Abstract
Glucose-regulated protein 94 (GRP94) is an endoplasmic reticulum (ER) chaperone for which only few client proteins and no cofactors are known and whose mode of action is unclear. To decipher the mode of GRP94 action in vivo, we exploited our finding that GRP94 is necessary for the production of insulin-like growth factor (IGF)-II and developed a cell-based functional assay. Grp94−/− cells are hypersensitive to serum withdrawal and die. This phenotype can be complemented either with exogenous IGF-II or by expression of functional GRP94. Fusion proteins of GRP94 with monomeric GFP (mGFP) or mCherry also rescue the viability of transiently transfected, GRP94-deficient cells, demonstrating that the fusion proteins are functional. Because these constructs enable direct visualization of chaperone-expressing cells, we used this survival assay to assess the activities of GRP94 mutants that are defective in specific biochemical functions in vitro. Mutations that abolish binding of adenosine nucleotides cannot support growth in serum-free medium. Similarly, mutations of residues needed for ATP hydrolysis also render GRP94 partially or completely nonfunctional. In contrast, an N-terminal domain mutant that cannot bind peptides still supports cell survival. Thus the peptide binding activity in vitro can be uncoupled from the chaperone activity toward IGF in vivo. This mutational analysis suggests that the ATPase activity of GRP94 is essential for chaperone activity in vivo and that the essential protein-binding domain of GRP94 is distinct from the N-terminal domain.
Keywords: endoplasmic reticulum, peptide hormones, protein folding
The endoplasmic reticulum (ER) chaperone glucose-regulated protein 94 (GRP94) is structurally homologous to other members of the heat shock proteins 90 (HSP90) family that reside in the cytosol or in mitochondria. GRP94 contains four domains: the N-terminal (N) domain contains a nucleotide binding site (1, 2), a peptide binding site (3, 4), and unmapped sites responsible for interaction with dendritic cells during antigen presentation (5). The second domain, as in HSP90, is a charged linker between the N and middle (M) domains, but it is longer in all GRP94s and functionally important for binding of both nucleotide and calcium (6, 7). The middle domain interacts with the N-terminal domain (28) and the C-terminal domain mediates the dimerization of GRP94 (8), and ER localization via the C-terminal KDEL peptide.
In contrast to most other ER chaperones, GRP94's function has long remained enigmatic. First, despite being one of the most abundant ER chaperones (9), only a handful of proteins have been demonstrated to be clients of GRP94, including immunoglobulins (10), some integrins (11), some Toll-like receptors (12), plant CLAVATA proteins (13) and, recently, insulin-like growth factor II (IGF-II) (14). This is a surprisingly small number of clients relative to other general chaperones and considering the high expression levels of GRP94. Furthermore, the known clients bear no obvious structural similarities to each other, highlighting the unknown nature of GRP94's recognition mechanism. Second, the N-terminal nucleotide binding site of GRP94 is targeted by specific inhibitors, geldanamycin and radicicol that also bind to the homologous site in HSP90, and by a nucleotide analog, NECA, whose binding is specific for GRP94 (15, 16). However, whereas HSP90 function is intimately linked to ATP binding and hydrolysis, as shown by yeast growth assays (17, 18) and by in vitro tests (19–21), the role of ATP hydrolysis in the action cycle of GRP94 has so far not been explored. GRP94 clearly binds adenosine nucleotides in vitro (22), but the low ATPase activity reported by Li and Srivastava (23) has been controversial (6, 15, 24). Only recently has the ATPase activity of purified GRP94 been reliably measured (25, 26). These studies demonstrate that the conservation of amino acids involved in ATP binding and hydrolysis between HSP90 and GRP94 is the result of a similar, but not identical mechanism of nucleotide utilization in vitro.
The third enigmatic aspect of GRP94 is the absence of reported cochaperones, which contrasts with the many proteins that modulate the activities of the cytosolic Hsp90 chaperones. For example, proteins from the p23 family facilitate the release of peptides from Hsp90 (27–29) and another cochaperone, Aha-1, stimulates Hsp90 ATPase activity (27). No ER homologs have been identified for GRP94. The C-terminal peptide of HSP90, EEVD, which binds tetratricopeptide repeat proteins (30, 31), is distinct from the C terminus of GRP94, KDEL, which serves as an ER localization signal. The lack of cochaperones raises questions regarding the mechanism of regulating binding and release of client proteins from GRP94 in vivo.
Fourth, GRP94 is absent from most unicellular organisms (except the parasite Leishmania) and thus does not exist in the genetically amenable yeast that has been so instructive in deciphering the mode of action of Hsp90. It is no surprise, therefore, that the functional significance and the mode of action of GRP94 have been difficult to elucidate. Even the RNAi approach used in Caenorhabditis elegans (WormBase, release WS179) has not been incisive so far, because although knockdown of GRP94 expression causes larval arrest and showed that GRP94 is essential, this system is not readily amenable to scoring the phenotype of site-directed mutants.
To better understand how GRP94 functions, we devised a cellular assay that measures physiologically significant structure−function relationships, akin to the use of yeast for assessing mutants in HSP90. This cell-based assay reports on active GRP94 and measures the relative activity of mutants. We use this assay to show that ATP binding and hydrolysis are essential for the function of GRP94 in the cell, while the peptide-binding site in the N-terminal domain of GRP94 that is central to the immunological activity is not essential for the chaperone function measured in vivo.
