Abstract
Trachealess (Trh) is a PAS domain transcription factor regulating Drosophila tracheogenesis. No other Trh homolog has been associated with a respiratory phenotype. Seeking homolog(s) regulating lung development, we screened murine genomic DNA using trh oligonucleotides, identifying only Npas3. Npas3 mRNA peaks in lung from E10.5 to E13.5, verified by sequencing, with immunostaining in airway epithelial cells. Npas3-null mice have reduced lung branching morphogenesis but are viable prenatally. Npas3-null newborns die in respiratory distress, with diminished alveolarization, decreased Shh, Fgf9, Fgf10, and Bmp4 mRNAs, and increased Spry2, consistent with reduced FGF signaling. Exogenous FGF10 rescues branching morphogenesis in Npas3-null lungs. In promoter reporter assays, NPAS3 directly upregulates Shh and represses Spry2. Npas3+/− mice have a milder lung phenotype, surviving postnatally, but develop emphysema. Therefore, absence of a developmentally expressed transcription factor can alter downstream gene expression and multiple signaling pathways in organogenesis. NPAS3 haploinsufficiency may also lead to emphysema.
Keywords: branching morphogenesis, emphysema, fibroblast growth factors, sonic hedgehog, transcription factors
The respiratory organ of Drosophila is the tracheal system (1), a tubular network that begins in the larva, opening to the outside at the spiracles. The basic structure resembles the lung, with branching of epithelial tubules, which transport O2 directly to target sites (1). Like the lung, the gas exchange surface area increases with branching, and most gas exchange occurs in the distal epithelium.
Drosophila tracheogenesis occurs in 3 phases (1): embryonic branching morphogenesis; formation of fine terminal branches, driven by O2 demands (2); and remodeling by cell proliferation and migration to form the adult tracheal system during metamorphosis. Mutants have been identified with specific tracheal defects (1). trachealess (trh)-null embryos have no trachea (1, 3). Trh is a bHLH transcription factor with 2 PAS domains (PER-ARNT-SIM) (4, 5). PAS domains regulate heterodimerization (6), a requirement for functional activity.
By phylogram analysis and amino acid sequence alignment, NPAS3 is most similar to Trh, followed by NPAS1, SIM1, SIM2, HIF-1α, EPAS1, and PER1 [supporting information (SI) Fig. S1]. Mice lacking NPAS1 and/or NPAS3 have been previously reported to have neurological defects (7, 8). Similarly, mice lacking SIM1 have neurological defects and die shortly after birth, but with no lung defects (9, 10). Also, Sim2-null mice die shortly after birth with respiratory distress from chest wall defects; however, detailed analysis of these mice revealed normal lungs (11).
Other proteins regulate Drosophila tracheogenesis downstream from Trh, including Breathless (FGFR), Branchless (FGF), Sprouty, and Notch (12, 13). In breathless-null embryos tracheal precursor cells invaginate but stay at the invagination site, and the main branches fail to form (1). Additional neurogenic gene mutants having pleiotropic and tracheal defects include Enhancer of Split (1) and Notch or its ligand, Delta, which have severe branching defects. Thus, multiple genes interact synergistically or additively to strongly favor normal development of the respiratory system. Multiple signaling pathways also permit fine-tuning of the structural parameters leading to function of the fully developed organ. We tested the hypothesis that a mammalian Trh homolog exists and that this homolog can activate multiple downstream signaling pathways involved in the lung development.
Results
Npas3 Expression in Developing Lungs.
We screened a murine genomic library using PCR primers specific for Drosophila trh. One clone was identified using a probe for the bHLH domain, confirmed with a PAS-A probe, and identified as murine Npas3.
