Abstract
Functional heterogeneity has been investigated for decades in the hippocampal region of the mammalian cerebral cortex, and evidence for vaguely defined “dorsal” and “ventral” regions is emerging. Direct evidence that hippocampal field CA1 displays clear regional, laminar, and pyramidal neuron differentiation is presented here, based on a systematic high-resolution analysis of a publicly accessible, genome-wide expression digital library (Allen Brain Atlas) [Lein et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176]. First, genetic markers reveal distinct spatial expression domains and subdomains along the longitudinal (dorsal/septal/posterior to ventral/temporal/anterior) axis of field CA1. Second, genetic markers divide field CA1 pyramidal neurons into multiple subtypes with characteristic laminar distributions. And third, subcortical brain regions receiving axonal projections from molecularly distinct spatial domains of field CA1 display distinct global gene expression patterns, suggesting that field CA1 spatial domains may be genetically wired independently to form distinct functional networks related to cognition and emotion. Insights emerging from this genomic–anatomic approach provide a starting point for a detailed analysis of differential hippocampal structure–function organization.
Keywords: genetics, genomics, hippocampus, learning and memory, neuroanatomy
The basic outlines of hippocampal architecture were established by the pioneering work of Ramón y Cajal (1) and his student Lorente de Nó (2), who named field CA3 (with large pyramidal neurons and mossy fibers), field CA2 (with large pyramids but no mossy fibers), and field CA1 (with small pyramids). The hippocampus and other interconnected medial temporal lobe cortical areas have been known for some time to be critical for learning and memory (3), and more recently the fact has become clear that other functions are subserved as well (4–6). Overall, the evidence suggests that the “dorsal” (septal or in humans posterior) hippocampus is involved in navigation and related spatial memory, whereas in contrast the “ventral” (temporal or in humans anterior) hippocampus influences stress responses and motivated or emotional behaviors. Experimental neuroanatomical work has established in rats and primates that different transverse levels along the dorsoventral or longitudinal axis establish extrinsic axonal connections in a highly differentiated and topographic way (7–11). Nevertheless, the exact spatial arrangement of the dorsal and ventral hippocampus has not, until now, been clearly established. Here, we directly parcel field CA1 into multiple, spatially distinct molecular domains and subdomains using robust gene markers selected from a comprehensive digital gene expression library (Allen Brain Atlas [ABA], www.brain-map.org) (12) combined with hippocampal cytoarchitectonic analysis. The evidence indicates that field CA1 is not a homogenous cortical area but instead displays clear regional and laminar specificity. These basic structural insights immediately suggest genetic, physiological, and behavioral experiments to clarify exactly how these molecular expression domains are differentially involved in two of the most complex cerebral functions—cognition and emotion.
Results
More than 4,000 genes are expressed in the hippocampal formation, based on data on the ABA web site. We first screened expression patterns of >2,000 genes in the hippocampal region, based on expression level, expression density, and gene clustering. From this, we carefully analyzed expression patterns of 48 genes in the category field CA1 pyramidal layer and 400 “return” genes when we interrogated points in dorsal, intermediate, ventral, and ventral tip parts of field CA1 (Fig. 1 A–C). Although these genes display very heterogeneous expression patterns, we found that many display consistent regional specificities in field CA1. Using these genes as molecular spatial markers, we have divided field CA1 into three distinct domains along the dorsoventral or longitudinal axis (Fig. 1D and Fig. S1): dorsal (CA1d), intermediate (CA1i), and ventral (CA1v) domains. They are visualized simultaneously in transverse mouse brain levels between 2.7 and 2.9 mm caudal to bregma where the maximal dorsoventral extent of field CA1 is displayed (Fig. 1 D–F and Fig. S1B). At these levels CA1d, CA1i, and CA1v occupy approximately the dorsal, intermediate, and ventral thirds of field CA1, respectively. The CA1d–CA1i border here is parallel to the ventral edge of the dentate gyrus lateral blade, whereas the CA1i–CA1v border is approximately level with the dorsal edge of the rhinal fissure (Fig. 1D and Fig. S1B). Rostral and dorsal to these transverse levels CA1d extends to the rostral (septal) tip of field CA1 (Fig. 1E and Fig. S1A), whereas caudally domain CA1d is gradually replaced by pyramidal neurons of the dorsal subiculum (Fig. S1 C and D), which is distinguished from domain CA1d by lack of a stratum oriens. In contrast, domain CA1v extends rostrally and ventrally for only a short distance (Fig. 1E). Dorsolaterally domain CA1v is progressively displaced by the ventral subiculum, until it merges with domain CA1i at the caudal end of field CA1 (Fig. 1E and Fig. S1 C and D).