Results and Discussion
We previously discovered that grp94−/− embryonic stem (ES) cells are hypersensitive to serum withdrawal and growth factor removal, and undergo rapid apoptosis (14, 32). Unlike GRP94-containing cells (WT), grp94−/− (KO) ES cells cannot respond to the stress by producing the growth factor IGF-II (14). The requirement for GRP94 is explained by a transient interaction of the chaperone with biosynthetic intermediate(s) of pro-IGF-II, without which the mature hormone is not produced and the intermediate is targeted to ER-associated degradation (14). The difference between grp94+/+ and grp94−/− cells was used to devise a 96-well plate assay that measures cell viability after the shift to low serum medium (Fig. 1A). The half-life of grp94−/− cells under these conditions was <12 h, whereas wild-type cells showed only marginal reduction in viability.
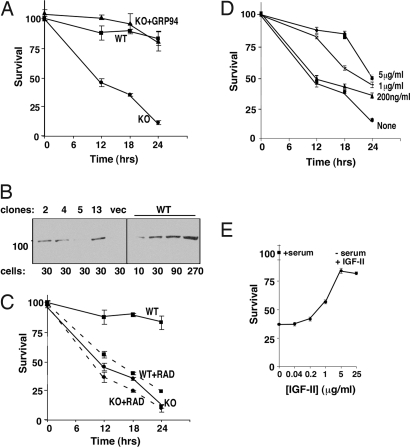
Fig. 1.
The survival of grp94−/− cells in serum-free medium is dependent on IGF-II. (A) Grp94−/− cells are hypersensitive to serum withdrawal because they lack active GRP94. Confluent cells in triplicate wells of a 96-well plate were washed and shifted to growth in ES cell medium lacking FCS (t = 0). At the indicated times the viability of the cells was assessed by the XTT assay. KO, survival curve of grp94−/− cells. WT, survival curve of grp94-sufficient cells. KO + GRP94, survival curve of grp94−/− cells that stably express wild-type GRP94 (clone 2). (B) Expression level of GRP94 in stably transfected grp94−/− clones. Shown are total cell lysates of clones 2, 4, 5, 13, expressing wild-type GRP94 or grp94−/− clones transfected with empty vector (vec), or WT cells. The lysates were separated by SDS/PAGE and analyzed by Western blotting with the anti-GRP94 monoclonal antibody 9G10 (all lanes are from the same gel, spliced together). The numbers below the gel indicate a number of cell equivalents in thousands, loaded in each lane. (C) Treatment with radicicol leads to similar sensitivity to serum withdrawal as ablation of the grp94 gene. Normal (WT) and grp94−/− cells (KO) were grown as in A, shifted to serum-free medium with or without 5 μM radicicol (RAD), and their survival quantified as above. (D) Survival of grp94−/− cells in serum-free medium in the presence of various concentrations of recombinant IGF-II, added at t = 0. (E) The survival of grp94−/− cells in serum-free medium supplemented with each concentration of IGF-II at the 18-h time point is plotted as a function of the concentration of IGF-II.
When wild-type cells were treated with 5 μM radicicol (Fig. 1C) or 17AAG (data not shown), the 2 pan-HSP90 inhibitors, cell viability was reduced to the level of grp94−/− cells. Treatment of grp94−/− cells with these inhibitors did not accelerate their apoptosis significantly. These observations suggest that GRP94 is targeted by these inhibitors in vivo.
Stable cell lines derived from the grp94−/− cells, in which GRP94 was reexpressed at near endogenous levels (Fig. 1B), showed survival similar to grp94+/+ cells (Fig. 1A). They also displayed similar sensitivity to radicicol or 17AAG (data not shown). We conclude that the hypersensitivity of knockout cells to serum withdrawal results directly from the absence of functional GRP94. Previously (14), we identified a growth factor whose secretion requires the activity of GRP94: insulin-like growth factor II. Indeed, supplying exogenous IGF-II rescued knockout cell survival in serum-free medium in dose-dependent fashion (Fig. 1 D and E). This result verified that the underlying cause for the hypersensitivity of knockout cells is inability to secrete IGF-II in the absence of GRP94. Exogenous IGF-I was just as effective as IGF-II (14), consistent with the shared receptor and signaling pathway for these 2 growth factors.
