Abstract
Numerous studies focus on the tumor suppressor p53 as a protector of genomic stability, mediator of cell cycle arrest and apoptosis, and target of mutation in 50% of all human cancers. The vast majority of information on p53, its protein-interaction partners and regulation, comes from studies of tumor-derived, cultured cells where p53 and its regulatory controls may be mutated or dysfunctional. To address regulation of endogenous p53 in normal cells, we created a mouse and stem cell model by knock-in (KI) of a tandem-affinity-purification (TAP) epitope at the endogenous Trp-53 locus. Mass spectrometry of TAP-purified p53-complexes from embryonic stem cells revealed Tripartite-motif protein 24 (Trim24), a previously unknown partner of p53. Mutation of TRIM24 homolog, bonus, in Drosophila led to apoptosis, which could be rescued by p53-depletion. These in vivo analyses establish TRIM24/bonus as a pathway that negatively regulates p53 in Drosophila. The Trim24-p53 link is evolutionarily conserved, as TRIM24 depletion in human breast cancer cells caused p53-dependent, spontaneous apoptosis. We found that Trim24 ubiquitylates and negatively regulates p53 levels, suggesting Trim24 as a therapeutic target to restore tumor suppression by p53.
Keywords: apoptosis, bonus, TAP, ubiquitylation
Although intensively studied in tumor cells, the mechanisms that regulate tumor suppressor p53 in normal cells remain poorly understood. Biochemical analysis of endogenous p53 poses a challenge, as p53 is held at low levels in non-transformed cells in the absence of stress (1). Mutant p53 generally exhibits increased protein stability in tumor-derived cells (2, 3). This offers opportunities for molecular analysis, although of a protein that may display loss- or gain-of-function (4). Indeed, several assumptions about p53 regulation, including the relative importance of specific post-translational modifications, sites of mutation, and interactions with regulatory proteins, have been challenged in mouse models where in vivo functions of p53 can be determined (3, 5–7).
In addition to its principle role in maintenance of genomic stability and tumor suppression, p53 regulates specific functions in normal cells (1). These include regulation of gene expression during development (8, 9), cellular progression to senescence and aging (10), and metabolic functions (6). Although a p53ERTAM mouse model, where p53 function and response may be temporally regulated (11), has been invaluable, questions regarding endogenous regulation and control of p53, before stress stimuli, transformation, or tumorigenesis, remain unanswered. To elucidate mechanisms that regulate endogenous p53, we developed a mouse and stem cell model based on TAP-tag fusion with the ORF of Tpr53, which allows TAP-purification and analysis of p53 protein partners and post-translational modifications by mass spectrometry. Here, we describe a protein-interaction partner of p53 that negatively regulates p53 protein stability by ubiquitylation. We addressed the in vivo significance of Trim24/bonus-p53 interaction in Drosophila, and uncovered a negative regulator of p53 levels in this organism. Human tumor-derived cancer cells, which retain wild-type TP53, respond to siRNA-targeted depletion of TRIM24 by undergoing apoptosis. This identification of a previously unknown, negative regulator of p53 suggests that potential therapeutic targets to restore p53 functions remain to be identified.
Results and Discussion
Creation of a Mouse and Stem Cell Model for p53 Analysis.
