Abstract
Most kinases are capable of recognizing and phosphorylating peptides containing short, linear sequence motifs. To measure the activation state of many kinases from the same cell lysate, we created a multiplexed, mass-spectrometry-based in vitro kinase assay. Ninety chemically synthesized peptides derived from well-characterized peptide substrates and in vivo phosphorylation sites with either known or previously unidentified upstream kinases were reacted individually in a plate format with crude cell lysates and ATP. Phosphorylation rates were directly measured based on the addition of 90 same-sequence, site-specific phosphopeptides enriched in stable isotopes to act as ideal quantitative internal standards for analysis by liquid chromatography coupled to tandem mass spectrometry. This approach concurrently measured up to 90 site-specific peptide phosphorylation rates, reporting a diagnostic fingerprint for activated kinase pathways. We applied this unique kinome-activity profiling strategy in a variety of cellular settings, including mitogen stimulation, cell cycle, pharmacological inhibition of pathways, and to a panel of breast cancer cell lines. Finally, we identified the source of activity for a peptide (derived from a PI3K regulatory subunit) from our library. This peptide substrate demonstrated mitotic and tyrosine-specific phosphorylation, which was confirmed to be a novel Src family kinase site in vivo.
Reversible protein phosphorylation is a ubiquitous process throughout eukaryotes, and dysregulation of protein kinase signaling pathways is a hallmark of most human cancers (1). Hyperactivation of signaling pathways occurs during tumor pathogenesis as a result of over-expression of signal activators (growth factor receptors, Ras, PI3K, and so forth), structural alteration of kinases (Src and BCR-Abl, and so forth), or loss of negative mediators (PTEN, LKB1 and SHP2, and so forth) (1–4). As a consequence, the signaling network is rewired and a new equilibrium is established that can involve retuning sensitivity to upstream signals, bypassing routes, and creating additional nodes and connections. Eventually, cells may acquire self-sufficiency in growth signals and limitless replicative potential, becoming insensitive to growth inhibition and apoptosis signals (1, 5).
Chemically synthesized peptides of optimized sequence have long been used as substrates for in vitro phosphorylation using both purified kinases and cell lysates (6, 7). Several methods have been proposed for detection of the phosphorylated product (8–10). However, these approaches are unable to resolve the actual site of phosphorylation on peptides, which may lead to off-target events. In addition, they often require substantial sample manipulation to ultimately detect the phosphorylated product, which reduces both precision and sensitivity. Despite the breadth of techniques available, highly quantitative and direct measurement methods are necessary to address the diverse clinical manifestations of aberrant cellular signaling events.
Because of its specificity and precise quantitative nature, mass spectrometry represents an ideal platform to quantify products formed from enzymatic reactions (11, 12). Here we report the development of an integrated method termed “KAYAK” (Kinase ActivitY Assay for Kinome profiling) for multiplexed, large-scale kinase activity profiling. We quantitatively measured site-specific phosphorylation activities toward 90 different peptides using high-resolution MS. The KAYAK approach faithfully reported the activation state of cellular signaling pathways in various settings. We demonstrate that a collection of kinase activities can serve as a tractable marker for dissecting highly heterogeneous signaling networks in cancer cells from a molecular level, which could inform future design of targeted therapeutic interventions.
Results
The KAYAK Strategy for Parallel Measurement of Kinase Pathway States.
We began by synthesizing 90 peptides and an additional 90 same-sequence reference “heavy” phosphopeptides [supporting information (SI) Table S1]. Because we sought to apply this KAYAK strategy toward aberrant signaling in cancer, we chose a dozen sequences to first cover two critical cancer-signaling pathways (PI3K and MAPK). The remainder was based on sites ascribed to known pathways in the literature, peptides with known specificities, and ≈30 uncharacterized sites from existing large-scale in vivo phosphoproteomics studies (13, 14). All synthetic peptides contained an additional C-terminal extension tripeptide of -Pro-Phe-Arg for ease of synthesis, isotope incorporation, and purification.