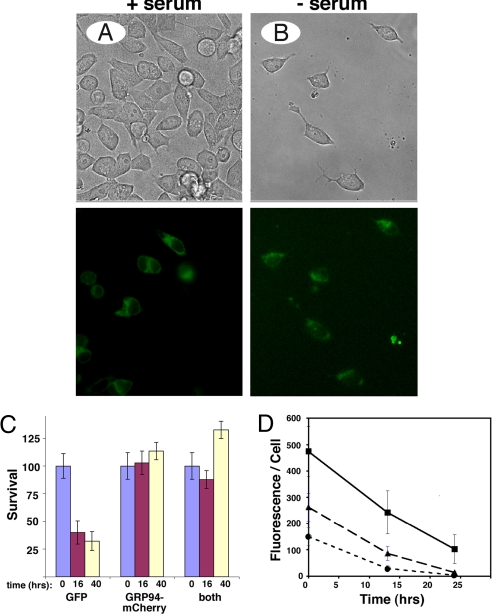
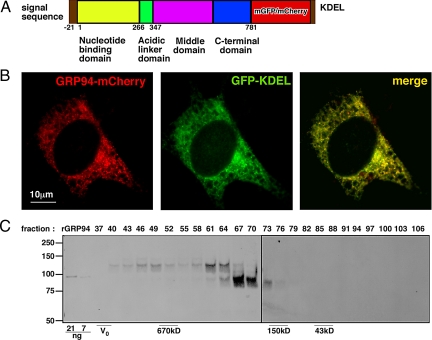
To develop complementation of cell viability as a test of the functionality of various GRP94 mutants, we used transiently transfected cells. However, the mediocre transfection efficiency of grp94−/− mouse embryonic fibroblasts (MEFs) (10–30%, see Fig. 3) meant that a majority of cells in each culture died when serum was removed, complicating accurate assessment of complementation. To circumvent this difficulty, we engineered fluorescent reporters for transfected grp94−/− MEFs. Monomeric GFP (mGFP) or mCherry were fused in frame after codon 778 of canine GRP94 cDNA and the ER retrieval tetrapeptide KDEL was added to the C terminus of the fusion protein, for correct subcellular localization (Fig. 2A). When expressed in grp94−/− cells, GRP94-mCherry displayed the expected reticular distribution, including both the nuclear envelope and the peripheral ER (Fig. 2B) and colocalized with the ER marker, ER-GFP. As seen in Fig. 2B, there was a very high degree of colocalization between the two (yellow color). To further characterize the fusion protein, we expressed GRP94-mCherry stably in C2C12 cells and determined the level of expression and dimerization status by size exclusion chromatography followed by gel electrophoresis and immunoblotting. C2C12 was chosen as a host that expresses endogenous GRP94 for comparison, because stable MEF lines expressing GRP94-mCherry could not be obtained. As shown in Fig. 2C, the endogenous GRP94 migrated as a dimer, as expected (8). The major peak of GRP94-mCherry migrated as a slightly larger dimer, consistent with its being approximately 60 kDa larger than the endogenous protein. No monomeric species was observed for either the endogenous GRP94 or the fusion protein. The ability of the GRP94 fusion proteins to heterodimerize with endogenous GRP94 is currently under investigation. In addition, a minor peak of the fusion protein migrated as a species 700 kDa or larger (although clearly included in the column), possibly representing microaggregates (Fig. 2C). The total level of expression of GRP94-mCherry was one-sixth of that of the endogenous GRP94. We conclude that the GRP94-mCherry and, presumably also GRP94-mGFP, are mostly dimeric, like the endogenous protein, and are properly expressed in the cell. The expression of the GRP94-mGFP fusion protein allowed direct visualization of transiently expressing cells and thus obviated the difficulty of scoring viability of a mixed cell population. In cultures transfected with GRP94-mGFP and then shifted to serum-free medium, there was preferential survival of fluorescent cells as the nontransfected cells died, and after 48 h, all of the surviving cells expressed GRP94-mGFP (Fig. 3 A and B). The preferential survival of GRP94-expressing cells was also demonstrated by coexpression of cytosolic GFP alongside GRP94-mCherry. As shown in Fig. 3C, survival of red cells was much better than that of green (GFP-transfected) cells. When doubly transfected cells were scored, their survival matched that of GRP94-mCherry-expressing cells and not that of cells expressing cytosolic GFP (Fig. 3C). We conclude that GRP94-mGFP and GRP94-mCherry are each functional and complement the inability of grp94−/− cells to respond to serum deprivation. Addition of exogenous IGF-II rescued the survival of transfected cells in the absence of serum (Fig. 1 D and E), suggesting that the underlying cause for the cell death is inability to secrete IGF-II in the absence of GRP94 (see also Table 1).
Fig. 3.
Rescue of cell viability after withdrawal of growth factors. (A and B) Cells transfected with GRP94-mGFP shown before (A, Left panels) and 40 h after serum withdrawal (B, Right panels). Bright field and fluorescence images of the same fields, showing transfected and untransfected cells under both conditions. Magnification 200×. (C) Quantitation of survival of cells cotransfected with cytosolic GFP and GRP94-mCherry. Cells expressing GFP alone, or GRP94-mCherry alone, or both constructs were scored at the indicated times after the shift to serum-free medium, and their percentage of viability is plotted. (D) Quantitation of total fluorescence per transfected cell. Grp94−/− cells expressing WT GRP94-mGFP were incubated in serum-free medium and photographed at the indicated times. For quantitation, cells were binned into brightly fluorescent at t = 0 (squares), medium fluorescent (triangles), and dull fluorescent cells (circles). Total pixel intensity per cells was calculated with the SlideBook software and is presented as means ± SD of 10–21 cells per time point.
Fig. 2.
Proper expression of GRP94-fluorescent fusion proteins. (A) Schematic representation of the fusion proteins. Yellow, the N-terminal domain of GRP94; green, the charged linker; purple, the M domain; blue, the C-terminal domain; red, monomeric green fluorescence protein (mGFP) or monomeric cherry fluorescence protein (mCherry); brown, the N-terminal leader sequence and the C-terminal KDEL ER targeting signal. The amino acids boundaries of the domains are indicated by the numbers beneath the scheme, as are the putative functional attributes of the domains. (B) Fluorescence image of a grp94−/− cell coexpressing GRP94-mCherry together with the ER marker GFP-KDEL. Note the reticular staining, characteristic of the ER. (C) Detergent lysate of a C2C12 clone expressing GRP94-mCherry (3.4 mg total protein) was fractionated on a Sephacryl S-300 size exclusion column. Every third fraction was analyzed by reducing SDS/PAGE (fraction numbers are indicated above each lane) and immunoblotted with monoclonal anti-GRP94 antibody to detect the endogenous and fusion proteins simultaneously. Recombinant GRP94 was loaded on lanes 1 and 2 (21 and 7 ng, respectively) to serve as loading and size standards. The excluded volume, V0, was measured with blue dextran and the size markers were thyroglobulin (670 kDa), IgG (150 kDa) and ovalbumin (43 kDa). Their migration is indicated beneath the gel.
