Abstract
Because heme is a major iron-containing molecule in vertebrates, the ability to use heme-bound iron is a determining factor in successful infection by bacterial pathogens. Until today, all known enzymes performing iron extraction from heme did so through the rupture of the tetrapyrrol skeleton. Here, we identified 2 Escherichia coli paralogs, YfeX and EfeB, without any previously known physiological functions. YfeX and EfeB promote iron extraction from heme preserving the tetrapyrrol ring intact. This novel enzymatic reaction corresponds to the deferrochelation of the heme. YfeX and EfeB are the sole proteins able to provide iron from exogenous heme sources to E. coli. YfeX is located in the cytoplasm. EfeB is periplasmic and enables iron extraction from heme in the periplasm and iron uptake in the absence of any heme permease. YfeX and EfeB are widespread and highly conserved in bacteria. We propose that their physiological function is to retrieve iron from heme.
Keywords: deferrochelation activity, Dyp peroxydase, heme iron extraction, new bacterial function, heme permease
Heme is ubiquitous, abundant, and vitally necessary as a cofactor in oxidoreduction and gas transport. Most microorganisms display a complete heme biosynthetic pathway, but are able to acquire the essential ferrous iron from exogenous heme (1). Free heme or heme arising from hemoproteins is internalized intact and subsequently degraded in the cytosol.
Diverse mechanisms for heme uptake have been identified in bacteria. They involve extracellular hemoproteins (hemophores) that capture heme and deliver it to bacteria (2, 3) and cell surface receptors that bind heme, hemoproteins, and/or hemophores. Surface receptors of Gram-positive bacteria are cell-wall-anchored proteins that scavenge heme and relay it to specific ABC transporters involved in heme internalization (4).
Gram-negative bacterial surface receptors are outer membrane proteins that actively transport heme through this membrane. Although the Escherichia coli K12 laboratory strain is missing its own heme outer membrane receptor, it acquires the ability to use heme as an iron source owing to the expression of a foreign heme receptor such as the Serratia marcescens HasR receptor. Once in the periplasm, heme is caught by heme permease which consists of a periplasmic heme-binding protein and an ABC transporter sharing similarities with Gram-positive heme ABC transporters (1). An alternative heme permease has been identified in E. coli K12. It comprises the dipeptide ABC transporter DppBCDF functioning with 2 interchangeable periplasmic peptide/heme-binding proteins, either DppA or MppA (5).
In the cytoplasm, iron is extracted through the action of heme-degrading enzymes that cleave the tetrapyrrol ring. For this purpose, some bacteria use orthologs of the mammalian HO heme oxygenases. This class of enzymes degrades heme into biliverdin, CO, and iron (6). Furthermore, non-HO homolog heme degrading enzymes have been reported for some bacteria (7–9). Although heme degradation by this class of weakly similar enzymes has been confirmed by CO production, the precise nature of the resulting products remains controversial (10).
However, genome BLAST searches fail to identify orthologs of any heme-degrading enzymes in many species including Shigella dysenteriae, Salmonella typhi, and E. coli pathogenic strains, despite their ability to use heme as an iron source. This is also the case of the E. coli K12 laboratory strain expressing a foreign heme receptor.
In this study, we identified 2 E. coli paralogs, YfeX and EfeB without previously known cellular functions that surprisingly release iron from heme without tetrapyrrol degradation, a novel enzymatic reaction corresponding to the deferrochelation of the heme.
We demonstrate here the role of YfeX and EfeB in the recovery of exogenous heme iron.
Isolation of E. coli Genes Potentially Involved in Heme Degradation.
E. coli features no orthologs of known heme-degrading enzymes. Yet E. coli K12 expressing a heterologous heme outer membrane receptor such as the S. marcescens HasR receptor, in combination with its native heme/dipeptide inner membrane permease DppABCDF, is able to use exogenous heme as an iron source. We therefore hypothesized that it might be equipped with enzyme(s) displaying iron capturing activity. We developed a screening method based on the fluorescence properties of heme metabolites to find such enzymes (11, 12). A strain expressing P. aeruginosa heme oxygenase (PigA) was used as a positive control. C600 (pBAD24-pigA) colonies could be easily distinguished from colonies carrying the empty vector pBAD24 on LB Ara plates by their color and by their fluorescence under near-UV light irradiation (405 nm). A genomic library of E. coli K12 MG1655 in pBAD24 was screened for its ability to confer such a phenotype upon strain C600 on LB Ara plates. Out of the 100,000 clones screened, one recombinant plasmid, carrying the yfeX gene, was responsible for a bright red fluorescent colony phenotype. YfeX is a protein of unknown function belonging to the family of dye-decolorizing peroxidases (Dyp-peroxidase) identified in fungi (13). E. coli has another protein belonging to the Dyp family, EfeB (14). Both proteins, YfeX and EfeB share only a low level of sequence similarity and are thus distantly related paralogs. The E. coli K12 MG1655 efeB gene was cloned into pBAD24. Strain C600 (pBAD24-efeB) also gave rise to fluorescent colonies upon arabinose induction. Additionally, bacterial cell cultures expressing either PigA, YfeX, or EfeB showed differing colors under visible light. The strain expressing PigA was dark green, whereas strains expressing either YfeX or EfeB were colored red. (supporting information (SI) Fig. S1). The fluorescence as well as the red color is characteristic of strains accumulating porphyrins.
The Major Fluorescent Compound Produced by Strains Overexpressing YfeX or EfeB is Protoporphyrin IX (PPIX).
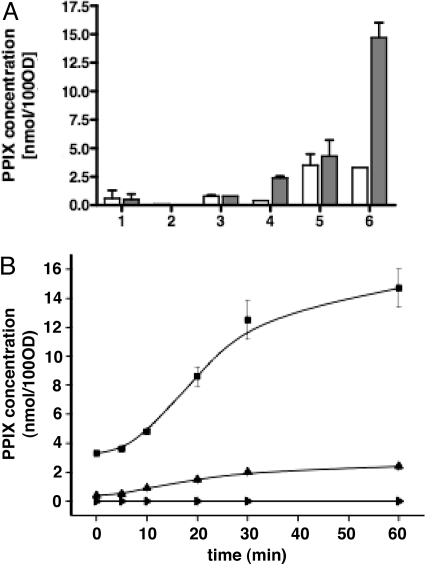
To determine the chemical nature of compounds accumulated by strains overexpressing either YfeX or EfeB, intracellular porphyrins of cell soluble fractions of these strains incubated without addition of exogenous hemin were extracted and separated by fluorescence-based HPLC. Using naturally occurring porphyrins as standards, the main peak of the cell extracts eluted at the same time as PPIX. Mass spectrometry confirmed that the chemical nature of this compound was PPIX (Fig. S2 A and B). Both strains contained large amounts of PPIX (3 and 10 times higher than the control strain, respectively) (Fig. 1A, lanes 1, 3, 5). Besides a very low level of coproporphyrin (COP) III, no other pigments including biliverdin were found either in pellets or soluble fractions of cells overexpressing YfeX or EfeB. Thus, strains overproducing either YfeX or EfeB accumulated PPIX, which was presumably formed from intracellular heme.
Fig. 1.
PPIX concentrations in cell soluble fractions incubated with and without hemin. Soluble fractions of JP313 carrying pBAD24 (empty vector), pBAD24-yfex, and pBAD24-efeB cultures were prepared as described in Methods. Porphyrin concentrations were normalized so as to represent the porphyrin amount in cell lysates corresponding to a bacterial suspension with an optical density of 100 at 600 nm (OD600 100). (A) PPIX concentrations in cell lysates incubated at room temperature for 0 min (white bar) and 60 min (gray bars) without or with addition of hemin at a final concentration of 50 μM. (1) JP313 (pBAD24) incubation without added hemin; (2) JP313 (pBAD24) incubation with added hemin; (3) JP313 (pBAD24-efeB) incubation without added hemin; (4) JP313 (pBAD24-efeB) incubation with added hemin; (5) JP313 (pBAD24-yfeX) incubation without added hemin; (6) JP313 (pBAD24-yfeX) incubation with added hemin. (B) Time dependent PPIX formation in the same cell lysates as in (A) after incubation with added hemin (50 μM). PPIX concentration was measured over time with the starting point at hemin addition to cell soluble fraction of the following strains: JP313 (pBAD24) (filled inverted triangle), JP313 (pBAD24-efeB) (filled triangle) and JP313 (pBAD24-yfeX) (filled square). The data points represent the means of duplicates experiments and the error bars represent the standard deviation.
Exogenous Heme and Meso-Heme Are Converted to PPIX and Meso-PPIX (MPPIX) by Cell Lysates Producing YfeX or EfeB.
We hypothesized that PPIX could also be formed from exogenous heme upon the action of either YfeX or EfeB. To test this, exogenous hemin (50 μM) was added to soluble cell fractions and incubated for 60 min; PPIX concentration increased over the incubation time only in extracts from YfeX or EfeB overproducing cells (Fig. 1A, lane 2, 4, 6). The respective PPIX accumulation time profiles show a steady increase of PPIX during the first 30 min of incubation and a plateau (Fig. 1B) for longer incubation times. This strongly suggests that exogenous heme was transformed into PPIX under the action of these 2 proteins.
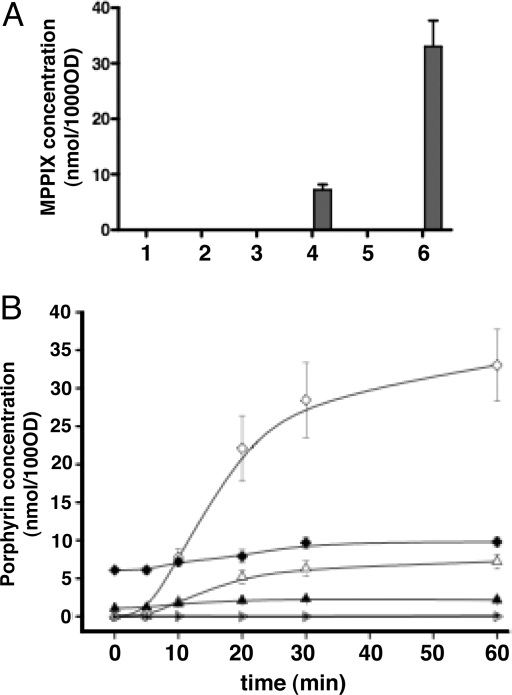
To further demonstrate that YfeX and EfeB have an effect on PPIX formation from exogenous sources we used as substrate a nonnaturally occurring heme, meso-heme, that is not produced by bacterial heme biosynthesis (15). Standard PPIX and meso-PPIX (MPPIX) could be separated by fluorescence-based HPLC (Fig. S3 A–C). Meso-heme (50 μM) or buffer was added at room temperature to soluble cell fractions of control strain or strains overproducing YfeX and EfeB, respectively, and accumulation of MPPIX and PPIX was monitored as a function of time after extraction from cell lysates and separation by fluorescence based HPLC (Fig. S3D). No detectable amount of MPPIX was found in the absence of meso-heme as substrate, confirming that MPPIX is not a natural byproduct of heme biosynthesis (Fig. 2A, lane 1, 3, 5). However, in the presence of meso-heme, in soluble cell fractions of strains expressing either YfeX or EfeB but not of the control strain (Fig. 2A, lanes 2, 4, 6), a steadily increasing accumulation of MPPIX over 30 min of incubation passing into a plateau was observed thereafter (Fig. 2B). At the plateau, 8 to 30 times more MPPIX accumulated in strains overexpressing EfeB and YfeX, respectively, than in the control strain (Fig. 2B). We also measured PPIX accumulation in the cell lysates incubated with meso-heme (Fig. 2B). PPIX, as well as COPIII, levels remained unchanged upon addition of meso-heme, indicating that meso-heme does not impair the natural heme biosynthesic pathway. Thus, YfeX and EfeB are involved in the conversion of exogenous heme into PPIX and are expected to bind heme and PPIX. We tested this hypothesis on purified YfeX.
Fig. 2.
PPIX and MPPIX concentration in cell soluble fractions incubated with and without meso-heme. The procedures used are the same as in Fig. 1. (A) MPPIX concentrations in cell lysates incubated at room temperature for 0 min (white bars) and 60 min (gray bars) without or with meso-heme addition at a final concentration of 50 μM. (1) JP313 (pBAD24) incubation without meso-heme; (2) JP313 (pBAD24) incubation with meso-heme; (3) JP313 (pBAD24-efeB) incubation without meso-heme; (4) JP313 (pBAD24-efeB) incubation with meso-heme; (5) JP313 (pBAD24-yfeX) incubation without meso-heme; (6) JP313 (pBAD24-yfeX) incubation with meso-heme. No MPPIX was detected at t = 0 in either of the samples. (B) MPPIX and PPIX production as a function of time in the same cell lysates as in (A), after meso-heme (50 μM) addition. PPIX (solid symbols) and MPPIX (open symbols). Concentrations were measured over time with the starting point at meso-heme addition to cell soluble fractions of the following strains: JP313 (pBAD24) (filled inverted triangle), JP313 (pBAD24-efeB) (filled triangle) and JP313 (pBAD24-yfeX) (filled square). The data points represent the mean and the error bars represent the standard deviation.
YfeX Binds Heme and PPIX in Vitro.
Addition of PPIX or hemin to purified apo-YfeX (Fig. S4) led to different Soret band at 407 nm for PPIX and 404 nm for hemin and very distinctive features in the 450–800 nm part of the spectra. (Fig. S5 A and B). Hemin and PPIX affinities to apo-YfeX (1 μM) were measured by monitoring the Soret bands with absorption spectroscopy. Titration curves shown in Fig. S6 A and B indicate that YfeX interacts with both hemin and PPIX at a molar ratio of 1 and with a Kd of 3.9 nM ±1.6 nM for hemin and of 4.8 nM ± 2.8 nM for PPIX. Unexpectedly, the saturation curve presents a slightly sigmoidal shape for PPIX titration that might indicate a cooperative effect that was not further investigated.
PPIX loaded YfeX was incubated with an excess of heme for 2 h. After elimination of unbound tetrapyrrols, UV-visible spectra showed a hemin loaded profile, demonstrating that hemin displaced PPIX from YfeX (Fig. S5C).
DyP peroxidase structural and sequence comparisons have revealed the conservation of the residues surrounding heme, including the histidine axial iron ligand (13). Structure modeling of YfeX identified H215 as the corresponding iron binding histidine residue (Fig. S7). After mutation to alanine, intracellular porphyrins of cell soluble fraction of strain overexpressing YfeX H215A were extracted and analyzed as described above. The observed basal porphyrin level was similar to that of the control strain carrying the empty vector (Fig. S2C). Subsequently, the YfeX H215A mutant protein was produced in amounts similar to the wild type protein. It was stable and purified according to the protocol for wild type YfeX (Fig. S4). As expected, the affinity for hemin was strongly reduced and more surprisingly, PPIX binding as well with Kd higher than 10−5 M (data not shown), suggesting that heme and PPIX share at least partially a common ligand pocket.
YfeX Is Loaded with PPIX in Vivo.
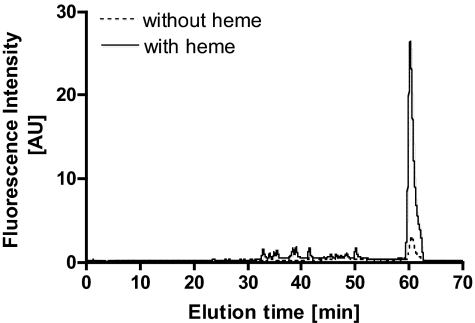
In addition to the apo-YfeX fraction used in the experiments described above, we were able to collect a second YfeX containing fraction during YfeX purification that exhibited an absorption peak at 407 nm. YfeX containing fractions were purified from cell lysates incubated with or without hemin for 30 min at room temperature. After incubation with hemin, the apoprotein fraction decreased with a concomitant increase in the fluorescent fraction. HPLC separation of the pigments bound to the YfeX fluorescent fraction showed that Yfex was loaded with PPIX (Fig. 3). These results indicate that PPIX produced from hemin through YfeX action partly remains bound to YfeX.
Fig. 3.
PPIX binding to YfeX in vivo. The fluorescent YfeX fraction was purified from C600 (pBAD 24-yfex) cell lysates incubated with and without hemin (50 μM) for 30 min at room temperature by anion exchange chromatography and exhibited an absorption peak at 406 nm. Pigments bound to the pure protein were extracted separated by HPLC and identified as in Fig. S2. The peak at 60.3 min corresponds to PPIX. Dotted line corresponds to purification of cell lysate without added hemin. Solid line corresponds to purification of cell lysate with added hemin.
YfeX and EfeB Are Required for Exogenous Heme Iron Acquisition.
Taken together, our in vitro results indicate that YfeX and EfeB are PPIX and heme-binding proteins and that they are involved in the heme transformation into PPIX. To test whether this activity is related to the use of exogenous heme as an iron source, the yfeX and efeB genes replaced by a Km cassette were transduced into strain FB827 (pAM238-hasR). This strain is lacking enterobactin, the major E. coli siderophore, and, owing to HasR, is able to acquire exogenous heme. The strains were tested for growth on minimal iron chelated plates (M63* Gly Ara Dip) around wells containing various hemoglobin concentrations. Both FB827 yfeX ::Km (pAM238-hasR) and FB827 efeB ::Km (pAM238-hasR) grew less efficiently than the wild-type strain around the hemoglobin-containing wells: significant growth was only observed around wells containing 50 μM hemoglobin, whereas the wild-type FB827 (pAM238-hasR) strain still formed a significant growth halo around wells containing a 50 times lower hemoglobin concentration of 1 μM (Table 1 and Fig. S8). The double mutant FB827 Δ yfeX efeB ::Km (pAM238-hasR) was unable to grow around the hemoglobin-containing wells at any tested concentration (Table 1). This demonstrates that the 2 proteins are the sole proteins able to retrieve iron from heme. It also suggests that the 2 YfeX and EfeB paralogs fulfill similar functions in heme iron utilization.
Table 1.
yfeX and efeB role in heme-iron acquisition
| FB827 (pAM-hasR) carrying the relevant mutation and/ or plasmids | Growth on M63* Dip around wells containing Hb (mM): |
|||
|---|---|---|---|---|
| 50 | 10 | 5 | 1 | |
| None | +++ | ++ | + | + |
| yfeX::Km | + | +/− | − | − |
| efeB::Km | + | +/− | − | − |
| D yfeX efeB::Km | − | − | − | − |
| yfeX::Km pBAD 24-yfeX | +++ | ++ | + | + |
| yfeX::Km pBAD 24-yfeXH215A | + | +/− | − | − |
| yfeX::Km pBAD 24- efeB | +++ | ++ | + | + |
| yfeX::Km pBAD 24-pigA | +++ | ++ | + | + |
| efeB::Km pBAD 24-efeB | +++ | ++ | + | + |
| efeB::Km pBAD 24-yfeX | +++ | ++ | + | + |
| efeB::Km pBAD 24-pigA | +++ | ++ | + | + |
| dppF::Km pBAD 24 | − | − | − | − |
| dppF::Km pBAD 24- efeB | − | − | − | − |
| dppF::Km pBAD 24- yfeX | − | − | − | − |
| dppF::Km pBAD24-efeUOBO157:H7 | +++ | ++ | + | + |
| dppF::Km pBAD24-efeUBO157:H7 | − | − | − | − |
The relevant chromosomal mutations carried by strain FB827 (pAM-hasR) and the plasmid-borne complementing genes carried on pBAD 24 are indicated in the first column. Cultures of strain FB827 (pAM 238-hasR) carrying the various mutations and plasmids were grown in M63 Gly media at 37 °C to an OD 600 of 1, and 100 μl aliquots were mixed with 3.5 ml of M63* soft agar 0.7% and poured onto M63* Dip plates containing 100 μM Dip to chelate any residual iron and 0.02% l-arabinose to induce the pBAD24-encoded genes. Aliquots of 50 μl of bovine hemoglobin at various concentrations calculated on the basis of the heme monomer were provided in wells punched in the solidified agar. Plates were incubated for 48 h at 37 °C and the growth halo radius around the wells was measured. All experiments were reproduced 3 times. The second column indicates the various strain growth around wells containing various Hb concentrations. The plates were incubated 48 h at 37 °C, and the radius of the growth around each well was measured. +++, radius of 10 mm; ++, radius of 6 mm; +, radius of 2 mm; −, no growth around the wells. All experiments were repeated 3 times.
YfeX and EfeB Can Complement Each Other and Both Are Complemented by the P. aeruginosa Heme Oxygenase PigA.
To test whether yfeX and efeB can complement each other, FB827 (pAM238-hasR) carrying either yfeX or efeB mutations was transformed with either pBAD24-yfeX or pBAD24-efeB. Each mutant having a defect in heme iron acquisition, this was fully complemented by overexpression of the corresponding wild-type allele or the wild-type paralog (Table 1). This intergenic complementation demonstrated that the 2 genes, yfeX and efeB, have at least partially redundant functions. The pBAD24-yfeX H215A expressing the mutated YfeX protein affected in heme- and PPIX-binding did not complement the yfeX mutation (Table 1). Addition of iron instead of hemoglobin (FeCl3 or FeSO4 0.1 mM) to the wells restored growth of all 3 mutants (the 2 single and the double yfeX efeB ::Km) (data not shown).
Expression of (pBAD24-pigA) in FB827 yfeX ::Km (pAM238-hasR) and FB827 efeB ::Km (pAM238-hasR) mutants fully restored the ability of the strain to grow around hemoglobin-containing wells (Table 1). The complementation for heme iron use by a gene encoding heme oxygenase strongly suggests that YfeX and EfeB are involved in intracellular iron release from heme.
Inactivation of Dpp Permease Abolishes Heme Iron Acquisition in the E. coli K12 Strain Overexpressing Either YfeX or EfeB.
EfeB is exported to the periplasm via the twin arginine translocation system (TAT) (14). The periplasmic localization of EfeB suggests that it could retrieve iron from heme in that cellular compartment. Periplasmic iron released from heme would be picked up by inner membrane iron transporters such as Feo or Fec (16). However, we have shown previously that Dpp permease is essential for heme iron transport, suggesting that only cytoplasmic heme can provide iron (5). Nevertheless, this was not done in conditions of overexpression of YfeX or EfeB. To address this question, FB827 dppF::Km (pAM238-hasR) was transformed with either pBAD24-yfeX or pBAD24-efeB. Overproduction of these proteins failed to restore wild-type growth on M63* Gly Ara Dip plates around wells containing hemoglobin (Table 1). Thus, in E. coli K12, despite a periplasmic localization of EfeB, only cytoplasmic heme was able to provide iron, suggesting that EfeB is at least partly active in the cytoplasm.
The efeUOB Operon of E. coli O157:H7 Allows Heme Iron Acquisition in the dppF Mutant.
efeB belongs to the iron-regulated efeUOB operon involved in Fe2+ uptake under acidic conditions (17). EfeU is homologous to the yeast high affinity iron permease Frtp1 (17, 18). This iron uptake system is defective in E. coli K12 due to a frameshift mutation interrupting the efeU reading frame after amino acid 37 (18). Unlike E. coli K12, many other E. coli strains, such as enterohemorrhagic O157:H7, have an intact efeU sequence and their efeUOB genes expressed in E. coli K12 stimulate iron uptake. All 3 genes were also shown to be necessary for Fe2+ uptake (17). We tested whether expression of a nonmutated efeUOB operon would allow heme iron acquisition from the periplasm, i.e., in the absence of the Dpp permease. efeUOB genes of E. coli O157 ::H7 were amplified and cloned into pBAD24. While FB827 dppF::Km (pAM238-hasR) (pBAD24) could not grow around the hemoglobin-containing wells (Table 1), FB827 dppF::Km (pAM238-hasR) (pBAD24-efeUOBO157 :H7)) was able to use heme iron as well as the wild-type strain. Thus, O157:H7 efeUOB genes were epistatic over the dppF mutation for heme iron utilization. The in-frame deletion of efeO was constructed and the corresponding plasmid was tested for heme iron acquisition. FB827 dppF::Km (pAM238-hasR) (pBAD24-efeUBO157 :H7)) could not grow around the hemoglobin-containing wells (Table 1). These results indicate that iron can be extracted from heme in the periplasm by EfeB and that EfeU and EfeO proteins are required for iron internalization.
Discussion
To identify new bacterial proteins involved in iron extraction from heme, we developed a screening method based on the fluorescent properties of tetrapyrrollic molecules lacking their iron. We succeeded in finding 2 such E. coli paralogs, YfeX and EfeB. Either protein, when overexpressed, promotes PPIX accumulation, an intermediate in the heme biosynthetic pathway. This could be the result of a higher heme biosynthetic activity due to a reduced intracellular heme pool (19, 20). To challenge this hypothesis, YfeX and EfeB activities were tested with meso-heme, a nonnatural porphyrin. Exogenously added meso-heme is converted into meso-PPIX by YfeX or EfeB producing cell extracts. Thus, unambiguously, the increased PPIX pool is issued from the YfeX and EfeB activities on the exogenous heme. YfeX binds both heme and PPIX with a stoichiometry of one for each and with comparable affinities for heme and for PPIX. We found a displacement of PPIX by heme suggesting that they bind to the same or overlapping sites. Moreover, the effect of the YfeX H215A mutation on both heme and PPIX affinities for YfeX also indicates that PPIX and heme share a common binding site. Spectroscopic analysis of pigments which are in vivo bound to YfeX before purification showed that part of the protein is loaded with PPIX and that incubation with heme strongly increases the PPIX loaded protein.
Although YfeX is involved in heme conversion into PPIX in cell lysates, this conversion of heme into PPIX was not observed with purified YfeX protein. It is likely that the reaction requires a cofactor which is not present on the purified protein.
Our genetic studies on the E. coli K12 strain which has been made competent for heme uptake by the expression of the heme outer membrane HasR show experimental evidence that the yfeX, efeB double mutant is unable to use heme as an external iron source and thus unquestionably establish that YfeX and EfeB are the sole proteins that can provide heme iron to E. coli K12. Each single yfeX or efeB gene deletion mutant has a partial defect in using heme as an exogenous iron source, indicating that the corresponding proteins have limiting activities when chromosomally encoded and that they might have complementary functions. In addition, when plasmid encoded, YfeX and EfeB fully complement the deletion of the other gene. Thus, YfeX and EfeB have at least partially redundant functions and can function independently from each other.
YfeX is cytoplasmic, whereas EfeB which has a double arginine signal peptide is exported to the periplasm (14). How can proteins that belong to different cellular compartments complement each other? One reason for such complementation could be that iron produced in the periplasm by EfeB is transported to the cytoplasm by an inner membrane iron transporter. This is not the case in E. coli K12 because inactivation of the DppABCDF heme permease prevents heme iron acquisition showing that only cytoplasmic heme is an iron source. As a matter of fact, the aerobic iron permease EfeU, encoded by the efeUOB operon is nonfunctional in E. coli K12 owing to a frame shift mutation. Unlike E. coli K12, many other E. coli strains, such as the enterohemorrhagic O157:H7, have an intact efeU sequence (17). When the complete efeUOB operon of E. coli O157:H7 is provided to E. coli K12, it allows heme iron acquisition from the periplasm, i.e., in the absence of the DppABCDF heme permease. All 3 genes are necessary for heme iron acquisition in the absence of the heme permease. Consistent with its role in providing iron from heme, the efeUOB operon is induced in iron restricted conditions and is not cryptic in E. coli strains having heme receptor genes.
To explain the EfeB activity in E. coli K12 in the absence of iron transport, it is more likely that some EfeB is active in the cytoplasm. EfeB is translocated to the periplasm by the TAT system, a pathway dedicated to prefolded proteins, some of which carrying a cofactor. EfeB binds heme in the cytoplasm, and the mature heme loaded protein is detected in the cytoplasm (14). This cytoplasmic form is likely to be active in heme-iron extraction.
Taken together, our data indicate that YfeX and EfeB are capable of extracting iron from heme while preserving the protoporphyrin ring intact, an enzymatic reaction which has not been previously described and which corresponds to a demetallation of the heme. In 2 cases, ferrochelatases from meat (21) and from Haemophilus influenzae (22) have been shown to catalyze the reverse reaction in vitro in the presence of reductants. However, YfeX and EfeB do not have significant sequence similarities to any known ferrochelatase and thus most likely have evolved separately to achieve the reaction by a mechanism distinct from ferrochelatases. The new enzymatic activity described here is referred to as deferrochelation activity.
YfeX and EfeB belong to the Dyp peroxidase family which differ from other peroxidases in their fold and their heme ligands. Many hemoproteins including hemoglobin have been shown to have peroxidase activity in the presence of peroxide (23). This in vitro peroxidase activity of the Dyp proteins may not correspond to their physiological activity. The molecular mechanism allowing iron extraction from heme is presently unknown. Spontaneously, iron is removed from heme by treatment with HCl (24). The enzymatic reaction could involve a distortion of the tetrapyrrol ring, as it is achieved by ferrochelatase during metal insertion (25). However, in the heme–TyrA structure (a YfeX ortholog), the protoporphyrin ring is plane (26).
YfeX and EfeB proteins are widespread and highly conserved in Gram-positive and Gram-negative bacteria. We propose that the deferrochelation activity represents their physiological function, enabling heme iron acquisition without internalization of heme, a potentially hazardous substance in the case of EfeB orthologs and without production of CO, an antibacterial gas. Porphyrins and heme efflux pumps have been identified in several organisms (27, 28). In addition, EfeB orthologs are present in many Gram-positive species. This could allow the use of iron from heme directly at the cell surface.
On the other hand, they are absent in higher eukaryotic organisms, making them potential targets for new antibacterial drugs, especially since there is growing evidence that heme utilization systems are required for bacterial virulence.
Methods
Bacterial Strains and Plasmids.
E. coli K12 strains FB8 (wild type, F−), FB827 (entF::Tn10), MG1655, JP313, and C600 are from laboratory collection. Strains JW2424 (yfeX::Km) and JW1004 (efeB ::Km) were obtained from the National BioResource Project (E. coli Hub) by means of the E. coli database “GenoBase” (http://ecoli.aist-nara.ac.jp/). E. coli O157:H7 EDL 933 was a gift of C. Lebouguenec (Institut Pasteur, Paris, France).
pAM238, pBAD24, pAM238-hasR, and pCP20 are from the laboratory collection.
Media and Growth Conditions.
Hemin (>90%, pure), bovine hemoglobin (Hb), and 2,2′-dipyridyl (Dip) were obtained from Sigma. Protoporphyrin IX (PPIX), meso-heme, and meso-protoporphyrin (MPPIX) were purchased from Frontier Scientific.
Bacteria were grown aerobically at 37 °C or 30 °C in LB medium, M63 or M63 without added iron salt (M63*). All minimal media were supplemented with 0.4% glucose (glu) or glycerol (gly). For arabinose induction, 0.2% l-arabinose (Ara) was added to induce the pBAD24 promoter. When required, Dip was added to a final concentration of 80 μM to M63*. Antibiotics were added at the usual concentrations for E. coli. Dip and Ara inducer concentrations are not indicated in the text.
Growth promotion assays were done as described in ref/ 29.
Genetic and Molecular Biology Techniques Were Done by Standard Methods.
The double Δ yfeX efeB ::Km mutant was constructed by P1 transduction after efeB::Km excision by specific recombination mediated by FLP recombinase encoded by the pCP20 plasmid. Verification of the Km cassette excision was done by PCR.
MG1655 chromosomal DNA fragments partially digested with Sau3A were ligated into pBAD24 plasmid and introduced into strain C600. The transformed cells were plated on LB Amp, Ara agar to induce the pBAD promoter. The plasmid DNA inserts conferring the screened phenotype were sequenced to identify the cloned gene.
Plasmid Constructions.
Plasmids carrying efeB from E. coli K12 and efeUOB from E. coli 0157:H7 were constructed by amplification of MG1655 genomic DNA and E. coli O157:H7, respectively, using complementary oligonucleotides (sequences available on demand). Amplified fragments were inserted into pBAD24. In-phase efeO deletion and site directed mutagenesis of H215A of yfeX were performed using the quick-change site-directed mutagenesis kit Stratagene with complementary oligonucleotides (sequences available on demand). Amplified mutant and wild type genes were checked by DNA sequencing.
The NdeI-XhoI 700 bp DNA fragment carrying the P. aeruginosa pigA gene in PET21 plasmid was purified and cloned into pBAD 24 digested with XbaI-SalI.
Membrane and Soluble Fraction Preparations.
Cultures of JP313 (pBAD24-yfeX) and JP313 (pBAD24-efeB) were grown overnight at 30 °C in M63 Gly Ara medium. Cells were harvested by centrifugation for 15 min at 8,000 × g at 4 °C. Each cell pellet was resuspended in Bug Buster buffer (Novagen) for a concentration of 1 g of bacterial dry weight in 5 ml of buffer (100 OD600/ml). The mixtures were incubated 30 min at room temperature to lyse the cells and centrifuged again for 10 min at 10,000 × g at 4 °C. Supernatants contained the soluble fractions (cytoplasm + periplasm), and the pellet contained all membranes. The soluble fractions were dialyzed overnight against 50 mM Tris·HCl, pH 8, at 4 °C.
Porphyrin Extraction Procedure, HPLC Separation and Mass Determination by Mass Spectrometry.
Quantitative extraction of porphyrins from soluble fractions, membrane pellets and whole cell pellets was achieved by adding 300 μl of extraction solvent (ethanol/DMSO/acetic acid; 80/20/1; vol/vol/v) to 200 μl of soluble fraction and 1 ml of extraction solvent to the membrane, respectively. Samples were subjected to 5 sonication cycles of 5 sec at 0 °C, amplitude 10% using a sonicator probe (Branson Digital Sonifier). They were then centrifuged at 12,500 g for 5 min and the porphyrin containing supernatant was used for subsequent HPLC analysis. Re-extraction of the samples was performed until no further fluorescence of the extract could be observed under UV light (excitation wavelength: 405 nm, lamp: Karl Storz endoscope).
Porphyrin separation was achieved as described before (30) and detailed procedures are given in Fig. S2 legend.
YfeX and EfeB Activities in Soluble Fractions.
Time course studies of PPIX and MPPIX formation were performed with 200 μl samples of soluble fractions of JP313 (pBAD24-yfeX), JP313 (pBAD24-efeB), and JP313 (pBAD24) cell cultures resuspended for an OD600 of 20. Reactions were initiated by the addition of either 50 μM hemin, or meso-heme or buffer alone to the samples and incubation at room temperature. At times t = 0, t = 5 min, t = 10 min, t = 20 min, t = 30 min, and t = 60 min, total porphyrins in the samples were extracted by the addition of 300 μl of extraction solvent (ethanol/DMSO/acetic acid; 80/20/1; vol/vol/v) and analyzed by HPLC.
YfeX Purification.
A 1 ml sample of C600 (pBAD24-yfeX) soluble fraction was incubated with hemin 50 μM or without hemin at room temperature for 30 min. After centrifugation for 5 min at 10,000 × g to discard aggregates, the mixtures were loaded on anion exchange chromatography (mono Q) preequilibrated with 50 mM Tris·HCl pH 8. Elution was performed with a 0 to 1 M NaCl gradient in the same buffer. Fractions were collected and their YfeX content and purity evaluated by SDS/PAGE. Purified YfeX with an apparent molecular mass of 33 kDa was eluted into 2 fractions with different UV/visible absorption spectra. One fraction had only a 280 nm protein absorption peak which corresponded to apo-YfeX. The N-terminal amino acid sequence was determined by the “Plateforme d'Analyze et de Microséquence des Protéines” of the Institut Pasteur. It corresponded to the N terminus of YfeX: SQVQSG. Apo-YfeX was used in the binding experiments. YfeX H215A was purified following the same protocol.
YfeX Binding Studies.
PPIX and hemin binding studies were carried out at room temperature. ApoYfeX was in Tris·HCl 50 mM pH 8. Concentration was evaluated from absorbance at 277 nm with a calculated ε = 32,500 M−1 cm−1. Apo-YfeX was diluted to 1 μM, a concentration which gives a measurable signal. Hemin and PPIX solutions were diluted in Tris·HCl 50 mM pH 8 for a 20 μM final. Aliquots of 1 to 5 μl of either hemin or PPIX were successively added to cell containing 1 ml of the apoprotein. This corresponds to PPIX or hemin range of final concentrations from 100 nM up to 1.5 μM for hemin and 100 nM up to 2.5 μM for PPIX. Absorption spectra were recorded 5 min after each hemin or PPIX addition in a 0.2–1 cm path length cells on a Beckman DU 800 spectrophotometer and were followed by measuring the absorbance from 250 nm to 700 nm. Absorbance at the Soret bands (404 nm for hemin and 407 nm for PPIX) were reported as a function of hemin and protoporphyrin IX concentration, corrected for absorbance of free heme and PPIX.
To determine the Kd for hemin and PPIX, titration curves could be fitted according to a one-site binding model using FitP software. Heme and PPIX binding to apo-YfeX H215A were done following the same protocol.
Supplementary Material
Acknowledgments.
We thank Dr. Daniel Béal IBPC for precious advices in fluorescence colony screening under near-UV light, Dr. Chantal Le Bouguennec and Dr. Alain Chaffotte for strains and helpful discussions and Dr. Simon Andrews for constructive advice. Furthermore, we thank Dr. Karine Ndjoko Ioset and Dr. Jean Luc Wolfender for their valuable help with the HPLC-MS analysis. We are grateful to Dr. Elie Dassa for careful reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903842106/DCSupplemental.
References
- 1.Wandersman C, Delepelaire P. Bacterial iron sources: From siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 2.Maresso A, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLOS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cescau S, et al. Heme acquisition by hemophores. BioMetals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 4.Reniere M, Torres V, Skaar EP. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. BioMetals. 2007;3-4:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 5.Létoffé S, Delepelaire P, Wandersman C. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc Natl Acad Sci USA. 2006;103:12891–12896. doi: 10.1073/pnas.0605440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frankenberg-Dinkel N. Bacterial heme oxygenases. Antioxid Redox Signaling. 2004;5:825–834. doi: 10.1089/ars.2004.6.825. [DOI] [PubMed] [Google Scholar]
- 7.Skaar E, Gaspar A, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puri S, O'Brian M. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J Bacteriol. 2006;188:6476–6482. doi: 10.1128/JB.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu R, et al. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–2846. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee W, Renière ML, Skaar EP, Murphy ME. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme degrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilks A, Ortiz de Montellano P, Rabsch W. Rat liver heme oxygenase. High level expression of a truncated soluble form and nature of the meso-hydroxylating species. J Biol Chem. 1993;268:22357–22362. [PubMed] [Google Scholar]
- 12.Frustaci J, O'Brian M. Characterization of a Bradyrhizobium japonicum ferrochelatase mutant and isolation of the hemH gene. J Bacteriol. 1992;174:4223–4229. doi: 10.1128/jb.174.13.4223-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugano Y. DyP-type peroxidases comprise a novel heme peroxidase family. Cell Mol Life Sci. 2008;66:1387–1403. doi: 10.1007/s00018-008-8651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sturm A, et al. YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. J Biol Chem. 2006;281:13972–13978. doi: 10.1074/jbc.M511891200. [DOI] [PubMed] [Google Scholar]
- 15.Perttila U, Sievers G. The heme environment of leghemoglobin. Absorption and circular dichroism spectra of artficial leghemoglobins and myoglobins. Biochim Biophys Acta. 1980;624:316–328. doi: 10.1016/0005-2795(80)90250-0. [DOI] [PubMed] [Google Scholar]
- 16.Koster W. Cytoplasmic membrane iron permease systems in the bacterial cell envelope. Front Biosci. 2005;10:462–477. doi: 10.2741/1542. [DOI] [PubMed] [Google Scholar]
- 17.Cao J, et al. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol Microbiol. 2007;65:857–875. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- 18.Grosse C, et al. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol. 2006;62:120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- 19.Woodard S, Dailey H. Regulation of heme biosynthesis in Escherichia coli. Arch Biochem Biophys. 1995;316:110–115. doi: 10.1006/abbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 20.Verderber E, et al. Role of the hemA gene product and delta-aminolevulinic acid in regulation of Escherichia coli heme synthesis. J Bacteriol. 1997;179:4583–4590. doi: 10.1128/jb.179.14.4583-4590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taketani S, et al. Heme synthase (ferrochelatase) catalyzes the removal of iron from heme and demetalation of metalloporphyrins. Biochemistry. 2007;46:15054–15061. doi: 10.1021/bi701460x. [DOI] [PubMed] [Google Scholar]
- 22.Loeb M. Ferrochelatase activity and protoporphyrin IX utilization in Haemophilus influenzae. J Bacteriol. 1995;177:3613–3615. doi: 10.1128/jb.177.12.3613-3615.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper C, et al. Peroxidase activity of hemoglobin towards ascorbate and urate: A synergistic protective strategy against toxicity of Hemoglobin-Based Oxygen Carriers (HBOC) Biochim Biophys Acta. 2008;1784:1415–1420. doi: 10.1016/j.bbapap.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Dawson RMC, Elliot DC, Elliott WH, Jones KM. Data for Biochemical Research. Oxford: Clarendon; 1986. [Google Scholar]
- 25.Karlberg T, et al. Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues. J Mol Biol. 2008;378:1074–1083. doi: 10.1016/j.jmb.2008.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zubieta C, et al. Identification and structural characterization of heme binding in a novel dye-decolorizing peroxidase, TyrA. Proteins. 2007;69:234–243. doi: 10.1002/prot.21673. [DOI] [PubMed] [Google Scholar]
- 27.Stauff D, et al. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J Bacteriol. 2008;190:3588–3596. doi: 10.1128/JB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatsumi R, Wachi M. TolC-dependent exclusion of porphyrins in Escherichia coli. J Bacteriol. 2008;190:6228–6233. doi: 10.1128/JB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Létoffé S, Delepelaire P, Wandersman C. Functional differences between heme permeases: Serratia marcescens HemTUV permease exhibits a narrower substrate specificity (restricted to heme) than the Escherichia coli DppABCDF peptide-heme permease. J Bacteriol. 2008;190:1866–1870. doi: 10.1128/JB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fotinos N, et al. Effects on gram-negative and gram-positive bacteria mediated by 5-aminolevulinic Acid and 5-aminolevulinic acid derivatives. Antimicrob Agents Chemother. 2008;52:1366–1673. doi: 10.1128/AAC.01372-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.