To test substrate suitability in a multiplexed assay, we measured phosphorylation rates individually using 100 μM of each substrate peptide, 6-μg lysate in kinase buffer from serum-starved HEK293 cells, and 5 mM ATP in a plate format. Reactions proceeded for 60 min followed by acidification and addition of isotope-labeled internal standard (IStd) peptides. After pooling 45 samples, phosphopeptides were enriched and analyzed by nano-scale liquid-chromatography (LC) separation coupled to on-line peptide detection by high-resolution MS (Fig. 1A). Many peptides derived from known phosphorylation sites contain additional Ser, Thr, and Tyr residues in their flanking sequences, sometimes leading to formation of additional phosphorylation position isomers. However, these site isomers were generally resolved by HPLC, and the phosphorylation site was subsequently confirmed by MS/MS analysis (Fig. 2). Each phosphopeptide and its IStd peptide co-eluted, facilitating site-specific quantification by direct ratio to the IStd peptide abundance (Fig. 1B and see Fig. 2).
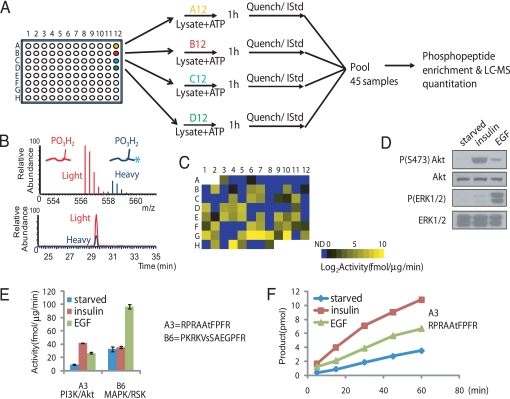
Fig. 1.
General scheme of the KAYAK strategy. (A) Overview of the procedure, where 90 synthetic peptides are used as substrates for in vitro kinase assays. (B) Example of high-resolution MS and elution chromatogram for a light and heavy (internal standard; IStd) pair of phosphopeptides. Asterisk indicates incorporation of a proline residue containing 6 additional Daltons of heavy isotopes. (C) Intensity map representation of substrate phosphorylation activities (average of triplicate experiments) from 6 μg starved HEK293 cell lysates toward each of the 90 peptides in their respective plate positions. ND, not detected, indicates the ones below the threshold. (D) Immunoblot analysis of the lysates of insulin- and EGF-stimulated HEK293 cells using the indicated antibodies. (E) Examples of phosphorylation rates (performed from 3 separate experiments) for a preferred Akt (A3; RPRAAtFPFR) and RSK (B6; PKRKVsSAEGPFR) substrate, respectively, using the lysates in (D). (F) Time course of the Akt-specific substrate, A3, phosphorylation reaction using the same lysates in D.
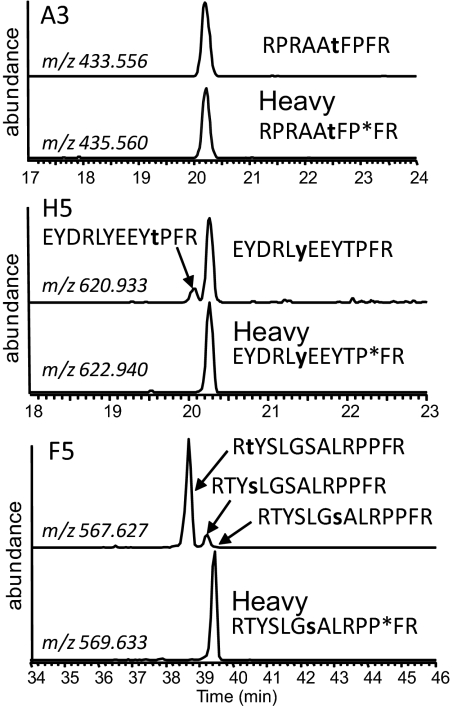
Fig. 2.
Site-specific measurement of peptide phosphorylation rates. Peptides phosphorylated at different Ser/Thr/Tyr residues were resolved by LC and phosphorylation sites were localized by concurrent tandem MS analysis (see Fig. S3 for an example). Perfect co-elution of the internal standard and product facilitated the determination of a site-specific phosphorylation rate. Extracted ion chromatograms were produced using a ± 10 ppm tolerance surrounding the predicted mass-to-charge (m/z) ratio for each phosphopeptide and each internal standard (contains heavy proline residue, indicated by asterisk). Confirmed phosphorylation sites are bold and lowercase. Peptide A3 has a single acceptor site residue, and its phosphorylation product perfectly co-eluted with the internal standard. In contrast, the phosphorylation products of H5 and F5 both contained multiple position isomers. In the case of F5, no site-specific phosphorylation toward the site in the IStd was detected.