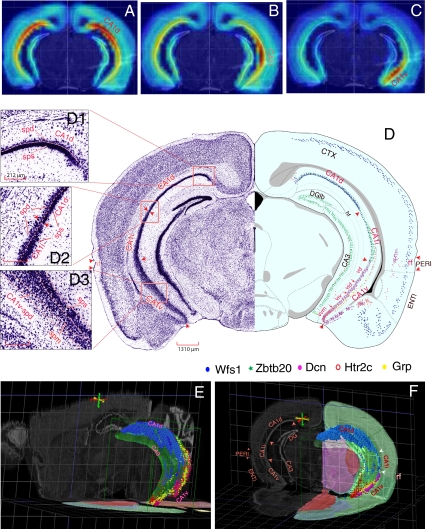
Fig. 1.
(A–C) Overall gene expression “heat maps” for “seed voxel” points in dorsal, intermediate, ventral domains of field CA1. (D) One transverse level of the ARA (15) displaying the maximal dorsoventral scope of field CA1 and its corresponding Nissl-stained histological images. Distributions of five marker genes, Wfs1 (blue), Zbtb20 (green), Dcn (purple), Htr2c (red), and Grp (yellow) are plotted on this level to reveal three molecular domains of field CA1 (CA1d, CA1i, and CA1v). (D1, D2, D3) High-resolution Nissl images that contain CA1d, CA1i, and CA1v, respectively (see text for details). (E) A three-dimensional model of Ammon's horn (in the context of the whole mouse brain). The overall shape of CA1 and its molecular domains revealed by CA1 gene markers Wfs1 (blue), Dcn (purple), and Htr2c (red) occupy the outside surface of the C-shaped cylinder of Ammon's horn, with the dorsal end (CA1d) extending much more rostral than the ventral end (CA1v). These two domains merge at the caudalmost end of CA1. (F) Three-dimensional expression patterns of 4 representative genes, Wfs1 (blue), Grp (yellow), Dcn (purple), and Htr2c (red), in one transverse plane of field CA1. These genes show distinct regional specificities and clearly define the spatial extent of CA1d (Wfs1), CA1v (Grp, Htr2c, and Dcn), and CA1i (revealed here by lack of these gene markers). Three-dimensional images of CA1 were generated in BrainExplore (http://www.brain-map.org), one three-dimensional model of the ARA (15, 40). See Fig. S1 for more detailed mapping of these genes in 4 representative levels of the ARA. CTX, cerebral cortex; DGlb, dentate gyrus, lateral blade; ENTl, lateral entorhinal area; PERI, perirhinal cortical area. (Scale bars: D, 1,310 μm; D1, D2, D3, 212 μm to match the same magnifications of the same images on ABA web site.)
The extent of domain CA1d is uniquely defined by strong expression of Wfs1 (Figs. 1 D–F and 2 and Fig. S1 A–D) and a number of other gene markers including Nov, Kcnh7, Ndst4, and 2610017I09Rik, whereas domain CA1v is defined by strong and unique expression of many other gene markers, including Dcn, Grp, Htr2c, Col5a1, and Gpc3 (Figs. 1 D–F, 2, and 3 and Fig. S1 A–D). (The full names of these genes and image series numbers of their corresponding full sets of gene expression images in the ABA are listed in Table S1.) The middle third of field CA1 (CA1i) is characterized most obviously by the absence or much weaker expression of the above markers (Fig. 1 D–F and Fig. S1 A–D). However, many genes have expression patterns that help to distinguish domain CA1i more directly. For example, Zbtb20 and Gpt2 are expressed in both domains CA1d and CA1i but not in CA1v, with Zbtb20 expressed more strongly in CA1i than CA1d (Fig. S1 B–D) and the reverse occurring for Gpt2. Of fundamental importance, although the expression of many genes in field CA1 may not be restricted to CA1d, CA1i, or CA1v, the expression does consistently respect one of the boundaries between them. Because genes expressed in domain CA1i are more commonly also expressed in CA1d but not CA1v, CA1i is possibly simply a subdomain of CA1d.
Fig. 2.
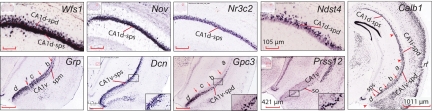
Digital images of six representative genes expressed specifically in three molecular domains of field CA1: CA1d (Wfs1), CA1i (Zbtb20), CA1v (Grp), and the CA1v subdomains, CA1vd, CA1vid, and CA1viv (Col5a1 and Tc1568100) and CA1vv (Gpr101 and Col5a1) at two different rostrocaudal levels (A, B) of field CA1. CA1v subdomains are labeled: a (CA1vd), b (CA1vid), c (CA1viv), and d (CA1vv). Images in (C) are gene expression heat maps of corresponding images in (B) (for definition of heat map, see ABA web site, http://www.brain-map.org) that display relative intensities of gene expression signals. All images were downloaded from the ABA web site. (Gene names and the image series numbers of their original digital images are listed in Table S1.) Overall expression patterns of Wfs1, Zbtb20, and Grp in the hippocampus are schematically mapped in more detail in Fig. S1. All images in the same panels have the same magnifications. Scale bars are displayed in the lower right corner of Gpr101 images.
Fig. 3.
Gene markers showing regional and laminar specificities of field CA1 pyramidal neurons. For high-resolution Nissl-stained cytoarchitecture and delineation of pyramidal neuron sublayers, see Fig. 1 and Fig. S1. CA1v subdomains are labeled: a (CA1vd), b (CA1vid), c (CA1viv), and d (CA1vv). All digital images were downloaded from the ABA gene expression library. (Gene names and the image series numbers of their original digital images are listed in Table S1.) Images in the same rows have the same magnifications.
The concept of subdomains is an important one, especially in domain CA1v where four are obvious (Figs. 1D and 2 and Fig. S1B): dorsolateral (CA1vd), intermediodorsal (CA1vid), intermedioventral (CA1viv), and ventromedial tip (CA1vv). The tiny CA1vv is very distinctive because of weak Grp expression (Figs. 1D and 2 and Fig. S1 B–D) and much stronger, localized Htr2c and Col5a1 expression (Figs. 1 D–F and 2 and Fig. S1 B–D). In addition, a number of other genes such as Gpr101 (Fig. 2) and Dlk1are expressed almost exclusively in CA1vv. Genes expressed in the other three subdomains display more or less overlapping or gradient-type patterns. For example, Tc1568100 and Loc432748 are weakly expressed in subdomain CA1vv, are heavily expressed in CA1viv, and become progressively more restricted to a single sublayer of pyramidal neurons in CA1vid and then CA1vd (Fig. 2). Many other genes, including Zdhhc7 and Pole4, are expressed progressively more strongly in CA1vid, CA1vd, and even CA1i—but not in CA1viv and CA1vv.
The size and shape of field CA1 pyramidal neurons and differences with field CA3 pyramids are firmly established (1, 2), and boundaries between these two hippocampal cortical areas are clearly visualized with molecular markers (13, 14). However, these pyramidal neuron populations have long been considered relatively uniform within an area. The expression pattern heterogeneity described above next led us to reexamine carefully field CA1 cytoarchitecture based on high-resolution Nissl-stained images presented in the Allen Reference Atlas (ARA) (15). We found that field CA1 cytoarchitecture is clearly heterogeneous both regionally and with respect to pyramidal neuron layers (Figs. 1D and 3 and Fig. S1 A–D). Following Swanson in rats (16), we first divided the pyramidal layer into superficial and deep sublayers. The cytoarchitecture of domain CA1d is characterized by a remarkably darkly stained, densely packed superficial pyramidal layer (CA1d-sps) and a thin deep layer with many fewer pyramids (CA1d-spd; Fig. 1D1 and Fig. S1 A–D), as originally described by Lorente de Nó (2). The CA1d-spd is considerably thinner rostrally, with mostly only one row of pyramids (Fig. S1 A and B), but progressively thickens caudally into 3 or 4 rows of loosely arranged pyramids (Fig. S1 C and D). Nevertheless, expression patterns for most genes (such as Nov and Ndst4) are very consistent from rostral to caudal in transverse sections of field CA1d, although a few genes (including Lct) show much stronger expression in rostral levels of CA1d than in more caudal levels, and a few others (including Kit) present the reverse gradient. Wfs1 and Nr4a1 are expressed in both CA1d-sps and CA1d-spd sublayers, Nov and Nr3c2 tend to express more specifically in CA1d-sps, and Ndst4 and Astn2 are expressed preferentially in CA1d-spd (Fig. 3). Some neurons in the stratum oriens also express Astn2. Compared with domain CA1d, the superficial pyramidal layer of CA1i (CA1i-sps) is much less dense and more lightly stained, whereas the deep layer (CA1i-spd) is relatively thicker with 3 or 4 rows of pyramids (Fig. 1D2 and Fig. S1B).
CA1v displays even more complex cytoarchitecture. In addition to superficial (CA1v-sps) and deep (CA1v-spd) pyramidal layers, a middle sublayer (CA1v-spm) containing smaller pyramids appears to be sandwiched between the other layers in subdomains CA1vd and CA1vid (Fig. 1D3 and Fig. S1B). In the progressively more ventral CA1viv, the middle pyramidal sublayer disappears, and pyramidal neuron morphology becomes less distinguishable between the superficial and the deep layers (Fig. 1 D and D3 and Fig. S1B). On reaching subdomain CA1vv—the ventral tip of field CA1—pyramids align in an apparently uniform, evenly stained, thick layer of approximately 7–9 neuronal rows (Fig. 1D and Fig. S1B). This cytoarchitectonic sublamination pattern in CA1v is substantiated by gene expression patterns. First, Grp is expressed strongly in both CA1v-sps and CA1v-spd but weakly in CA1v-spm (Figs. 2 and 3 and Fig. S1B); Dcn is expressed strongly in CA1v-sps but weakly in CA1v-spd and CA1v-spm (Fig. 3 and Fig. S1 B–D), whereas Htr2c and Gpc3 are strongly expressed in CA1vv, but only in the deep layer of CA1viv, CA1vid, and CA1vd (Figs. 1 D–F and 3 and Fig. S1 B–D). Second, Tc1568100 and Loc432748 are expressed strongly in the CA1viv-sps, but only the CA1vid-spm and CA1vd-spm (Fig. 2), whereas Prss23 is expressed specifically in the deep layer of CA1viv, CA1vid, and CA1vd and in CA1i (Fig. 3). And third, Prss12 is expressed specifically only in the striatum oriens of domain CA1v (Fig. 3). Interestingly, some genes display laminar differences in different domains. For example, Calb1 is expressed selectively in the superficial layer of CA1d and CA1viv but is expressed more strongly in the deep layer of CA1i, CA1vd, and CA1vid (Fig. 3).
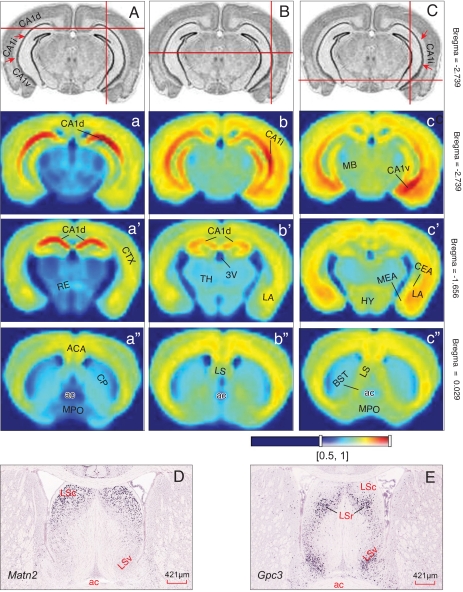
Next, we examined global spatial gene expression pattern correlations between these field CA1 molecular domains and other brain regions (Fig. 4; all correlation coefficients provided in Table S2). We found that domain CA1d exhibits a high level of intrastructural correlation between different transverse planes of CA1d, but lower correlations with domain CA1v, confirming their molecular specificities across the dorsoventral rather than the rostrocaudal axis. As expected from earlier results (above), domain CA1i expression patterns are correlated more with those of CA1d than with CA1v. Most interestingly, probabilistic coexpression correlations of domains CA1d and CA1v with other brain regions are very distinctive. For example, although both domains exhibit very high correlation values with the cortical mantle as a whole, domain CA1v shows higher correlation values than CA1d with cortical amygdalar areas and the basolateral amygdalar complex, both of which share direct bidirectional axonal connections with CA1v (10, 11, 17, 18). In terms of subcortical regions, CA1v displays strikingly higher coexpression correlations than CA1d with neuron populations associated with autonomic, neuroendocrine, and affective behaviors, including the central and medial amygdalar nuclei, intermediate and ventral parts of the lateral septal nucleus, bed nuclei of the stria terminalis, and hypothalamus—all of which share massive direct and indirect neuronal connectivity with CA1v (9, 10, 17–19). These obvious correlations strongly suggest that CA1d and CA1v are genetically wired independently with different functional specificities.
Fig. 4.
Global gene coexpression “energy” maps result from setting seed voxel points in domain CA1d (A), CA1i (B), and CA1v (C) at three representative transverse levels of the mouse brain at approximately bregma −2.739 (a, b, c), −1.656 (a′, b′, c′), and 0.029 (a″, b″, c″). The heat map energy definition is shown at the bottom. The coexpression correlation coefficients of these CA1 molecular domains with other brain structures are listed in Table S2. For a detailed description of AGEA spatial gene expression correlation map, see Ng et al. (24). (D and E) Correlated gene expression patterns of domain CA1d marker gene Matn2 and CA1v marker gene Gpc3 in the lateral septal nucleus. (Scale bars: D and E, 421 μm.)
Finally, Risold and Swanson (9, 16) proposed the existence of a series of structure–function domains along the longitudinal axis of field CA1 and adjacent subiculum that send topographically organized projections to the three major parts of the lateral septal nucleus that in turn share bidirectional connections with hypothalamic structures mediating the expression of different classes of innate motivated behavior. To explore potential genetic mechanisms underlying this topographic wiring pattern, we compared coexpression patterns of CA1 domain-specific genes with those of the lateral septal nucleus. We found that a number of CA1 domain-specific genes are also expressed in the lateral septal nucleus and that their coexpression correlations respect the topologically ordered dorsal-to-dorsal and ventral-to-ventral organization of their axonal projection connectivity. For example, genes that show strong expression in domain CA1d, such as Wfs1 and Matn2, are also strongly expressed in the caudal end of the caudal and rostral lateral septal nucleus (LSc, LSr), which are topologically dorsal (Fig. 4D). In contrast, Dlk1 and Gpr101—the CA1vv (and ventral subicular) marker genes—also show strong expression patterns in the ventral lateral septal nucleus (LSv). Genes that show significant expression in more dorsal subdomains of CA1v are most likely also to coexpress in ventral levels of the LSr. Thus, Loc432748, a specific marker gene for CA1viv, presents specific signal only in a specific zone of the LSr. Gpc3 and Htr2c, which are strongly expressed in CA1vv and the rest of domain CA1v, are strongly expressed in the LSv and LSr but not in LSc (Fig. 4E). Man1a and Zdhhc7, which are expressed throughout field CA1 (except in CA1vv), express strongly in the LSc and LSr (but not LSv). By comparison of the topology and topography of these gene expression correlations with the hippocampal functional domains defined by Risold and Swanson in rat (9, 16), CA1vv defined here appears to correspond well to their Domain 5—the ventral tip of field CA1 and subiculum, which sends projections specifically to the LSv (9)—one of the primary sources of cerebral inputs to the hypothalamic neuroendocrine zone and region controlling feeding behavior. The progressively more dorsally located CA1viv, CA1vid, and CA1vd may correspond to their Domains 2–4. These CA1v subdomains send topographic projections to different zones of the LSr, which in turn project to the hypothalamic medial column that controls two basic classes of social behavior—reproduction and defense (9). Finally, CA1d and CA1i together may correspond to their Domain 1, which presumably innervates LSc and LSr levels that in turn project to the theta-rhythm-controlling supramammillary nucleus (8). The densest outputs of dorsal CA1 have been shown to be to the dorsal subiculum (11)—the major source of the postcommissural fornix pathway innervating the medial and lateral mammillary nuclei and anterior thalamic complex. As a matter of fact, this dorsal CA1 to subiculum projection is the major hippocampal component that processes spatial memory (20–22).
Discussion
Many gene markers have been used as reproducible landmarks for defining nervous system divisions at various stages of development (23) and in the adult (15). This is an extension of the revolutionary chemical neuroanatomy approach that began in the 1950s with cholinesterase histochemistry, followed by histo-fluorescence and then immunohistochemistry (see ref. 15). The resulting chemoarchitectonic data greatly refined classical cytoarchitectonics based on Nissl staining. The present study, combined with two recent reports (24, 25), supply proof-of-principle examples of how the genomic–neuroanatomic approach can further refine architectonic maps and identify neuronal phenotypes amenable to selective experimental genetic manipulation. Although raw data about these expression patterns (digitized images) are freely available on the ABA web site and powerful online informatics tools enable users conveniently to identify regionally specific gene expression patterns, synthesis and interpretation of this information relative to existing structural, functional, and behavioral knowledge is still required. Specifically, we provide here a testable model to guide and stimulate further analysis of hippocampal structure–function in health and disease.
Our genetic–anatomic analysis and the current axonal connectivity literature (7–11, 17–19, 26) suggest that domains CA1d and CA1i lie in approximately the dorsal half of the hippocampus, and they display strong global gene expression correlations primarily with other parts of cerebral cortex and subcortical regions innervated by dorsal hippocampus and related to theta-rhythm modulation and navigation. In contrast, domain CA1v lies in approximately the ventral half of the hippocampus, and this domain shares strong gene coexpression patterns and projections with cerebral cortex and subcortical regions mediating neuroendocrine, autonomic, and goal-oriented or emotional behavioral responses. The natural boundary between CA1i and CA1v in mice coincides with the rhinal fissure's dorsal border, a boundary shown by certain CA1 domain-specific genes and by other cortical marker genes, including Col5a1, which is strongly expressed in isocortical layers 6a and 6b (Fig. 2). The evidence suggests that the dorsal and ventral halves of field CA1 are genetically wired independently to support cognitive and emotional responses, respectively (4–6).
The model of three major CA1 molecular domains presented here shows interesting correlations with the 9 even more striking molecular regions recently reported along the longitudinal axis of mouse field CA3 (25). The observations of Thompson et al., together with our own in fields CA1 and CA3, suggest that field CA3—and thus Ammon's horn as a whole—is also divided into three major molecular domains: (i) a dorsal domain (CA3d) including their regions 1–3 that is accompanied by domain CA1d; (ii) an intermediate domain (CA3i) including their regions 4 and 5 that is accompanied by domain CA1i; and (iii) a ventral domain (CA3v) including their regions 6–7 that is accompanied by domain CA1v. The domains CA3d and CA3i (regions 1–5) form approximately the dorsal half of field CA3, whereas the domain CA3v (regions 6 and 7) forms approximately the ventral half.
Ramón y Cajal first noted structural differences along the hippocampal longitudinal axis, related to what he called the superior and inferior perforant paths (1) and what would later be referred to loosely as the dorsal and ventral hippocampus, respectively (9, 27). Lorente de Nó (2) vaguely divided the “Ammonic system” into three longitudinal segments based on now discredited connectional data and also divided field CA3 and field CA1 into three parallel, longitudinally running zones (a, b, and c for both). Lorente de Nó's (2) subfields CA3a, CA3b, and CA3c appear to correspond well with three molecular regions identified by Thompson et al. (25): region 3 (Mas1 expression) with CA3a, region 2 (Fmo1 expression) with CA3b, and region 1 (Ttn1 expression) with CA3c. In field CA1, at least some genes (such as Lct and Kit) were found here to show differential expression patterns across the topological medial to lateral (transverse) axis, but correspondences with Lorente de No's subfields CA1a, CA1b, and CA1c are not yet clear.
Extensive work since the early 1970s established that intrahippocampal circuitry, typified by the classical trisynaptic circuit from entorhinal area to dentate gyrus to field CA3 to field CA1, runs transverse to the longitudinal axis and that specific extrinsic inputs and outputs are associated with specific regions along the longitudinal axis of the hippocampal formation (8–11, 17–19, 28–33). The dorsal half of field CA1 has a high concentration of place neurons (e.g., ref. 34) and sends most of its extrinsic cortical projections, either directly or indirectly via dorsal subiculum, to the cortical retrosplenial area (11, 35), which is involved in cognitive processing of visual sensory information at least in part for spatial and contextual memory. The dorsal subiculum receives its strongest input from dorsal CA1 and sends massive projections to the mammillary and anterior thalamic nuclei, two regions containing abundant navigation neurons (36). These two diencephalic regions in turn project back to the dorsal hippocampus and retrosplenial area (35). This and other evidence (see refs. 4–6, 20, and 21) suggest that the dorsal half of field CA1 (domains CA1d and CA1i) is critically involved in cognitive spatial memory processing, at least in rodents.
The most distinguishing feature of connectivity associated with the ventral half of field CA1 (domain CA1v and its 4 subdomains) is dense bidirectional connections with amygdalar regions implicated in emotional behaviors (10, 11, 17, 18). Ventral but not dorsal field CA1 also projects directly (19) to the hypothalamic periventricular region and medial zone, which integrate neuroendocrine, autonomic, and somatic motor responses associated with three basic classes of motivated behaviors common to all animals: ingestive (feeding and drinking), reproductive (sexual and parental), and defensive (fight or flight) (26). These direct descending projections from ventral not dorsal CA1 are reinforced by multiple indirect pathways involving relays in the ventral subiculum, amygdalar region, lateral septal nucleus, and bed nuclei of the stria terminalis (9–11, 17–19, 26, 33, 37), all of which display gene coexpression patterns correlated with those of CA1v as shown here.
The observation that the dorsal half and ventral half of field CA1 display distinct global gene expression correlations with brain structures that they project to either directly or indirectly is striking. Although the mechanistic significance of this relationship between CA1 molecular domains and their extrinsic circuitry is not yet clear, from the above the dorsal and ventral halves are obviously genetically wired independently for different functional specializations. The former is selectively involved in cognitive aspects of the learning and memory associated with navigation, exploration, and locomotion, whereas in contrast the latter is part of the temporal lobe associated most directly with motivational and emotional aspects of the classic Klüver–Bucy Syndrome. In fact, at least two CA1v marker genes, Grp and Htr2c, have been implicated in psychiatric disorders (38, 39).
This and similar work (25) are part of a systematic examination of relationships between gene expression patterns and neuronal networks to clarify genetic contributions to brain structure and function. However, the detection sensitivity and specificity for many genes, especially those of low abundance, probably were compromised in the large-scale, high-throughput process of ABA data production (12). This would make the list of domain-specific markers assembled here rather incomplete, and many genes critical for determining adult brain structure and connections likely are expressed during development not in the adult (23). Even for the domain-specific markers listed here (Table S1), information in Gene Ontology suggests many diverse (and unknown) functions whose possible role in neuronal development and adult structure–function remain to be investigated.
Relative to isocortex, very little is known about laminar and regional differences in field CA1 pyramidal cell subtypes. Both Ramón y Cajal (1) and Lorente de Nó (2) observed deep and superficial pyramids and noticed that they establish unique relationships with basket cells. However, the addition of multiple genetic markers for differentiation of field CA1 pyramids opens many new possibilities for establishing analytical distinctions.
Materials and Methods
This study was based on a systematic, high-resolution analysis of >4,000 genes expressed in the hippocampal formation through a publicly accessible online gene expression digital library—the ABA. We first manually screened expression patterns of >2,000 genes under the Anatomic Search tool category “hippocampal region,” based on expression level, expression density, and gene clustering, and carefully analyzed expression patterns of 48 genes in the category “field CA1 pyramidal layer” under “fine structure search.” We then used the Gene Finder tool of the Anatomic Gene Expression Atlas (AGEA) application (Fig. 1 A–C) to filter candidate genes showing regional specificities. Detailed methodology of the AGEA has been described in a recent report (24). In brief, because all ABA image-based, in situ hybridization data are spatially registered to the common anatomic framework of the ARA (15) with a standard coordinate system and hierarchical ontology, the AGEA in principle enables users to view a comprehensive list of genes expressed in any given three-dimensional voxel (“seed voxel”) of the mouse brain atlas using the Gene Finder application (24). With this strategy, we carefully analyzed the expression of ≈400 “return” genes when we set seed voxel points in dorsal, intermediate, and ventral parts of field CA1. AGEA also allows users to view global expression correlations associated with any seed voxel. With this tool, we have also analyzed gene expression correlations of different domains of the CA1.
Supplementary Material
Acknowledgments.
This work was supported in part by National Institutes of Health Grants MH083180 (to H.-W.D.), RR013642 (to A.W.T.), NS16686 (to L.W.S.), and MH62122 (to M.S.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812608106/DCSupplemental.
References
- 1.Ramón y, Cajal S. Estudios sobre la corteza cerebral humana. Trab Inst Cajal Invest Biol. 1901–1902;1:1–227. [Google Scholar]
- 2.Lorente de Nó RJ. Studies on the structure of the cerebral cortex. II. Continuation of the study of the ammonic system. J Psychol Neurol. 1934;46:113–177. [Google Scholar]
- 3.Squire LR. Memory and the hippocampus: A synthesis from findings with rats, monkeys, and humans. Psychol Rev. 1992;99:195–231. doi: 10.1037/0033-295x.99.2.195. [DOI] [PubMed] [Google Scholar]
- 4.Moser MB, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8:608–619. doi: 10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Anagnostaras SG, Gale GD, Fanselow MS. The hippocampus and Pavlovian fear conditioning: Reply to Bast et al. Hippocampus. 2002;12:561–565. doi: 10.1002/hipo.10071. [DOI] [PubMed] [Google Scholar]
- 6.Kjelstrup KG, et al. Reduced fear expression after lesions of the ventral hippocampus. Proc Natl Acad Sci USA. 2002;99:10825–10830. doi: 10.1073/pnas.152112399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson LW, Cowan WM. Hippocampo-hypothalamic connections: Origin in subicular cortex, not ammon's horn. Science. 1975;189:303–304. doi: 10.1126/science.49928. [DOI] [PubMed] [Google Scholar]
- 8.Swanson LW, Cowan WM. An autoradiographic study of the organization of the efferent connections of the hippocampal formation in the rat. J Comp Neurol. 1977;172:49–84. doi: 10.1002/cne.901720104. [DOI] [PubMed] [Google Scholar]
- 9.Risold PY, Swanson LW. Structural evidence for functional domains in the rat hippocampus. Science. 1996;272:1484–1486. doi: 10.1126/science.272.5267.1484. [DOI] [PubMed] [Google Scholar]
- 10.Witter MP, Amaral DG. Hippocampal formation. In: Paxinos G, editor. The Rat Nervous System. Amsterdam: Elsevier; 2004. pp. 637–704. [Google Scholar]
- 11.Cenquizca LA, Swanson LW. Spatial organization of direct hippocampal field CA1 axonal projections to the rest of the cerebral cortex. Brain Res Rev. 2007;56:1–26. doi: 10.1016/j.brainresrev.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 13.Lein ES, Callaway EM, Albright TD, Gage FH. Redefining the boundaries of the hippocampal CA2 subfield in the mouse using gene expression and 3-dimensional reconstruction. J Comp Neurol. 2005;485:1–18. doi: 10.1002/cne.20426. [DOI] [PubMed] [Google Scholar]
- 14.Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong H-W. The Allen Reference Atlas: A Digital Color Brain Atlas of the C57BL/6J Male Mouse. Hoboken, NJ: Wiley; 2007. [Google Scholar]
- 16.Swanson LW. Brain Maps: Structure of the Rat Brain. 3rd Ed. Amsterdam: Elsevier; 2004. [Google Scholar]
- 17.Pitkanen A, Pikkarainen M, Nurminen N, Ylinen A. Reciprocal connections between the amygdala and the hippocampal formation, perirhinal cortex, and postrhinal cortex in rat. Ann N Y Acad Sci. 2000;911:369–391. doi: 10.1111/j.1749-6632.2000.tb06738.x. [DOI] [PubMed] [Google Scholar]
- 18.Petrovich GD, Canteras NS, Swanson LW. Combinatorial amygdalar inputs to hippocampal domains and hypothalamic behavior systems. Brain Res Rev. 2001;38:247–289. doi: 10.1016/s0165-0173(01)00080-7. [DOI] [PubMed] [Google Scholar]
- 19.Cenquizca LA, Swanson LW. Analysis of direct hippocampal cortical field CA1 axonal projections to diencephalon in the rat. J Comp Neurol. 2006;497:101–114. doi: 10.1002/cne.20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moser E, Moser MB, Andersen P. Spatial learning impairment parallels the magnitude of dorsal hippocampal lesions, but is hardly present following ventral lesions. J Neurosci. 1993;13:3916–3925. doi: 10.1523/JNEUROSCI.13-09-03916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunsaker MR, Fieldsted PM, Rosenberg JS, Kesner RP. Dissociating the roles of dorsal and ventral CA1 for the temporal processing of spatial locations, visual objects, and odors. Behav Neurosci. 2008;122:643–650. doi: 10.1037/0735-7044.122.3.643. [DOI] [PubMed] [Google Scholar]
- 22.Gigg J. Constraints on hippocampal processing imposed by the connectivity between CA1, subiculum and subicular targets. Behav Brain Res. 2006;174:265–271. doi: 10.1016/j.bbr.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Puelles L, Martínez S, Martínez-de-la-Torre M, Rubenstein JLR. Gene maps and related histogenetic domains in the forebrain and midbrain. In: Paxinos G, editor. The Rat Nervous System. Amsterdam: Elsevier; 2004. pp. 3–25. [Google Scholar]
- 24.Ng L, et al. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- 25.Thompson CL, et al. Genomic anatomy of the hippocampus. Neuron. 2008;60:1010–1021. doi: 10.1016/j.neuron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886:113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- 27.Gloor P. The Temporal Lobe and Limbic System. New York: Oxford Univ Press; 1997. pp. 325–589. [Google Scholar]
- 28.Witter MP. Anatomical organization of the parahippocampal-hippocampal network. Ann N Y Acad Sci. 2000;911:1–24. doi: 10.1111/j.1749-6632.2000.tb06716.x. [DOI] [PubMed] [Google Scholar]
- 29.Witter MP, Van Hoesen GW, Amaral DG. Topographical organization of the entorhinal projection to the dentate gyrus of the monkey. J Neurosci. 1989;9:216–218. doi: 10.1523/JNEUROSCI.09-01-00216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolorfo CL, Amaral DG. Entorhinal cortex of the rat: Organization of intrinsic connections. J Comp Neurol. 1998;398:49–82. doi: 10.1002/(sici)1096-9861(19980817)398:1<49::aid-cne4>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Van Groen T, Miettinen P, Kadish I. The entorhinal cortex of the mouse: Organization of the projection to the hippocampal formation. Hippocampus. 2003;13:133–149. doi: 10.1002/hipo.10037. [DOI] [PubMed] [Google Scholar]
- 32.Swanson LW, Wyss JM, Cowan WM. An autoradiographic study of the organization of intrahippocampal association pathways in the rat. J Comp Neurol. 1978;181:681–715. doi: 10.1002/cne.901810402. [DOI] [PubMed] [Google Scholar]
- 33.Swanson LW, Köhler C, Björklund A. The limbic region. I: The septohippocampal system. In: Hökfelt T, Björklund A, Swanson LW, editors. Integrated Systems of the CNS, Part I. Handbook of Chemical Neuroanatomy. Vol 5. Amsterdam: Elsevier; 1987. pp. 125–277. [Google Scholar]
- 34.Jung MW, Wiener SI, McNaughton BL. Comparison of spatial firing characteristics of units in dorsal and ventral hippocampus of the rat. J Neurosci. 1994;14:7347–7356. doi: 10.1523/JNEUROSCI.14-12-07347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risold PY, Thompson RH, Swanson LW. The structural organization of connections between hypothalamus and cerebral cortex. Brain Res Brain Res Rev. 1997;24:197–254. doi: 10.1016/s0165-0173(97)00007-6. [DOI] [PubMed] [Google Scholar]
- 36.Taube JS. The head direction signal: Origins and sensory-motor integration. Annu Rev Neurosci. 2007;30:181–207. doi: 10.1146/annurev.neuro.29.051605.112854. [DOI] [PubMed] [Google Scholar]
- 37.Dong H-W, Petrovich GD, Swanson LW. Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Res Rev. 2001;38:192–246. doi: 10.1016/s0165-0173(01)00079-0. [DOI] [PubMed] [Google Scholar]
- 38.Roesler R, Henriques JA, Schwartsmann G. Gastrin-releasing peptide receptor as a molecular target for psychiatric and neurological disorders. CNS Neurol Disord Drug Targets. 2006;5:197–204. doi: 10.2174/187152706776359673. [DOI] [PubMed] [Google Scholar]
- 39.Drago A, Serretti A. Focus on HTR2C: A possible suggestion for genetic studies of complex disorders. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30864. in press. [DOI] [PubMed] [Google Scholar]
- 40.Lau, et al. Exploration and visualization of gene expression with neuroanatomy in the adult mouse brain. BMC Bioinformatics. 2008;18:9:153. doi: 10.1186/1471-2105-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.