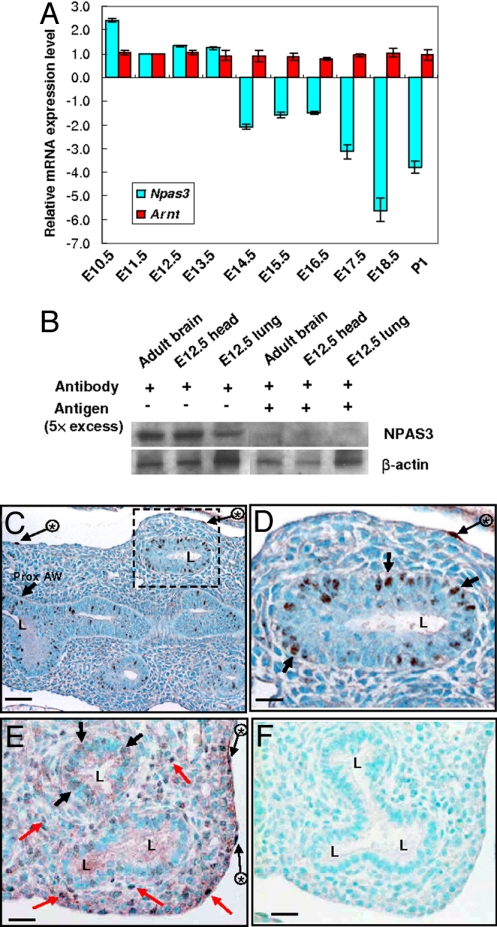
Using QRT-PCR we measured mRNA levels of Npas3 in developing lungs (primers given in Table S1). Arnt mRNA levels were also measured, because Tango, the Drosophila ortholog of ARNT, is required for Trh function via formation of heterodimer between Tango and Trh (4, 5). As shown in Fig. 1A, Npas3 expression is highest in E10.5 embryonic lungs, drops by ≈50% at E11.5, but remains high during E11.5−E13.5. Npas3 expression further decreases to ≈30%−60% of the E11.5 levels from E14.5 to E17.5, then to ≈20% of the E11.5 levels at E18.5 and postnatal day 1 (P1). Arnt mRNA levels have no significant change during E10.5–P1. To assess NPAS3 protein in developing lungs, Western blotting was carried out at E12.5, using adult brain and E12.5 head as positive controls. The 92-kDa NPAS3 band is present, and preabsorption of NPAS3 antibody with a 5-fold excess of peptide antigen abrogates the specific band (Fig. 1B). This peptide is entirely specific for NPAS3 (with no sequence similarities to NPAS1, NPAS2, or other PAS proteins).
Fig. 1.
Developmental expression of Npas3 mRNA and protein in murine lungs. (A) QRT-PCR analysis of Npas3 and Arnt mRNA in lungs from E10.5 to P1. (B) Western blot analysis of NPAS3 in E12.5 lungs, using adult brains and E12.5 heads as positive controls, and preabsorbing NPAS3 antibody with excess antigen for specificity. (C–F) NPAS3 immunohistochemistry in E13.5 (C and D) and E15.5 (E and F) lungs. Some NPAS3-positive epithelial nuclei (black arrows), mesothelial cells (long black arrows with asterisks), and mesenchymal cells (red arrows) are indicated. D is the higher magnification of dashed line box in C; F is a section immediately serial to E, running in parallel using antigen-preabsorbed NPAS3 antibody. L, airway lumen; Prox. AW, proximal airway. (Scale bars: C, 50 μm; D–F, 25 μm.)
We have also performed QRT-PCR analysis of Npas3 mRNA in postnatal lungs from 1 to 10 months of age (Fig. S2). Npas3 expression in adult lungs drops to ≈20% of the newborn levels and remains constant during 1–10 months. Thus, Npas3 continues to be expressed in adult lungs, albeit at lower levels than in embryonic and P1 lungs.
We then localized NPAS3 protein in developing lungs using immunohistochemistry. At E13.5, NPAS3 immunostaining is observed in airway epithelial cell nuclei (Fig. 1 C and D). At E15.5, NPAS3 immunostaining is weaker in epithelial cells, but multiple mesenchymal cells are NPAS3 positive (Fig. 1E). NPAS3 positivity also occurs in mesothelial nuclei on the pleural surface (Fig. 1 C–E). All NPAS3 immunostaining is removed by preabsorption of NPAS3 antiserum with a 5-fold excess of peptide antigen (Fig. 1F).
Analyses of Npas3-Null Mice.