Only two LC-MS runs were required to analyze the entire plate. We observed robust activities toward many peptides, even under the serum-starved conditions, with more than half of the substrate peptides (49 out of 90) phosphorylated at rates of at least 1 fmol/μg lysate/min (Fig. 1C). The peptide showing the highest phosphorylation rate (position G10 in the 96-well plate, KKKRFsFKKSPFR, lowercase s/t/y in peptide sequences denotes the phosphorylation site) corresponded to the conversion of only 18% of the substrate, suggesting our reaction scheme resided within the linear portion of the kinase reaction. Notably, the activity measurements were highly reproducible across biological replicates and encompassed a range of more than 3 orders of magnitude. This wide dynamic range ensured that variations in kinase activities were easily distinguishable, providing a tractable index of kinase-mediated cellular networks and pathways.
Profiling the Activities of Kinase Mediated-Signaling Networks After Mitogen Stimulation.
We examined the ability of these peptides to report specific changes in kinase activation after pathway stimulation. Lysates from HEK293 cells were collected after insulin or EGF treatment and compared to their activities in the serum-starved state using the KAYAK approach. Fig. 1D shows the Western blot analysis of these lysates where the PI3K and MAPK pathways were activated, as indicated by elevated phospho-Akt and phospho-ERK1/2 levels, respectively. Phosphorylation of a derivative of a known Akt substrate peptide, Aktide (15) (plate position: A3, RPRAAtFPRF) was in good agreement with immunoblot results for Akt activation, showing a strong increase in phosphorylation after a 10-min insulin treatment (4.6-fold) and a weaker but substantial increase (2.9-fold) after EGF stimulation for 5 min (Fig. 1E). In contrast, peptide B6 reported increased phosphorylation activity after EGF (3-fold) but not insulin treatment of the cells. Peptide B6 corresponds to PKRKVsSAEGPFR, which was derived from sequences spanning Ser-6 of nonhistone chromosomal protein HMG-14. This site has been shown to be phosphorylated in vivo as a result of EGF stimulation by MAPK downstream effectors, RSK and MSK (16). We also confirmed the linearity of the product formation in a time-course experiment for the Akt peptide substrate, A3 (Fig. 1F). These assays were extremely sensitive; kinase activities toward several peptides could be confidently measured using as little as 50 ng of crude lysate per reaction (Fig. S1).
The average measured activities from 3 separate experiments using all 90 peptides are shown in Fig. 3A for serum-starved, insulin- and EGF-stimulated lysates. While the same peptides were generally phosphorylated by these lysates, we observed differences in the absolute activity levels for 8 peptides (Table S2, see Fig. 3). For example, a peptide derived from tuberin (E11, RKRLIsSVEDPFR, S1798) showed up-regulated phosphorylation after both insulin (1.7-fold) and EGF (2.3-fold) stimulation. This is in agreement with the report that this site is phosphorylated in vivo upon activation of either the PI3K or MAPK pathways (17). In contrast, phosphorylation of several peptides (e.g., B6, C6, C11, and G5) (see Table S2) increased only under EGF-stimulated but not insulin-treated conditions, making them putative activity reporters of the MAPK pathway. Although the substrate library contained several EGFR-derived peptides that contained sites known to be phosphorylated upon receptor activation in vivo, we did not observe an increase of phosphorylation of these peptides in the EGF-stimulated (or any other) cell lysate, indicating that a correct context was critical for their phosphorylation. Nevertheless, the KAYAK method supported at least 8 peptides (see Table S2 and Fig. S2) capable of distinguishing quiescent from activated PI3K and MAPK signaling pathways.
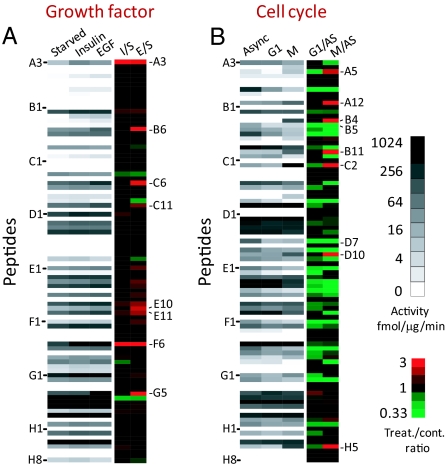
Fig. 3.
