Abstract
Many cell–cell adhesive events are mediated by the dimerization of cadherin proteins presented on apposing cell surfaces. Cadherin-mediated processes play a central role in the sorting of cells into separate tissues in vivo, but in vitro assays aimed at mimicking this behavior have yielded inconclusive results. In some cases, cells that express different cadherins exhibit homotypic cell sorting, forming separate cell aggregates, whereas in other cases, intermixed aggregates are formed. A third pattern is observed for mixtures of cells expressing either N- or E-cadherin, which form distinct homotypic aggregates that adhere to one another through a heterotypic interface. The molecular basis of cadherin-mediated cell patterning phenomena is poorly understood, in part because the relationship between cellular adhesive specificity and intermolecular binding free energies has not been established. To clarify this issue, we have measured the dimerization affinities of N-cadherin and E-cadherin. These proteins are similar in sequence and structure, yet are able to mediate homotypic cell patterning behavior in a variety of tissues. N-cadherin is found to form homodimers with higher affinity than does E-cadherin and, unexpectedly, the N/E-cadherin heterophilic binding affinity is intermediate in strength between the 2 homophilic affinities. We can account for observed cell aggregation behaviors by using a theoretical framework that establishes a connection between molecular affinities and cell–cell adhesive specificity. Our results illustrate how graded differences between different homophilic and heterophilic cadherin dimerizaton affinities can result in homotypic cell patterning and, more generally, show how proteins that are closely related can, nevertheless, be responsible for highly specific cellular adhesive behavior.
Keywords: binding affinities, cadherins, cell adhesion, differential adhesion hypothesis, surface plasmon resonance
Expression of different cadherins has been associated with the sorting of cells into distinct layers or compartments (1, 2). This behavior is often viewed as a manifestation of homotypic cell-sorting behavior—like cells adhere to one another. However, cell layers characterized by the expression of different cadherins sometimes remain in contact with one another, suggesting that heterotypic adhesion may also be of physiological relevance. Consistent with in vivo observations, in vitro aggregation assays have shown that cells expressing different classical cadherins can adhere to one another (3, 4). In some such instances, cells form distinct aggregates that possess a common interface, whereas in others, cells are completely mixed. Thus, cells expressing cadherins can exhibit homotypic and/or heterotypic adhesive properties, albeit for reasons that remain to be explained. Here, we probe the molecular basis of this behavior.
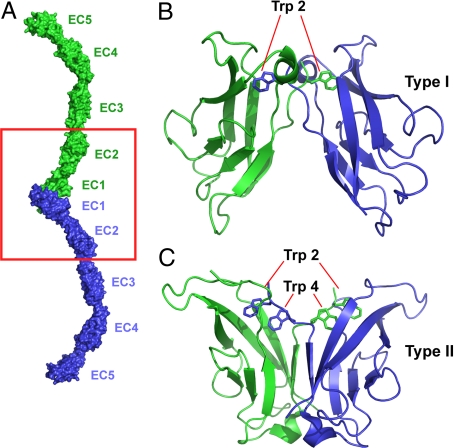
Cadherins constitute a large family of cell surface adhesion receptors that can be grouped into numerous subfamilies (5). The type I and type II “classical cadherins” are found only in vertebrates. Classical cadherins (Fig. 1A) contain 5 extracellular cadherin domains (EC1–EC5; Fig. 1A), a single transmembrane segment, and a cytoplasmic domain that contains highly conserved binding sites for catenin proteins, which provide indirect links to the cytoskeleton (1, 2). Wild-type classical cadherins dimerize through an interface formed solely between their EC1 domains (Fig. 1 B and C) (6). This interface (known as the “strand dimer”), which has been well-characterized in numerous structural studies (7–10), is formed in part through the swapping of an N-terminal β-strand. There are 6 type I cadherins and 13 type II cadherins in vertebrates; differences between them play a crucial role in the sorting of cells into separate tissues.
Fig. 1.
Structure of cadherin molecules. (A) The structure of the adhesive dimeric complex formed by ectodomains of type I C-cadherin (7) is shown in molecular surface representation. Note that the adhesive interface is encompassed entirely within the EC1 domain. The red boxed region includes the interacting EC1–2 regions corresponding to the cadherin constructs used in this study. (B) Expanded ribbon diagram view of the strand-exchanged adhesive interface between the N-terminal EC1 domains of the type I E-cadherin (9). The side chains of the Trp anchor residues are shown. (C) A ribbon diagram view of the strand-exchanged adhesive interface between the N-terminal EC1 domains of the type II cadherin 8 (15). The side chains of the Trp-2 and Trp-4 anchor residues are indicated.
We have focused our analysis on the interactions of 2 type I cadherins, N-cadherin and E- cadherin, that have been implicated in morphogenetic processes involving both heterotypic and homotypic adhesive interactions (1, 2). E- and N-cadherins are very similar both in sequence and structure (6, 11). The 2 EC1 domains superimpose geometrically almost perfectly (11), and their interfacial residues are very similar. How do the small differences between them mediate highly specific cell-patterning behavior (12)?
To begin to address this question, we carried out aggregation assays with cells expressing either N- or E-cadherin and, in parallel, used equilibrium analytical ultracentrifugation (AUC) and surface plasmon resonance (SPR) to determine the homophilic and heterophilic binding affinities of the individual cadherin molecules. Relating the observed molecular and cellular behaviors requires the forging of a theoretical link between cadherin dimerization affinities and cell–cell adhesive strengths. We established this link in 2 sequential steps. By using the measured binding affinities, we first calculated the work, W(I, J), associated with separating 2 cells of types I and J. The values of W(I, J) for cells expressing N- and E-cadherins are then used in conjunction with the differential adhesion hypothesis (DAH) to predict the behavior of mixtures of such cells. Our experimental results and theoretical analysis provide a conceptual basis for understanding the sorting and adhesive behaviors of cadherin-expressing cells as derived from the molecular properties of closely related cadherin proteins.
Results
Cell Aggregation Assays.
Aggregation assays using transfected L and CHO cells have been widely used to study the cellular adhesive properties of different cadherins; however, the observations have not generally lent themselves to unambiguous interpretation. Steinberg and coworkers (4, 13) in particular have emphasized that the assays are often carried out under different shear forces that can affect the outcome of the experiments. This is perhaps not unexpected. After initial cell–cell contact, a variety of processes, such as junction formation and intercellular signaling, are initiated, and the extent to which they compete kinetically with the equilibration of cell mixtures may well influence in vitro behavior. Here, we report a series of cell assays carried out under different shear force conditions that pertain directly to the results of this work.
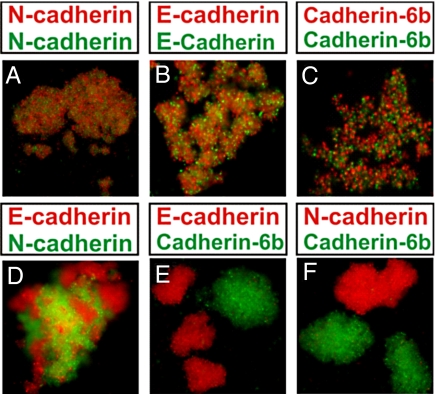
Cell aggregation assays were carried out with CHO cells expressing either N- or E-cadherins, or cadherin-6b, a type II cadherin. In mixing assays, cell suspensions were permitted to aggregate with agitation at 70–80 rpm, whereas in hanging-drop assays (Fig. S1), no agitation was applied, thus minimizing shear force. Cells expressing the same cadherin formed intermixed aggregates (Fig. 2). In contrast, mixtures of N-cadherin-expressing and E-cadherin-expressing cells formed distinct homotypic aggregates that adhered to one another (Fig. 2D), whereas mixtures of cells expressing cadherin 6b and N- or E-cadherins formed separate aggregates that did not adhere to one another (Fig. 2 E and F).
Fig. 2.
Mixing cell aggregation assays with cadherin-expressing CHO cells. Mixing aggregation assays using 2 identical CHO cell lines expressing the same cadherin—(A) N-cadherin with N-cadherin, (B) E-cadherin with E-cadherin, and (C) cadherin-6b with cadherin-6b—result in aggregates composed of an interspersed mixture of each cell line. (D) Dissociated cells expressing E- and N-cadherins form separate homotypic aggregates that adhere to one another. In contrast, cells that express type II cadherin-6b form aggregates that do not adhere to cells expressing either E-cadherin (E) or N-cadherin (F). (Magnification: 20×.)
The results obtained with the hanging-drop assays (Fig. S1) are qualitatively similar. In assays on cell lines expressing the same cadherin, the expected intermixing was clearly observed. In contrast, mixtures of cells expressing N- and E-cadherins formed separate yet mutually adhesive, homotypic aggregates that produced a mosaic pattern. Finally, as was observed for the mixing assay, cells expressing cadherin-6b formed separate aggregates that did not adhere to cells expressing either N- or E-cadherin.
Patterns such as those displayed in Fig. 2D have been observed previously (3, 14) and have been interpreted in terms of homotypic cell-sorting segregation (3). On the other hand, the pattern seen in Fig. S1D can be interpreted easily in terms of complete intermixing. However, we are suggesting here that a more nuanced interpretation of cell aggregation data is appropriate. Specifically, Fig. 2 and Fig. S1 illustrate examples of (i) complete mixing of cells expressing the same cadherin (Fig. 2 A–C and Fig. S1 A–C); (ii) the formation of separate aggregates that adhere to one another (Fig. 2D and Fig. S1D); and (iii) the formation of separate aggregates that do not adhere to one another (Fig. 2 E and F and Fig. S1 E and F). There are obvious differences in the patterns observed in all 3 cases that can, in principle, be related to the molecular properties of the cadherins that are expressed on the cell surface.
Indeed, despite the ambiguities associated with the cell assays, the patterns observed clearly reflect the adhesive properties of cadherin molecules. For example, the identity of the specificity-determining EC1 domain affects cell-patterning behavior, as observed, for example, in both in vitro and in vivo studies with chimeras where the EC1 domain of one cadherin has been replaced with that of another (14, 15). In addition, the W2A mutant, which disrupts the EC1–EC1 interface, is known to abrogate in vitro cell–cell adhesion (14). On the other hand, cells expressing some cadherins (e.g., P- and E-cadherins) intermix completely (4). Why is this? It seems apparent that these issues cannot be addressed without quantitative measurements of the dimerization properties of individual cadherin molecules.
AUC Measurements of Homophilic Binding Affinities.
Dissociation constants (Kd) were measured for the homodimerization of 2-domain constructs (EC1–2) of N- and E-cadherins from mouse, human, and chicken (Table 1; see also SI Methods). These constructs include the entire dimerization interface and binding sites for 3 calcium ions between the EC1 and EC2 domains (Fig. 1). In all cases, and at 2 temperatures, the Kd for the dimerization of N-cadherin was significantly lower than that of E-cadherin. For the mouse proteins, the Kds at 37 °C were 22.6 ± 1.7 μM and 160.0 ± 21.3 μM for N- and E-cadherin, respectively (Fig. S2 and Table 1). The Kd for a 3-domain construct (EC1–3) of mouse E-cadherin (141.0 ± 10.3 μM at 37 °C) was essentially identical to that of the 2-domain construct, consistent with crystallographic observations that cadherin dimerization only involves contacts between EC1 domains. These Kds are all significantly lower than the value of 720 μM obtained from an NMR titration using a refolded E-cadherin EC1–EC2 construct (16), but that protein has a β-strand in a different orientation than seen in other cadherins and may have been affected by denaturation in its urea-based purification or during the NMR experiment. Indeed, recent single-molecule force measurements on live cells determined a binding energy for E-cadherin (17) virtually identical to that reported here and, in addition, our results are in the range of a previous report for the Kd of a complete ectodomain (5-domain) construct of C-cadherin (64 μM) (18). That N- and E-cadherins would differ in their Kds by almost an order of magnitude is an unanticipated finding.
Table 1.
Kds for the homodimerization of N-cadherin and E-cadherin EC1–2 proteins from mouse, human, and chicken
| Species | Kd 25 °C, μM |
Kd 37 °C, μM |
|||
|---|---|---|---|---|---|
| N-cadherin | E-cadherin | Cadherin-6 | N-cadherin | E-cadherin | |
| Mouse | 25.8 ± 1.5 | 96.5 ± 10.6 | 3.13 ± 0.12 | 22.6 ± 1.7 | 160.0 ± 21.3 |
| Human | 24.6 ± 5.0 | 156.0 ± 10.0 | NA* | 22.1 ± 6.5 | 217 ± 30 |
| Chicken | 19.7 ± 2.0 | 62.0 ± 9.5 | NA* | 20.4 ± 1.4 | 110.0 ± 6.8 |
*Binding of cadherin-6 was tested only for mouse.
Type II cadherins form homodimeric interfaces with larger, buried hydrophobic surface areas than observed in type I cadherins (Fig. 1) (15), suggesting that they might have greater dimerization affinities. Consistent with this expectation, the measured Kd of the type II cadherin-6, 3.13 μM (Table 1), was almost an order of magnitude stronger than that of even N-cadherin.
SPR Measurements of Heterophilic Binding Affinities.
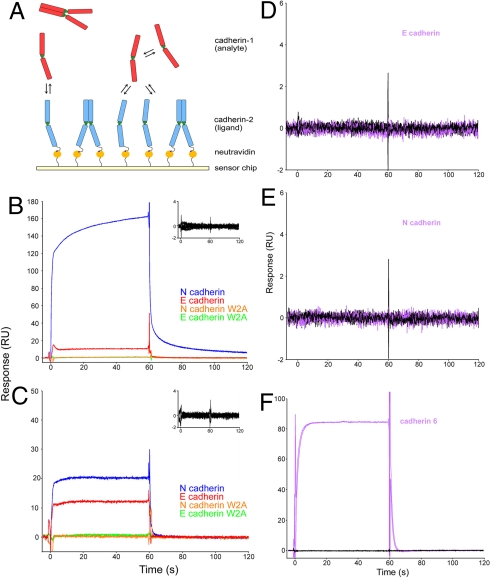
The measurement of heterophilic Kds for cadherins presents a technical challenge due to the presence of homophilic binding. To overcome this problem, we combined AUC, which yields precise values for homophilic Kds, with SPR, which allows assessment of relative homophilic and heterophilic Kds. SPR experiments involved flowing an “analyte” over a chip upon which a “ligand” was immobilized (Fig. 3A). Because cadherins homodimerize, some fraction of both the analyte and the ligand will be in a dimeric state, and therefore unavailable for binding. To determine the amount of available monomer in the analyte, the homophilic Kd for each analyte (measured by AUC) was used to calculate the “effective monomer concentration.” The same procedure cannot be used for the ligand because the extent of dimerization on the chip is unknown, preventing the determination of absolute binding affinities. Nevertheless, we were able to determine relative affinities by comparing the response of different analytes flowed over the same ligand surface (Fig. 3 and Materials and Methods).
Fig. 3.
SPR binding experiments using N- and E-cadherins. (A) Biotinylated cadherin of a given type (ligand, shown in blue) was tethered over a neutravidin-coated sensor chip. Another cadherin of a given type (analyte, shown in red) was injected independently over the surface. The interaction between an analyte monomer (red) and the ligand monomer (blue) produces a binding signal. (B) N-cadherin (blue traces) and E-cadherin (red traces) were injected over a surface containing N-cadherin at 30 μM and 17 μM, respectively, which correspond to 13.1 μM free monomer. Each sample was injected in duplicate. N-cadherin W2A (orange traces) and E-cadherin W2A (green traces) mutants, which were also injected at the same monomer concentrations, show no binding. Inset shows an overlay of buffer blank injections performed throughout the experiment over the same surface. (C) During the same experiment, N- and E-cadherins were also injected over a surface containing E-cadherin, under the same conditions as described in B. Mouse cadherin-6 (purple traces) was injected at 121 μM (effective monomer concentration, 13.1 μM) over the same E-cadherin (D) and N-cadherin (E) surfaces. Black traces represent buffer blanks that were performed throughout the experiment. (F) To confirm that lack of binding was not due to an inactive protein, binding of cadherin-6 was tested against a cadherin-6 surface.
SPR experiments were performed by using EC1–2 cadherin constructs from mouse, human, and chicken as both ligands and analytes at 37 °C (Fig. 3 and Fig. S3). With proteins from all 3 species, flowing an N-cadherin analyte over an N-cadherin surface produced a much larger SPR response than did an identical monomer concentration of E-cadherin flowing over the same surface (with the ratio between the two ranging from about 15:1 to 6:1). With E-cadherin as ligand, the heterophilic response obtained by using N-cadherin as analyte was somewhat larger than when E-cadherin was used (but with a ratio ranging between about 2:1 and 1.5:1). Thus, in contrast to results with the N-cadherin surface, the 2 responses are of comparable magnitude. Taken together, the AUC and SPR results demonstrate that vertebrate cadherins exhibit a rank order of affinities of N/N > N/E > E/E. Thus, these measurements of the interactions of E- and N-cadherins reveal a second, unanticipated result: the heterophilic Kd is intermediate between the 2 homophilic values.
The SPR results obtained for cadherin-6 reveal a very different pattern. Despite the much higher homophilic binding affinity as measured by AUC (see above), SPR experiments (Fig. 3 D and E) detected no heterophilic binding between cadherin-6 and either N- or E-cadherin. This is consistent with the shape of the respective EC1 domains (compare Fig. 1 B and C), whose surfaces are not compatible with the formation of a common interface (15).
Relating Cadherin Dimerization Affinities to Cellular Behavior.
In this section, we use the experimentally determined binding affinities to calculate values of W(I, J) for cells expressing different amounts of N- and E-cadherins. W(I, J), the work required to separate 2 cells, is a measure of adhesive strength and provides a direct link to theories, such as Steinberg's DAH (19, 20), that predict cell-patterning behavior based on cellular properties. As we have discussed previously, we assume that upon the initial encounter of 2 cells of types I and J, the adhesive strength is proportional to the number of trans-cadherin dimers formed and to the binding free energy of each dimer (12).
Thus,
where Ndimer(I, J) is the number of trans-i-j dimers linking cell types I and J; i and j denote the types of cadherin on each cell (where, for clarity, lowercase is used to denote molecules and uppercase used to denote cells); and Δg(i, j) is the transdimerization free energy between cadherins i and j. If cadherins are laterally mobile and randomly distributed in the plane of the cell membrane, the number of dimers formed will depend on the 2D cadherin densities (12). These can be related to the 3D concentrations of their corresponding interacting (EC1) domains, Ci and Cj. The 3D concentration of trans-dimers (expressed here in moles/liter), Cij, is given by Cij = CiCj/Kd(i, j) so that Ndimer = νLCij, where L is Avogadro's number and ν is the volume of the region between interacting cells that is accessible to the EC1 domains (12).
Eq. 1.1 can be rewritten in a form that provides an explicit relationship between adhesive strengths, 3D concentrations, and Kds.
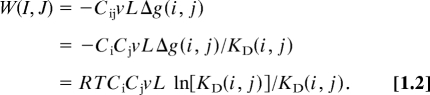 |
Eq. 1.2 is derived from elementary considerations by assuming that a local equilibrium is established in the interface between interacting cells, with junction formation being viewed here as a later event. The concentrations that appear in the equation can be obtained by solving the quadratic equation that relates Ci, Cj, and Cij to Kds and total cadherin concentrations. Eq. 1.2 now enables calculation of W(I, J) for cells expressing N- and E-cadherins and prediction of their aggregation behavior.
We apply Eq. 1.2 to the case of a mixture of 2 cell types, each of 10-μm diameter and expressing either E- or N-cadherin, with a total of 25,000 cadherin molecules presented on the cell surface (in the range determined by Steinberg and coworkers for transfected L cells; ref. 4). These expression levels correspond to 2D densities of about 80 cadherins per square micrometer, which can be transformed into 3D concentrations if we know the range of fluctuations available to an EC1 domain on the cell surface (12). Based on our estimate of this value of about 12 nm, each cell will have an effective total EC1 concentration of about 10 μM (12). Introduction of the Kds at 37 °C for E- and N-cadherins reported here yields dimer concentrations of CE = 0.56 μM and CN = 2.5 μM, corresponding to a monomer to dimer ratio of about 17:1 for E-cadherin and about 3:1 for N-cadherin. Thus, for a cell–cell interface containing 1,000 cadherins from each cell, upon initial intercellular contact about 56 dimers will be formed between 2 cells expressing E-cadherin and 250 dimers formed between 2 cells expressing N-cadherin. Because the Kds for E- and N-cadherins translate into values of Δg(E, E) and Δg(N, N) of −5.3 kcal/mol and −6.5 kcal/mol, respectively, the corresponding values for the adhesive strength at the cellular level would be W(E, E) ≈ 305 kcal/mol and W(N, N) ≈ 1,540 kcal/mol. W(E, N) would have an intermediate value closer to the value of W(E, E) than to W(N, N). For 2 interacting cells, the corresponding values of W(I, I) are 5.1 × 10−19 and 2.6 × 10−18 calories for I = E-cadherin and I = N-cadherin, respectively (Fig. S4).
Although these numbers are approximations, they show that measured differences in molecular binding affinities will result in substantial differences in adhesive strengths between 2 cells. Moreover, the separation in Kds between N- and E-cadherins implies that W(N, N) > W(E, E) for a range of cell surface concentrations, even including cases where E-cadherin concentrations are greater than those of N-cadherin (Fig. S4).
The effect of these cell–cell adhesive strengths, W(I, J), on cell patterning can be evaluated in the context of the DAH (13, 19, 20), which posits an analogy between cell-sorting behavior and the interfacial properties of binary mixtures of immiscible liquids (21). The predictions of the DAH are related to the value of the parameter Φ—the difference between the average of the 2 homophilic adhesive strengths and the heterophilic adhesive strength.
When I = J so that the 2 cell types are identical, Φ = 0, which corresponds to a case of complete mixing. When I ≠ J (i.e., the cell types are different), a number of possibilities arise. (i) The strength of the heterophilic interaction is greater than the average of the homophilic interactions, so that Φ < 0. In such cases, there will be a tendency for I and J cells to mix. (ii) When Φ > 0 (i.e., when the average strength of homophilic interactions is greater than the strength of the heterophilic interaction), I and J cell types will form separate aggregates. When heterophilic interactions are weak [i.e., W(I, J) ≈ 0], there will be no contact between the pure I and pure J aggregates. (iii) When Φ > 0 but heterophilic interactions are significant [i.e., W(I, J) > 0], the separate aggregates will adhere to one another. As discussed by Foty and Steinberg (13), these relationships can be used to predict cell aggregation behavior if the relevant values of W are known.
Based on the values derived here for cells expressing N- and E-cadherins, at comparable expression levels, W(N, N) is significantly larger than both W(E, E) and W(N, E), so that the average of the homophilic adhesive strengths will be greater than the heterophilic adhesive strength. That is, because W(N, N) > W(E, E) and W(E, E) ≈ W(E, N), then Φ > 0. Because W(N, E) is significant, corresponding to case iii described in the previous paragraph, the expected behavior is that mixtures of cells expressing N- and E-cadherins will form separate aggregates that adhere to one another, as observed previously (3) and in our cell assays (Fig. 1 and Fig. S1). For mixtures of cells expressing cadherin-6b and either N- or E-cadherin, the average of the homophilic interactions will be significant while heterophilic interactions are very weak; that is, Φ > 0, but W(6b, N) and W(6b, E) ≈ 0. The predicted behavior is then the formation of homotypic aggregates that do not adhere to one another, as also observed in the cell assays reported above.
Discussion
It is widely accepted that upon encounter of 2 cells, the initial recognition event involves the formation of trans-dimers between cadherin monomers (22, 23) located on apposing surfaces, and that this is followed by the clustering of these dimers into larger structures (see refs. 17 and 23). It has been shown in in vitro and in vivo assays that cell–cell adhesive specificity is determined by the identity of the interacting EC1 domains, and it has been natural to assume a connection between dimerizaton affinities and adhesive propensities. However, such a connection has never been established clearly, in part because relative affinities for the homodimerization and heterodimerization of different cadherins have not been reported. The determination of these affinities for N- and E-cadherins and for cadherin-6 now enables a more quantitative discussion of the adhesive properties of cadherin-expressing cells than has previously been possible.
The major results of this study are the determination with AUC of the homodimerization affinities of N- and E-cadherins (22.6 and 160.0 μM, respectively), and the demonstration with SPR that the heterodimeric affinity is intermediate between these 2 values. The corresponding binding free energies, −6.5 kcal/mol for N-cadherin and −5.3 kcal/mol for E-cadherin, are in one sense quite close, as expected from the similarity in sequence and structure between the 2 EC1 domains. However, as discussed above, such small differences can be amplified by the presence of many cadherins on the cell surface so as to yield significant differences in cell–cell adhesive strengths. The DAH provides a framework for understanding how these differences can be exploited in cell-patterning behavior and explains how homotypic cell aggregation can occur in the presence of strong heterotypic interactions.
There have been previous reports of significant heterophilic interactions between N- and E-cadherins. For example, Geiger and coworkers (24) described the formation of adherens junctions between liver cells expressing N-cadherin and retinal cells expressing E-cadherin. By using a laminar flow assay, Niessen and Gumbiner (3) found evidence that the N–E affinity is stronger than the E–E affinity, consistent with the results of this work. Molecular force measurements have also detected both homophilic and heterophilic affinities for N- and E-cadherins; however, the bond energies that were reported for the EC1–EC1 interaction (25) (corresponding to Kds in the 10–100 mM range) are unusually weak and are inconsistent with all reported solution data. Moreover, interpretation of the measurements is complicated by the fact that the densities that were used in the experiments (≈104 cadherins per square micrometer) (21) were such that the cadherins were closely packed on each surface, corresponding to an effective radius per cadherin molecule of only ≈55 Å. In contrast, as mentioned above, our results are in excellent agreement with single molecule force measurements on live cells containing E-cadherin (17).
Although this work establishes a connection between cadherin dimerization affinities and cellular behavior, the relationship is not unambiguous. Cell–cell adhesive strengths are also determined by cadherin expression levels (for example, Eq. 1.2) (4), so that both affinities and concentrations can, in principle, be exploited in vivo. Moreover, the process of trans-dimer formation may be coupled to the formation of junctions (26), including interactions with catenins, which have been shown to affect adhesive forces (17). Nevertheless, the existence of evolutionarily conserved differential affinities between N- and E-cadherins offers a direct resolution to the apparent paradox posed by the observed homotypic and heterotypic affinities between E-cadherin-expressing and N-cadherin-expressing cells and, more generally, provides a new basis for interpreting cadherin-mediated cell-patterning behavior. Specifically, we have shown that cell layer separation is expected in the presence of strong heterophilic binding affinities if at least one of the homophilic affinities is significantly stronger than the heterophilic interaction. There is persuasive biological logic to this design. A balance between homotypic and heterotypic cellular affinities, the origins of which lie in molecular dimerization affinities, promotes the formation of separate tissue layers, yet it still permits these layers to adhere to one another. We note that many tissues appear to display this behavior (27–29).
Methods
Cell Aggregation Assays.
To distinguish between the cell lines, each CHO cell line of the type I cadherins, E-cadherin or N-cadherin, or the type II cadherin, cadherin-6b, was labeled with either 15 μg/mL DiI (1,1-dioctadecyl-3,3,3,3-tetramethylindocarbocyanine; Vibrant cell-labeling solution; Molecular Probes) or 15 μg/mL DiO (3,3′-dioctadecyloxacarbocyanine perchlorate; Vibrant cell-labeling solution; Molecular Probes) in DMEM with 10% FCS at 37 °C in a humidified atmosphere comprising 5% CO2 for 12 h. The cells were washed extensively with Hanks Balanced Salt Solution (HBSS) to avoid cross-contamination. A complete dispersed cell suspension was obtained by treating confluent CHO cells with enzyme-free cell dissociation solution (Sigma) or 0.25% Trypsin-EDTA solution (Invitrogen) at 37 °C for 10 min.
We performed 2 aggregation assays that use different levels of shear force. Mixing assays were performed with a slight modification from that described by Shimoyama et al. (30). The cells were resuspended in DMEM containing 10% FCS and 70 units of DNase I. A total of 5 × 104 cells per 0.5 mL was added to 24-well Ultra low cluster plates (Corning Costar) and allowed to aggregate for 24 h at 37 °C on a rotary shaker with 70–80 rpm in a humidified atmosphere comprising 5% CO2/95% air. The parental CHO cell line showed no aggregation in these conditions, strongly suggesting that the observed aggregation is cadherin-dependent.
Hanging-drop assays were modified slightly from that described by Kim et al. (31). After labeling and dissociation, 5 × 103 cells of each type were placed in a 28-μL drop on the lid of a 6-cm Petri dish above a reservoir of 5 mL of culture medium. This configuration was left for 6–8 h at 37 °C in a humidified atmosphere comprising 5% CO2/95% air.
SPR Binding Analysis.
Binding experiments were performed by using a Biacore T100 biosensor equipped with a low-charge Series S CM4 sensor chip (Biacore). Neutravidin was immobilized over all flow cells in HEPES Buffered Saline (HBS) running buffer (10 mM Hepes, pH 7.4; and 150 mM NaCl) at 32 °C. A mixture of 200 mM N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide and 50 mM N-hydroxysuccinimide was injected over the flow cells for 7 min at 20 μL/min. Immunopure neutravidin (Pierce) at 50 μg/mL in 10 mM sodium acetate, pH 4.5, was flowed over the activated surfaces for 7 min at 20 μL/min. Excess activated groups were blocked by using a 7-min pulse of 1.0 M ethanolamine-HCl at the same flow rate, resulting in immobilization levels of 10,000–13,000 response units (RU).
Biotinylated mouse N- and E-cadherins were captured over flow cells 1 and 2, respectively, at 37 °C in a running buffer of 20 mM Tris·HCl, pH 8.0; 150 mM NaCl; 10 mM CaCl2; 3 mM Tris [2-carboxy-ethyl] phosphine; 0.2 mg/mL BSA; and 0.005% (vol/vol) surfactant P20. Protein stocks were diluted 50-fold in running buffer and injected in consecutive 10-s pulses at 20 μL/min. Capture levels were 1,000 RU of N-cadherin and 265 RU of E-cadherin, which correspond to 63.6 μM free monomer on each flow cell available for interaction with the analyte. For the binding analysis at 25 °C, the capture levels were adjusted accordingly (890 RU of N-cadherin and 350 RU of E-cadherin) to yield 63.6 μM unbound monomer on each surface. (These numbers were derived by using the Kds reported in Table 1 and the relationship 1,000 RU = 1 ng/mm2.) Flow cell 3 was used as a reference surface to remove bulk refractive index changes and injection noise from the binding data. In a separate experiment, 530 RU of cadherin-6 was captured adjacent to the N- and E-cadherin surfaces, to confirm the binding activity of mouse cadherin-6.
In each binding cycle, the association phase was monitored at 50 μL/min for 60 s, followed by a 60-s dissociation phase at the same flow rate. At the end of the dissociation time, the binding responses returned back to baseline; therefore, the regenerations step was omitted. Buffer was injected at 50 μL/min for 60 s to wash out the flow path and minimize sample carryover into the next cycle. Eight buffer cycles were introduced at the beginning of each experiment to minimize temperature fluctuations. Binding of each cadherin was tested in duplicate to verify the reproducibility of the assay. Buffer binding cycles were performed between sample injections to allow for proper referencing of the binding responses. At 37 °C, mouse N- and E-cadherins were injected at 28.3 μM and 15.3 μM, respectively, which correspond to 13.1 μM unbound monomer in each sample. N- and E-cadherins carrying the W2A mutation were injected at 13.1 μM of total protein because they can only exist as monomers in solution, as confirmed by AUC. At 25 °C, 13.1 μM free monomer was flowed over the surface for each N- and E-cadherin (26.4 μM N-cadherin and 16.7 μM E-cadherin). Similarly to the 37 °C binding experiment, N-cadherin W2A and E-cadherin W2A were injected at 13.1 μM of total protein because they are both monomers. Mouse cadherin-6 was tested at 121 μM, which yields a free-monomer concentration of 13.1 μM (Kd = 3.13 ± 0.12 μM). Binding data were processed by using Scrubber 2.0 (BioLogic Software). The results were confirmed in at least 2 independent experiments.
Supplementary Material
Acknowledgments.
We thank Drs. A. Palmer and R. Axel for their insightful comments. This work was supported by National Science Foundation Grant MCB-0416708 (to B.H.H.) and National Institutes of Health Grant R01 GM062270–07 (to L.S.). T.M.J. is supported by a grant from National Institute of Neurological Disorders and Stroke. The financial support of the US-Israel Binational Science Foundation is gratefully acknowledged (Grant No. 2006-401, to A.B.-S., B.H., and L.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905349106/DCSupplemental.
References
- 1.Takeichi M. Cadherins: A molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- 2.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 3.Niessen CM, Gumbiner BM. Cadherin-mediated cell sorting not determined by binding or adhesion specificity. J Cell Biol. 2002;156:389–399. doi: 10.1083/jcb.200108040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duguay D, Foty RA, Steinberg MS. Cadherin-mediated cell adhesion and tissue segregation: Qualitative and quantitative determinants. Dev Biol. 2003;253:309–323. doi: 10.1016/s0012-1606(02)00016-7. [DOI] [PubMed] [Google Scholar]
- 5.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 6.Patel SD, et al. Cadherin-mediated cell-cell adhesion: Sticking together as a family. Curr Opin Struct Biol. 2003;13:690–698. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 8.Haussinger D, et al. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: Changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324:823–839. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- 9.Parisini E, et al. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373:401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro L, et al. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 11.Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: Do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:954–968. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CP, et al. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci USA. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foty RA, Steinberg MS. The differential adhesion hypothesis: A direct evaluation. Dev Biol. 2005;278:255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Shan WS, et al. The adhesive binding site of cadherins revisited. Biophys Chem. 1999;82:157–163. doi: 10.1016/s0301-4622(99)00115-5. [DOI] [PubMed] [Google Scholar]
- 15.Patel SD, et al. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 16.Haussinger D, et al. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajpai S, et al. {alpha}-Catenin mediates initial E-cadherin-dependent cell-cell recognition and subsequent bond strengthening. Proc Natl Acad Sci USA. 2008;105:18331–18336. doi: 10.1073/pnas.0806783105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappuis-Flament S, et al. Multiple cadherin extracellular repeats mediate homophilic binding and adhesion. J Cell Biol. 2001;154:231–243. doi: 10.1083/jcb.200103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinberg MS. On the mechanism of tissue reconstruction by dissociated cells, III. Free energy relations and the reorganization of fused, heteronomic tissue fragments. Proc Natl Acad Sci USA. 1962;48:1769–1776. doi: 10.1073/pnas.48.10.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinberg MS. Adhesion in development: An historical overview. Dev Biol. 1996;180:377–388. doi: 10.1006/dbio.1996.0312. [DOI] [PubMed] [Google Scholar]
- 21.Evans DF, Wennerstrom H. The Colloidal Domain: Where Physics, Chemistry, Biology, and Technology Meet. New York: VCH Publishers; 1994. p. 515. [Google Scholar]
- 22.Troyanovsky S. Cadherin dimers in cell-cell adhesion. Eur J Cell Biol. 2005;84:225–233. doi: 10.1016/j.ejcb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc Natl Acad Sci USA. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volk T, Cohen O, Geiger B. Formation of heterotypic adherens-type junctions between L-CAM-containing liver cells and A-CAM-containing lens cells. Cell. 1987;50:987–994. doi: 10.1016/0092-8674(87)90525-3. [DOI] [PubMed] [Google Scholar]
- 25.Prakasam AK, Maruthamuthu V, Leckband DE. Similarities between heterophilic and homophilic cadherin adhesion. Proc Natl Acad Sci USA. 2006;103:15434–15439. doi: 10.1073/pnas.0606701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Troyanovsky RB, Laur O, Troyanovsky SM. Stable and unstable cadherin dimers: Mechanisms of formation and roles in cell adhesion. Mol Biol Cell. 2007;18:4343–4352. doi: 10.1091/mbc.E07-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doi Y, et al. Development of complementary expression patterns of E- and N-cadherin in the mouse liver. Hepatol Res. 2007;37:230–237. doi: 10.1111/j.1872-034X.2007.00028.x. [DOI] [PubMed] [Google Scholar]
- 28.Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- 29.Prozialeck WC, Lamar PC, Appelt DM. Differential expression of E-cadherin, N-cadherin and beta-catenin in proximal and distal segments of the rat nephron. BMC Physiol. 2004;4:10. doi: 10.1186/1472-6793-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimoyama Y, Tsujimoto G, Kitajima M, Natori M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem J. 2000;349:159–167. doi: 10.1042/0264-6021:3490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JB, et al. N-Cadherin extracellular repeat 4 mediates epithelial to mesenchymal transition and increased motility. J Cell Biol. 2000;151:1193–1206. doi: 10.1083/jcb.151.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.