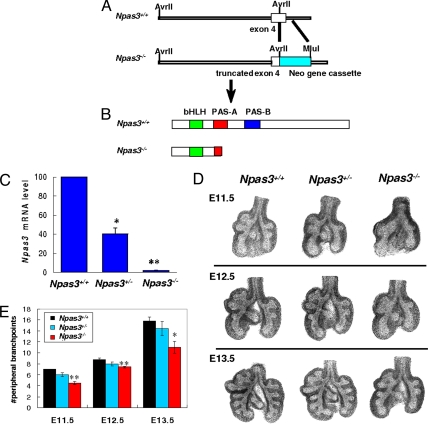
Knowing that Npas3 is expressed in embryonic and postnatal lungs, we generated Npas3-null mice by targeting the PAS-A domain, which is needed for heterodimerization and transactivation by NPAS3 (5), similar to Trh (14). PAS-A also determines target gene specificity (15). A part of exon 4 and intron 4 of Npas3 gene was replaced by Neo gene cassette (Fig. 2A), resulting in a truncated PAS-A domain with premature termination (Fig. 2B). By QRT-PCR, Npas3-null mice lack Npas3 mRNA, whereas heterozygote (Het) littermates have ≈50% of wild-type (WT) levels (Fig. 2C).
Fig. 2.
Npas3 targeting and branching morphogenesis in Npas3-deficient lungs. (A and B) To generate Npas3-null mice, a 1.4-kb DNA fragment in exon 4 and intron 4 of murine Npas3 gene was replaced by Neo gene cassette, leading to a frameshift mutation and premature termination. (C) QRT-PCR analysis of Npas3 mRNA from Npas3-deficient vs. WT littermate lungs at E12.5. *P < 0.01, ** P < 0.001, n = 4. (D) Representative WT vs. Npas3-deficient lung buds at E11.5, E12.5, and E13.5. (E) Pooled results of numbers of peripheral branch points in lung buds, with 4–5 litters per timepoint. Branching of Npas3−/− lung buds significantly reduced compared with Npas3+/+ littermates. *P < 0.05, **P < 0.01.
We analyzed in vivo lung branching morphogenesis in Npas3-deficient embryos at E11.5, E12.5, and E13.5 (Fig. 2 D and E). Npas3-null mice have significantly reduced branching morphogenesis at all these time points, similar to inducible trh-null Drosophila (3).
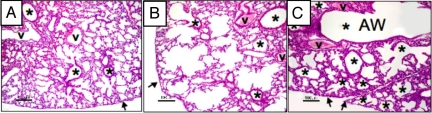
Within hours after birth, most of the Npas3-null pups become gasping and cyanotic (Fig. S3). By 48 h, 95% of Npas3-null mice die, whereas all of the WT and Npas3-Hets survive. Lung histopathology on P1 shows normal alveolar structure in WT pups (Fig. 3A), at the saccular stage of development, just preceding true alveolarization, which begins at ≈P3. However, Npas3-Hets have widespread defects in alveolar septation, resulting in lungs that resemble centrilobular emphysema (Fig. 3B). Npas3-null pups are most severely affected, with only patches of future alveolar tissue interspersed with dilated airways, giving the appearance of bronchiectasis (Fig. 3C).
Fig. 3.
Histopathological features of Npas3-deficient lungs at P1. Lung histopathology at P1 for (A) WT, (B) Npas3-Het, and (C) Npas3-null littermates. Compared with WT, Npas3-deficient lungs have dilated airways with few alveoli. Npas3-Hets have defects in the primitive alveolar parenchyma alternated with segments of more normal alveoli (saccular stage) and many airways extend near the pleura (arrows). Npas3-null pups have reduced alveolar parenchyma. The conducting airways are massively dilated, similar to bronchiectasis. v, blood vessel; *, airway (AW). (Scale bar: 100 μm.)