Peptide phosphorylation rates accurately report pathway activation states. (A) Intensity map of kinase activities of starved (S), insulin-stimulated (I), and EGF-stimulated (E) HEK293 cells (average of triplicate experiments). Fold change over the starved state is shown. Peptides with signals below detection threshold were not included for ratio calculation. (B) Intensity map of kinase activities of asynchronously growing (AS) HeLa cells, and cells arrested in either G1/S or G2/M phase of the cell cycle. Fold change over the asynchronous state is shown. Peptides were ordered according to their positions in the 96-well plate. Sequences of all peptides can be found in Table S1.
Profiling the Activities of Kinase-Mediated Signaling Networks During Cell Cycle.
To identify peptides specifically targeted by additional pathways activated in cancer, phosphorylation rates from cell cycle-arrested lysates were determined for the entire library. Asynchronously growing HeLa cells were compared with cells synchronized in G1/S or G2/M phases of the cell cycle (see Fig. 3B). Phosphorylation of many peptides containing Pro at the + 1 position of S/T was now dramatically increased in G2/M phase (see Table S2 and Fig. 3B), which was verified by Western blot experiments (see Fig. S2E). This was in accordance with the mitotic activation of proline-directed kinases such as the cyclin-dependent kinases (18). For example, peptide C2 (IPTGTtPQRKPFR, derived from kinesin-like protein kif11, Thr-927) showed a 19-fold increase in phosphorylation during the G2/M phase compared with asynchronously growing or G1/S cells (see Fig. S2D). This was in agreement with a previous in vivo quantitative phosphoproteomics study, where the same site showed up-regulated mitotic phosphorylation (13).
Although tyrosine-specific phosphorylation was detected on several target peptides, their levels remained largely unchanged or decreased after nocodazole arrest (see Fig. 3B and Fig. S2 B and F). One exception was the phosphorylation rate of H5 (EYDRLyEEYTPFR) derived from phosphoinositide 3-kinase (PI3K) regulatory subunit p85α(Tyr-467)/p55γ(Tyr-199). Unexpectedly, H5 showed a dramatic increase in phosphorylation (13-fold) in the mitotic lysate. Retention time comparison and tandem MS experiments using both collision-induced dissociation and electron transfer dissociation confirmed that the indicated Tyr rather than a more C-terminal TP motif was phosphorylated (Fig. S3).
KAYAK Peptides as Reporters of Pathway Inhibition.
We next applied the KAYAK method to measure the effect of pharmacological inhibitors or siRNA-mediated knockdown of kinases (Figs. S4). EGF-stimulated cell lysates once again phosphorylated several peptides including B6, C6, C11 and G5 (see Table S2), and these effects were blocked by pretreating cells with a MEK-specific inhibitor, U0126. In contrast, phosphorylation levels of these peptides were not changed as a result of insulin stimulation. Peptides B6, C11, and G5 were confirmed as specific targets of RSK by siRNA-mediated knockdown of RSK1/2 (see Fig. S4) and purified kinase (Fig. S5).
PI3 Kinase Regulatory Subunit p55 Shows Src-Dependent Tyrosine Phosphorylation During Mitosis.
Because of the striking increase in mitotic phosphorylation of H5 peptide (EYDRLyEEYTPFR), we sought to identify its activity source (Fig. 4A). The tyrosine residue of interest is conserved among various members of the PI3K regulatory subunit (i.e., Tyr-197 of p55α, Tyr-199 of p55γ, Tyr-467 of p85α, and Tyr-464 of p85β) (Fig. S6A). Using a phospho-specific antibody toward this site, we found the in vivo phosphorylation level on p55 was also dramatically increased during G2/M phase (Fig. 4B). Increased phosphorylation was not detected at 85 kDa. At this stage, we cannot differentiate the relative contribution of p55α and p55γ, and herein we use Tyr-199 of p55 to designate the site. To rule out the possibility that this mitotic phosphorylation event was an artifact of nocodazole treatment, we synchronized HeLa cells in early S-phase and then monitored phospho-p55 (Tyr-199) levels as cells underwent a natural mitosis (Fig. 4C). Phospho-PI3K regulatory subunit p55 (Tyr-199) levels increased during cell progression from G1/S through G2/M.