Table 1.
GRP94-dependent IGF-II secretion
| Clone name | IGF-II (ng/mL) |
|---|---|
| 42.1 | 1.7 ± 0.2 |
| 14.1 | <0.25 |
| 14.1.2 | 1.5 ± 0.1 |
| 14.1.13 | 1.3 ± 0.3 |
| ΔK clone 19 | <0.25 |
| ΔK clone 23 | <0.25 |
ELISA measurements of IGF-II in supernatants of cells, 24 h after serum withdrawal. Wild-type MEF (42.1) and MEF deficient in GRP94 (14.1) that had been stably transfected with wild-type GRP94 (14.1.2 and 14.1.13, see also Fig. 1B) secreted the indicated levels of IGF-II in the absence of serum. Transfectants expressing the ΔK construct (clones 19 and 23) failed to secrete IGF-II in the absence of serum. The detection limit of this assay is 0.25 ng/mL. Means + SD are shown for 2–4 independent experiments for each clone.
The main reason that even fluorescent-GRP94-containing cells die during this assay is decreased expression of the fusion protein with time. The average intensity of fluorescent cells 24 h after the shift to serum-free medium was only 31% of the average fluorescence immediately after the shift (which was typically done 18–20 h after initiating the transfection) (Fig. 3D). Placing the GRP94 gene under the control of the E1α promoter, which is more efficient in MEF than the CMV promoter, gave rise to higher and more persistent expression of the chimeric proteins, but only improved the survival of wild-type cells by about 12 h (data not shown).
We used the ability of GRP94-fluorescent proteins to improve the survival of serum-deprived grp94−/− cells to perform structure–function analysis, by testing the ability of various GRP94 mutants to rescue viability. To construct a nonfunctional chaperone as a negative control, we deleted amino acids 144–488 in frame (ΔK-mGFP). This construct removes part of the N-terminal domain, all of the charged linker domain, and much of the middle domain and therefore was expected to ablate the chaperone function of GRP94. As shown in Fig. 4A, the ΔK mutant indeed could not rescue cell viability. Under all conditions tested and in all experiments (n > 20), the viability of ΔK-mGFP-expressing cells was no better than nontransfected cells and was 2- to 3-fold worse than the viability of cells expressing wild-type GRP94-mGFP (Fig. 4A and Table 2). When the secretion of IGF-II was measured directly, MEF cells expressing either endogenous or transfected GRP94 responded to serum deprivation by secreting the hormone. On the other hand, grp94−/− cells stably expressing the ΔK-mGFP protein secreted undetectable levels of IGF-II (Table 1). Furthermore, when exogenous IGF-II was added to the cells in serum-free medium, survival of cells expressing both wild-type GRP94-mGFP and ΔK-mGFP was improved in dose-dependent fashion, and the difference in their survival was abolished, confirming again that the main underlying mechanism is the ability of GRP94 to support endogenous IGF production (data not shown).
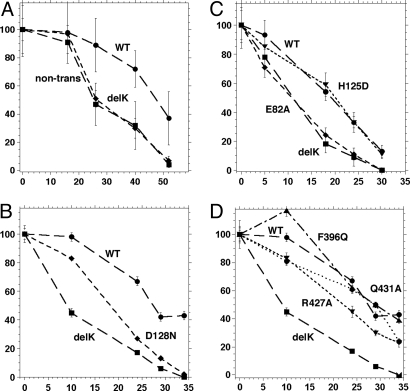
Fig. 4.
Functionality of GRP94 mutants. (A) GRP94delK-mGFP is an inactive mutant of GRP94. Grp94−/− cells were transfected with wild-type fusion construct or with the delK fusion construct, containing an in-frame deletion of amino acids 144–488. The cells were shifted to serum-free growth medium 18–24 h later and viability was scored at the indicated times thereafter. Viability of nontransfected cells at the same time points was scored in parallel. Viable cells per field were counted, averaged over 8–15 fields per time point, and presented as percentage of the viable cells at the time of serum shift. Circles, survival curve of WT GRP94-mGFP expressing cells; squares, survival curve of GRP94delK-mGFP expressing cells; diamonds, survival of nontransfected cells. (B) Activity of ATP binding mutant. Survival curve of the D128N mutant (diamonds), which is unable to bind nucleotide, radicicol, or geldanamycin, is compared to the survival of wild type (WT, circles) and delK mutant (squares). A representative experiment out of 3. (C) Activities of nonhydrolyzing and of peptide nonbinder mutants. Survival curves of E82A (diamonds), which cannot hydrolyze ATP, and of H125D (triangles), which does not bind peptides in vitro, are compared to those of wild type and delK, as indicated. Shown is a representative experiment out of 2 for E82A and 5 for H125D. (D) Activity of M domain mutants. R427A (downward triangles) and Q431A (diamonds) are substitutions in the catalytic loop and F396Q (upward triangles) is a mutant in the hydrophobic patch in the M domain (see text). Wild type (circles) and delK (squares) are the positive and negative controls, respectively. The experiment shown is representative of 3–5 for the various mutants.