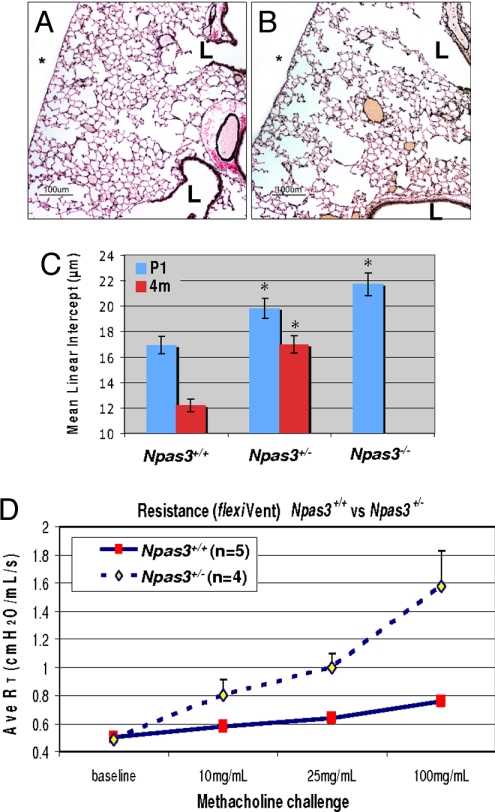
To determine if Npas3-deficient mice can repair these lung defects by adulthood, we performed histopathology and pulmonary function testing at 4 months of age (4 mo). Compared with WT littermates (Fig. 4A), Npas3-Hets have decreased alveolarization (Fig. 4B), similar to defects in primitive alveoli observed at P1 (Fig. 3 B and C), with the appearance of centriacinar emphysema. To quantify these differences, mean linear intercepts (MLI) were determined by computerized image analysis at P1 and 4 mo. MLI is widely used to measure mean alveolar diameter, which in the absence of injury reflects alveolar development. As shown in Fig. 4C, the normal MLI in WT mice decreased from ≈17 μm at P1 to ≈12 μm at 4 mo, a 28% decrease in mean alveolar airspace diameter. In contrast, the MLI of Npas3-Hets decreased from ≈20 μm at P1 to ≈17 μm at 4 mo, a 14% decrease. Both Npas3-null and -Het mice at P1 or 4 mo have significantly increased MLI compared with WT littermates, consistent with defective distal lung development and/or alveolar homeostasis both pre- and postnatally.
Fig. 4.
Lung histopathology of adult Npas3-deficient mice. (A and B) Lung histopathology from WT (A) and Npas3-Het littermates (B) at 4 months (4 mo) of age (Voerhoff's elastic tissue stain). L, airway lumen; *, pleural surface; bar, 100 μm. (C) Mean linear intercepts of lungs from P1 and 4-mo mice (3 litters each): *P < 0.0001 compared with WT. Note that there were no 4-mo-old Npas3-null mice because pups rarely survived beyond 48 h.
Considering the similarity of lung histopathology between Npas3-null mice and human disorders with reduced alveolarization, such as emphysema, we carried out pulmonary function testing of Npas3-Hets and WT littermates using the flexiVent apparatus. At 3, 4, 6, and 7 mo, Npas3-Hets do not have altered baseline airway resistance. Neither airway inflammation nor consistent evidence of pulmonary neuroendocrine cell hyperplasia was observed in Npas3-deficient mice.
Altered Downstream Gene Expression in Npas3-Null Lungs.
To explore mechanisms underlying the pulmonary phenotype of Npas3-deficient mice, we carried out QRT-PCR analyses for a panel of 22 different genes implicated in lung development. At E11.5, no significant difference was observed in any of 22 mRNAs between WT and Npas3-null lungs. At P1, however, there are multiple differences in gene expression (Table 1). In Npas3-null lungs, Shh mRNA is decreased most significantly (<30% of the WT levels). There is also a 50% decrease in Gli2 gene expression, which functions downstream from SHH to mediate its effects on transcription. Expression of other FGF signaling mediator genes Fgf9, Fgf18, and Ffg10 is decreased approximately one-third to one-half, whereas expression of sprouty2 (Spry2), an antagonist of FGF signaling, is increased ≈3-fold. Bmp4, which interacts with both SHH and FGF signaling pathways, has significantly reduced gene expression in lungs of Npas3-null mice.
Table 1.
QRT-PCR analyses of altered gene expression in P1 lungs from Npas3-null mice compared with WT littermates
| Gene | Fold change (null vs. WT) | P value |
|---|---|---|
| Shh | −3.2 | 0.0028 |
| Fgf9 | −1.9 | 0.0011 |
| Gli2 | −1.9 | 0.0364 |
| Bmp4 | −1.6 | 0.0125 |
| Fgf18 | −1.5 | 0.0460 |
| Fgf10 | −1.4 | 0.0426 |
| Spry2 | 2.7 | 0.0289 |
| Fgf7 | 1.4 | 0.0399 |
To assess the abundance of some of the corresponding proteins, we immunostained E15.5 lungs from Npas3-null embryos and their WT littermates (Fig. S4). WT lungs have strong SHH immunostaining in proximal airway epithelium and throughout the mesenchyme (Fig. S4A). In Npas3-null lungs, there is a similar distribution of SHH, but the immunostaining is weaker and there are fewer positive cells (Fig. S4B). In contrast, WT lungs have SPRY2 (Fig. S4C) present in scattered mesenchymal cells and proximal epithelial cells, whereas Npas3-null lungs have the increased numbers of SPRY2-positive cells in distal airway epithelium (Fig. S4D). FGF10 immunostaining occurs in WT lung mesenchymal cells, most strongly in the subpleural mesenchyme (Fig. S4E), whereas Npas3-null lungs have reduced FGF10 immunostaining, almost entirely in the subpleural region (Fig. S4F).