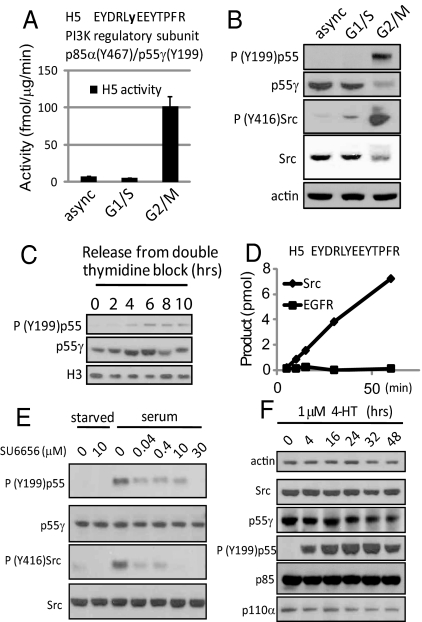
Fig. 4.
Identification and validation of Src family kinase activity toward Tyr-199 of PI3K regulatory subunit p55. (A) Activity toward substrate peptide H5 using lysates of asynchronously growing HeLa cells and cells arrested in G1/S and G2/M phase. (B) Immunoblot analysis of phospho-PI3K regulatory subunit p55 (Tyr-199) levels in these same lysates. (C) Phospho-PI3K regulatory subunit p55 (Tyr-199) immunoreactivity in HeLa cells undergoing a natural mitosis. (D) In vitro phosphorylation of peptide H5 using 2 ng of purified Src or EGFR. (E) Treatment of asynchronously growing HEK293 cells using a Src family kinase (SFK) specific inhibitor, SU6656. (F) Immunoblot analysis of vSrc-ER expressing MCF10A cells treated with 1 μM 4-HT to activate vSrc.
It has been reported that Src, a soluble tyrosine kinase, is transiently activated during mitosis (19). We therefore investigated whether increased phospho-p55 levels during mitosis were a result of activated Src. Indeed, Src-activating phosphorylation (Tyr-416) increased during G2/M (see Fig. 4B), correlating with elevated phospho-p55 (Tyr-199). H5 could also be phosphorylated in vitro by purified Src but not EGFR (Fig. 4D). To further confirm the site's Src-dependent nature in vivo, we treated asynchronously growing HEK293 cells with a Src family kinase (SFK) inhibitor, SU6656 (20). The levels of both phospho-Src (Tyr-416) and phospho-p55 (Tyr-199) diminished after treatment (Fig. 4E). In another experiment, MCF10A cells expressing v-Src:estrogen receptor (vSrc-ER) (21) were treated with 1 μM 4-hydroxytamoxifen (4-HT) to activate v-Src (Fig. 4F). Increased phosphorylation of p55 at Tyr-199 was observed within 4 h and persisted whenever v-Src was activated. This was not an artifact of 4-HT treatment because MCF10A cells incubated with 1 μM 4-HT displayed unaltered phosphorylation at this site (see Fig. S6). The consistent correlation between Src activity and phospho-p55 (Tyr-199) in different cells suggests that this tyrosine residue is a general Src-dependent phosphorylation site in vivo. Interestingly, we also observed that the protein level of p55γ decreased after prolonged Src activation (see Fig. 4 B and F). These findings demonstrate that even without prior knowledge of the kinase, its kinetics, or specificity, in vitro peptide phosphorylation can lead to the discovery of both the responsible kinase in vivo and even the site's biological context.
KAYAK Profiling of Kinome Activities in Cancer Cell Lines.
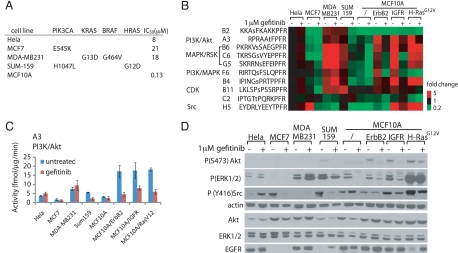
In tumors, activating mutations are often found in several core signaling pathways (2). To assess the ability of the KAYAK method to accurately identify differences in the signaling pathway activities, we compared phosphorylation of the peptides by the lysates of 7 asynchronously growing breast cancer cell lines and also after treatment with an EGFR inhibitor, gefitinib (Fig. 5). Cell lines were selected to represent the highly heterogeneous nature of breast cancer (22). A summary of activating mutations is presented in Fig. 5A. For example, MDA-MB231 is a highly invasive cell line that contains the mutant form of K-Ras (G13D) and B-Raf (G464V) (23). Sum159 cell line also contains a mutation within the MAPK pathway (H-Ras G12D) (24). We also included MCF10A cells, which are nontumorigenic epithelial cells, and MCF10A cells over-expressing ErbB2, insulin-like growth-factor receptor (IGFR) and H-RasG12V.