Table 2.
Survival index of GRP94 point mutants
| Mutant | Survival index | N |
|---|---|---|
| WT | 1.00 | 9 |
| ΔK | 0.33 ± 0.03 | 9 |
| D128N | 0.51 ± 0.12 | 3 |
| D128NG132A | 0.40 ± 0.07 | 2 |
| E82A | 0.43 ± 0.03 | 2 |
| F396Q | 0.74 ± 0.24 | 3 |
| R427A | 0.68 ± 0.19 | 4 |
| Q431A | 0.64 ± 0.19 | 5 |
| H125D | 0.88 ± 0.16 | 4 |
The survival index was calculated at the time point within each experiment where survival of the ΔK transfected cells was one-third the value of the WT-transfected cells. This normalized the activity of each GRP94 mutant with reference to these 2 internal controls and accounted for the time axis variability of survival curves from experiment to experiment.
One of the major mechanistic questions about GRP94 is the role of ATP in its action cycle. To address this question, we initially used the mutant D128N-G132A, containing 2 substitutions in the nucleotide-binding pocket of GRP94 (16). When tested as a recombinant protein in vitro, D128N-G132A fails to bind ATP or the nucleotide site inhibitors radicicol and geldanamycin, but is still active in peptide binding (4). When tested as an mGFP fusion protein, D128N-G132A was inefficient in supporting cell survival, although it was still marginally better than the global mutant ΔK (Table 2). To confirm this effect we also prepared the single point mutant D128N and showed that it also was inefficient in rescuing grp94−/− cells from serum deprivation, even if at early time points (up to 18 h) D128N consistently showed intermediate survival between ΔK and WT GRP94 (Fig. 4B; Table 2). We conclude from the comparison of activities in vitro and in vivo that the nucleotide/inhibitor-binding site of GRP94 is important for the chaperone activity assayed here in cells. Residue D128 is homologous to D79, a conserved residue in all cytoplasmic Hsp90s. D79 is essential for nucleotide binding by Hsp90, and the D79N Hsp90 cannot support yeast growth (17). Thus, our data show a conserved need for nucleotide binding in vivo across all Hsp90 family members.
We next investigated the role of ATP hydrolysis in the activity of GRP94. Structure analysis methods, including x-ray crystallography, mass spectrometry, and optical techniques show that the ATP- and ADP-bound forms of GRP94 have equivalent conformations (15, 33, 34). Yet, most attempts to measure significant ATP hydrolysis by purified GRP94 have failed (22, 24). Nonetheless, all of the amino acids that are involved in ATP hydrolysis in other members of the family are conserved precisely in all GRP94s from lower and higher eukaryotes, including plants. Therefore, we systematically tested the importance of specific conserved amino acids for GRP94 function. First, we investigated the importance of Glu-82, the homolog of Glu-33 in HSP90 that contacts the γ phosphate of ATP and is essential for ATP hydrolysis by HSP90 in vitro and in yeast (17, 18, 29, 35, 36). Recently, this conserved amino acid was shown to be essential for the ATPase activity measured for recombinant GRP94 (25). When Glu-82 was mutated to alanine and tested in the IGF-dependent cell survival assay, E82A was incapable of supporting cell survival during serum withdrawal (Fig. 4C), similar to the ATP-nonbinding mutants. These data strongly suggest that ATP hydrolysis is important for GRP94 function in cells.
Proteins of the GHKL family have an unusual split ATPase structure, where residues that are needed for nucleotide hydrolysis reside in both the N and M domains and contact the bound nucleotide across the N/M domains interface. In the middle domain of Hsp90, residues R380 and Q384 are essential for ATP hydrolysis in vitro because they are involved in positioning the γ phosphate for hydrolysis (36). When tested in yeast cells, these Hsp90 mutants fail to support growth (36), consistent with the importance of ATP hydrolysis. We assessed the roles of the homologous amino acids, R427 and Q431, in the M domain of GRP94. When alanine mutants of each of these residues were tested in grp94−/− cells, each displayed an intermediate activity between the wild-type GRP94-mGFP and ΔK-mGFP (Fig. 4D and Table 2). The partial rescue phenotype was observed, even when the double mutant R427A-Q431A was tested (data not shown). Thus, these catalytic loop residues are not essential in vivo and reflect the ATPase activity in vitro (25). We posit the existence of a cellular factor(s), which partly compensates for R427A, but not for E82A. We were unable to determine whether the partial rescue of viability correlated with intermediate levels of IGF-II secretion, because the transient expression conditions did not yield sufficient levels of IGF-II for robust measurements.
These in vivo results are best understood in light of the structural data (25) and enzymatic analysis of GRP94 (26). Both studies demonstrated that recombinant GRP94 does, in fact, possess ATPase activity, no slower than that of human HSP90β, and that this activity is inhibited by radicicol. Dollins et al. (25) abolished the ATPase activity by substituting Glu-82 with Ala. In addition, R427A retained only 15% of the ATPase activity in vitro even though R427 is not adjacent to the bound nucleotide in the crystal structure. Using the same mutants in cells, we observed that ATP hydrolysis is important for the chaperone activity of GRP94. We hypothesize that even partial ATPase activity is sufficient in vivo, perhaps because the slow hydrolytic activity is augmented in cells by interaction with factors that enhance and regulate it, analogous to Aha-1, which enhances the ATPase activity of HSP90 200-fold (27, 37). Such a hypothetical factor would act to rotate the catalytic loop and better position it, leading to a higher rate of hydrolysis. A prediction of this hypothesis is that under conditions of diminished ATP supply to the ER, the chaperone activity of GRP94 may be inhibited and therefore the production of IGF under such conditions should be reduced.