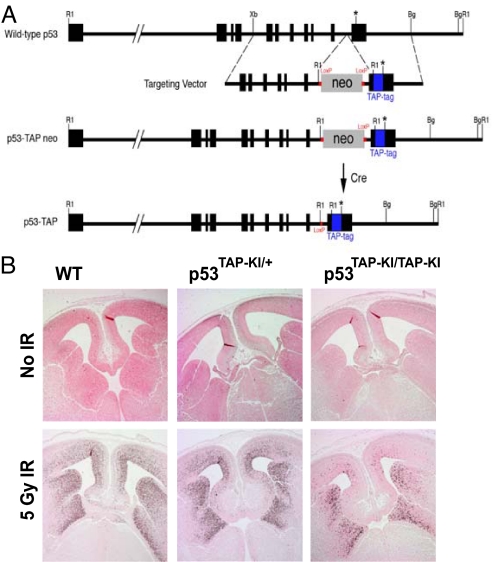
To facilitate analysis of endogenous p53 in normal cells, we performed gene targeting to create mouse embryonic stem (ES) cells and mice (12), which express endogenously regulated p53 protein fused with a C-terminal TAP tag (p53-TAPKI, Fig. 1A) (13). Germline transmission of the p53-TAPKI allele was confirmed by Southern hybridization and allele-specific PCR ( Fig. S1A). p53-TAPKI remains entirely responsive to exogenous and endogenous stimuli in p53TAP-KI/+ ES cells and mice. The induction of apoptosis (Fig. S1B) and expression levels of p53-regulated genes, Cdkn1a (p21/CIP1), Mdm2, and Perp (Fig. S1C), were virtually identical over a time course of doxorubicin exposure in p53-TAPKI and wild-type ES cells. Chromatin immunoprecipitation (ChIP) showed that p53-TAPKI and p53 proteins are identical in their interactions with endogenous p53-response elements of specific genes (Fig. S1D).
Fig. 1.
Knock-in of a C-terminal TAP-tag does not disrupt regulatory functions of p53. (A) Targeting strategy: Homologous recombination and targeting, as shown, generated a C-terminal TAP tag fusion of p53 by inserting a loxP-flanked PGK-neo cassette into intron 10 of p53 and a TAP-tag cassette in frame with full-length p53 into exon 11. After selection of targeted ES cells, the neo cassette was removed by introduction of a cre-expressing plasmid to create p53TAP-KI/+ ES cells (see SI Text for details). (B) Apoptosis in vivo: IHC of e13.5 CNS (WT, p53TAP-KI/+, p53TAP-KI/TAP-KI) for cleaved caspase 3 to indicate apoptosis, without (no IR) or with exposure to γ-IR (5 Gy IR).
In ES cells, p53-mediated apoptosis is independent of transcription, as it is primarily driven by p53-mitochondrial interactions (14). Therefore, we performed analyses in vivo to assess p53-TAPKI response to stress, where downstream functions of p53 are primarily driven by p53-regulated transcription. p53TAP-KI/+ mice were intercrossed to obtain viable, homozygous p53-TAPKI (p53TAP-KI/TAP-KI) mice, which were born at the expected ratio of genotypes and show no tumor development 1 year after birth (Fig. S1E). We exposed embryonic day 13.5 (e13.5) embryos, of wild-type, heterozygous, and homozygous p53/TAPKI genotype, to γ-irradiation to induce an apoptotic response that is p53-dependent (15). Sections of the central nervous system (CNS) and immunohistochemistry (IHC) for activated, cleaved caspase 3 showed that p53-TAPKI mounted an apoptotic response in vivo, indistinguishable from wild-type p53 (Fig. 1B).
To regulate apoptosis and multiple downstream pathways, p53 forms a tetrameric complex (16). Endogenous p53-TAPKI and wild-type p53 expressed in p53TAP-KI/+ ES cells formed mixed hetero-tetramer p53-TAPKI/wild-type p53 protein complexes, fractionating between 200 kDa and >1 MDa by size-exclusion chromatography (Fig. S1F). Taken together, these data show that TAP-fusion to p53 and formation of heteromeric, p53 protein complexes did not disrupt p53-functions in transcription and apoptosis.
Identification of Trim24 as a p53 Partner.
Extracts of p53TAP-KI/+ and p53+/+ ES cells were purified by TAP-chromatography (13), before analysis by mass spectrometry (Fig. S2A). Multiple peptides confirmed the identity of p53, as well as 2 known p53-interacting partners: Mdm2 (17, 18) and 53BP1 (19). Additionally, we recovered proteins not previously known as p53-partners, for example, Trim24 (20). Originally named transcription intermediary factor 1-alpha (TIF1α), Trim24 was identified as a co-regulator of retinoid signaling (21). Trim24 is highly expressed in a tissue-specific profile of tumors, and is a known translocation target of the Braf (T18) and Ret oncogenes in acute promyelocytic leukemia and papillary thyroid carcinomas, respectively (22–24).