Fig. 5.
Peptide phosphorylation rates accurately report activating mutations in kinase pathways. Seven breast cancer cell lines and HeLa cells were examined. (A) Summary of the activating mutations within the PI3K and MAPK pathways and IC50 of these cells toward gefitinib treatment (22, 25, 26). (B) KAYAK activities (average of duplicates, shown as fold difference normalized to asynchronously growing HeLa cells) in cancer cell lines with or without 1 μM treatment of gefitinib for 24 h. Peptide sequences with their reported kinases pathways/kinases are shown. (C) Example of activity profile from (B) for A3 peptide (PI3K/Akt pathway) across these cell lines. (D) Western blot analysis of the lysates for the cell lines using the indicated antibodies.
Basal kinase activities varied among these cell lines suggesting differences in the underlying pathway activation state (see Fig. 5 B and C). For example, MDA-MB231 and Sum159 cells displayed substantially higher MAPK activities (indicated by B6, C6, and G5) (see Table S2) compared with MCF7 and MCF10A cells. These results were in agreement with Western blot analysis (see Fig. 5D). Because MDA-MB231 and MCF10A cells express similar levels of EGFR, higher MAPK activities observed in MDA-MB231 cells were likely driven by the activating mutations of Ras and Raf. In addition, over-expression of ErbB2, IGFR and H-RasG12V in MCF10A cells led to higher basal activities in both the PI3K/Akt and MAPK pathways.
Cells displayed further diversification in their response to gefitinib treatment. Growth of HeLa cells is resistant to gefitinib (IC50 = 8 μM) (25) and activities of PI3K or MAPK in these cells were not affected by 1 μM gefitinib treatment. A breast cancer cell line with a high IC50 (21 μM) (22), MCF7, also showed minimal activity decrease in both PI3K and MAPK pathway. In contrast, MCF10A cells are sensitive to gefitinib, with a cell growth IC50 of 0.13 μM (26). PI3K and MAPK activities in normal MCF10A cells and MCF10A/ErbB2, MCF10A/IGFR were strongly inhibited after 1 μM gefitinib treatment. MAPK activity of MCF10A cells over-expressing H-RasG12V showed gefitinib-resistance. Because Ras signals downstream of EGFR and upstream of MAPK, the mutant form of Ras could lead to disengagement of MAPK from EGFR. However, whether Ras mutation can convey resistance of MAPK activity to EGFR inhibition is also cell-context dependant. Although both MDA-MB231 and Sum159 cells harbor Ras mutations, MAPK activity in MDA-MB231 cells was completely refractory to EGFR inhibition. Accordingly, growth in MDA-MB231 cells is resistant to gefitinib treatment, with an IC50 of 18 μM (22). In contrast, MAPK activity in Sum159 cells showed some sensitivity toward gefitinib treatment. We also observed a differential response of Src activity (as reported by H5 peptide and corroborated by Western blot) toward gefitinib treatment. Src was inhibited in MCF7, Sum159, MCF10A/IGFR, and MCF10A/H-RasG12V cells, whereas Src activity in HeLa and MCF10A cells was resistant to gefitinib inhibition. Overall, phosphorylation activity measured from the KAYAK approach correlated with the activating mutations within the pathways in diverse cell lines.
Discussion
Although large-scale genome profiling methods have been used to determine the genomic alterations of many kinases (2), activity measurements are a more direct method to characterize the activation status of a kinase and its downstream effectors in the signaling network. Chemically synthesized peptides of optimized sequences have been used for more than 30 years as in vitro phosphorylation substrates using both purified kinases and cell lysates (6, 7). However, a sensitive, multiplexed, and site-specific detection method has been lacking. The method reported herein measured kinase activities toward up to 90 different substrate peptides using phosphopeptide enrichment and high-resolution MS.