Adjacent to the catalytic loop of Hsp90 there is a small cluster of hydrophobic residues centered on Val-348-Phe-349 (36). The side chains of these residues are postulated to contact residues in the N-terminal domain that are exposed upon nucleotide binding and position the catalytic loop residues near the critical glutamate residue (E82 in GRP94) (36). The F349Q substitution in HSP90 lowers ATP hydrolysis in vitro and results in a growth defect in vivo (36). We tested the analogous mutation in GRP94, F396Q. Surprisingly, this mutant exhibited only a mild phenotype and was able to support cell survival reasonably well (Fig. 4D and Table 2). In light of the different phenotypes for the analogous HSP90 mutants when tested in yeast, we hypothesize the equivalent catalytic loop and hydrophobic patch mutations in the M domain still allow partial nucleotide hydrolysis in GRP94 in vivo.
A different in vivo activity long associated with GRP94 is its ability to boost T cell responses, by providing a route for presentation of peptides to the T cell receptor (38). We showed previously that the portion of GRP94 relevant for this activity is the N-terminal and charged linker domains that comprise a peptide-binding domain (5). The β sheet of the N-terminal domain is a peptide-binding site and His-125, within this sheet, is essential for peptide binding in vitro (3, 4). His-125 contacts the bound peptide and substitution with aspartate abolished peptide binding (4). Peptide binding to this N-terminal site is also inhibited when the nucleotide pocket at the other lobe of the N-terminal domain is occupied by radicicol or geldanamycin (8). Because the peptide-binding site in vitro is also the protein-binding site in vivo in most chaperones, we tested whether the mutant H125D supports growth of grp94−/− cells under the serum deprivation stress. Unexpectedly, the in vivo results dramatically ran contrary to prediction. As shown in Fig. 4C, H125D rescues cell survival almost as efficiently as wild-type GRP94 (Table 2). Thus, the peptide-binding site in the N-terminal domain, whose activity is measured by binding to the purified chaperone, is not essential for GRP94's function measured in this cell-based assay. Rather than using the β sheet site in the N-terminal domain to bind client proteins in vivo, GRP94 must employ another domain for pro-IGF interactions. The various crystal structures of GRP94 and Hsp90 do not ascribe a clear function to the N-terminal β sheet. For example, there is no obvious conformational change in the β sheet when apo-GRP94 and nucleotide-bound GRP94 are compared (16, 25, 39). Yet, indirect biophysical evidence suggests that the β sheet binding site is regulated by both the nucleotide-binding site in the N-terminal domain and by the charged domain that links the N and the M domains (3, 7). Possibly, the peptide-binding site is used in vivo for binding a regulatory protein, which is yet to be identified.
It is important to note that the mutants do not rescue viability to different extents simply because of different levels of expression. All of the mutants are expressed at equivalent levels, and in fact, ΔK, which does not rescue at all, is consistently expressed somewhat better than WT or the mutants that do rescue viability (Table S1). An additional potential explanation for the variable rescue ability is that the effective cellular pools of mutants vary because of differential misfolding/aggregation of mutants. To investigate this possibility, we measured the mobility of GRP94 mutants in the ER, using fluorescence recovery after photobleaching (FRAP) experiments. Because of increases in size, aggregated proteins exhibit little or no diffusional mobility (e.g., refs. 40, 41). All of the versions of GRP94 used here behaved as soluble proteins that readily diffuse into photobleached regions of the ER (Fig. S1A). We saw no evidence of aggregation, which would have been discerned as an immobile fraction of molecules, nor for any subcompartmentalization of mutants within the ER (data not shown). The D values for wild-type GRP94, the inactive E82A mutant, or the active H125D protein were rather similar, although the diffusion of H125D was statistically slower than that of wild type. Only the ΔK mutant exhibited considerably faster diffusional mobility. All of the GRP94 versions were considerably slower than an inert ER luminal marker, ER-GFP (Fig. S1B). The lack of correlation of D values with rescue of viability supports the interpretation that the differential rescue ability of point mutants in our assay are the result of differential activity and not of expression differences or misfolding. The FRAP data are consistent with engagement of GRP94 mutants in distinct protein–protein interactions, whose natures, and sensitivity to ER stress conditions, such as serum withdrawal, are currently being pursued.
