Abstract
The prion protein (PrP) is responsible for a group of neurodegenerative diseases called the transmissible spongiform encephalopathies. The normal function of PrP has not yet been discovered, but indirect evidence suggests a linkage to its ability to bind copper. In this article, low-copper-concentration bindings of Cu2+ to PrP are investigated by using a recently developed hybrid density functional theory (DFT)/DFT method. It is found that at the lowest copper concentrations, the binding site consists of 4 histidine residues coordinating the copper through ε imidazole nitrogens. At higher concentrations, 2 histidines are involved in the binding, one of them in the axial position. These results are in good agreement with existing experimental data. Comparison of free energies for all modes of coordination shows that when enough copper is available, the binding sites will spontaneously rearrange to accommodate more copper ions, despite the fact that binding energy per copper ion decreases with concentration. These findings support the hypothesis that PrP acts as a copper buffer in vivo, protecting other proteins from the attachment of copper ions. Using large-scale classical molecular dynamics, we also probe the structure of full-length copper-bound PrP, including its unfolded N-terminal domain. The results show that copper attachment leads to rearrangement of the structure of the Cu-bonded octarepeat region and to development of turns in areas separating copper-bound residues. These turns make the flexible N-terminal domain more rigid and thus more resistant to misfolding. The last result suggests that copper binding plays a beneficial role in the initial stages of prion diseases.
Keywords: copper attachment, hybrid DFT, misfolding, neurodegenerative diseases, metalloprotein
Transmissible spongiform encephalopathies (TSEs) are neurodegenerative diseases in which infectious agents called prions are exclusively composed of an abnormally folded form of the protein PrP (1, 2). The TSEs include bovine spongiform encephalopathy or “mad cow disease” in cattle, scrapie in sheep, chronic wasting disease in deer and elk, and kuru and Creutzfeldt–Jakob disease in humans. Prions represent a new type of infectious particle different from other known infectious agents such as viruses or bacteria.
The PrP is a normal component of many types of tissues and is found at highest levels in the central nervous systems in presynaptic membranes (3), where it is attached through a glycolphosphatidylinositol anchor (4). Excluding a signal peptide, it consists of 209 amino acids and has a folded C-terminal domain and a flexible and unstructured N-terminal domain. In TSEs, the PrP misfolds from its normal cellular form, PrPC, to the abnormal pathological scrapie form, PrPSc, creating an infectious prion. This conversion also changes the prion protein from being sensitive to proteinase K (PK) digestion to being highly resistant toward it. The misfolded proteins accumulate forming amyloidic deposits that are hallmarks of the neurodegenerative diseases. The detailed structure of the PrPSc has not been resolved yet, but it was found that the amyloidic deposits are β-sheet rich (5).
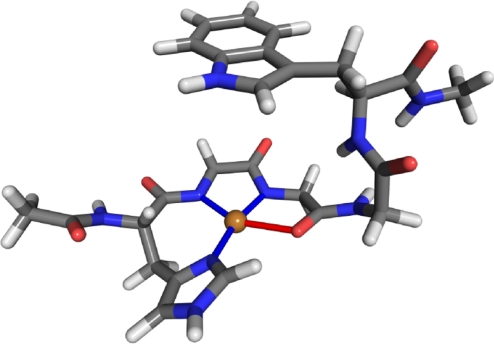
The normal function of the PrPC in healthy tissue is still unknown. However, it is now well established (6, 7) that the PrPC efficiently binds Cu2+ and further studies (8) link this ability to its function. Current research also indicates that the PrPC can act as a copper ion buffer protecting cells from effects of uncomplexed Cu2+. For example, comparison between normal and PrP knockout mice shows (9) that the knockout mice had extensive oxidative damage not present in the normal ones. Furthermore, neurons containing PrP are more resistant to copper toxicity than cells lacking the protein (10–12). Other PrP functions have also been suggested. These include superoxide dismutase activity (13) and transporting copper ions through membranes. (14–16) To determine PrP's function, the mechanisms and properties of Cu2+ binding have to be understood. A range of experimental techniques, including EPR (17–19), CD (20), X-ray crystallography (16), and NMR (21, 22) have been used to investigate Cu2+ binding to PrP. These studies determined that, at maximum copper occupancy, the full-length prion protein can bind 5 or 6 copper ions. Four of these binding sites are located in the octarepeat domain, residues 60–91, which consists of 4 tandem repeats of the amino acid sequence PHGGGWGQ. In this case, the copper ions are coordinated by a single histidine residue, deprotonated backbone amide nitrogens, and a backbone carboxyl oxygen. This mode of binding has been confirmed by theoretical studies (23, 24) and the copper geometry is displayed in Fig. 1. In addition, experimental evidence (28, 29) indicates that the structured C terminus is capable of binding Cu2+ as well. These binding sites were very recently investigated (30) theoretically by using QM/MM Car–Parinello molecular dynamics. More recent experiments (25, 26) have shown that this picture of copper binding to the PrP is incomplete and that at low copper concentrations, which are physiologically the most relevant, the octarepeat domain binds Cu2+ in 2 different modes. At the lowest Cu2+ concentrations, a single copper is coordinated by multiple histidine imidazoles without involvement of the backbone. This type of coordination is also found (27) to exist at mildly acidic pH of 5.5, which disfavors the deprotonation of backbone nitrogens occurring at high concentration Cu2+ binding. At intermediate concentrations, the copper ion is coordinated by 1 or 2 histidines, 2 water molecules, and a deprotonated backbone nitrogen. Further increase in copper ion concentration yields the previously mentioned maximum copper occupancy binding geometry.