Considering the role of FGF signaling in angiogenesis and the link between alveolarization and development of the pulmonary capillary bed, we assessed the pulmonary vasculature by laminin immunostaining. Compared with WT lungs, which have many subpleural laminin-positive capillaries and larger vascular channels (Fig. S4G), Npas3-null lungs have only a few dysmorphic subpleural capillaries, despite multiple larger vascular structures (Fig. S4H). These observations suggest that NPAS3 could regulate angiogenesis in the lung and brain, possibly via FGF signaling molecules and SHH.
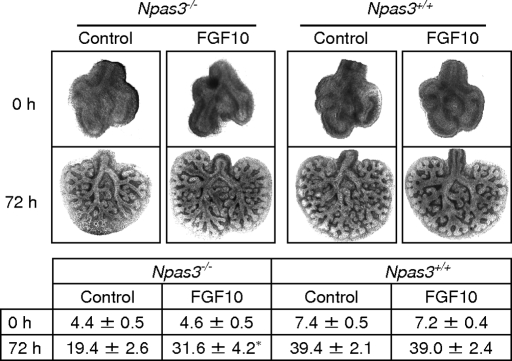
To determine which altered proteins might contribute to the Npas3-null pulmonary phenotype, we cultured WT and Npas3-null E11.5 lung buds with exogenous FGF10 in the growth medium. FGF10 restored branching morphogenesis in Npas3-null lung buds (Fig. 5). However, although the peripheral branching of Npas3-null lung buds significantly increased with exogenous FGF10, it did not reach the level of branching of WT littermate controls, suggesting the deficiency of other regulators of branching morphogenesis in Npas3-null lungs.
Fig. 5.
FGF10 restores branching of Npas3-null embryonic lungs in vitro. Littermate Npas3+/+ and Npas3−/− E11.5 lungs were cultured in growth medium alone (control) or plus 500 ng/mL recombinant human FGF10 for 72 h. (Upper) Representative lung buds cultured in medium or medium plus recombinant FGF10 at 0 h and 72 h. (Lower) Pooled data of lung peripheral branch-point number (mean ± SD) at 0 h and 72 h. Asterisk (*) indicates branching of Npas3−/− lung buds was significantly increased with FGF10 at 72 h (P < 0.01, n = 5), but was less than littermate WT control (P < 0.05, n = 5).
We then assessed whether FGF10 can rescue a cell migration defect in E11.5 Npas3-null lungs. The mesenchyme was removed from distal tips of lung buds. Neither WT nor Npas3-null epithelial cells migrated toward the negative control bead (Fig. S5). In contrast, WT epithelial cells migrated rapidly toward the FGF10-coated bead, but Npas3-null epithelial cells did not migrate toward FGF10. Thus, FGF10 can rescue defective branching but not defective migration in Npas3-null lungs.
These cumulative observations indicate that NPAS3 plays an important role in early lung morphogenesis by regulating expression of multiple genes that play major roles in organogenesis. To determine whether NPAS3 might directly regulate expression of any of these potential target genes, we carried out luciferase reporter assays using a Shh promoter construct (16) and a Spry2 promoter construct (17) that had been previously characterized. A NPAS3 expression vector was constructed by cloning murine Npas3 cNDA into pcDNA3.1. A pcDNA1-ARNT expression construct was also obtained (18). These constructs were used to transfect into a murine lung type II cell line, MLE-12 (19). NPAS3 alone induces <2-fold increase of Shh promoter transactivation, whereas NPAS3 plus ARNT synergistically increase Shh luciferase activity ≈5-fold (Fig. S6A). In contrast, NPAS3 alone represses Spry2 promoter transactivation to ≈10% of baseline levels, and NPAS3 plus ARNT synergistically decrease Spry2 luciferase activity to <2% of baseline levels (Fig. S6B). Western blots confirmed that NPAS3 levels are comparable between the experimental groups for both reporter constructs (data not shown).
Discussion
The present study provides definitive evidence for NPAS3 being the functional mammalian homolog of Trh. NPAS3 has highest amino acid sequence homology to Trh, especially in the consensus domains. Our observations support the hypothesis that NPAS3 is essential for normal lung development. In addition, NPAS3 plays a significant role in maintaining lung homeostasis, because haploinsufficient adults develop alveolar loss. During development, NPAS3 functions by transactivating genes important for lung development and/or regeneration, such as Shh, and many of the same genes could be implicated in maintaining lung homeostasis. Nonetheless, the spectrum of targets of NPAS3 must differ in part from those of Trh because Npas3-null mice do have normal tracheal development. These differences support the concept of NPAS3 as being an ortholog of Trh, despite structural similarities in branching morphogenesis for mice and fruit flies.
Impaired lung structure and function in Npas3-deficient mice lead to pulmonary lethality shortly after birth. This is at least partly due to altered gene expression of multiple FGF signaling molecules, similar to Trh (4). Several laboratories have validated the importance of FGF signaling in lung development (20–22). Vascular development is an intrinsic part of alveolarization, and multiple growth factors contribute to this vital process, including FGFs and VEGFs (23).
The mechanism by which NPAS3 controls lung development is complex, with upregulation of FGF signaling at multiple levels, decreased gene expression of Spry2 as an FGF inhibitor, and activation of Shh gene expression (Fig. 6). Addition of FGF10 can rescue branching morphogenesis of lung buds from Npas3-null embryos. However, FGF10 does not rescue the defect in epithelial cell migration. Cumulatively, these observations are consistent with NPAS3 activating or suppressing multiple distinct signaling pathways in lung development and repair. A single gene target may be implicated in only part of the Npas3-deficient phenotype, as we demonstrated for FGF10. NPAS3 is not only a proximal regulator of lung development, but also appears to have a significant influence on postnatal lung pathology.
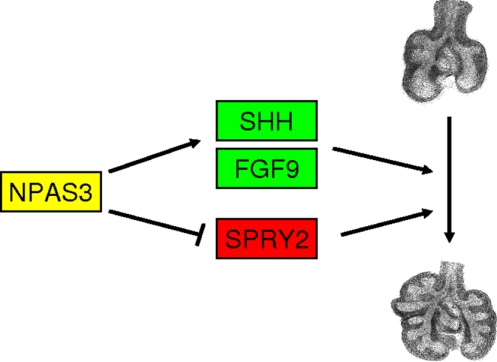
Fig. 6.
Working hypothesis. Schematic drawing of how NPAS3 might function in epithelial and mesenchymal cells in developing lungs. NPAS3 regulates transcription of Shh and Fgf9, which would lead to increased SHH signaling via PATCHED with activation of Gli2 and the cascade of genes activated by SHH signaling. NPAS3 activation of FGF signaling includes FGF9 and FGF10, which reciprocally activate both epithelial and mesenchymal cells. FGFR2 signaling is inhibited by SPRY2, but NPAS3 represses Spry2 expression, and thus facilitates FGF signaling during lung development.
In conclusion, investigation of how NPAS3 deficiency might be leading to abnormal lung development and emphysema will open new roads for understanding the fetal origins of adult disease. Emphysema is a highly prevalent and debilitating chronic lung disease that is initiated or exacerbated by cigarette smoking or air pollution. We have demonstrated that Npas3 continues to be expressed in adult lungs, albeit at lower levels than in embryonic or P1 lungs. Therefore, the progressive postnatal phenotype of Npas3-deficient mice may be related to both structural defects from developmental dysregulation in utero and/or a reduction in ongoing adult expression that is needed for normal homeostasis. In genomic analyses, associations have been identified between human chromosome 14q12–13 (where NPAS3 maps) and inflammatory lung diseases (24–26). Thus, Npas3-deficient individuals could be at high risk for free radical injury due to an apparent defect in alveolar repair. Even in adults, new onset secondary septation can prevent and/or repair the alveolar destruction of emphysema (27). NPAS3 could also play a role in bronchopulmonary dysplasia (BPD) because lungs from the newborn Npas3-null mice have diminished alveolarization, similar to infants with BPD. This suggests that NPAS3-induced responses during postnatal lung injury are likely to be similar to pathways triggered during lung development, such as SHH and FGF signaling cascades.
Materials and Methods
See details in SI Materials and Methods.
Genomic Library Screening.
A murine genomic BAC library was screened using 2 pairs of PCR primers specific for Drosophila trh gene. Hybridizing restriction fragments from positive clones were sequenced.
RNA Isolation and QRT-PCR.
Genomic DNA-free total RNA isolated from murine lungs was reverse transcribed. Quantitative real-time PCR was performed using TaqMan or SYBR technology with ABI-PRISM 7300 detection system.
Npas3 Gene Targeting and Generation of Knockout Mice.
A 1.4-kb DNA fragment in exon 4 and intron 4 of murine Npas3 gene was replaced by Neo gene cassette. The targeting vector was transfected into 129Sv/Ev embryonic stem (ES) cells by electroporation. Correctly targeted ES cells were microinjected into C57BL/6J blastocysts to generate chimeras and germline transmission of the Npas3-null mice. The founders were inbred on C57BL/6 genetic background for over 10 generations.
Western Blot Analyses.
Proteins were fractionated on 7.5% SDS-PAGE and transferred to PVDF membrane. Immunoblotting was performed using anti-NPAS3 antibody, HRP-conjugated secondary antibody, and the enhanced chemiluminescent reagent. Antibody preabsorbed with excess antigenic peptide was used as the negative control.
Histopathology and Immunohistochemistry.
Tissues were fixed in 4% PFA and embedded in paraffin. Immunostaining of paraffin sections was performed using the primary antibodies, biotylated secondary antibodies, and ABC method as previously described (28).
Promoter Assay.
The Npas3 cDNA (154–2778) was cloned into pcDNA3.1/V5-His vector. This NPAS3 construct contains the functional bHLH, PAS-A, and PAS-B domains. pGL3-Shh promoter vector was from M. Kadakia (Wright State University, Dayton, OH) (16). pGL2-Spry2 reporter construct was from D. Warburton (University of Southern California, Los Angeles, CA) (17). pcDNA1-ARNT was from O. Hankinson (University of California, Los Angeles, CA) (18). MLE-12 cells were transfected using Lipofectamine 2000, and luciferase activity was measured 48 h after transfection.
Embryonic Lung Explant Culture.
E11.5 lungs were cultured as described (29). In rescue experiments, recombinant human FGF10 was added to growth medium at 500 ng/mL, and lung explant cultures were maintained at 37 °C, 5% CO2 for 72 h.
FGF10-Coated Bead Cultures.
Heparin beads were washed with medium alone or further soaked with FGF10. Mesenchyme-free epithelium from E11.5 lung bud tips and a FGF10 coated bead (or medium washed bead) were embedded into growth factor-reduced Matrigel and cultured at 37 °C, 5% CO2 for 72 h.
Supplementary Material
Acknowledgments.
We thank Dr. David Warburton for providing the Sprouty2 reporter construct, Dr. Madhavi Kadakia for the Shh promoter construct, and Dr. Oliver Hankinson for the ARNT expression construct. We thank Dr. John Shannon and Mike Burhans for technical instruction on FGF10-coated bead culture experiments. We appreciate the excellent administrative assistance of Karen Hunt. This work was supported by National Institutes of Health (NIH) Grant 2R01 HL44984 (to M.E.S.) and an Established Investigator Award from the American Asthma Foundation. The Mouse Physiology Core was also supported by NIH Grants ES 011961 and ES 016347 (to W.M.F. and E.N.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902426106/DCSupplemental.
References
- 1.Manning G, Krasnow MA. Development of Drosophila Melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. Development of the Drosophila tracheal system; pp. 609–685. [Google Scholar]
- 2.Affolter M, et al. Tube or not tube: Remodeling epithelial tissues by branching morphogenesis. Dev Cell. 2003;4:11–18. doi: 10.1016/s1534-5807(02)00410-0. [DOI] [PubMed] [Google Scholar]
- 3.Wilk R, Weizman I, Shilo B-Z. Trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- 4.Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284:1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- 5.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 6.Pongratz I, Antonsson C, Whitelaw ML, Poellinger L. Role of the PAS domain in regulation of dimerization and DNA binding specificity of the dioxin receptor. Mol Cell Biol. 1998;18:4079–4088. doi: 10.1128/mcb.18.7.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erbel-Sieler C, et al. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci USA. 2004;101:13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunskill EW, et al. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur J Neurosci. 2005;22:1265–1276. doi: 10.1111/j.1460-9568.2005.04291.x. [DOI] [PubMed] [Google Scholar]
- 9.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogan BL. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 11.Goshu E, et al. Sim2 mutants have developmental defects not overlapping with those of Sim1 mutants. Mol Cell Biol. 2002;22:4147–4157. doi: 10.1128/MCB.22.12.4147-4157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelzer E, Shilo BZ. Cell fate choices in Drosophila tracheal morphogenesis. Bioessays. 2000;22:219–226. doi: 10.1002/(SICI)1521-1878(200003)22:3<219::AID-BIES3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.Rosin D, Shilo BZ. Branch-specific migration cues in the Drosophila tracheal system. Bioessays. 2002;24:110–113. doi: 10.1002/bies.10041. [DOI] [PubMed] [Google Scholar]
- 14.Shilo BZ, et al. Branching morphogenesis in the Drosophila tracheal system. Cold Spring Harb Symp Quant Biol. 1997;62:241–247. [PubMed] [Google Scholar]
- 15.Zelzer E, Wappner P, Shilo BZ. The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caserta TM, et al. p63 overexpression induces the expression of Sonic Hedgehog. Mol Cancer Res. 2006;4:759–768. doi: 10.1158/1541-7786.MCR-05-0149. [DOI] [PubMed] [Google Scholar]
- 17.Ding W, Bellusci S, Shi W, Warburton D. Functional analysis of the human Sprouty2 gene promoter. Gene. 2003;322:175–185. doi: 10.1016/j.gene.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Reisz-Porszasz S, Probst MR, Fukunaga BN, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT) Mol Cell Biol. 1994;14:6075–6086. doi: 10.1128/mcb.14.9.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacDonald JI, Possmayer F. Stimulation of phosphatidylcholine biosynthesis in mouse MLE-12 type-II cells by conditioned medium from cortisol-treated rat fetal lung fibroblasts. Biochem J. 1995;312:425–431. doi: 10.1042/bj3120425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Izvolsky KI, Qian J, Cardoso WV. Identification of FGF10 targets in the embryonic lung epithelium during bud morphogenesis. J Biol Chem. 2005;280:4834–4841. doi: 10.1074/jbc.M410714200. [DOI] [PubMed] [Google Scholar]
- 21.Hyatt BA, Shangguan X, Shannon JM. FGF-10 induces SP-C and Bmp4 and regulates proximal-distal patterning in embryonic tracheal epithelium. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1116–L1126. doi: 10.1152/ajplung.00033.2004. [DOI] [PubMed] [Google Scholar]
- 22.Volpe MV, Ramadurai SM, Pham LD, Nielsen HC. Hoxb-5 down regulation alters Tenascin-C, FGF10 and Hoxb gene expression patterns in pseudoglandular period fetal mouse lung. Front Biosci. 2007;12:860–873. doi: 10.2741/2108. [DOI] [PubMed] [Google Scholar]
- 23.Galambos C, et al. Defective pulmonary development in the absence of heparin-binding vascular endothelial growth factor isoforms. Am J Respir Cell Mol Biol. 2002;27:194–203. doi: 10.1165/ajrcmb.27.2.4703. [DOI] [PubMed] [Google Scholar]
- 24.Demoly P. Respiratory allergic disease genes (Translated from French) Rev Pneumol Clin. 2003;59:67–75. [PubMed] [Google Scholar]
- 25.Blumenthal MN, et al. A genome-wide search for allergic response (atopy) genes in three ethnic groups: Collaborative Study on the Genetics of Asthma. Hum Genet. 2004;114:157–164. doi: 10.1007/s00439-003-1030-5. [DOI] [PubMed] [Google Scholar]
- 26.CSGA. A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15:389–392. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 27.Massaro GD, Massaro D. Retinoic acid treatment abrogates elastase-induced pulmonary emphysema in rats. Nat Med. 1997;3:675–677. doi: 10.1038/nm0697-675. [DOI] [PubMed] [Google Scholar]
- 28.Sunday ME, Yoder BA, Cuttitta F, Haley KJ, Emanuel RL. Bombesin-like peptide mediates lung injury in a baboon model of bronchopulmonary dysplasia. J Clin Invest. 1998;102:584–594. doi: 10.1172/JCI2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levesque B, et al. NPAS1 regulates branching morphogenesis in embryonic lung. Am J Respir Cell Mol Biol. 2007;36:427–434. doi: 10.1165/rcmb.2006-0314OC. [DOI] [PMC free article] [PubMed] [Google Scholar]