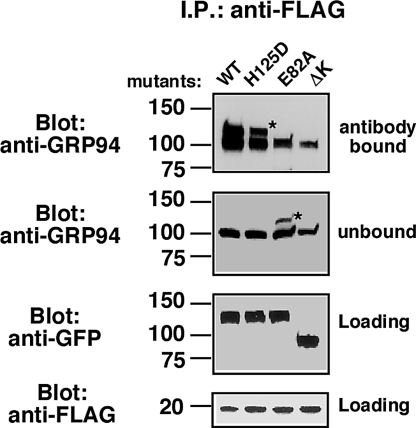
To determine whether the survival promoting activity of GRP94 is the result of physical interaction with pro-IGF-II, we tested whether GRP94 associates with pro-IGF-II by immunoprecipitation experiments. Cells (293T) were transfected with both FLAG-tagged pro-IGF-II and either WT GRP94-GFP, H125D-GFP, or E82A-GFP. After immunoisolation on anti-FLAG beads, the coprecipitated proteins were probed with monoclonal anti-GRP94. As shown in Fig. 5, the endogenous GRP94 (100-kDa band) was associated with the tagged pro-IGF-II in all cases. The 2 forms of GRP94-GFP, which rescued viability, WT- and H125D-GFP, also coassociated with pro-IGF-II and were distinguished from endogenous GRP94 by their higher apparent size. On the other hand, E82A-GFP, which did not support viability, did not coimmunoprecipitate with pro-IGF-II (Fig. 5). This result provides further evidence that the functional readout directly reflects chaperone activity toward IGF-II.
Fig. 5.
Physical association of GRP94 mutants with pro-IGF. Pro-IGF-II tagged with FLAG was expressed in 293T cells together with either wild-type GRP94-GFP or the H125D, E82A, or ΔK mutants of the fusion protein. Detergent lysates were immunoprecipitated using anti-FLAG. Fifty percent of the eluted material, 10% of the antibody-unbound material, and 5% of the input material were resolved on the same SDS gel. The top half of the gel was probed with 9G10 anti-GRP94, or with anti-GFP, as indicated. The bottom half of the gel was probed with anti-FLAG. The unbound fraction indicates the efficiency of immunodepletion of the total cell lysate. *, denotes the migration of GRP94-GFP. ΔK serves as specificity control in this experiment, because the epitope for the anti-GRP94 monoclonal is deleted in this construct and it is only detectable with anti-GFP antibodies. n = 2.
In conclusion, we describe an assay predicated on making cells dependent on the function of GRP94, as a prerequisite for production and secretion of IGF, which is necessary for survival under some stress conditions. Enabling this assay is a fluorescent fusion protein of GRP94, which is active and thus allows assessment of functional complementation on a cell-by-cell basis. Using this assay, we have uncoupled the immunological activity of GRP94 from its IGF chaperone activity and show that ATP binding and hydrolysis are essential for chaperone activity in cells. These findings resolve long-standing questions about this ubiquitous ER chaperone and indicate that GRP94 uses an action cycle similar to that of the cytosolic and bacterial HSP90s. Our finding concerning the putative GRP94 peptide-binding domain requires a new investigation to define how GRP94 binds its client proteins.
Materials and Methods
Plasmids.
The cDNA for canine GRP94 was cloned into pCDNA3.1 and fused after codon 777 to monomeric Green or Cherry fluorescent proteins [mGFP (42) or mCherry (43)] followed by a KDEL C-terminal sequence. Site-directed mutagenesis was performed by the QuikChange method (Stratagene). The ΔK-mGFP mutant was constructed by in-frame deletion of residues 144–488.
Cells and Transfections.
The line 14.1 is a grp94−/− mouse embryonic fibroblast (32). The cells were transfected on fibronectin-coated 35-mm glass bottom culture dishes (MatTek). Twenty hours later, the cells were scored microscopically; the plates were washed and incubated in serum-free medium. Cell numbers were scored at intervals of 4–10 h for the next 2 days.
Quantitation of Cell Survival.
Because of the inherent variability in the time course of cell survival, each experiment included both WT-mGFP and the ΔK-mGFP mutant, to serve as positive and negative controls, respectively. The time after the shift to serum-free medium when only 50% of the cells survived (T1/2) was estimated from each survival curve and the survival because of mutant clone expression was compared to that of the wild-type GRP94 within each experiment.
For further information, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Drs. J. Burkhardt, S. Seeholzer and R. Kalb for many useful suggestions and R. Rubenstein for generous provision of equipment. This work was supported by grants from the W.W. Smith Foundation, the Commonwealth of Pennsylvania, and the National Institutes of Health (to Y.A.). O.O. was supported by the Juvenile Diabetes Research Foundation Grants AG18001 and NS-059367 and the Arthritis Foundation. E.L.S. is an Ellison Medical Foundation New Scholar in Aging and is supported by National Institute on Aging 1R21AG032544–01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902626106/DCSupplemental.