This strategy represents a significant improvement over other current approaches. The major advantage rests on the direct measurement by MS of the enzymatic products. This obviates the need for background subtraction of radioactivity or calibration of antibody-based detection-using standards. The quantitative measurement is based on the addition of 90 phosphopeptide internal standards. Each has the exact sequence as its substrate counterpart, but is enriched in stable isotopes and phosphorylated at a known site. Phosphorylated substrate peptides co-elute perfectly with their respective heavier IStd phosphopeptide, and the direct ratio of detector response for light-to-heavy pairs measures product amount. Site-specific profiling of kinase activities is achieved (see Fig. 2). Despite largely unknown kinase specificities and kinetics, faithful pathway reporting was clearly observed for many peptides in different settings, including mitogen stimulation (see Figs. 1 and 3), cell cycle arrest (see Fig. 3), pathway inhibition (see Fig. S4), and activating mutations (see Fig. 5).
For peptides with unfavorable kinetics or specificities, two method improvements can be attempted. First, the substrate concentration can be “tuned” in our assay, exploiting differences in affinity or kinetics between competing kinases. An overview of phosphorylation of the peptides using purified kinases is shown in Fig. S7. Peptide (F6) derived from nitric oxide synthase Ser-1176 can be phosphorylated by both Akt and RSK1 at the same site using purified kinases (see Fig. S5D). Because the Km values for each kinase-peptide pair differed, reducing the substrate concentration to 5 μM resulted in the preferential phosphorylation by Akt, the kinase reported for this site (27). Second, the 90-peptide kinase assay could be performed as a single reaction. Besides increasing throughput and a 90-fold decrease in sample consumption, competition between peptides would be expected to increase specificity because contending kinases should be occupied by their preferred substrates.
The KAYAK assay identified a unique mitosis-specific activity for Src family kinases toward PI 3-kinase regulatory subunit p55. Crystallography studies of the PI3 kinase p110α/p85α complex show that Tyr-467/p85α (homologous to Tyr-199/p55γ) is localized at the interface between the inter-SH2 domain (iSH2) of p85α and the C2 domain of p110α (28). Specifically, Tyr-467 is 2.7 Å away from His-450 of the catalytic subunit, within the distance for potential hydrogen bond formation (see Fig. S6D). This interaction and even the interface will likely be disrupted by phosphorylation of Tyr-467/Tyr-199. We speculate that phosphorylation of Tyr-467/Tyr-199 on the regulatory subunit (p85/p55) within the iSH2 domain could be a mechanism for SFK-dependent regulation of PI3 kinase activity during mitosis or more generally. Functional studies are ongoing.
While the EGFR inhibitor gefitinib has been approved for treatment in non-small cell lung cancer, growth and proliferation of many breast cancer cell lines are resistant to EGFR inhibition (22). Breast cancer is highly heterogeneous, often having mutations or over-expression of different signaling molecules within several key pathways. For example, MDA-MB231 cells have a very high basal MAPK activity, potentially because of mutation of the 2 key components, KRAS and BRAF, within the pathway. As a result, MAPK is constituatively active, which is resistant to EGFR inhibition. Interestingly, using KAYAK and immunoblot experiments, we found over-expression of IGFR in MCF10A cells activated both PI3K and MAPK pathways, which was blocked by gefitinib treatment. Previous reports have shown that IGF induces heterodimerization of IGFR and EGFR to activate ERK1/2 (29). Because gefitinib specifically inhibits EGFR but not IGFR (29), our findings suggest that one action of IGFR in activating downstream signaling pathways is mediated through a direct effect on EGFR tyrosine kinase activity.
Although a deregulated PI3K-, MAPK-, Src-, or the cyclin-dependent kinase-mediated signaling pathway is often an obligatory event in tumorgenisis (2, 4), we are currently expanding our peptide substrate catalog to target kinases implicated in other pathophysiological processes, including casein-kinases, GSK3, ATM/ATR, mTOR, and so forth. Future studies could also use KAYAK to investigate the activities of signaling pathways as a net effect of both active kinases and phosphatases present in cells. Moreover, the strategy behind the KAYAK approach could be applied to additional enzyme classes. Specifically, protease activities from plasma samples may represent an untapped resource for disease biomarkers. We predict that multiplexed peptide-based activity assays, exploiting high-resolution MS, will become a mainstay of clinical diagnosis, rational drug design, and disease prognosis.