Fig. 1.
Binding of copper ion to a HGGGW fragment. The copper ion is shown in gold, carbon atoms are colored gray, nitrogen atoms blue, oxygen atoms red, and hydrogens light gray.
Despite these advances, the copper binding to the PrPC is still not fully understood. The 3D binding geometry is only known for the high-occupancy binding, which has been studied the most, whereas for the other types of binding, important aspects, such as the number of histidine residues and water molecules involved in binding, have not been determined. The binding energies are unknown, and for the multiple-histidine coordination it is unclear which N atoms in the imidazole group bind to the copper ion.
In this article, we theoretically investigate the physiologically important low copper-concentration binding modes of Cu2+ to the prion protein. We determine the precise geometries of the bindings and consider the structure of copper-bound full-length PrPC. Analysis of energetics of all of the binding modes shows that the binding strength decreases with the number of coordinated copper ions, which supports the notion that PrPC functions as a copper buffer. Calculations on the full-length PrPC reveal that that binding of copper leads to development of new structural features in the unfolded part of the protein, increasing the stability of this entire region. Because the stabilized protein has a lesser tendency to misfold, this suggests a protective role of copper in the initial stages of prion diseases.
Results and Discussion
Low-Concentration Copper Binding to PrP.
Experiments (25, 26) have shown that at low copper concentration the copper ion is coordinated by multiple histidine imidazoles. However, the 3D geometry has not been determined, and it is not known which nitrogen atoms in the imidazole group participate in the binding. Furthermore, experiments (25) could not distinguish between alternative copper-ion coordinations involving either 4 histidine residues or 3 histidines and a water molecule.
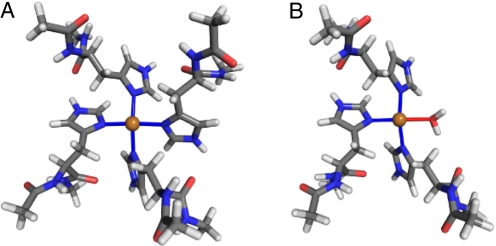
The 2 alternatives are investigated theoretically by using the hybrid density functional theory (DFT) method. This method was recently used (23) to evaluate the high-concentration copper binding in PrPC, and the results were in agreement with structures obtained from experiments (31) and other theoretical studies (24, 32). In fact, compared with other theoretical studies, our results provided refinement of the binding-site geometry, because of the use of explicit solvation instead of an implicit one (24, 32). We first investigate the binding through δ-nitrogens, Nδ, on imidazole rings with ε-nitrogens being protonated. The optimized geometry with Cu coordinated to 4 histidines is shown in Fig. 2A. The binding is mostly planar, with all 4 Cu-N bonds being 2.00 ± 0.02 Å. The angles between the imidazole ring planes and the binding plane are also alike, 55° to 60°.
Fig. 2.
Relaxed δ-binding geometries of a copper ion to 4 histidine residues (A) and 3 histidine residues and a water molecule (B). For clarity, water molecules not coordinating directly with the copper ion are not shown. All Cu-N bond distances are between 1.98 and 2.02 Å and the N-Cu-N bond angles are between 88° and 95°. The atom colors are the same as in Fig. 1.
When one of the histidines is replaced by a water molecule (Fig. 2B), the bonding geometry remains similar, with Cu-bonding being still planar and the Cu-N distances ≈2.00 Å. However, the Cu-O bond to the water molecule is longer, 2.29 Å, indicating weaker bonding than to the histidine residues. Comparison of energetics of the 2 configurations confirm this: The binding through 4 histidines is favored by 0.62 eV over that through 3 histidines and a water molecule.
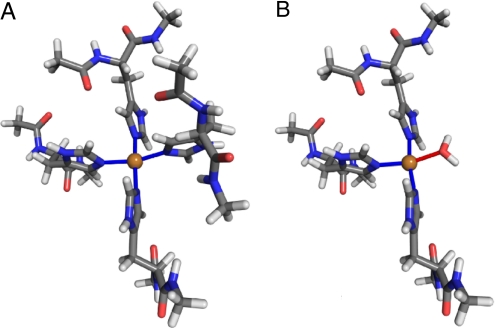
The relaxed structures for the ε binding are shown in Fig. 3. The geometries are similar to those in the δ configurations: The binding is planar, and the bond distances between copper and nitrogen atoms are close to 2.00 Å. However, the angles between the binding plane and the imidazole rings are 85° to 95°, compared with 55° to 60° in the δ case. The configuration with 3 histidines and a water molecule is unfavorable by a similar energy difference of 0.73 eV.
Fig. 3.
Relaxed ε-binding geometries of a copper ion to 4 histidine residues (A) and 3 histidine residues and a water molecule (B). All Cu-N bond distances are between 1.98 and 2.02 Å and N-Cu-N bond angles are between 88° and 95°. The atom colors are the same as in Fig. 1.