References
- 1.Wearsch PA, Voglino L, Nicchitta CV. Structural transitions accompanying the activation of peptide binding to the endoplasmic reticulum Hsp90 chaperone GRP94. Biochemistry. 1998;37:5709–5719. doi: 10.1021/bi9801006. [DOI] [PubMed] [Google Scholar]
- 2.Schulte TW, et al. Interaction of radicicol with members of the heat shock protein 90 family of molecular chaperones. Mol Endocrinol. 1999;13:1435–1448. doi: 10.1210/mend.13.9.0339. [DOI] [PubMed] [Google Scholar]
- 3.Vogen SM, et al. Radicicol-sensitive peptide binding to the N-terminal portion of GRP94. J Biol Chem. 2002;277:40742–40750. doi: 10.1074/jbc.M205323200. [DOI] [PubMed] [Google Scholar]
- 4.Gidalevitz T, et al. Identification of the N-terminal peptide binding site of glucose-regulated protein 94. J Biol Chem. 2004;279:16543–16552. doi: 10.1074/jbc.M313060200. [DOI] [PubMed] [Google Scholar]
- 5.Biswas C, et al. The N-terminal fragment of GRP94 is sufficient for peptide presentation via professional APCs. Int Immunol. 2006;18:1147–1157. doi: 10.1093/intimm/dxl049. [DOI] [PubMed] [Google Scholar]
- 6.Simen BB. Chicago: Univ of Chicago; 2002. Gene targeting and biochemical analysis of the endoplasmic reticulum chaperone GRP94. PhD thesis. [Google Scholar]
- 7.Biswas C, et al. The peptide binding activity of GRP94 is regulated by calcium. Biochem J. 2007;405:233–241. doi: 10.1042/BJ20061867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wearsch PA, Nicchitta CV. Endoplasmic reticulum chaperone GRP94 subunit assembly is regulated through a defined oligomerization domain. Biochemistry. 1996;35:16760–16769. doi: 10.1021/bi962068q. [DOI] [PubMed] [Google Scholar]
- 9.Gilchrist A, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 11.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, et al. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishiguro S, et al. SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 2002;21:898–908. doi: 10.1093/emboj/21.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrovsky O, Ahmed NT, Argon Y. GRP94 protects cells grown without serum from apoptosis by regulating the secretion of insulin-like growth factor II. Mol Biol Cell. 2009;20:1855–1864. doi: 10.1091/mbc.E08-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wassenberg JJ, Reed RC, Nicchitta CV. Ligand interactions in the adenosine nucleotide binding domain of the Hsp90 chaperone, GRP94. II. Ligand-mediated activation of GRP94 molecular chaperone and peptide binding activity. J Biol Chem. 2000;275:22806–22814. doi: 10.1074/jbc.M001476200. [DOI] [PubMed] [Google Scholar]
- 16.Soldano KL, Jivan A, Nicchitta CV, Gewirth DT. Structure of the N-terminal domain of GRP94: Basis for ligand specificity and regulation. J Biol Chem. 2003;278:48330–48338. doi: 10.1074/jbc.M308661200. [DOI] [PubMed] [Google Scholar]
- 17.Obermann WM, et al. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakob U, et al. Assessment of the ATP binding properties of Hsp90. J Biol Chem. 1996;271:10035–10041. doi: 10.1074/jbc.271.17.10035. [DOI] [PubMed] [Google Scholar]
- 20.Scheibel T, Weikl T, Buchner J. Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc Natl Acad Sci USA. 1998;95:1495–1499. doi: 10.1073/pnas.95.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheibel T, et al. ATP-binding properties of human Hsp90. J Biol Chem. 1997;272:18608–18613. doi: 10.1074/jbc.272.30.18608. [DOI] [PubMed] [Google Scholar]
- 22.Rosser MFN, Nicchitta CV. Ligand interactions in the adenosine nucleotide binding domain of the Hsp90 chaperone, GRP94. I. Evidence for allosteric regulation of ligand binding. J Biol Chem. 2000;275:22798–22805. doi: 10.1074/jbc.M001477200. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Srivastava PK. Tumor rejection antigen gp96/grp94 is an ATPase: Implications for protein folding and antigen presentation. EMBO J. 1993;12:3143–3151. doi: 10.1002/j.1460-2075.1993.tb05983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wearsch PA, Nicchitta CV. Interaction of endoplasmic reticulum chaperone GRP94 with peptide substrates is adenine nucleotide-independent. J Biol Chem. 1997;272:5152–5156. doi: 10.1074/jbc.272.8.5152. [DOI] [PubMed] [Google Scholar]
- 25.Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frey S, Leskovar A, Reinstein J, Buchner J. The ATPase cycle of the endoplasmic chaperone Grp94. J Biol Chem. 2007;282:35612–35620. doi: 10.1074/jbc.M704647200. [DOI] [PubMed] [Google Scholar]
- 27.Siligardi G, et al. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J Biol Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 28.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young JC, Obermann WM, Hartl FU. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 31.Scheufler C, et al. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 32.Wanderling S, et al. GRP94 is essential for mesoderm induction and muscle development because it regulates IGF secretion. Mol Biol Cell. 2007;18:3764–3775. doi: 10.1091/mbc.E07-03-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dollins DE, Immormino RM, Gewirth DT. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. J Biol Chem. 2005;280:30438–30447. doi: 10.1074/jbc.M503761200. [DOI] [PubMed] [Google Scholar]
- 34.Chu F, et al. Identification of novel quaternary domain interactions in the Hsp90 chaperone, GRP94. Protein Sci. 2006;15:1260–1269. doi: 10.1110/ps.052065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- 36.Meyer P, et al. Structural and functional analysis of the middle segment of hsp90: Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 37.Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 38.Tamura Y, et al. Immunotherapy of tumors with autologous tumor-derived heat shock protein preparations. Science. 1997;278:117–120. doi: 10.1126/science.278.5335.117. [DOI] [PubMed] [Google Scholar]
- 39.Immormino RM, et al. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279:46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, et al. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 41.Nehls S, et al. Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- 42.Snapp EL, et al. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 44.Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc Natl Acad Sci USA. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siggia ED, Lippincott-Schwartz J, Bekiranov S. Diffusion in inhomogeneous media: Theory and simulations applied to whole cell photobleach recovery. Biophys J. 2000;79:1761–1770. doi: 10.1016/S0006-3495(00)76428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snapp EL, Altan N, Lippincott-Schwartz J. Measuring protein mobility by photobleaching GFP chimeras in living cells. Curr Protoc Cell Biol. 2003 doi: 10.1002/0471143030.cb2101s19. Chapter 21:Unit 21.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.