Materials and Methods
Unphosphorylated peptides (containing natural abundance of stable isotopes) and same-sequence phosphorylated peptides (internal standard, contained 13C515N-Proline, ΔM = 6.0201 Da) were synthesized by Cell Signaling Technology. All in vitro kinase reactions were performed using 6-μg cell lysate (unless otherwise noted). Samples were analyzed with an LTQ-FT or LTQ-orbitrap mass spectrometer (ThermoFisher). Description of additional experimental procedures can be found in the SI Experimental Procedures.
Supplementary Material
Acknowledgments.
We thank J. Blenis, L. Cantley, and J. Brugge (Harvard Medical School, Boston) and their group members for helpful discussion and for generously providing the reagents, R. Baserga (Kimmel Cancer Institute, Thomas Jefferson University, Philadelphia) for providing the IGFR expression plasmids, and J. Knott, C. Hunter, S. Rogers, and J. Reynolds for help with peptide synthesis. This work was supported in part by Grants HG3456 and GM67945 from the National Institutes of Health and an industry-sponsored research project to S.P.G. from Merck and ThermoFisher.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905165106/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 4.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 5.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Kuenzel EA, Krebs EG. A synthetic peptide substrate specific for casein kinase II. Proc Natl Acad Sci USA. 1985;82:737–741. doi: 10.1073/pnas.82.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuda I, et al. A synthetic peptide substrate for selective assay of protein kinase C. Biochem Biophys Res Commun. 1990;166:1220–1227. doi: 10.1016/0006-291x(90)90996-z. [DOI] [PubMed] [Google Scholar]
- 8.Diks SH, et al. Evidence for a minimal eukaryotic phosphoproteome? PLoS ONE. 2007;2:e777. doi: 10.1371/journal.pone.0000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janes KA, et al. A high-throughput quantitative multiplex kinase assay for monitoring information flow in signaling networks: application to sepsis-apoptosis. Mol Cell Proteomics. 2003;2:463–473. doi: 10.1074/mcp.M300045-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Shults MD, et al. A multiplexed protein kinase assay. Chembiochem. 2007;8:933–942. doi: 10.1002/cbic.200600522. [DOI] [PubMed] [Google Scholar]
- 11.Gao H, Leary JA. Multiplex inhibitor screening and kinetic constant determinations for yeast hexokinase using mass spectrometry based assays. J Am Soc Mass Spectrom. 2003;14:173–181. doi: 10.1016/S1044-0305(02)00867-X. [DOI] [PubMed] [Google Scholar]
- 12.Pi N, Armstrong JI, Bertozzi CR, Leary JA. Kinetic analysis of NodST sulfotransferase using an electrospray ionization mass spectrometry assay. Biochemistry. 2002;41:13283–13288. doi: 10.1021/bi020457g. [DOI] [PubMed] [Google Scholar]
- 13.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutillas PR, et al. Ultrasensitive and absolute quantification of the phosphoinositide 3-kinase/Akt signal transduction pathway by mass spectrometry. Proc Natl Acad Sci USA. 2006;103:8959–8964. doi: 10.1073/pnas.0602101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim JH, et al. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Roux PP, et al. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- 19.Zheng XM, Shalloway D. Two mechanisms activate PTPalpha during mitosis. EMBO J. 2001;20:6037–6049. doi: 10.1093/emboj/20.21.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blake RA, et al. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reginato MJ, et al. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrer-Soler L, et al. An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa) -induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands (Review) Int J Mol Med. 2007;20:3–10. [PubMed] [Google Scholar]
- 23.Thompson EW, et al. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol. 1992;150:534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- 24.Hollestelle A, et al. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol Cancer Res. 2007;5:195–201. doi: 10.1158/1541-7786.MCR-06-0263. [DOI] [PubMed] [Google Scholar]
- 25.Giocanti N, et al. Additive interaction of gefitinib (‘Iressa’, ZD1839) and ionising radiation in human tumour cells in vitro. Br J Cancer. 2004;91:2026–2033. doi: 10.1038/sj.bjc.6602242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Normanno N, et al. The MEK/MAPK pathway is involved in the resistance of breast cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol. 2006;207:420–427. doi: 10.1002/jcp.20588. [DOI] [PubMed] [Google Scholar]
- 27.Dimmeler S, et al. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 28.Huang CH, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad T, Farnie G, Bundred NJ, Anderson NG. The mitogenic action of insulin-like growth factor I in normal human mammary epithelial cells requires the epidermal growth factor receptor tyrosine kinase. J Biol Chem. 2004;279:1713–1719. doi: 10.1074/jbc.M306156200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.