To determine which of the 4-histidine configurations is more advantageous energetically, one needs to account for the fact that the total energy of the histidine depends on its protonation state. Our calculations show that protonation at the ε nitrogen is preferable by 0.06 eV. Because the number of atoms in both systems is the same, the energy difference can be calculated as
where Eδ and Eε are the total energies of δ and ε bindings, respectively, and EHisδ and EHisε are the energies of histidine fragments with δ and ε protonations. The calculated ΔEδε shows that the ε binding is preferred by 0.21 eV. This result is consistent with experiment (33), which showed that when the copper ion is coordinated to multiple histidine residues, the binding involves ε nitrogens.
Medium-Concentration Copper Binding to PrPC.
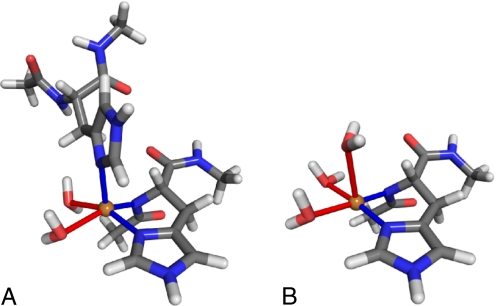
Copper binding to PrPC at medium concentrations was experimentally investigated by Chattopadhyay et al. (25) and Wells et al. (26). These workers proposed similar models of Cu2+ binding with 2 histidine imidazoles involved in binding, one in equatorial and the other in axial position, although the presence of the axial histidine could not be conclusively proven in Chattopadhyay's work. Starting from their model, we constructed atomic structures with the copper ion coordinated by deprotonated imidazole δ-N, 2 water molecules in an equatorial plane, and either an axial histidine or an axial water molecule. The relaxed structures for both cases are shown in Fig. 4. Comparing the energies of the 2 systems, we find that the axial histidine is energetically favored over the axial water by 0.26 eV, confirming histidine's participation in the binding. The Cu distance to the axial δ-N is 2.19 Å, whereas the axial water is located at 2.61 Å, indicating weaker binding. The equatorial binding is mostly planar in both cases, although with the axial histidine present, Cu is slightly shifted out of the plane toward the coordinating axial nitrogen. The Cu-O distances for equatorial waters are 2.25 and 2.35 Å for axial histidine and 2.13 and 2.19 Å for axial water. The Cu-N equatorial distances are 2.00 Å.
Fig. 4.
Relaxed medium concentration copper bindings with axial histidine present (A) and with axial histidine replaced by a water molecule (B). The bond distances between Cu and axial nitrogens are 2.00 Å, whereas the equatorial nitrogen is located at 2.35 Å. The Cu-O distances between the copper ion and the axial water molecules are 2.25 and 2.35 Å for A and 2.13 and 2.19 Å for B. The additional axial water in B is located at 2.61 Å. The equatorial bond angles starting from N-Cu-N and continuing clockwise are 89°, 90°, 79°, and 94° for A and 93°, 92°, 85°, and 91° for B. The atom colors are the same as in Fig. 1.
PrP as a Copper Buffer.
At present, the leading hypothesis for PrP's function in vivo is that it acts as a copper buffer (34), binding uncomplexed extracellular Cu2+ and protecting other proteins from its deleterious effects. In this study, we have examined 2 of the 3 binding modes of copper occurring in the octarepeat domain of the PrPC. The third type of binding was explored in our previous work (23) as well as by other workers (24). Using insights from these studies, we now analyze the plausibility of the copper buffer hypothesis.
For each of the 3 binding modes, we calculate the binding energies of Cu2+ to PrPC relative to Cu2+ solvated in water. The low concentration, multiple-histidine binding has the energy of 5.22 eV, which is the largest. The medium concentration coordination of copper is less strong, with a binding energy of 2.75 eV. The high occupancy, high concentration binding is the weakest, with an energy gain of 1.76 eV. These results are consistent with recent experimental values for PrPC's copper affinity (26, 35), where the same ordering was found, with low concentration binding being in the nanomolar range, whereas the high coordination concentration reached micromoles.
Both the experimental and the theoretical results thus show that the strength of binding decreases with copper concentration. This may seem contradictory to the PrPC's suggested buffering role, which requires binding of increasing amount of copper with increasing concentration, but the interplay between occupancy and the binding strength enables PrPC to perform the buffering function. In the low concentration regime, the copper ion is coordinated by all 4 histidines in the octarepeat domain, and thus only 1 ion can be bound to the PrPC, at an energy gain of 5.22 eV. As the concentration increases, an additional ion will be bound, but now each copper ion is coordinated to 2 histidines, allowing 2 copper ions per PrPC. Because the medium concentration binding energy is 2.75 eV, the energy gain becomes 2 × 2.75 eV − 5.22 eV = 0.28 eV. An energy gain also occurs for the transition from the medium-concentration binding to the high-concentration one. In this case, 2 copper ions are replaced by 4 more weakly bound ones. This replacement is exothermic by 4 × 1.76 eV − 2 × 2.75 eV = 1.54 eV. Our results thus show that with increasing presence of copper in its environment, PrPC will sequester more copper ions, therefore functioning as a copper buffer, in accordance with earlier hypotheses (34).
The ordering above, which considered only the total (internal) energy, is the same also when the free energies are calculated, because the entropy of the Cu-containing PrP increases when the number of Cu ions bound to PrP increases. This is because the attachment of a single Cu ion, while having the largest binding energy, imposes severe constraints on the internal degrees of freedom of the PrP, bonding to 4 histidine residues in the octarepeat region. In the intermediate binding mode, only 2 histidine residues are coordinated to each Cu ion, allowing for increased thermal motion, whereas in the high concentration case, each Cu ion binds to a single residue, leaving the PrP even less constrained. This qualitative argument is confirmed by our classical molecular dynamics simulations on full-length models for all 3 modes of Cu-bound PrP and calculating entropies by using Schlitter's method (36, 37). The T*S values at 300 K increase by ≈0.16 eV when the number of Cu ions is increased from 1 to 2 and, additionally, by ≈0.23 eV when 2 more Cu ions are added. Combining the energetic and entropic contributions at 300 K, the binding of the second Cu ion releases 0.44 eV and the attachment of additional 2 ions gains 1.77 eV.
Structural Properties of Full-Length Copper-Bound Prion Protein.
We also investigate the effect of copper binding on the structure of full-length PrPC. In this study, we only consider the low concentration 4-histidine coordination, which is the strongest bound and likely to be the most physiologically relevant, because its copper affinity is similar to that of other copper-binding proteins (26, 38, 39). Molecular mechanics is used in this study, because of the large number of atoms required in the simulations.
The N-terminal domain of PrPC, which contains the copper-binding region, is unfolded and flexible. Therefore, its structure in either free or copper-bound states cannot be revealed by current experiments. Even with the use of computer simulation, determining the structure of the full-length PrPC is challenging, because no experimental data can be used to form an initial structure. Furthermore, because the N-terminal domain is unfolded, even the equilibrium structure is highly flexible, instead of being well defined, as in the case of folded proteins. In addition, the unfolded part of the PrPC takes up significantly more space than a folded protein of the same size and therefore requires a substantially larger simulation cell than a typical folded protein with the same number of residues. To overcome these difficulties, a massively parallel, long-time simulation is used to explore the structural properties of full-length PrPC.
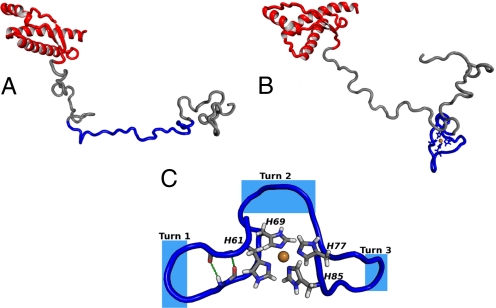
The initial structure of the free full-length PrPC is set up as described in Materials and Methods. An equilibration of ≈30 ns is followed by a 20-ns molecular dynamics run. A snapshot of the equilibrated structure is shown in Fig. 5A. The folded C-terminal region is largely conserved, with the exception of residues 192–194, which convert from a 3–10 helix to an ordinary α-helix. The unfolded N-terminal region develops only turns and random coils; no higher-order structural features are present. The octarepeat region, containing histidine residues that bind the copper ion, is mostly extended with only a few transient turns developed there.
Fig. 5.
Snapshots of equilibrated structures of PrPC. (A) Free PrPC. The octarepeat region is shown in blue. Note that this region is extended and only few turns are developed. (B) Copper-bound PrPC. (C) Close-up of the octarepeat region. The intermediate segments between copper-bound histidines form turns that reverse the direction of the backbone. Several types of turns are observed. Turn 1 is a β-turn, with β-bridge between residues 62 and 67 just after the turn. In this case, the intermediate segment forms a short β-hairpin. Turn 2 is wide and cannot be classified as a turn based on the standard criteria. Turn 3 is similar to 1, with the exception that no β-bridge is formed within the segment. The atom colors are the same as in Fig. 1.
In the case of copper-bound PrPC, the copper is assumed to bind to histidine residues in the octarepeat domain, which extend from residue 60 to 91 and consist of 4 tandem repeats of the 8-residue sequence PHGGGWGQ. Copper binding to octarepeat histidines, His 61, 69, 77, and 85, creates 3 intermediate segments, 62–68, 70–76, and 78–84, separating the copper-bound histidines with identical sequences of GGGWGQP. The initial structure of the copper-bound PrPC is based on the equilibrated structure of the free PrPC, on which the geometry of the copper-bound fragment obtained from quantum calculations is imposed. The copper-bound protein is first equilibrated for 28 ns, after which a molecular dynamics run of 20 ns is performed to examine the dynamic stability of the equilibrated structure. A snapshot of the equilibrated copper-bound PrPC is shown in Fig. 5B, whereas a close-up of the octarepeat region is displayed in Fig. 5C. The figures show that the structure of the copper-bound octarepeat region is characterized by turns formed in the intermediate segments. These turns revert the direction of the protein chain so that it returns back to the binding site. However, the segments remain highly flexible and change shape throughout the calculation.
The types of turns are determined by distances between the α-carbons of neighboring copper-bound histidines. Because only side-chain imidazoles are directly involved in copper binding, the histidine α-carbons are well movable, and distances between them fluctuate between 5 and 10 Å. For distances shorter than ≈8 Å, the turns formed by the intermediate segments are regular β-turns, composed of either GWGQ or GGWG residues. These segments can develop internal hydrogen bonds, just outside the turn region. In such cases, the segments form short β-hairpin structures, which are stable for several nanoseconds. However, when distances between the histidine α-carbons are greater than ≈8 Å, the turns are wide, irregular, and cannot be classified as turns by using STRIDE (40) or DSS (41), although they still facilitate the change in the direction of the protein backbone. Outside of the octarepeat region in the N-terminal domain, the structure is similar to that found for the free PrPC, i.e., only unstable turns and random coils exist there. The C-terminal domain is the same as in the case of the free protein and the initial folded structure is conserved, showing that the influence of the bound copper does not extend to its C-terminal domain. Within the length of our simulation (20 ns), there is no indication of β-sheets formation or transformation to PrPSc, which is rich in β-sheets.
The stabilities of structural features in the N-terminal domain are also examined for both free and copper-bound PrPC. We count the number of MD steps over which noncoil structural features (turns, occasional β-bridges), developed in the N-terminal domain, persist without being destroyed. Secondary structure assignment is made by using STRIDE (40). In the copper-bound protein the structural features persist 17% longer than in the case of the free protein. Calculating this quantity only for residues outside the octarepeat domain, we find almost the same value of 15%, showing that binding of copper enhances the structural stability of the whole N-terminal domain and not only the atoms near the copper ion. A possible reason for this is that the octarepeat region contains a high concentration of glycines, which are highly flexible and therefore contribute to the pliability of the unfolded part of PrPC. When this region is stabilized by copper binding, a stabilizing effect on the whole N-terminal domain can be expected.
Despite numerous studies, it is still unclear whether copper plays a beneficial or a deleterious role in prion diseases. It was found earlier that the presence of copper greatly slows amyloid formation (42) and that the addition of copper reduces the accumulation of PrPSc (43). On the other hand, copper is present in PrPSc deposits (44), and its removal by chelation delays the onset of prion diseases (45). Copper was also found to promote conversion of recombinant PrPC into the PK-resistant form, but this conversion required aging, and the effect on fresh prion protein is small (46). Our finding that copper binding stabilizes the structure of the N-terminal domain indicates that copper has a mildly protective role against prion diseases when bound in the low-concentration structure, because a stabilized N-terminal domain should have lesser tendency for misfolding.
Summary and Conclusions
Hybrid DFT/DFT calculations were used to study the geometry and energetics of copper ion attachment to the prion protein, which is responsible for a group of neurodegenerative diseases. Several different binding geometries were examined, corresponding to different copper concentrations. At low copper concentrations it was found that the copper ion is bound to 4 histidine imidazole groups through Nε atoms, whereas at higher concentrations, 2 histidine imidazoles, one of them in axial position, coordinate the copper. These results are in good agreement with the existing experimental data. An evaluation of the energetics of all of the copper coordination modes shows that interplay between the number of bound ions and the binding strength is such that when enough copper is available, the binding site(s) will spontaneously rearrange to accommodate more copper ions with a net energy gain, despite the fact that the binding strength per copper ion decreases with the number of ions per PrP. The strong affinity for Cu and the multiplicity of concentration-dependent Cu binding sites supports the suggested copper buffering role of the prion protein, as storing excess Cu2+ and protecting other proteins from the effects of uncomplexed copper.
The effect of copper binding in the physiologically most relevant low-concentration configuration on the conformation of the prion protein was also investigated through very large scale molecular dynamics simulation. This is the only way to investigate the effect of Cu on the conformation of the unstructured part of the prion protein, which cannot be determined experimentally because of its dynamic flexibility. Our results show that copper binding alters the structure of the unfolded N-terminal domain, creating flexible turns in the octarepeat region that coordinates the copper ion. Furthermore, Cu binding increases the stability of the entire N-terminal domain, increasing its resistance to misfolding. The last results suggest that copper binding in the low concentration mode, which is the dominant one in vivo, delays the onset of prion diseases.
Materials and Methods
Hybrid DFT Calculations.
Solvation effects are important for biomolecules, and therefore the solvent has to be included in biosimulations. Because a full quantum-mechanical description of a large amount of solvent is prohibitively expensive, various hybrid embedding schemes were developed, such as the QM/MM (47–49), which describes the solvent by using molecular mechanics. This article employs a recently developed massively parallel hybrid DFT/DFT method (23) in which the chemically active part of the system, including its first solvation shells, is treated at an ab initio DFT level, whereas an approximate orbital-free DFT is used for the remainder of the solvent. Thousands of solvent molecules can be included in the calculations at a low computational cost, because the solvent molecules in the orbital-free region have fixed internal structures and frozen electron densities. The advantage of this method is that all interactions in the system are treated at the DFT level and that the flow of molecules across the DFT/orbital-free-DFT interface is allowed, which preserves the fidelity of the simulations for the reactive part of the system.
In the chemically active ab initio part of the system, the DFT equations are solved by using the highly parallel real-space multigrid method (RMG) (50, 51) code. The calculations use a grid with spacing of 0.32 bohr, corresponding to the kinetic energy cutoff of 48 Ry, ultrasoft pseudopotentials (52), and the generalized gradient approximation (53). The simulations are performed on up to 512 CPUs of the Cray XT4 supercomputer.
Structural Model of Copper Binding to PrPC.
The prion protein is represented by fragments of histidine residues to which neutral ends are applied, resulting in 29 atoms in each fragment. The fragments, the copper ion, and the water molecules closest to the copper ion (6–9 water molecules, depending on the type of binding) are treated by the full Kohn–Sham (KS) DFT. Up to 4 fragments are included in the calculations, resulting in up to 143 KS DFT atoms including 9 water molecules. The remainder of the simulation cell, with dimensions of 44.45 Å, is filled with up to 2m644 water molecules at liquid density. The water molecules not included in the KS DFT are treated by the orbital-free DFT.
In the calculations, several initial structures matching the experimental data are used. They are first equilibrated by performing 1,000 molecular dynamics steps at 300 K to remove the bias of the initial configuration and escape from local energy minima. They are then relaxed, and configurations with the lowest energies are identified. The final structures are discussed in Results and Discussion, where the different copper-binding modes are described.
Structural Model and Simulations of the Full-Length PrPC.
The structural model of the full-length PrPC is constructed in several steps: First, the flexible N-terminal domain is built according to its amino acid sequence for residues 23–124 as a β-strand by using PyMOL (54). The residues 1–22, which are part of the signal peptide, are not included in the calculation. The peptide is partially equilibrated by running molecular dynamics for 6 ns, which leads to the destruction of the initial structure and creation of random coils and turns. For the C-terminal domain, an entry with PDB ID code 1QLX (55) from the Protein Data Bank database, which gives the structure of residues 125–228, is used. The 2 parts are joined together to form a model of the full-length PrPC, consisting of 206 amino acids.
The initial structure of the copper-bound PrPC is based on the structure of equilibrated free PrPC, on which the copper binding geometry determined from DFT calculations is imposed. The histidines participating in the binding are those in the octarepeat domain, His-61, His-69, His-77, and His-85.
The stabilities of the full-length free and copper-bound PrPC structures are investigated by classical molecular dynamics using the package NAMD (56) with the CHARMM27 (57) force-field parameters. Periodic boundary conditions are used, and the protein is immersed in a large water box in which the distance between any protein atom and an edge of the box is at least 10 Å. The model PrPC is positively charged, +7e, and therefore 7 Cl− counter ions are added to the simulation cell at random positions to neutralize the system. The total number of atoms is 195,748 for the free PrPC and 164,371 for the copper-bound PrPC. The temperature is maintained at 310 K by means of Langevin dynamics and the pressure is kept constant at 1 atm. The constant pressure is maintained by the NosŁ–Hoover Langevin Piston algorithm (58, 59). The time steps in the Verlet-I/r-RESPA method (60, 61) are: 4 fs for the long-range electrostatic forces, 2 fs for the short-range nonbonded forces, and 2 fs for the bonded forces. Bonds involving hydrogen are constrained with SHAKE (62) to enable large time steps.
As is well known, copper and other transition metals are difficult to describe with force fields, because of the importance of quantum effects originating in their d shell that cannot be neglected. Therefore, the copper ion is not directly present in the simulation. Instead, its effect on the copper-bound protein is modeled by constraining the binding-site geometry to that determined from hybrid DFT calculations. To mimic the free rotation of the coordinating imidazole rings, which is observed during hybrid DFT molecular dynamics simulations, only the 2 nitrogen atoms in the imidazole rings are fixed.
After these calculations were completed, an improved description of the Cu-binding site was developed. In this model, the Cu is connected to the coordinating atoms through springs with appropriate stiffness, so that the amplitudes of bond vibrations are the same in classical and quantum calculations. Parts of calculations were repeated with this method and there was little impact on the quantities discussed in this work, including the flexibility of the copper-bound PrP. The rest of the protein is free to move.
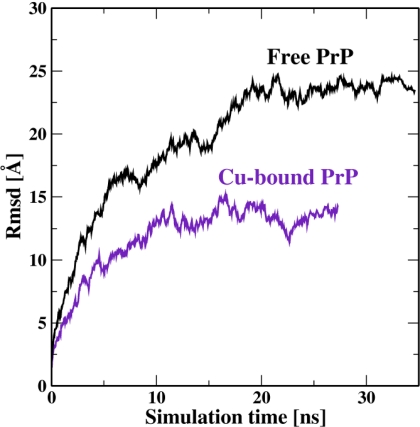
Both the free and copper-bound proteins are equilibrated until the RMSD of the backbone flattened out. Because of the flexibility of the N-terminal domain and the fact that it is built without detailed knowledge of its structure, it takes 20–30 ns for the RMSD to reasonably flatten out. A complete equilibration may require an even longer simulation, but this would be prohibitively expensive computationally. The RMSD as a function of the simulation time is displayed in Fig. 6. Other quantities, such as various energy components and the solvent-accessible surface of the protein are already well equilibrated within the simulation time.
Fig. 6.
RMS deviations of the protein backbone as a function of simulation time for free and Cu-bound PrP.
Acknowledgments.
We are indebted to Dr. Nicholas M. Glykos for his CARMA entropy program and help, and to Dr. Jason Vertrees for his help with generating graphics by using PyMOL. This work was mainly supported by Department of Energy (DOE) Grant DE-FG02-98ER45685, with the petascale code development supported by DOE Grant DE-FG05-08OR23331 and National Science Foundation (NSF) Grant OCI-0749320. R.C. was supported by the NSF Research Experiences for Undergraduates Program. The calculations were carried out at the National Center for Computational Sciences at Oak Ridge National Laboratory.
Footnotes
The authors declare no conflict of interest.
References
- 1.Prusiner S. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herms J, et al. Evidence of presynaptic location and function of the prion protein. J Neurosci. 1999;19:8866–8875. doi: 10.1523/JNEUROSCI.19-20-08866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caughey B, Chesebro B. Prion protein and the transmissible spongiform encephalopathies. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 5.Baxa U, Cassese T, Kajava A, Steven A. Structure, function, and amyloidogenesis of fungal prions: Filament polymorphism and prion variants. Adv Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- 6.Hornshaw MP, Mcdermott JR, Candy JM, Lakey JH. Copper-binding to the n-terminal tandem repeat region of mammalian and avian prion protein—Structural studies using synthetic peptides. Biochem Bioph Res Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 7.Hornshaw MP, Mcdermott JR, Candy JM. Copper-binding to the N-terminal tandem repeat regions of mammalian and avian prion protein. Biochem Bioph Res Commun. 1995;207:621–629. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 8.Brown D, et al. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 9.Klamt F, et al. Imbalance of antioxidant defense in mice lacking cellular prion protein. Free Radical Biol Med. 2001;30:1137–1144. doi: 10.1016/s0891-5849(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 10.Vassallo N, Herms J. Cellular prion protein function in copper homeostasis and redox signalling at the synapse. J Neurochem. 2003;86:538–544. doi: 10.1046/j.1471-4159.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 11.Rachidi W, et al. Expression of prion protein increases cellular copper binding and antioxidant enzyme activities but not copper delivery. J Biol Chem. 2003;278:9064–9072. doi: 10.1074/jbc.M211830200. [DOI] [PubMed] [Google Scholar]
- 12.Zeng FN, Watt NT, Walmsley AR, Hooper NM. Tethering the N-terminus of the prion protein compromises the cellular response to oxidative stress. J Neurochem. 2003;84:480–490. doi: 10.1046/j.1471-4159.2003.01529.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown DR, et al. Normal prion protein has an activity like that of superoxide dismutase. Biochem J. 1999;344:1–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Whittal R, Ball H, Cohen F, Prusiner A, Baldwin M. Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci. 2000;9:332–343. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauly P, Harris D. Copper stimulates endocytosis of the prion protein. J Biol Chem. 1998;273:33107–33110. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 16.Burns CS, et al. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aronoff-Spencer E, et al. Identification of the Cu 2 binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistryistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns CS, et al. Copper coordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–6803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonomo R, Impellizzeri G, Pappalardo G, Rizzarelli E, Tabbi G. Copper (II) binding modes in the prion octapeptide PHGGGWGQ: A spectroscopic and voltammetric study. Chemistry. 2000;6:4195–4202. doi: 10.1002/1521-3765(20001117)6:22<4195::aid-chem4195>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Viles J, et al. Copper binding to the prion protein: Structural implications of four identical cooperative binding sites. Proc Natl Acad Sci USA. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson G, et al. Location and properties of metal-binding sites on the human prion protein. Proc Natl Acad Sci USA. 2001;98:8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CE, Klewpatinond M, Abdelraheim SR, Brown DR, Viles JH. Probing copper(2+) binding to the prion protein-using diamagnetic nicke(2+) and H-1 NMR: The unstructured N terminus facilitates the coordination of six copper(2+) ions at physiological concentrations. J Mol Biol. 2005;346:1393–1407. doi: 10.1016/j.jmb.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 23.Hodak M, Lu W, Bernholc J. Hybrid ab initio Kohn–Sham density functional theory/frozen-density orbital-free density functional theory simulation method suitable for biological systems. J Chem Phys. 2008;128 doi: 10.1063/1.2814165. 014101. [DOI] [PubMed] [Google Scholar]
- 24.Pushie M, Rauk A. Computational studies of Cu(II)[peptide] binding motifs: Cu[hggg] and cu[hg] as models for Cu(II) binding to the prion protein octarepeat region. J Biol Inorg Chem. 2003;8:53–65. doi: 10.1007/s00775-002-0386-7. [DOI] [PubMed] [Google Scholar]
- 25.Chattopadhyay M, et al. The octarepeat domain of the prion protein binds cu(II) with three distinct coordination modes at pH 7.4. J Am Chem Soc. 2005;127:12647–12656. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wells M, et al. Multiple forms of copper (II) co-ordination occur throughout the disordered N-terminal region of the prion protein at pH 7.4. Biochem J. 2006;400:501–510. doi: 10.1042/BJ20060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells MA, et al. A reassessment of copper(II) binding in the full-length prion protein. Biochem J. 2006;399:435–444. doi: 10.1042/BJ20060458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cereghetti GM, et al. Electron paramagnetic resonance evidence for binding of Cu2+ to the C-terminal domain of the murine prion protein. Biophys J. 2001;81:516–525. doi: 10.1016/S0006-3495(01)75718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cereghetti GM, et al. Stability and Cu (II) binding of prion protein variants related to inherited human prion diseases. Biophys J. 2003;84:1985–1997. doi: 10.1016/S0006-3495(03)75007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colombo MC, et al. Copper binding sites in the C-terminal domain of mouse prion protein: A hybrid (QM/MM) molecular dynamics study. Proteins. 2008;70:1084–1098. doi: 10.1002/prot.21604. [DOI] [PubMed] [Google Scholar]
- 31.Millhauser G. Copper binding in the prion protein. Acc Chem Res. 2004;37:79–85. doi: 10.1021/ar0301678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji H, Zhang H. A theoretical study on Cu (II) binding modes and antioxidant activity of mammalian normal prion protein. Chem Res Toxicol. 2004;17:471–475. doi: 10.1021/tx034232y. [DOI] [PubMed] [Google Scholar]
- 33.Miura T, Hori-i A, Mototani H, Takeuchi H. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. doi: 10.1021/bi9909389. [DOI] [PubMed] [Google Scholar]
- 34.Millhauser G. Copper and the prion protein: Methods, structures, function, and disease. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter ED, Chattopadhyay M, Millhauser GL. The affinity of copper binding to the prion protein octarepeat domain: Evidence for negative cooperativity. Biochemistry. 2006;45:13083–13092. doi: 10.1021/bi060948r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlitter J. Estimation of absolute and relative entropies of macromolecules using the covariance matrix. Chem Phys Lett. 1993;215:617–621. [Google Scholar]
- 37.Glykos NM. CARMA: A molecular dynamics analysis program. J Comput Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 38.Linder M. Biochemistry of Copper. New York: Plenum; 1991. Extracellular copper substituents and mammalian copper transport; pp. 73–134. [Google Scholar]
- 39.Masuoka J, Hegenauer J, Van Dyke B, Saltman P. Intrinsic stoichiometric equilibrium constants for the binding of zinc (II) and copper (II) to the high affinity site of serum albumin. J Biol Chem. 1993;268:21533–21537. [PubMed] [Google Scholar]
- 40.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins Struct Funct Genet. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 42.Bocharova O, Breydo L, Salnikov V, Baskakov I. Copper (II) inhibits in vitro conversion of prion protein into amyloid fibrils. Biochemistry. 2005;44:6776–6787. doi: 10.1021/bi050251q. [DOI] [PubMed] [Google Scholar]
- 43.Hijazi N, Shaked Y, Rosenmann H, Ben-Hur T, Gabizon R. Copper binding to PrPC may inhibit prion disease propagation. Brain Res. 2003;993:192–200. doi: 10.1016/j.brainres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Wadsworth J, et al. Strain-specific prion-protein conformation determined by metal ions. Nat Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 45.Sigurdsson E, et al. Copper chelation delays the onset of prion disease*. J Biol Chem. 2003;278:46199–46202. doi: 10.1074/jbc.C300303200. [DOI] [PubMed] [Google Scholar]
- 46.Qin K, et al. Copper (II)-induced conformational changes and protease resistance in recombinant and cellular PrP effect of protein age and deamidation. J Biol Chem. 2000;275:19121–19131. doi: 10.1074/jbc.275.25.19121. [DOI] [PubMed] [Google Scholar]
- 47.Warshel A, Levitt M. Theoretical studies of enzymic reactions—Dielectric, electrostatic and steric stabilization of carbonium-ion in reaction of lysozyme. J Mol Biol. 1976;102:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 48.Singh UC, Kollmann PA. A combined ab initio quantum-mechanical and molecular mechanical method for carrying out simulations on complex molecular-systems—Applications to the ch3cl + cl- exchange-reaction and gas-phase protonation of polyethers. J Comput Chem. 1986;7:718–730. [Google Scholar]
- 49.Field MJ, Bash PA, Karplus M. A combined quantum-mechanical and molecular mechanical potential for molecular-dynamics simulations. J Comput Chem. 1990;11:700–733. [Google Scholar]
- 50.Briggs EL, Sullivan DJ, Bernholc J. Real-space multigrid-based approach to large-scale electronic structure calculations. Phys Rev B. 1996;54:14362–14375. doi: 10.1103/physrevb.54.14362. [DOI] [PubMed] [Google Scholar]
- 51.Hodak M, Wang S, Lu W, Bernholc J. Implementation of ultrasoft pseudopotentials in large-scale grid-based electronic structure calculations. Phys Rev B. 2007;76 085108. [Google Scholar]
- 52.Vanderbilt D. Soft self-consistent pseudopotentials in a generalized eigenvalue formalism. Phys Rev B. 1990;41:7892. doi: 10.1103/physrevb.41.7892. [DOI] [PubMed] [Google Scholar]
- 53.Perdew JP, Burke K, Ernzerhof M. Generalized gradient approximation made simple. Phys Rev Lett. 1996;77:3865. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 54.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 55.Zahn R, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips JC, et al. Scalable molecular dynamics with namd. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feller SE, MacKerell AD. An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 58.Tu K, Tobias DJ, Klein ML. Constant pressure and temperature molecular dynamics simulation of a fully hydrated liquid crystal phase dipalmitoylphosphatidylcholine bilayer. Biophys J. 1995;69:2558–2562. doi: 10.1016/S0006-3495(95)80126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-pressure molecular-dynamics simulation—The Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 60.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular-dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 61.Grubmüller H, Heller H, Windemuth A, Schulten K. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol Simul. 1991;6:121–142. [Google Scholar]
- 62.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]