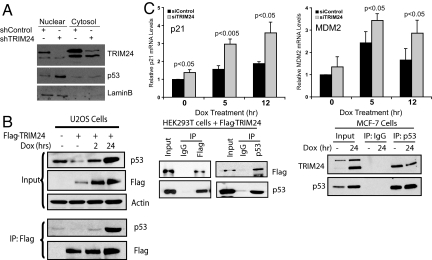
In contrast to high levels of Trim24 expression in specific tumors, decreased levels of Trim24 in normal ES cells led to greatly increased levels of nuclear-localized p53 (Fig. 2A), while ectopic expression of Trim24 in human cell lines decreased p53 levels (Fig. 2B). Trim24-p53 interactions were verified by co-immunoprecipitation (co-IP) in ES cells (Fig. S2B) and in multiple, human cell lines (Fig. 2B). In parallel with RNAi-mediated decreases in Trim24 protein and RNA levels (Fig. 2A, Fig. S2C, and Fig. S3), specific endogenous, p53-target genes Cdkn1a (p21/CIP1) and Mdm2 were significantly activated before and after doxorubicin treatment (Fig. 2C and Fig. S3A). Decreased Trim24 and increased p53 had no statistically significant effect on other p53-regulated genes, for example, Perp, or Trp-53 itself (Fig. S2C and Fig. S3). In MEFs, isolated from e13.5 embryos, Trim24 depletion led to similar trends in p21 and Mdm2 expression, as well as Perp (Fig. S2D). Cells depleted (Fig. S3A) or lacking (Fig. S2D) p53 did not show significant effects of Trim24 depletion, supporting the p53-dependence of Trim24-mediated regulation.
Fig. 2.
Trim24 is a negative regulator of p53. (A) RNAi-mediated depletion of Trim24 led to increased p53 protein in parallel with decreased Trim24 protein in ES cells (shTrim24 and shControl). (B) TRIM24 interacts with p53 in human cells. Cell lysates of U2OS, HEK293T, or MCF7 cells (Input) were used for IP of endogenous TRIM24, Flag-TRIM24 (Flag) and/or p53 after Dox (0.5 μg/mL) or without Dox induction of stress for the indicated time period (hr). Flag-TRIM24 was ectopically expressed (+ Flag-TRIM24) in U2OS and HEK293T cells and analyses performed 24 h later. Immunoblotting was conducted with indicated antibodies after SDS/PAGE to detect interacting proteins, as indicated. (C) Loss of Trim24 led to activation of specific p53-target genes in ES cells. Significant changes in RNA expression, determined by real-time RT-PCR, are indicated by P values less than 0.05 (Student's t test). Values are expressed as fold-change in comparison to siControl-treated cells at t = 0.
An In Vivo Analysis of the Trim24-p53 Regulatory Pathway.
Mammalian Trim24 is 1 of 3 members of the TIF1 subfamily within the larger TRIM group of 70 family members (25). Protein-protein interactions between TRIM family members are known, and siRNA depletion of Trim24, Trim28, and Trim33 (TIF1α, TIF1ß, and TIF1γ, respectively) revealed compensation, which may alter Trim24-regulated responses. Mammalian Trim28 (Kap1, TIF1ß) has known roles in epigenetic silencing, E3-SUMO-ligase activity and interacts with several proteins, including Mdm2 and E2F1 (26–28). To determine if Trim24 specifically regulates p53 in vivo, we turned to Drosophila where the gene bonus is the single, evolutionarily conserved homologue of the TIF1 subfamily (29), and most similar to TRIM24 and TRIM33.
