Abstract
A number of common mutations in the hemoglobin β (HBB) gene cause β-thalassemia, a monogenic disease with high prevalence in certain ethnic groups. As there are 30 HBB variants that cover more than 99.5% of HBB mutant alleles in the Thai population, an efficient and cost-effective screening method is required. Three panels of multiplex primer extensions, followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry were developed. The first panel simultaneously detected 21 of the most common HBB mutations, while the second panel screened nine additional mutations, plus seven of the first panel for confirmation; the third panel was used to confirm three HBB mutations, yielding a 9-Da mass difference that could not be clearly distinguished by the previous two panels. The protocol was both standardized using 40 samples of known genotypes and subsequently validated in 162 blind samples with 27 different genotypes (including a normal control), comprising heterozygous, compound heterozygous, and homozygous β-thalassemia. Results were in complete agreement with those from the genotyping results, conducted using three different methods overall. The method developed here permitted the detection of mutations missed using a single genotyping procedure. The procedure should serve as the method of choice for HBB genotyping due to its accuracy, sensitivity, and cost-effectiveness, and can be applied to studies of other gene variants that are potential disease biomarkers.
To date, 739 point mutations in the hemoglobin, β (HBB) gene causing β-thalassemia (MIM# 141900) have been reported in HbVar: A Database of Human Hemoglobin Variants and Thalassemias (http://globin.cse.psu.edu/globin/hbvar/menu.html, accessed March 2009), but each ethnic group has a limited number of common mutations and a considerable number of rarer mutations.1 The c.79G>A (also known as CD26G>A or Hb E) is the most frequent HBB variant in Southeast Asia including Thailand.2 “Thai” generally refers to speakers of Thai (Tai) languages. The ethnic groups of Thailand comprise Thais (constituting 85% of the population) and Hill Peoples living primarily in the north, as well as other groups including the Chinese and minorities in the south.3 In the Thai population, approximately 40 HBB mutations have been identified,4 of which 30 variants account for more than 99.5% of all mutant HBB alleles (Table 1). Thus, an efficient, cost-effective, and highly accurate screening method is required for their detection.
Table 1.
Thirty Common and Rare HBB Mutations of Patients with β-Thalassemia and Carriers from the Siriraj-Thalassemia Program Project, Faculty of Medicine Siriraj Hospital, Mahidol University during 2003–2004
| Common HBB mutations (13) |
HBB mutations causing abnormal Hb (10) |
Rare HBB mutations (7) |
|||
|---|---|---|---|---|---|
| Common name | HGVS nomenclature | Common name | HGVS nomenclature | Common name | HGVS nomenclature |
| CD26G>A (Hb E) | c.79G>A* | CD147+AC (Hb Tak) | c.441_442insAC*†‡ | CD43G>T | c.130G>T* |
| CD41/42-TTCT | c.124_127delTTCT*† | CD126T>G (Hb Dhonburi) | c.380T>G* | CD123/125 (−8 bp) | c.370_377delACC CCACC† |
| CD17A>T | c.52A>T*†‡ | CD136G>A (Hb Hope) | c.410G>A* | −87C>A | c.−137C>A† |
| −28A>G | c.−78A>G* | CD6G>A (Hb C) | c.19G>A*† | CD15-T | c.46delT† |
| IVS2#654C>T | c.316−197C>T* | CD56G>A (Hb J-Bangkok) | c.170G>A* | CD8/9+G | c.27_28insG† |
| IVS1#5G>C | c.92 + 5G>C* | CD83G>A (Hb Pyrgos) | c.251G>A* | CD27/28+C | c.84_85insC† |
| CD19A>G (Hb Malay) | c.59A>G* | CD6A>C (Hb G Makassar) | c.20A>C*† | CD41-C | c.126delC*† |
| CD71/72 + A | c.216_217insA* | CD6A>T (Hb S) | c.20A>T*†‡ | ||
| IVS1#1G>T | c.92 + 1G>T† | CD121G>C (Hb D Punjab) | c.364G>C* | ||
| −31A>G | c.−81A>G† | CD1T>C (Hb Raleigh) | c.5T>C† | ||
| −30T>C | c.−80T>C* | ||||
| CD35C>A | c.108C>A† | ||||
| CD0T>G | c.2T>G* | ||||
Each column is listed in order of decreasing frequency.
HGVS, Human Genome Variation Society.
HBB mutations detectable by Panel 1 Multiplex SBE.
HBB mutations detectable by Panel 2 Multiplex SBE.
HBB mutations detectable by Panel 3 Multiplex VSET.
Many simple methods for genotyping HBB mutations have been used, including restriction fragment length polymorphism analysis,5 reverse dot-blot hybridization,6,7,8 amplification refractory mutation system,9 single strand conformation polymorphism analysis,4,10 denaturing gradient gel electrophoresis (DGGE),11,12 and direct DNA sequencing.13 Recent advances in genotyping technologies have enabled high sample-throughput screening of several mutations in a large number of samples. Allele-specific arrayed primer extension has been designed for the simultaneous detection of 15 nondeletion α-globin gene defects and 23 β-globin gene mutations commonly found in Southeast Asian countries to overcome the need to use multiple reverse dot-blot analyses.14 Multiplex minisequencing also has been widely applied as a basic molecular technique, with subsequent detection using a variety of different platforms, including capillary electrophoresis,15 denaturing high performance liquid chromatography16,17 and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS).18,19
MALDI-TOF MS has been developed as a genotyping tool based on the differences in mass of variant DNA sequences.20 This technique provides highly accurate identification due to its ability in a single run to detect directly the absolute masses of multiple variant sites.21 MALDI-TOF MS in combination with multiplex minisequencing has proven to be a cost-effective and efficient procedure for high-throughput genotyping of a number of disease-causing genes or of single nucleotide polymorphisms (SNPs).19,22,23,24
Nevertheless, bottlenecks in multiplex genotyping using MALDI-TOF MS include optimization of highly multiplex-primer extension (PE) reactions25 and the need to completely remove contaminating salt adducts that can compromise spectral quality and reduce accuracy of mass assignments.25,26,27 For genotyping of HBB, there is the additional problem of the very close proximity and partial overlapping of the mutations to one another. Recently, a successful single analysis of the eight most common HBB mutations in Taiwanese population has been achieved by using eight parallel minisequencing reactions and pooling of the minisequencing reaction products for subsequent sequential desalting and multiplex MALDI-TOF analysis.19
To reduce analysis time and cost of HBB genotyping, this study aimed to maximize multiplexing in both PCR and PE steps, based on having well-designed primers and well-optimized reaction conditions that give best yields for every possible allele in each multiplex reaction. We have developed an alternative approach for genotyping the 30 specific HBB mutations in the Thai population, which comprises tetraplex PCR to amplify four fragments spanning all 30 mutations, multiplex PE reaction of the PCR products, desalting with magnetic bead separation, and analysis of PE products by MALDI-TOF MS. Three separate panels of multiplex PE reactions were developed for 21 mutations, 16 variants including nine additional mutations and seven mutations identical to the first panel, and an optional third panel for confirmation of c.20A>T, c.52A>T, or c.441_442insAC (Hb Tak). Using this approach, the 30 HBB mutations were reliably and unambiguously detected and the technique was validated using a total of 162 randomly selected β-thalassemia samples previously genotyped by DGGE, restriction fragment length polymorphism, and/or direct sequencing techniques.
Materials and Methods
Collection of DNA Samples Carrying HBB Mutations
DNA samples were anonymously obtained from DNA bank of Siriraj-Thalassemia Program Project, Faculty of Medicine Siriraj Hospital, Mahidol University during 2005, with patients' informed consent at the time of blood collection conducted as part of a routine medical examination, and the process conforming to institutional and national ethical guidelines. Genomic DNA was isolated from whole blood using Puregene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN). These samples have been analyzed previously in our laboratory for HBB genotypes using DGGE, restriction fragment length polymorphism, or direct DNA sequencing.
For method development of multiplex genotyping with MALDI-TOF MS, 40 DNA samples of known HBB genotypes from a normal control, 30 β-thalassemia carriers with 30 different HBB mutations (Table 1), either single heterozygotes or compound heterozygotes, as well as nine different homozygotes were tested. Subsequently, 162 DNA samples randomly selected from β-thalassemia patients (compound heterozygote or homozygote) and carriers were tested for method validation.
Multiplex PCR Amplification and Purification
Four pairs of PCR primers, BGPL/BGEX1R, BGEX2.1L/BGEX2.2R, BGInt2L/BGInt2R, and BGEX3L/BGEX3R,4 were used in a single PCR reaction to generate tetraplex-PCR amplicons of 323, 276, 194, and 230 bp, respectively, which encompass all of the 30 HBB mutations, as shown in Figure 1. A 25-μl volume of multiplex PCR reaction comprised 1.5 mmol/L MgCl2, 200 μmol/L dNTPs, 0.5 U of Immolase DNA polymerase (Bioline USA Inc.), 100 ng of genomic DNA, and 200 nmol/L PCR primers, except for BGEX2.1L and BGEX2.2R, which required 600 nmol/L. Thermal cycling followed in a GeneAmp PCR System 2400 (PE Applied Biosystems, Foster City, CA) at 95°C for 7 minutes, followed by 35 cycles of 94°C for 30 seconds, 57°C for 30 seconds, and 72°C for 20 seconds, and an additional extension step at 72°C for 10 minutes.
Figure 1.
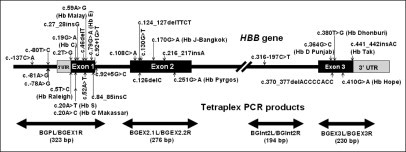
Schematic diagram of HBB gene showing distribution of 30 HBB mutations of interest and positions of tetraplex PCR products.
Excess PCR primers and dNTPs in the reaction were removed by incubating 5 μl of PCR mixture with 2 μl of “ExoSAP-IT” mix (U.S.B. Corporation, Cleveland, OH) at 37°C for 30 minutes followed by 20 minutes at 80°C.
Primer Design for Multiplex Primer-Extension Reaction
To economize on time and cost, multiplex PE was designed to cover all 30 mutations using at most three reactions. PE reaction can be a single-base extension (SBE) (PinPoint assay)28 or a very short extension (VSET).21 For SBE, the primer anneals immediately next to the mutation site and is extended with a single nucleotide, using terminating dideoxynucleoside triphosphate (ddNTP), whereas in VSET, the primer is extended by only one base from one allele and by two bases from the other allele using three ddNTPs and a fourth nucleotide in the deoxy form. Due to a high capacity of multiplexing, SBE was performed in our study while VSET was used to distinguish clearly the 9-Da mass difference between extension product containing dideoxyadenosine monophosphate and that of dideoxythymidine monophosphate.
Primers used throughout this study were synthesized by BioService Unit, National Center for Genetic Engineering and Biotechnology, Bangkok, Thailand. Design of the extension primers was based on the positions of individual mutations, taking into account the molecular mass range of primers or products from both alleles that can be reliably and unequivocally differentiated from one another by MALDI-TOF MS (4000 to 9000 Da). The masses of the primers and their corresponding extension products were calculated using Mongo Oligo Mass Calculator v2.05 (http://library.med.utah.edu/masspec/mongo.htm, accessed November 2006) and adding the mass of a particular ddNTP to that of the primers. Primers were grouped into three panels for multiplexing as shown in Table 2.
Table 2.
Expected Molecular Mass of Extension Primers and Products of Multiplex Single-Base Extension (SBE) and Very Short Extension (VSET) Assay Employed for Detection of HBB Mutations
| Primer No | Mutation site in HBB gene | Primer sequence | Primer strand | Primer conc. (nmol/L) | Primer mass (Da) | Normal product mass (Da) | Mutant product mass (Da) | Mass difference |
|---|---|---|---|---|---|---|---|---|
| Panel 1: Multiplex SBE–for detection of 21 HBB mutations (using 19 extension primers) | ||||||||
| 1.1 | c.19G>A (Hb C) | 5′-GCATCTGACTCCT-3′ | Sense | 425 | 3885.6 | +ddG = 4198.8 | +ddA = 4182.8 | −16 |
| 1.2 | c.20A>T (Hb S) | 5′-CCGGCAGACTTCTCC-3′ | Antisense | 200 | 4489.0 | +ddT = 4777.2 | +ddA = 4786.2 | +9 |
| c.20A>C (Hb G Makassar) | +ddG = 4802.2 | +25 | ||||||
| 1.3 | c.216_217insA | 5′-TGCTCGGTGCCTTTA-3′ | Sense | 300 | 4550.0 | +ddG = 4863.2 | +ddA = 4847.2 | −16 |
| 1.4 | c.410G>A (Hb Hope) | 5′-GAGGGCATTAGCCACA-3′ | Antisense | 150 | 4915.3 | +ddC = 5188.5 | +ddT = 5203.5 | +15 |
| 1.5 | c.79G>A (Hb E) | 5′-CCACCTGCCCAGGGCCT-3′ | Antisense | 100 | 5092.3 | +ddC = 5365.6 | +ddT = 5380.5 | +15 |
| 1.6 | c.52A>T | 5′-GTACTGCGCTGTGGGGC-3′ | Sense | 385 | 5258.4 | +ddA = 5555.6 | +ddT = 5546.6 | −9 |
| 1.7 | c.-78A>G | 5′-ATGGCTCTGCCCTGACTT-3′ | Antisense | 225 | 5441.6 | +ddT = 5729.8 | +ddC = 5714.8 | −15 |
| 1.8 | c.170G>A (Hb J-Bangkok) | 5′-CTCCTGATGCTGTTATGG-3′ | Sense | 175 | 5496.6 | +ddG = 5809.8 | +ddA = 5793.8 | −16 |
| 1.9 | c.59A>G (Hb Malay) | 5′-ACCACCAACTTCATCCACG-3′ | Antisense | 100 | 5661.7 | +ddT = 5949.9 | +ddC = 5934.9 | −15 |
| 1.10 | c.-80T>C | 5′-CATTAGCCAGGGCTGGGCA-3′ | Sense | 150 | 5853.8 | +ddT = 6142.0 | +ddC = 6127.0 | −15 |
| 1.11 | c.441_442insAC (Hb Tak) | 5′-GCAAGAAAGCGAGCTTAGTG-3′ | Antisense | 100 | 6215.1 | +ddA = 6512.3 | +ddT = 6503.3 | −9 |
| 1.12 | c.380T>G (Hb Dhonburi) | 5′-ACTTTCTGATAGGCAGCCTGC-3′ | Antisense | 87.5 | 6397.2 | +ddA = 6694.4 | +ddC = 6670.4 | −24 |
| 1.13 | c.130G>T | 5′-GGGACAGATCCCCAAAGGACT-3′ | Antisense | 150 | 6449.0 | +ddC = 6722.2 | +ddA = 6746.2 | +24 |
| 1.14 | c.2T>G | 5′-ACTAGCAACCTCAAACAGACACCA-3′ | Sense | 150 | 7252.8 | +ddT = 7541.0 | +ddG = 7566.0 | +25 |
| 1.15 | c.251G>A (Hb Pyrgos) | 5′-TGGGTCACCTGGACAACCTCAAGG-3′ | Sense | 125 | 7362.8 | +ddG = 7676.0 | +ddA = 7660.0 | −16 |
| 1.16 | c.316–197C>T | 5′-ACAGTGATAATTTCTGGGTTAAGG-3′ | Sense | 150 | 7446.9 | +ddC = 7720.1 | +ddT = 7735.1 | +15 |
| 1.17 | c.126delC | 5′-GTTGCGTACCCTTGGACCCAGAGGTT-3′ | Sense | 350 | 7978.2 | +ddC = 8251.4 | +ddT = 8266.4 | +15 |
| c.124_127delTTCT | +ddG = 8291.4 | +40 | ||||||
| 1.18 | c.364G>C (Hb D Punjab) | 5′-CTCTCTCCTCGCCCATCACTTTGGCAAA-3′ | Sense | 450 | 8395.5 | +ddG = 8708.7 | +ddC = 8668.7 | −40 |
| 1.19 | c.92 + 5G>C | 5′-TCCTTAAACCTGTCTTGTAACCTTGATA-3′ | Antisense | 225 | 8488.6 | +ddC = 8761.8 | +ddG = 8801.8 | +40 |
| Panel 2: Multiplex SBE–for detection of 16 HBB mutations (using 14 extension primers) | ||||||||
| 2.1 | c.19G>A (Hb C) | 5′-GCATCTGACTCCT-3′ | Sense | 425 | 3885.6 | +ddG = 4198.8 | +ddA = 4182.8 | −16 |
| 2.2 | c.52A>T | 5′-ATCCACGTTCACCT-3′ | Antisense | 385 | 4158.8 | +ddT = 4447.0 | +ddA = 4456.0 | +9 |
| 2.3 | c.20A>T (Hb S) | 5′-CCGGCAGACTTCTCC-3′ | Antisense | 140 | 4489.0 | +ddT = 4777.2 | +ddA = 4786.2 | +9 |
| c.20A>C (Hb G Makassar) | +ddG = 4802.2 | +25 | ||||||
| 2.4 | c.27_28insG | 5′-GCTCCTGAGGAGAAG-3′ | Sense | 250 | 4642.1 | +ddT = 4930.3 | +ddG = 4955.3 | +25 |
| 2.5 | c.126delC | 5′-CCTTGGACCCAGAGGTT-3′ | Sense | 125 | 5186.4 | +ddC = 5460.0 | +ddT = 5475.0 | +15 |
| c.124_127delTTCT | +ddG = 5500.0 | +40 | ||||||
| 2.6 | c.108C>A | 5′-AGGCTGCTGGTGGTCTA-3′ | Sense | 125 | 5257.5 | +ddC = 5530.7 | +ddA = 5554.7 | +24 |
| 2.7 | c.441_442insAC (Hb Tak) | 5′-GAGAAAGCGAGCTTAGTG-3′ | Antisense | 75 | 5612.7 | +ddA = 5909.9 | +ddT = 5900.9 | −9 |
| 2.8 | c.46delT | 5′-CATTGCCGTTACTGCCCTG-3′ | Sense | 125 | 5730.7 | +ddT = 6018.9 | +ddG = 6043.9 | +25 |
| 2.9 | c.−81A>G | 5′-GGCAGGAGCCAGGGCTGGGC-3′ | Sense | 125 | 6249.1 | +ddA = 6546.3 | +ddG = 6562.3 | +16 |
| 2.10 | c.84_85insC | 5′-GATGAAGTTGGTGGTGAGGCCC-3′ | Sense | 150 | 6871.5 | +ddT = 7159.7 | +ddC = 7144.7 | −15 |
| 2.11 | c.−137C>A | 5′-AGACCTCACCCTGTGGAGCCACAC-3′ | Sense | 100 | 7267.8 | +ddC = 7541.0 | +ddA = 7565.0 | +24 |
| 2.12 | c.370_377del ACCCCACC | 5′-ACTTTCTGATAGGCAGCCTGCACTG-3′ | Antisense | 150 | 7633.0 | +ddG = 7946.2 | +ddA = 7930.2 | −16 |
| 2.13 | c.92 + 1G>T | 5′-CTCCTGTCTTGTAACCTTGATACCAA-3′ | Antisense | 125 | 7856.2 | +ddC = 8129.4 | +ddA = 8153.4 | +24 |
| 2.14 | c.5T>C (Hb Raleigh) | 5′-ACTAGCAACCTCAAACAGACACCATGG-3′ | Sense | 200 | 8215.4 | +ddT = 8503.6 | +ddC = 8488.6 | −15 |
| Panel 3: Multiplex VSET–for confirmation of three HBB mutations (using three extension primers) | ||||||||
| 3.1 | c.20A>T (Hb S) | 5′-TGCATCTGACTCCTG-3′ | Sense | 150 | 4519 | +ddA = 4816.2 | +dT+ ddG = 5136.4 | 320.2 |
| 3.2 | c.52A>T | 5′-GTACTGCGCTGTGGGGC-3′ | Sense | 375 | 5258.4 | +ddA = 5555.6 | +dT+ ddA = 5859.8 | 303.8 |
| 3.3 | c.441_442insAC (Hb Tak) | 5′-GCAAGAAAGCGAGCTTAGTG-3′ | Antisense | 100 | 6215.1 | +ddA = 6512.3 | +dT+ ddG = 6832.5 | 320.2 |
Bold bases are modifications for mass adjustment.
Underlined-bold bases are modifications for removal of self-complementary or hairpin formation.
Italic-bold bases are positions where there may be mismatches of bases due to the presence of other closely-located HBB mutations or SNPs.
Multiplex PE Reaction
Optimization of multiplex PE reactions was performed by testing with various combinations of extension primers until maximum multiplexing was obtained. Primer concentrations were adjusted to obtain balanced intensities of MALDI-TOF MS signals. Finally, three panels, which included 19, 14, and 3 primers per reaction for detection of 21, 16 (with 7 repeated tests), and 3 (as confirmation) HBB mutations, respectively, were obtained as Panel 1 and Panel 2 for multiplex SBE and Panel 3 for multiplex VSET assay (Table 2).
Multiplex SBE reaction (Panels 1 and 2) was performed in a 20-μl volume containing 5 μl of ExoSAP-IT treated PCR product, 75 to 450 nmol/L extension primers (Table 2), 50 μmol/L ddNTP (Amersham Biosciences UK Ltd, Buckinghamshire, UK), 1 μl of Thermo Sequenase buffer, and 1 U of Thermo Sequenase DNA polymerase (Amersham Biosciences UK Ltd). For multiplex VSET reaction (Panel 3), the 20-μl reaction volume contained 5 μl of ExoSAP-IT treated PCR product, 100 to 375 nmol/L of extension primers (Table 2), 50 μmol/L each of ddATP, ddCTP, ddGTP, and dTTP (instead of ddTTP), 1 μl of Thermo Sequenase buffer, and 1 U of Thermo Sequenase DNA polymerase.
Thermal cycling for all PE reactions was performed in a GeneAmp PCR System 2400, consisting of an initial denaturation at 96°C for 1 minute, followed by 90 cycles of touch-down protocol using the same denaturation step at 96°C for 15 seconds and extension step at 60°C for 60 seconds in every cycle, but the annealing temperature for 15 seconds was reduced from starting 60°C to 42°C in 2°C decrement every two cycles. Annealing temperature of 42°C was then maintained for the subsequent 72 cycles.
Sample Preparation and MALDI-TOF MS Analysis
PE products (20 μl) were purified using 5 μl of magnetic beads (GenoPure) according to manufacturer's instructions (Bruker Daltonics GmbH, Bremen, Germany) to obtain single-stranded oligonucleotide products.
For preparation of matrix solution, a saturated solution of 3-hydroxypicolinic acid in 50% acetonitrile was mixed with 50 mmol/L di-ammonium hydrogen citrate at a 1:1 ratio (v/v). A 0.5 μl aliquot of matrix solution was spotted onto an AnchorChip target plate (Bruker–Daltonics) and dried at room temperature. Subsequently, a 0.5 μl aliquot of purified PE sample was applied on top of the dried matrix and dried at room temperature. Mass spectra were recorded in a linear Bruker reflex V delayed extraction MALDI-TOF mass spectrometer (Bruker Daltonics), using negative ion and linear mode. External calibration was performed using a standard oligonucleotide mixture (0.25 pmol/μL of Oligo 12 mer, average m/z = 3646.4; 1.25 pmol/μL of Oligo 20 mer, average m/z = 6118.0; and 5 pmol/μL Oligo 30 mer, average m/z = 9192.0). Data were analyzed using Flex Analysis software (Bruker Daltonics).
Genotype Determination
Genotype was manually assigned for each mutation based on mass difference between primer and extension peaks of the MALDI spectra by determining signal ratio of specific to nonspecific PE reaction products.
Results
Design of Extension Primer Sets
The extension primers were designed specifically for each of the 30 HBB mutations by dividing into two multiplex-SBE panels (Table 2). Very close proximity to each other in some of the 30 HBB mutations made it difficult to design extension primers that were interference-free. Base positions where there might be mismatches due to the presence of other closely located HBB mutations are shown in bold italics in Table 2.
Panel 1 multiplex SBE was designed to detect a total of 21 HBB mutations using 19 extension primers (two were shared by two variant sites). Detectable HBB mutations included ten common and two rare mutation types, as well as nine abnormal Hb variants (as classified in Table 1). Data obtained from Siriraj-Thalassemia Program Project, Faculty of Medicine Siriraj Hospital, Mahidol University during 2003–2004, have revealed that the eight most common HBB mutations account for about 90% of mutant alleles in the Thai population, namely, c.79G>A (Hb E), c.124_127delTTCT, c.52A>T, c.−78A>G, c.316−197C>T, c.92 + 5G>C, c.59A>G (Hb Malay), and c.216_217insA (Table 1). Thus Panel 1 multiplex SBE was capable of detecting 95% of mutant HBB alleles in Thailand.
Panel 2 multiplex SBE, using 14 extension primers, was designed to detect 16 HBB mutations. Nine variants other than those in Panel 1 could be detected in Panel 2, including three common mutations, one abnormal Hb variant, and five rare mutations (as classified in Table 1). The three common variants (c.−81A>G, c.92 + 1G>T, and c.108C>A) were grouped into this panel to avoid incompatibility with their closely located mutations in Panel 1. Seven mutations in Panel 1 were also detected in Panel 2 as confirmatory tests, namely, those having 9-Da mass difference between wild-type and mutant alleles causing difficulty in interpretation (c.20A>T, c.52A>T, and c.441_442insAC); the smallest mass allele yielding poor signal (c.19G>A); the second most frequent variant in Thais (c.124_127delTTCT) and its extension-primer sharing variant (c.126delC); as well as the one (c.20A>C) coupled with c.20A>T.
In Panel 3, instead of multiplex SBE, multiplex VSET was established as an optional procedure for resolving ambiguous results obtained from the three HBB mutations, namely, c.20A>T, c.52A>T, and c.441_442insAC, having 9-Da mass differences found in the first two panels.
Optimization of the Three Multiplex PE Reactions
To evaluate the minisequencing performance of the extension primer for each HBB mutation before incorporating it into the multiplex SBE set, each primer was individually used in a single-plex SBE reaction and the result was analyzed by MALDI-TOF MS. Using various different combinations of extension primers, successful set of 19-plex and 14-plex reaction was obtained for multiplex Panels 1 and 2 respectively. Optimal PE conditions were further tested by varying the amounts of extension primers to adjust the signal balance of MALDI-TOF MS peaks in the same multiplex set and by adjusting thermal profiles of PE reactions to obtain the highest yields of every possible extension products.
To reduce the possibility of primer interactions in the multiplex SBE panels, as well as interference of unextended primers in MALDI-TOF MS analysis, an optimized minimum concentration of each extension primer was used. However, this could still yield very high signal-to-noise ratio of mass spectra. An optimal thermal cycling profile was achieved by the use of a modified touchdown thermal-cycling minisequencing, with an initial annealing temperature at 60°C followed by gradual reduction to 42°C. With amplification of up to 90 cycles, more primers were consumed, resulting in higher product yields and lower peak intensities of the primers. As a result, more extension products could be clearly determined.
Genopure magnetic bead separation kit for purification of oligonucleotides was used to purify PE products. Good peak intensity of ionization, high signal-to-noise ratio, and negligible salt-adduct peaks were observed from the purified PE products.
Standardization of Multiplex SBE and VSET
Multiplex SBE, optimized for Panels 1 and 2, was used for genotyping of a normal control and heterozygotes (single and compound) carrying 30 HBB mutations (Figure 2 and Table 1). Mass differences between each pair of wild-type and mutant allele of 9, 15, 16, 24, 25, and 40 Da could be clearly distinguished by MALDI-TOF MS, except for the 9-Da difference between A and T of c.20A>T, c.52A>T, and c.441_442insAC, which appeared as broad peaks (Figure 2: 1.2, 1.7, and 1.12). Mass spectra of the extended products plus residual primers in Panels 1 and 2 multiplex SBE of representative samples are shown in Figure 3A–D. Inclusion in both multiplex SBE panels of the three mutations producing a 9-Da difference allowed confirmation of the results. In cases where there were high quality PCR and clean PE products, the three mutations could be unequivocally genotyped, and where inconclusive signals were obtained, Panel 3 multiplex VSET was performed to obtain the correct genotyping (Figure 4A–D).
Figure 2.
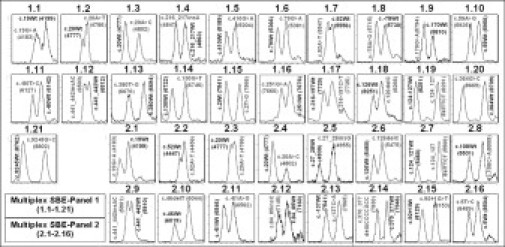
Mass spectra of HBB mutations detected by Panel 1 and Panel 2 multiplex SBE. Representative mass spectra obtained from 21 heterozygous HBB mutation sites detected by Panel 1 multiplex SBE (1.1–1.21) and 16 by Panel 2 (2.1–2.16) are shown. Genotype of the extension product is indicated above each peak and mass value is in parentheses. Wt = wild-type.
Figure 3.
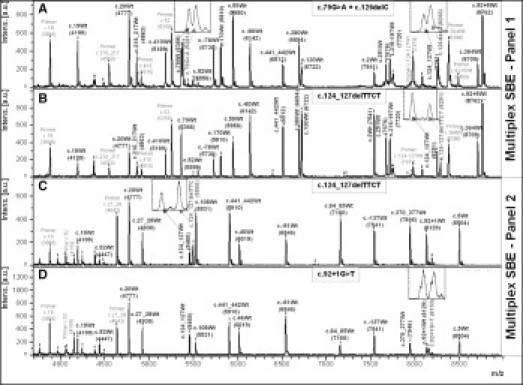
Mass spectra of 19-plex minisequencing reaction for genotyping of 21 HBB mutation sites in Panel 1 multiplex SBE and 14-plex minisequencing reaction for 16 HBB mutations in Panel 2 multiplex SBE. Panel 1 multiplex SBE: (A) c.79G>A + c.126delC (compound heterozygous) and (B) c.124_127delTTCT heterozygous. Panel 2 multiplex SBE: (C) c.124_127delTTCT heterozygous and (D) c.92 + 1G>T heterozygous. Genotype of extension product is indicated above each peak and mass value is in parentheses. Peaks labeled “Primer” indicate residual extension primers. Magnified views of heterozygote peaks are indicated by arrows and shown in insets.
Figure 4.
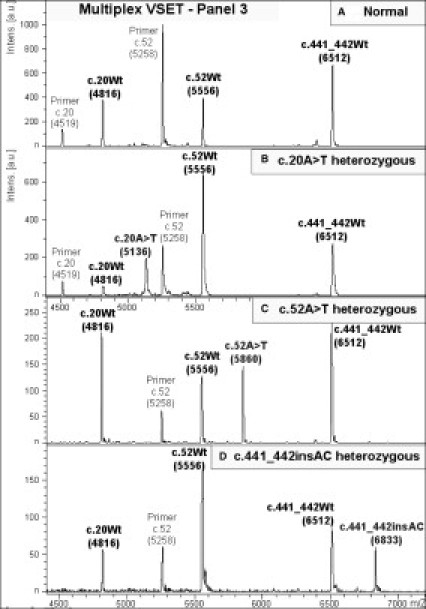
Mass spectra of 3-plex primer-extension reaction for genotyping in Panel 3 multiplex VSET: (A) normal, (B) c.20A>T heterozygote, (C) c.52A>T heterozygote, and (D) c.441_442insAC heterozygote. Genotype of extension product is indicated above each peak and mass value is in parentheses. Peaks labeled “Primer” indicate residual extension primers.
For discrimination of homozygotes from heterozygotes, we evaluated the performance of Panels 1 and 2 multiplex SBEs using nine (available) homozygous HBB mutations (Table 3). For Panel 1 multiplex SBE, eight homozygotes could be genotyped, giving the expected results. Five homozygotes, c.124_127delTTCT, c.52A>T, c.−81A>G, c.19G>A, and c.20A>T, could also be genotyped accurately by Panel 2 multiplex SBE. Moreover, Panel 3 multiplex VSET correctly identified homozygotes of c.52A>T and c.20A>T.
Table 3.
HBB Genotypes Tested
| Status of subject with known HBB genotype | ||||
|---|---|---|---|---|
| Single heterozygous (trait) | Compound heterozygous | Homozygous | ||
| c.−137C>A | c.27_28insG | c.124_127delTTCT | c.126delC + c.79G>A | c.−81A>G |
| c.−81A>G | c.46delT | c.130G>T | c.251G>A+ c.79G>A | c.−78A>G |
| c.−78A>G | c.52A>T | c.170G>A | c.410G>A+ c.79G>A | c.19G>A |
| c.−80T>C | c.59A>G | c.216_217insA | c.20A>T | |
| c.2T>G | c.79G>A | c.316−197C>T | c.52A>T | |
| c.5T>C | c.84_85insC | c.364G>C | c.59A>G | |
| c.19G>A | c.92 + 1G>T | c.370_377delACCCCACC | c.79G>A | |
| c.20A>T | c.92 + 5G>C | c.380T>G | c.92 + 5G>C | |
| c.20A>C | c.108C>A | c.441_442insAC | c.124_127delTTCT | |
However, due to the crowded features of the 30 HBB mutations, interferences or interactions among various HBB alleles and extension primers are of concern. Observations that need to be taken into account for screening unknown samples with the three Panels are listed in Table 4. For example, in Panel 1, homozygous c.−81A>G (which is not within the detection scope of Panel 1) results in the loss of detection of c.−80T>C because c.−81A>G is located at the 3′ end of an extension primer for c.−80T>C, leading to a strong mismatch between primer and template.
Table 4.
Allelic Loss Observed in Multiplex Panel
| Multiplex panel tested | Homozygous DNA sample tested | Observed allelic loss (of either allele) | Cause | Solution |
|---|---|---|---|---|
| Panel 1 | c.−81A>G (detectable by Panel 2) | c.−80T>C | Homozygous c.−81A>G causes a strong mismatch at the 3′ end of an extension primer for c.−80T>C | DNA sequencing focusing on c.−80T>C |
| Panel 2 | c.19G>A (Hb C) and c.20A>T (Hb S) (detectable by both Panels 1 and 2) | c.27_28insG | Homozygous c.19G>A and c.20A>T result in a mismatched site at the 9th and 8th base respectively from 3′ end of primer for c.27_28insG whose length is only 14 bases. The primer is furthermore at a disadvantage in competition for the same template region with the other two for c.19 and c.20 | DNA sequencing focusing on c.27_28insG or rechecking it by exclusion of primers for c.19G>A and c.20A>T from Panel 2 |
| c.92 + 5G>C (detectable by Panel 1) | c.92 + 1G>T | Homozygous c.92 + 5G>C has a base mismatch at the 4th base from 3′ end of a primer for c.92 + 1G>T | DNA sequencing focusing on c.92 + 1G>T | |
| Panel 3 | c.19G>A (Hb C) (detectable by both Panels 1 and 2) | c.20A>T (Hb S) | Homozygous c.19G>A has a mismatch at 3′ end of extension primer for c.20A>T (only in this panel) | c.20A>T (Hb S) can be detected in Panel 1 and 2 without this interference (different primer is used) |
Homozygous c.79G>A (Hb E) was also evaluated in Panel 2 multiplex SBE. This mutation site is located at the seventh base from the 3′ end of the c.84_85insC primer and has no effect on primer binding (Figure 5). Thus, similar results could be expected in Panel 1 multiplex SBE for the case of homozygous c.46delT and homozygous c.84_85insC when the effect on the binding of primer for c.52A>T and c.79G>A is considered respectively, as the mutation sites are at the sixth base from the 3′ end of both primers. However, the presence of base mismatch at the third position from 3′ end of an extension primer may inhibit PE reaction, as in the case of homozygous c.2T>G detected by c.5T>C primer in Panel 2 multiplex SBE (Figure 5).
Figure 5.

Mismatched bases generated by extension primers used in multiplex SBE reaction and by homozygous alleles of HBB mutations. Mismatched positions near the 3′ end are likely to cause allelic loss in multiplex SBE reaction, whereas those at sixth or seventh position are not. Mismatched bases are indicated in bold and italics in box.
Primers used for Panel 3 multiplex VSET could detect c.20A>T, c.52A>T, and c.441_442insAC mutations. The primer for detecting c.20A>T could also pick up c.20A>C mutation. However, it was observed that heterozygous c.20A>C gave no expected signal of mass 4792 Da from the mutant C allele (as SBE, not VSET). This may have been caused by a preferential incorporation of ddA for the wild-type allele (mass = 4816.2 Da) over that of the mutant and/or by poor ionization/desorption of oligonucleotide molecules of this mutant allele.
Validation of Multiplex SBE and VSET in Thalassemia Samples
One hundred and sixty-two randomly selected thalassemia subjects whose HBB mutations had previously been genotyped by DGGE, restriction fragment length polymorphism, and/or DNA sequencing were used to validate the technique. Samples consisted of 120 heterozygous healthy carriers having 15 different HBB mutations, and 42 patients (eight homozygotes and 34 compound heterozygotes) having two homozygous and 10 different compound heterozygous HBB mutations (Table 5). Every sample identified using a combination of Panels 1 + 2 (104 samples) or Panels 1 + 2 + 3 (29 samples) consistently demonstrated the expected genotype. Moreover, 10 samples determined by PE MALDI-TOF MS were shown to have been misidentified by conventional methods. For example, c.79G>A (Hb E) was frequently discovered in samples that previously had not been detected by DGGE.
Table 5.
Blinded Validation Analysis of Multiplex PE Panels Coupled with MALDI-TOF MS in 162 Randomly Selected Samples Carrying HBB Mutations Previously Genotyped by Conventional Methods
| Number of samples tested by PE panel that gave 100% concordant results compared with conventional methods |
|||||
|---|---|---|---|---|---|
| HBB mutation | Panel 1 alone | Panel 2 Alone | Panel 3 alone | Panel 1 + 2 (in same sample) | Panel 1 + 2 +3 (in same sample) |
| Homozygous mutation (2 types, 8 samples) | |||||
| c.−78A>G | 2 | 1 | — | 1 | — |
| c.79G>A | 6 | 6 | — | 5 | — |
| Compound heterozygous mutation (10 types, 34 samples) | |||||
| c.−81A>G/c.79G>A | 1 | 2 | — | 1 | — |
| c.−78A>G/c.59A>G | 1 | 1 | — | 1 | — |
| c.−78A>G/c.79G>A | 2 | 2 | — | 2 | — |
| c.52A>T/c.79G>A | 5 | 10 | 8 | 5 | 5 |
| c.59A>G/c.79G>A | 1 | 1 | — | 1 | — |
| c.92 + 1G>T/c.79G>A | 2 | 4 | — | 2 | — |
| c.124_127delTTCT/c.79G>A | 4 | 8 | — | 4 | — |
| c.364G>C/c.79G>A | 1 | 2 | — | 1 | — |
| c.441_442insAC/c.79G>A | 1 | 2 | 2 | 1 | 1 |
| c.216_217insA/c.441_442insAC | 1 | 2 | 2 | 1 | 1 |
| Heterozygous mutation (15 types, 120 samples) | |||||
| c.−81A>G | 1 | 1 | — | 1 | — |
| c.−78A>G | 10 | 9 | — | 8 | — |
| c.2T>G | 2 | 1 | — | 1 | — |
| c.19G>A | 2 | 1 | 1 | 1 | 1 |
| c.52A>T | 26 | 27 | 36 | 19 | 19 |
| c.59A>G | 5 | 2 | — | 2 | — |
| c.79G>A | 3 | 1 | — | 1 | — |
| c.92 + 1G>T | 3 | 4 | — | 3 | — |
| c.92 + 5G>C | 3 | 1 | — | 1 | — |
| c.124_127delTTCT | 33 | 35 | — | 27 | — |
| c.216_217insA | 3 | 3 | — | 3 | — |
| c.316−197C>T | 8 | 7 | — | 7 | — |
| c.370_377delACCCCACC | 1 | 1 | — | 1 | — |
| c.410G>A | 2 | 2 | — | 2 | — |
| c.441_442insAC | 3 | 4 | 5 | 2 | 2 |
| Total | 132 | 140 | 54 | 104 | 29 |
Discussion
We have developed a multiplex-PE method for genotyping using MALDI-TOF MS, based on mass difference between wild-type and mutant alleles generated by an addition of a single base. Using optimized minimum primer concentrations, a touchdown thermal cycling protocol, and only two multiplex panels, the method enabled efficient genotyping of 30 commonly found HBB mutations specific to the Thai population. The procedure was validated by correctly genotyping 162 thalassemia samples previously identified using conventional techniques of DGGE, restriction fragment length polymorphism, and/or direct DNA sequencing.
Due to the close proximity of the 30 mutation sites, primers used in genotyping HBB mutations were chosen so as to avoid false priming, self annealing, and primer-dimer artifact, genotyping errors that could arise in negative controls. Presence of any additional SNP sites in the extension primer sequence itself is an important factor when genotyping homozygous samples carrying such SNP sites as shown in Table 4. Other critical parameters determining success of multiplex-minisequencing protocols include high quantity and quality of amplicons, as well as high quality purified primers with appropriate sequences, mass ranges (about 4000 to 9000 Da) and thermal profiles.29 In this study, optimization of primer extension reaction was achieved by varying combinations of extension primers taking into account priority of HBB-mutation types and ensuring compatibility of all primers in the set, as well as uniqueness of individual masses. The optimized minimum concentration of primers used in the minisequencing reactions yielded a high signal-to-noise ratio, resulting in easier identification of the extended products. As excess primers can dimerize to form false peaks in the mass spectrum and can compete for the ion current, thereby reducing detection sensitivity of MALDI-TOF MS for the DNA fragments of interest,30 a modified touchdown minisequencing thermal-cycling program with a high cycle number of up to 90 cycles yielded sufficient products for subsequent MS detection, as previously demonstrated by Meyer et al.31
As shown in Figure 2, unequal signal intensities for heterozygous alleles are frequently observed in this study. Such uneven extension signals could be caused by differences in SBE efficiency due to preferential incorporation of one ddNTP over another, as well as by competitive ionization and desorption of oligonucleotide molecules in multiplex reactions. This phenomenon is acceptable unless the signals from both alleles are more than 10 times different and the low signal no longer satisfies the signal-to-noise criterion for genotype determination.32 Although low concentrations of primers were used, some primer signals still masked extension signals. Extension products of c.52A>T (either A or T) sometimes exhibited very low or no peak intensity. However, if a peak absence was observed in DNA samples whose genotypes could be assigned by the other two mutant alleles (in cases of homozygote or compound heterozygote), it could be ignored. Alternatively, problematic samples should be specifically rechecked by improving every step including multiplex-PCR amplification, primer extension, and purification, as well as sample preparation and MALDI-TOF MS analysis. If a repeat test still failed in one panel, the remaining panel could provide a back-up analysis for this mutation (viz. c.52A>T). Thus, false-negative results were not obtained from our protocol, resulting in 100% sensitivity.
In general, DNA analysis by MALDI-TOF MS is considered more complicated than protein analysis. Larger DNA fragments can give less intense and broader signals due to DNA instability or fragmentation (mainly depurination), contributing to the effect of a more complex isotope distribution of the poor mass spectrum.25 Higher sensitivity, resolution, and accuracy of the MALDI mass spectrum can only be achieved by using an appropriate TOF instrumental configuration.33 Linear-mode MALDI-TOF MS analysis, although unable to resolve isotopic peaks of parent and fragment ions, was used in our study to reduce fragmentation and to enhance signals from DNA molecules of mass larger than 3000 Da. Discrimination of monoisotopic ions of 1-Da mass difference, as obtained from the reflectron mode, cannot be seen in the linear mode, but only average masses were shown as a continuous single peak of each SBE allele due to the aggregation of ions of the individual isotopes, thereby generating the overlapping mass spectral peaks. This can lead to a high background and may result in false positives in those cases with low signal-to-noise spectra. In fact, only a potential false-positive result was generated by a Na+ salt adduct (21.98 Da) in the product of c.46delT wild-type allele (6019 Da), giving rise to a 6041-Da peak close to 6044 Da of the mutant allele. However, mutation c.46delT is rare, and when it is present, the mutant peak was always about twice that of the wild-type peak (see Figure 2, 2.10). Nevertheless, a mass difference of 25 Da in the mutant case should allow discrimination from the 22-Da difference of the salt adduct.
The c.79G>A (Hb E) mutation is often missed when the DGGE technique is used, as heteroduplex DNA species formed have very similar mobility in the gel. However, the PE reaction coupled with MALDI-TOF can readily discriminate such alleles based on differences in mass of the extended products.
Any individual DNA sample requested for HBB genotyping (regardless of relevant clinical information) should be screened first by Panel 1 multiplex SBE, which covers 95% of mutant HBB alleles in the Thai population (cost/mutation of $0.58). Panel 2 multiplex SBE, which detects an additional 4.5% of the mutations, should be performed on samples not identified by Panel 1. Panel 3 multiplex VSET is used only to confirm the results of three mutations, c.20A>T, c.52A>T, and c.441_442insAC, which constitute approximately 15% of Thai mutant HBB alleles. Even based on the use of up to three multiplex panels, unit cost of the protocol for detection of all 30 HBB mutations is $0.78 per genotype determination (average cost $0.69), while that using a combination of three conventional methods is $3.15. The major cost of MALDI-TOF genotyping is that for Thermo Sequenase DNA polymerase used in PE reactions. Other contributions to the cost include the purification steps using ExoSAP-IT and Genopure kit, as well as MALDI-TOF MS analysis. To reduce reagent costs further, the reaction volume for primer extension can be reduced to 10 μl, allowing the subsequent purification step to use only one half of the original protocol volume. A comparison of the costs involved in MALDI-TOF genotyping methods in this study and other conventional methods is summarized in Table 6. These estimates include only reagent costs. The costs of all plastic wares, consumables, and personnel were assumed to be similar for all of the methods. Nonetheless, the high levels of multiplexing and automation of MALDI-TOF genotyping likely should reduce personnel costs below those of other methods. Instruments can be amortized over a large number of tests for a rather long period of time, and thus the costs can be assumed to be negligible.
Table 6.
Cost Comparison between MALDI-TOF Genotyping and Conventional Methods Used for HBB Genotyping in this Study
| MALDI-TOF genotyping |
Conventional method |
|||||
|---|---|---|---|---|---|---|
| 1 Panel | 2 Panels | 3 Panels | DGGE | RFLP | DNA Sequencing | |
| Number of detectable mutations | 21 | 30 | 30* | 18 | 1 (Hb E) | 11† |
| Percent total HBB alleles in Thai population | 95% | 99.5% | 99.5* % | 65% | 30% | 4.5% |
| Analysis time (h) | 22 | 22 | 22 | 17 | 22 | 14 |
| Reagent cost ($) | ||||||
| DNA preparation | 5.00 | 5.00 | 5.00 | 5.00 | 5.00 | 5.00 |
| PCR | 1.25 | 1.25 | 1.25 | 7.50 (1.25 × 6) | 1.25 | 7.50 (1.25 × 6) |
| Post-PCR | 3.75 | 7.50 | 11.25 | − | 0.75 | 52.50 |
| Analysis step | 2.00 | 4.00 | 6.00 | 2.50 | − | 7.50 |
| Total cost | 12.00 | 17.80 | 23.50 | 15.00 | 7.00 | 72.50 |
| Cost ($) per mutation | 0.57 | 0.59 | 0.78 | 0.83 | 7.00 | 6.60 |
| Average for 30 mutations = 0.69 | Average for 30 mutations = 3.15 | |||||
Optional for specific confirmation of c.20A>T (Hb S), c.52A>T, and c.441_442insAC.
HBB mutations requiring DNA sequencing analysis are c.−137C>A, c.2T>G, c.5T>C (Hb Raleigh), c.19G>A (Hb C), c.20A>T (Hb S), c.20A>C (Hb G Makassar), c.46delT, C.251G>A (Hb Pyrgos), c.364G>C (Hb D Punjab), c.380T>G (Hb Dhonburi), and c.410G>A (Hb Hope).
Increasingly multiplex PE combined with MALDI-TOF MS has been applied to genotype mutations or SNPs in variety of genes.19,22,23,24,32,34,35 Solid phase capturable ddNTPs in SBE have been developed using biotinylated ddNTPs to generate 3′-biotinylated extension DNA products, which then are purified by streptavidin-coated magnetic beads before analysis by MS.27 This solid phase capturable-SBE method has been applied for simultaneous genotyping of 17 Y-chromosome SNPs,36 detection of 30 point mutations in p53 in a single tube,37 and for concurrent analysis of 40 SNPs of CYP2C9 and 50 SNPs of CYP2A13 genes.38 However, the cost-effectiveness of this approach has not been evaluated. The method developed in this study employs unmodified oligonuclotide primers, allowing the use of standard magnetic beads in the purification step, and thereby substantially reducing the unit cost when compared with that of the solid phase capturable-SBE procedure.
In conclusion, a reliable multiplex system for simultaneously genotyping 21 (using Panel 1 protocol) and 16 (Panel 2 protocol) HBB mutations involving 2 PE reactions and subsequent MALDI-TOF MS analysis has been successfully developed. The 9-Da mass difference between wild-type and mutant allele in 3 HBB mutations could be unambiguously confirmed using an additional PE set (Panel 3 protocol). Purification of PE products by a magnetic bead DNA purification system resulted in high quality MALDI-TOF MS spectra. Data analysis was a bottleneck for high-throughput genotyping using the current method, due to the high level of multiplexing. Software for automatic allele identification should be developed to calibrate each mass spectrum using extension primers in each reaction as internal references and to identify genotypes based on mass differences between primer and extension peaks. Nevertheless, this fast, accurate, and flexible multiplex genotyping system should prove useful for rapid and inexpensive screening of β-thalassemia carriers and patients, as well as for prenatal diagnosis and molecular screening of the Thai population harboring high frequencies of mutant HBB alleles. This approach also could be used as a model in the development of rapid and accurate genotyping of common point mutations in single-gene defects or of SNPs as markers in multigenic disorders, and for identification of potential markers for clinical diagnosis, monitoring and prognosis.
Acknowledgements
We thank Dr. Prapin Wilairat for valuable comments and suggestions.
Footnotes
Supported by the National Center for Genetic Engineering and Biotechnology, National Science and Technology for Development Agency (NSTDA), Thailand (BT-B-02-MM-SU-4904), Siriraj Chalermprakiat Fund (to W.T.) and Mahidol University Research Grant (to C.L.). P.Y. is a Senior Research Scholar of The Thailand Research Fund.
Contributor Information
Wanna Thongnoppakhun, