interesting observation of the rate acceleration of the first half-reaction by CoA requires further mechanistic exploration into potential explanations for this phenomenon.
Our first avenue of exploration was to examine the existing literature on enzymes that catalyze reactions similar to OSB-CoA synthetase as well as utilize Bi Uni Uni Bi ping-pong kinetic mechanisms. The enzyme pantothenate synthetase from M. tuberculosis (MtPS) catalyzes a Bi Uni Uni Bi ping-pong reaction in its transformation of ATP, pantoate and β-alanine into PPi, pantothene and AMP (15). In a series of steady-state and pre-steady-state kinetic studies on MtPS, Zheng and Blanchard demonstrated that in the absence of the third substrate, β-alanine, the rate of PPi formation and release for the first half-reaction is 1.3 sec−1. However, in the presence of β-alanine, the rate increases to 1.8 sec−1 as measured by the production of PPi, or to 3.4 sec−1 as measured by the formation of AMP (15). These results suggest that β-alanine is somehow capable of increasing the rate of the first half-reaction although it is predicted to bind only after the release of PPi. Unfortunately, further experimental studies or hypotheses to explain these observations have not yet been put forth.
The enzyme 4-chlorobenzoyl-CoA ligase (4CBA:CoA ligase) from Pseudomonas sp. Strain CBS3 catalyzes the transformation of 4-chlorbenzoate (4-CBA), ATP and CoA into PPi, 4-CBA-CoA and AMP (29). In a series of steady-state and pre-steady-state kinetic studies, Dunaway-Mariano and coworkers observe that the rate of production and release of PPi for the first half-reaction, the conversion of 4-CBA and ATP into 4-CBA-AMP and PPi, is 0.013 sec−1 in the absence of CoA. In the presence of CoA however, the rate increases about 3000-fold to 40 sec−1 (29). During the course of the reaction, the production of the intermediate, 4-CBA-AMP, was confirmed and its rate of formation measured (29). Unfortunately, it was not established in those studies whether or not 4CBA:CoA ligase utilizes a Bi Uni Uni Bi ping-pong mechanism. Subsequent experiments to do so have not yet been performed (Debra Dunaway-Mariano, personal communication).
Recent and extensive steady-state and pre-steady-state kinetic studies, in conjunction with X-ray structural studies, have been performed by the Dunaway-Mariano group on 4CBA:CoA ligase isolated from Alcaligenes sp. Strain AL3007 (34, 35). From their kinetic studies, it was rigorously established that 4CBA:CoA ligase utilizes a Bi Uni Uni Bi ping-pong kinetic mechanism and that PPi release must precede CoA binding (35). Interestingly, they found that the overall reaction is characterized by a burst-phase in the production of the product 4-CBA-CoA (60 sec−1) followed by a slower steady-state rate (10.2 sec−1). These data suggest that a step after the formation of the product 4-CBA-CoA is rate determining. To help further establish the mechanism for the rate liming step, the X-ray structures of two 4CBA:CoA ligase complexes, one with the reaction intermediate 4-CB-AMP bound, and the other with both AMP and 4-CP-CoA bound were determined (34). The structural data suggested that 4CBA:CoA ligase undergoes a significant conformational change in transforming between the 4-CB-AMP complex (designated conformer 2) to the substrate bound or 4-CB-AMP-Mg-PPi bound complex (designated conformer 1). The slow rate for isomerization of conformer 2 back to conformer 1 is proposed to be the rate limiting step in the 4CBA:CoA ligase kinetic mechanism (35). This step is indicated in by the green diagonal arrow in Scheme 2C in an analogous position of our proposed reaction scheme for OSB-CoA synthetase.
The observation of a rate limiting, conformational change in the 4CBA:CoA ligase catalyzed reaction, described as a 'domain alteration' mechanism (36), provides a compelling model that may apply to the OSB-CoA synthetase mechanism. By combining Schemes 2A and 2B, and then introducing potential conformational isomerization events between the various enzyme forms 'E' and 'F' that could occur along the reaction pathway, we propose a more general Bi Uni Uni Bi Isomerization (Iso) ping pong mechanism (Scheme 2C) that we believe applies to OSB-CoA synthetase specifically, and potentially to many other enzymes such as 4CBA:CoA ligase in general.
In the model proposed in Scheme 2C, in the absence of CoA, OSB-CoA synthetase could catalyze the first half-reaction solely with complexes involving enzyme form 'E' that are catalytically less active than complexes involving enzyme form 'F'. This path is indicated in Scheme 2A and would be considered a non-traditional ping-pong mechanism since it does not involve transformation to an 'F' form of the enzyme. It is also possible that in the absence of CoA, OSB-CoA synthetase could catalyze the first half-reaction in the classical ping-pong mechanism by converting the ternary E ·ATP·OSB complex directly to the F·OSB-AMP·PPi complex (represented by the black diagonal arrow in Scheme 2C) which would then release the product PPi forming F·OSB-AMP (yellow box in Scheme 2C). The kcat value of 3.9 min−1 would apply to this path. The F·OSB-AMP could then slowly isomerize back to an E·OSB-AMP complex which would then release OSB-AMP from E for another round of catalytic turnover at 1.1 min−1.
In the presence of CoA, once PPi is released from the F·OSB-AMP·PPi complex, CoA could rapidly combine with the F·OSB-AMP complex to then complete the formation and release of products from the 'F' enzyme complexes (indicated by the red shaded box in Scheme 2C). Once the free enzyme, F, is released at the end of this first reaction cycle, it would rapidly combine with substrates, ATP followed by OSB, to form the F·ATP·OSB complex that would then proceed to generate products. The overall rate of 134 to 155 min−1 would then apply to each round of the subsequent catalytic turnovers in steady-state. The isomerization of free F back to free E would be slow compared to the formation of the F ·ATP·OSB complex. This model would explain the kinetic results for the OSB-CoA synthetase reaction described above.
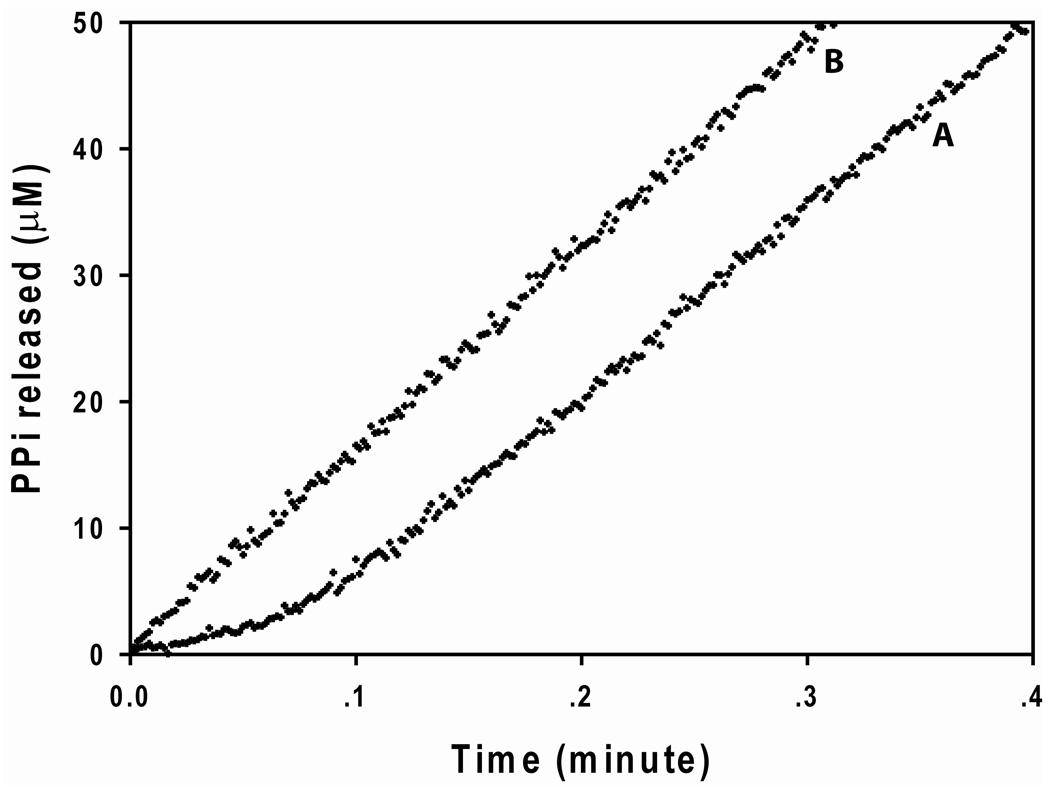
The model proposed in Scheme 2C would also then predict that a lag-phase may exist in the time course of the OSB-CoA synthetase catalyzed reaction. We therefore carefully performed a cuvette-based assay to more accurately capture the presence of any lag phase. In the presence of saturating concentrations of ATP, OSB and CoA, a lag phase is clearly evident within the first few seconds of the reaction (Figure 7) support a Bi Uni Uni Bi Iso ping pong mechanism whereby the rate of formation of the F·OSB-AMP complex from free E is slow in the initial turnovers of the enzyme, but once the enzyme complexes involving the F forms of the enzyme are produced, they are maintained during the steady-state turnover of the enzyme. Although the lag phase lends support that the F·OSB-AMP is produced, it does not rule out the possibility that CoA combines directly with the E·OSB-AMP complex to then produce the F·OSB-AMP·CoA complex (blue diagonal arrow in Scheme 2C).
Figure 7. Plot of PPi released as a function of time in the overall reaction of B. ANTHRACIS OSB-CoA synthetase.
In plot A, OSB-CoA synthetase was mixed with 256 µM OSB, 512 µM ATP and 1024 µM CoA and 2 µM MgCl2 and the release of PPi was monitored using the phosphate detection assay. The absorbance at 360 nm was converted into the amount of PPi released using a molar extinction coefficient of 11000 M−1 cm−1. A lag phase followed by a linear steady-state was observed for plot A. In plot B, OSB-CoA synthetase was first preincubated with 256 µM OSB, 512 µM ATP and 2 mM MgCl2 for 1 minute; a final concentration of 1024 µM CoA was then added and the release of PPi was monitored. A linear steady-state curve without a lag phase was observed for plot B. To ensure that the observed lag phase was not due to any inherent lag phase of the coupled assay, an excess amount of the coupling enzymes was added such that the inherent, theoretical lag phase would be at most 0.12 seconds (see Supporting Information on “Calculation of the inherent lag phase (τ) of the coupled assay in the pre-steady-state kinetic studies of E+ATP+OSB+CoA”). The lack of an observed lag phase in Figure 5 also supports that the observed lag phase is associated with the isomerization of OSB-CoA synthetase.
To test for which of the complexes (E·OSB-AMP or F·OSB-AMP) CoA combines most readily, we incubated OSB-CoA synthetase with saturating levels of ATP and OSB in the absence of CoA to attain the steady-state rate for the first half-reaction. We then initiated the second half-reaction by the addition of a saturating concentration of CoA to check for any lag phase before the steady-state phase of the overall reaction is achieved. The progress curve data in Figure 7 clearly indicate that the lag phase for the reaction has been abolished. This observation supports a mechanism whereby the F·OSB-AMP complex is generated and stabilized via the first half-reaction, and in the absence of CoA it slowly isomerizes back to the E·OSB-AMP complex releasing the intermediate. When CoA is present it readily combines with the F·OSB-AMP complex to complete the reaction cycle via the F-form enzyme complexes. Had a lag phase also been observed upon the addition of CoA, it would have suggested that the F·OSB-AMP-CoA complex is formed directly from the E·OSB-AMP (indicated by the blue diagonal arrow) but this is not the case for OSB-CoA synthetase.
In conclusion, the essential nature of menaquinone biosynthesis for the survival of bacteria, and the lack of this pathway in human, suggest that enzymes in this pathway may be potential targets for antibiotics discovery. This is the first report of the kinetic mechanism of OSB-CoA synthetase, a key enzyme for the bacteria menaquinone biosynthesis. The formation of the OSB-AMP intermediate in the OSB-CoA synthetase catalyzed reaction, as suggested by our kinetic mechanism, as well as our data on the TFMP-butyl-AMS, indicate that structural homologs of OSB-AMP can function as potential inhibitors of this enzyme. The ordered Bi Uni Uni Bi Iso ping-pong reaction mechanism proposed for the OSB-CoA synthetase reaction is a subset of a potentially more universal kinetic scheme for class of enzymes and others that catalyze similar reactions. The prediction of conformational isomerizations along the reaction pathways of enzymes that utilize Bi Uni Uni Bi ping pong mechanisms will help to provide a mechanistic framework whereby explanations can be provided to explain the different lag and burst phases associated with different steps of the reaction. The conformational isomerziation model also helps to explain how OSB-CoA synthetase accommodates two half-reactions using the same active site during the reaction. We are currently attempting to obtain the X-ray crystal structure of OSB-CoA synthetase in complex with various substrates, products and reaction intermediate mimics in order to reveal the structural mechanisms and isomerizations predicted for this two half-reaction mechanism.
Supplementary Material
Methods for the cloning, overexpression and purification of (his)6-tagged E. coli 1,4-dihydroxy-2-naphthoate-CoA (DHNA-CoA) synthetase, syntheses of Compounds 1 – 6, determination of pH optimum, and data on the effect of Mg2+ on B. anthracis OSB-CoA synthetase activity, calculation of the inherent lag phase (τ) of the coupled assay in the pre-steady-state kinetic studies of E+ATP+OSB+CoA, goodness-of-fit information of the experimental data to various equations in the bisubstrates initial velocity studies and schematics of the coupled enzyme assays utilized for the detection of B. anthracis OSB-CoA synthetase activity are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations
- OSB
o-succinylbenzoate
- OSB-CoA
O-succinylbenzoyl-coenzyme A
- TFMP-butyl-AMS
5’-O-(N-(2-trifluoromethylphenyl)-4-oxobutyl) adenosine sulfonamide
- IPP
inorganic pyrophosphatase
- PNP
purine nucleoside phosphorylase
- MESG
2-amino-6-mercapto-7-methylpurine ribonucleoside
- IPTG
isopropyl-beta-D-thiogalactopyranoside
Footnotes
This research is supported by a grant from NIH NIAID AI056575. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
REFERENCES
- 1.Bentley R, Meganathan R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol Rev. 1982;46:241–280. doi: 10.1128/mr.46.3.241-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JFML, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolonay JF, Beanan MJ, Dodson RJ, Brinkac LM, Gwinn M, DeBoy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pop M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang LX, Hance IR, Weidman JF, Berry KJ, Plaut RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Koehler TM, Hanna PC, Kolsto AB, Fraser CM. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 5.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Veldhoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Anal Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]
- 7.Bodem GB, Leete E. Biosynthesis of shihunine in Dendrobium pierardii. J. Am. Chem. Soc. 1976;98:6321. [Google Scholar]
- 8.Bryant RW, Jr, Bentley R. Menaquinone biosynthesis: conversion of o-succinylbenzoic acid to 1,4-dihydroxy-2-naphthoic acid and menaquinones by Escherichia coli extracts. Biochemistry. 1976;15:4792–4796. doi: 10.1021/bi00667a007. [DOI] [PubMed] [Google Scholar]
- 9.Kwon O, Bhattacharyya DK, Meganathan R. Menaquinone (vitamin K-2) biosynthesis: Overexpression, purification, and properties of o-succinylbenzoyl-coenzyme A synthetase from Escherichia coli. Journal of Bacteriology. 1996;178:6778–6781. doi: 10.1128/jb.178.23.6778-6781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meganathan R, Bentley R. Menaquinone (vitamin K2) biosynthesis: conversion of o-succinylbenzoic acid to 1,4-dihydroxy-2-naphthoic acid by Mycobacterium phlei enzymes. J. Bacteriol. 1979;140:92. doi: 10.1128/jb.140.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papas TS, Peterkofsky A. A random sequential mechanism for arginyl transfer ribonucleic acid synthetase of Escherichia coli. Biochemistry. 1972;11:4602–4608. doi: 10.1021/bi00774a029. [DOI] [PubMed] [Google Scholar]
- 12.Farrar WW, Plowman KM. Kinetics of acetyl-CoA synthetase-I. Mode of addition of substrates. Int J Biochem. 1975;6:537–542. doi: 10.1016/0020-711x(79)90019-3. [DOI] [PubMed] [Google Scholar]
- 13.Farrar WW, Plowman KM. Kinetics of acetyl-CoA synthetase-II. Products inhibition studies. Int J Biochem. 1979;10:583–588. doi: 10.1016/0020-711x(79)90019-3. [DOI] [PubMed] [Google Scholar]
- 14.Kim YS, Kang SW. Steady-State Kinetics of Malonyl-Coa Synthetase from Bradyrhizobium-Japonicum and Evidence for Malonyl-Amp Formation in the Reaction. Biochemical Journal. 1994;297:327–333. doi: 10.1042/bj2970327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng R, Blanchard JS. Steady-state and pre-steady-state kinetic analysis of Mycobacterium tuberculosis pantothenate synthetase. Biochemistry. 2001;40:12904–12912. doi: 10.1021/bi011522+. [DOI] [PubMed] [Google Scholar]
- 16.Cohn M, Hu A. Isotopic (18O) shift in 31P nuclear magnetic resonance applied to a study of enzyme-catalyzed phosphate–phosphate exchange and phosphate (oxygen)–water exchange reactions. Proc Natl Acad Sci U S A. 1978;75:200–203. doi: 10.1073/pnas.75.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grayson NA, Westkaemper RB. Stable analogs of acyl adenylates. Inhibition of acetyl- and acyl-CoA synthetase by adenosine 5'-alkylphosphates. Life Sci. 1988;43:437–444. doi: 10.1016/0024-3205(88)90523-1. [DOI] [PubMed] [Google Scholar]
- 19.Ferreras JA, Ryu J-S, Lello FD, Tan DS, Quadri LEN. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nature Chemical Biology. 2005;1:29. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 20.Truglio JJ, Theis K, Feng Y, Gajda R, Machutta C, Tonge PJ, Kisker C. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J Biol Chem. 2003;278:42352–42360. doi: 10.1074/jbc.M307399200. [DOI] [PubMed] [Google Scholar]
- 21.Akaike H. A new look at the statistical model identification. Automatic Control, IEEE Transactions on. 1974;19:716–723. [Google Scholar]
- 22.Menguy T, Chenevois S, Guillain F, le Maire M, Falson P, Champeil P. Ligand binding to macromolecules or micelles: use of centrifugal ultrafiltration to measure low-affinity binding. Anal Biochem. 1998;264:141–148. doi: 10.1006/abio.1998.2854. [DOI] [PubMed] [Google Scholar]
- 23.Wolfer GK, Jr, Rippon WB. Protocols for use of ultrafiltration in determination of free ligand concentration and of complexity of ligand/protein interactions. Clin Chem. 1987;33:115–117. [PubMed] [Google Scholar]
- 24.Grant BD, Adams JA. Pre-steady-state kinetic analysis of cAMP-dependent protein kinase using rapid quench flow techniques. Biochemistry. 1996;35:2022–2029. doi: 10.1021/bi952144+. [DOI] [PubMed] [Google Scholar]
- 25.Northrop DB. On the meaning of Km and V/K in enzyme kinetics. J. Chem. Edu. 1998;75:1153–1157. [Google Scholar]
- 26.Northrop DB. So what exactly is V/K, anyway? In: Frey PA, Northrop DB, editors. Biomedical and Health Research. 1999. pp. 250–263. [Google Scholar]
- 27.Sieweke HJ, Leistner E. o-Succinylbenzoate: coenzyme A ligase, an enzyme involved in menaquinone (vitamin K2) biosynthesis, displays broad specificity. Z Naturforsch [C] 1991;46:585–590. doi: 10.1515/znc-1991-7-814. [DOI] [PubMed] [Google Scholar]
- 28.Midelfort CF, Sarton-Miller I. The stereochemical course of acetate activation by yeast acetyl-CoA synthetase. J Biol Chem. 1978;253:7127–7129. [PubMed] [Google Scholar]
- 29.Chang KH, DunawayMariano D. Determination of the chemical pathway for 4-chlorobenzoate: Coenzyme a ligase catalysis. Biochemistry. 1996;35:13478–13484. doi: 10.1021/bi961284w. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Lee SW, Choi EC, Choi SY. Aminoacyl-tRNA synthetases and their inhibitors as a novel family of antibiotics. Appl Microbiol Biotechnol. 2003;61:278–288. doi: 10.1007/s00253-003-1243-5. [DOI] [PubMed] [Google Scholar]
- 31.Cassio D, Lemoine F, Waller JP, Sandrin E, Boissonnas RA. Selective inhibition of aminoacyl ribonucleic acid synthetases by aminoalkyl adenylates. Biochemistry. 1967;6:827–836. doi: 10.1021/bi00855a024. [DOI] [PubMed] [Google Scholar]
- 32.Vessey DA, Kelley M. Characterization of the reaction mechanism for the XL-I form of bovine liver xenobiotic/medium-chain fatty acid:CoA ligase. Biochem J. 2001;357:283–288. doi: 10.1042/0264-6021:3570283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley M, Vessey DA. Determination of the mechanism of reaction for bile acid: CoA ligase. Biochem J. 1994;304(Pt 3):945–949. doi: 10.1042/bj3040945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reger AS, Wu R, Dunaway-Mariano D, Gulick AM. Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase. Biochemistry. 2008;47:8016–8025. doi: 10.1021/bi800696y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R, Cao J, Lu X, Reger AS, Gulick AM, Dunaway-Mariano D. Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis. Biochemistry. 2008;47:8026–8039. doi: 10.1021/bi800698m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bandarian V, Pattridge KA, Lennon BW, Huddler DP, Matthews RG, Ludwig ML. Domain alternation switches B(12)-dependent methionine synthase to the activation conformation. Nat Struct Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods for the cloning, overexpression and purification of (his)6-tagged E. coli 1,4-dihydroxy-2-naphthoate-CoA (DHNA-CoA) synthetase, syntheses of Compounds 1 – 6, determination of pH optimum, and data on the effect of Mg2+ on B. anthracis OSB-CoA synthetase activity, calculation of the inherent lag phase (τ) of the coupled assay in the pre-steady-state kinetic studies of E+ATP+OSB+CoA, goodness-of-fit information of the experimental data to various equations in the bisubstrates initial velocity studies and schematics of the coupled enzyme assays utilized for the detection of B. anthracis OSB-CoA synthetase activity are provided. This material is available free of charge via the Internet at http://pubs.acs.org.