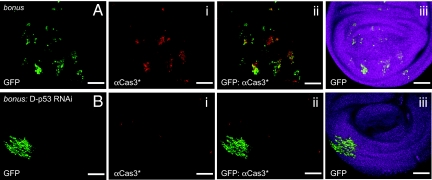
Bonus is a member of the Polycomb group of genes, and is required for male viability and metamorphosis (29). By mosaic analysis in imaginal discs, we found that clones of cells mutated in bonus, marked by green fluorescent protein (GFP, Fig. 3A), are small and highly apoptotic (Fig. 3Ai, cleaved caspase 3-positive). RNAi-mediated depletion of Drosophila p53 (D-p53) rescued the bonus apoptotic phenotype, resulting in considerably larger bonus clones and an absence of caspase 3 cleavage (Fig. 3B). Similar rescue was achieved by a dominant-negative form of mutant D-p53. These observations suggested that loss of bonus causes unrestrained D-p53 activity and induces apoptosis. Thus, bonus may be the principle negative regulator of D-p53 activity and cell death in tissues of Drosophila, which has no ortholog of Mdm2 or previously identified pathway regulating p53 protein stability (30).
Fig. 3.
Loss of Trim24 causes apoptosis in Drosophila. (A) bonus mutant clones [marked by GFP (A)] in Drosophila wing imaginal discs are small due to apoptosis (Cas3*-labeling). (B) RNAi-mediated depletion of D-p53 in bonus mutant clones (marked by GFP) suppresses apoptosis (lack of Cas3*-labeling) and leads to significant increase in clone size. Aiii and Biii show the outline of the disc. (Scale bar, 20 μm.)
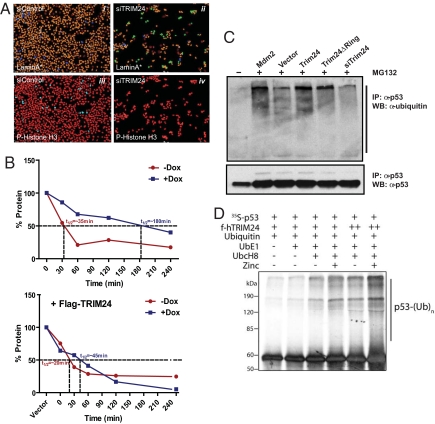
The link between TRIM24/bonus and p53-dependent cell death is conserved through evolution. Loss of TRIM24 in the human breast cancer cell line MCF7 (Fig. 4A) caused spontaneous apoptosis (ii, cleaved lamin A-positive), along with decreased mitosis (iv, phospho-H3-positive), compared to control cells (i and iii). Parallel siTRIM24/p53 studies in p53-positive MCF7, human lung carcinoma A549 and colon carcinoma HCT116 cells, as well as p53-depleted MCF7, p53-negative/mutant human prostate carcinoma PC3 and human breast carcinoma T47D, confirmed that p53-dependent apoptosis occurs with depletion of TRIM24 only in p53-positive cells (Fig. S4A and B). When TRIM24 is depleted in MCF7 cells, levels of p53 protein increase even in the absence of DNA damage (Fig. S4C).
Fig. 4.
TRIM24 negatively regulates p53 protein stability. (A) Depletion of TRIM24 in MCF7 cells increases apoptosis and decreases mitosis. Antibodies recognizing cleaved lamin-A (top) to detect apoptosis (green), or phospho-histone H3 (bottom) to detect cells in mitosis (blue), were used after transfection with siControl (left) and siTRIM24 (right) oligos. Cells were additionally stained with Hoescht followed by automated image acquisition and analysis. (B) p53-half-life is decreased by TRIM24 expression. MCF7 cells were treated with cycloheximide for 0, 15, 30, 60, 120, and 240 min (upper, vector only transfected) without DNA damage (–Dox) or with DNA damage (+Dox); and (lower, Flag-TRIM24) transfected with Flag-TRIM24 followed by similar treatment. p53, TRIM24 and Flag-TRIM24 protein levels were analyzed by immunoblotting, quantified by densitometry, and plotted against time to determine p53-half-lives. (C) Endogenous p53-ubiquitination assay: MCF7 cells were transfected with Mdm2, Flag-TRIM24, Flag-TRIM24ΔRing, siTRIM24, or vector control, as indicated. p53 was immunoprecipitated from lysates of transfected MCF7 cells, treated with MG132 (+) or untreated (-), and immunoblotted with anti-ubiquitin antibody to capture polyubiquitinated p53 and anti-p53 antibody to indicate levels of p53. (D) In vitro ubiquitination of p53: In vitro ubiquitination reactions were performed on in vitro translated 35S-p53 bound to hTRIM24 (+:20 μg, and ++:40 μg, estimated by staining) in the presence or absence of UbE1, UbcH8, and 25 μM zinc as indicated. Reactions were analyzed by autoradiogram.
TRIM24 Regulates p53 Protein Levels by Altering the Stability of p53.
Members of the TRIM family of proteins are defined by their conserved, N-terminal domains: a RING domain, 2 B-boxes, and a coiled-coil domain, while their C-terminal domains are more variable. Some members of the TRIM family are known E3-ubiquitin ligases, which ubiquitylate specific proteins for proteasome-mediated degradation (25). Specific negative regulators of p53, such as Mdm2, act via their RING domain in this manner to control p53 protein levels (7, 31).
We determined that the RING domain of TRIM24 functions as an E3-ubiquitin ligase to regulate p53 protein levels, similar to Mdm2 (Fig. 4C). Addition of an inhibitor of proteasomal-mediated degradation, MG132, led to increased p53 protein levels and detection of endogenously ubiquitylated p53. Ectopic expression of TRIM24 (Flag-TRIM24) or Mdm2 increased ubiquitylation of p53 and decreased p53 levels. When the RING domain of TRIM24 was deleted (TRIM24ΔRING), ubiquitylation of p53 decreased, which further declined when endogenous Trim24 in its entirety was depleted (siTrim24).
Ectopic expression of TRIM24 altered the kinetics of p53 protein decay (Fig. 4B and Fig. S5). Cessation of protein synthesis, by addition of cyclohexamide to MCF7 cells, led to a time-dependent loss of p53 protein, which was accelerated by ectopic expression of TRIM24. Stress-mediated induction of p53 (Dox) extended p53 protein half-life 5-fold, which is effectively opposed by ectopic TRIM24 (Fig. 4B). The role of TRIM24, as a direct regulator of p53 protein levels, is supported by in vitro ubiquitylation of p53 (Fig. 4D). Baculovirus-expressed, Flag-TRIM24 induced ubiquitylation of p53 in the presence of E2 UbcH8, in contrast to UbcH5 usage as an E2 by Mdm2 (32).
Our results suggest that Trim24 is a negative regulator of p53 that joins the ranks of Mdm2 and Cop1, as a RING-protein targeting p53 for degradation (33). Trim24 may lie at an intersection between networks of signaling pathways regulated by p53 and/or nuclear receptors. The dual roles of Trim24, as a co-repressor of specific nuclear receptors, for example, retinoic acid receptor (21), and regulator of p53 levels, likely dictate cell type and/or tissue-specific outcomes of Trim24-regulation (34). Mdm2 is clearly chief among the previously identified negative regulators of p53 levels, for example, PirH2, ARF-BP1, Cop1, and others (17, 18, 35). Whether these proteins negatively regulate p53 in specific cell types or under particular conditions awaits analysis of their functions in vivo.
Loss of p53's ability to serve as a tumor suppressor is the result of mutation of its gene and, more widely, dysfunction in pathways that modulate its stability, activation or downstream functions. Our identification of Trim24, as a p53-partner in cells of multiple origins, and its evolutionary conservation, as a negative regulator of p53, underscore the central nature of p53-control and its fine-tuning. Deregulation of E3-ubiquitin ligases, which target p53, is a common theme in human cancers (36). Therapeutic targeting of Trim24 protein to increase p53 protein levels may offer an avenue toward restoration of p53-functions and induced apoptosis of tumor cells.
Methods
Generation of p53TAP Mouse ES Cells and Mice.
A targeting vector to generate a C-terminal TAP tag fusion of p53 had a loxP-flanked PGK-neo cassette inserted into intron 10 of p53 and a TAP-tag cassette in frame with full-length p53 into exon 11, and contained approximately 3.5 kb 5′ and 2.5 kb 3′ homology arms with a MC1-TK negative selection marker. Details with regards to targeting and analysis of ES cells and generated mice are found in SI Text.
Drosophila Studies.
bonus mutant clones and RNAi-mediated inactivation of D-p53 were induced by the MARCM system (37), with the bonus21B allele (29). The UAS-D-p53 RNAi stock was provided by Clayton Morrison (Baylor College of Medicine).
Apoptosis Assays.
Apoptotic cells were counted by double staining with Annexin V (FITC) and propidium iodide as per manufacturer's instructions (BD PharMingen). Twenty thousand events were analyzed on Epics XL flow cytometer (Coulter) and % apoptosis was determined using System II software (Coulter). We performed immunohistochemistry for cleaved caspase-3 (Cell Signaling) on mouse embryonic brain sections as previously described (38).
TAP Purification, IP, Silver Stain, and Immunoblotting.
TAP-tag IP from mES cells was carried out using approximately 5–10 mg total protein whole cell lysate. The cells were lysed in lysis buffer 1 (LB1: 50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors Calbiochem, cat #539131), then clarified at 100,000 × g for 1 hour at 4°C. The supernatant was precleared with 125 μL (50% slurry) Protein A Sepharose beads (GE Healthcare) for 2 h rocking at 4°C. The beads were removed and anti-Protein A (P2921, Sigma) antibody was added to the precleared lysate overnight at 4°C. An identical approach was used with anti-TRIM24 (TIF1-α, A300–815A, Bethyl) for Trim24-IP. Beads were added as in preclearing and then collected in chromatography columns (Bio-Rad), followed by 1-mL washes for 10 min with LB1 (150 mM NaCl), LB2 (300 mM NaCl), LB3 (500 mM NaCl), LB2, and LB1. For TAP-IP with anti-Protein A, the protein complexes were cleaved away from the beads with 10 μg tobacco etch virus (TEV) protease in 20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 10% glycerol, 0.5 mM EDTA, 1 mM DTT, and protease inhibitors for 3 h at 4°C. IP-proteins were transferred to nitrocellulose membranes and immunoblotted with primary antibodies for p53 (CM5, Novocastra), TRIM24 (Bethyl), and β-actin (Genetex).
p53TAP Mice Survival Curve.
p53TAP/+ heterozygous mice were intercrossed to generate p53+/+, p53TAP/+, and p53TAP/TAP. Twenty mice from each genotype were monitored for tumor development, death, or signs of illness. The ages of the killed mice were recorded and Kaplan-Meier survival curves were calculated using Prism 5 (Graphpad).
Size-Exclusion Chromatography.
ES cells were collected, lysed and centrifuged as above. Approximately 0.5 mg mES whole cell lysate protein was subjected to size exclusion chromatography via Superose6 (10/25) (GE Healthcare) that was equilibrated with LB1 buffer. Details are found in SI Text.
Ubiquitination of p53.
MCF7 cells were transfected with plasmids, as indicated. Cells were treated with proteasomal inhibitor, MG132 for 6 h and lysed in RIPA buffer supplemented with 10 mM iodoacetamide (GE Healthcare) and protease inhibitors. Endogenous p53 was immunoprecipitated from 1 mg protein lysate using p53 (DO1) antibody and immunoblotted with anti-ubiquitin antibody (Sigma). The blot was reprobed with p53 to confirm the equal p53 pull down.
In Vitro Ubiquitination of p53.
N-terminal Flag-tagged human TRIM24 was expressed in a baculovirus system and purified, as previously described (39), but without elution from Flag-beads. In vitro ubiquitination was completed, as described previously (32), with the following modifications: 15 μL in vitro translated 35S-labeled p53 (TNT system, Promega) was incubated for 1 h at 4 °C with 20–40 μg bead-bound Flag-hTRIM24. The flag beads were washed 2 times, and incubated with 100 ng UbE1, 150 ng UbcH8, and 10 μg ubiquitin in the reaction buffer (BostonBiochem) for 1 h at 37 °C. Reactions were separated on 9% SDS/PAGE gels and the 35S-labeled p53 detected by autoradiogram.
Measurements of p53 Half Life.
MCF7 were transfected with Flag-TRIM24 or control DNA for 24 h and treated with 30 μg/mL protein synthesis inhibitor, cycloheximide (CHX), for different time points before analysis. To analyze p53 half life after doxorubicin (Dox), cells were pretreated with Dox for 4 h followed by treatment with CHX+Dox simultaneously. Protein levels were quantified as described above and plotted as percent protein left against time of CHX treatment to calculate p53 half life.
ChIP Assays and Additional Methods.
ChIP assays were preformed, as described previously (40), with modification as detailed in the SI Text. Cell Culture, siRNA depletion conditions and additional methods may be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank Cenix Bioscience, K. Vousden, S. Appikonda, G. Lozano, T. Terzian, R. Behringer, S. Dent, E. Koutelou, B. Atanassov, S. Stratton, and members of our laboratories for help with this work and H. Bellen (Howard Hughes Medical Institute, Baylor College of Medicine, Houston), G. Halder, and C. Morrison (University of Texas M.D. Anderson Cancer Center, Houston) for fly stocks. This work was supported by funds from the Kleberg Fund for Innovative Research, National Institutes of Health Grant GM081627, and CellCentric Ltd. (M.C.B.); the Kadoorie Foundation (R.L.J.); National Institutes of Health Grant GM068016 and the Welch Foundation (A.B.J.); and National Cancer Institute Cancer Center Support Grant to the University of Texas M.D. Anderson Cancer Center. A.K.J. is a Laura and John Arnold Foundation Odyssey Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11431.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813177106/DCSupplemental.
References
- 1.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 2.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 4.Levine AJ, Momand J, Finlay CA. The p53 tumor suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 5.Poyurovsky MV, Prives C. Unleashing the power of p53: Lessons from mice and men. Genes Dev. 2006;20:125–131. doi: 10.1101/gad.1397506. [DOI] [PubMed] [Google Scholar]
- 6.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 7.Clegg HV, Itahana K, Zhang Y. Unlocking the Mdm2–p53 loop: Ubiquitin is the key. Cell Cycle. 2008;7:287–292. doi: 10.4161/cc.7.3.5358. [DOI] [PubMed] [Google Scholar]
- 8.Hu W, Feng Z, Atwal GS, Levine AJ. p53: A new player in reproduction. Cell Cycle. 2008;7:848–852. doi: 10.4161/cc.7.7.5658. [DOI] [PubMed] [Google Scholar]
- 9.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 10.Feng Z, Hu W, Rajagopal G, Levine AJ. The tumor suppressor p53: Cancer and aging. Cell Cycle. 2008;7:842–847. doi: 10.4161/cc.7.7.5657. [DOI] [PubMed] [Google Scholar]
- 11.Christophorou MA, et al. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 12.Kokubo H, et al. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ Res. 2004;95:540–547. doi: 10.1161/01.RES.0000141136.85194.f0. [DOI] [PubMed] [Google Scholar]
- 13.Puig O, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 14.Han MK, et al. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 17.de Oca Luna RM, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:206–208. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 18.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 19.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Douarin B, et al. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 22.Le Douarin B, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong S, et al. A RA-dependent, tumour-growth suppressive transcription complex is the target of the PML-RARalpha and T18 oncoproteins. Nat Genet. 1999;23:287–295. doi: 10.1038/15463. [DOI] [PubMed] [Google Scholar]
- 24.Klugbauer S, Rabes HM. The transcription coactivator HTIF1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene. 1999;18:4388–4393. doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- 25.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Rauscher FJ, 3rd, Cress WD, Chen J. Regulation of E2F1 function by the nuclear corepressor KAP1. J Biol Chem. 2007;282:29902–29909. doi: 10.1074/jbc.M704757200. [DOI] [PubMed] [Google Scholar]
- 28.Zeng L, et al. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beckstead R, et al. Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit betaFTZ-F1-dependent transcription. Mol Cell. 2001;7:753–765. doi: 10.1016/s1097-2765(01)00220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006;13:909–912. doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- 31.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MH, Lozano G. Regulation of the p53-MDM2 pathway by 14–3-3 sigma and other proteins. Semin Cancer Biol. 2006;16:225–234. doi: 10.1016/j.semcancer.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Khetchoumian K, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- 35.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 37.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 38.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 40.Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J Biol Chem. 2005;280:24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.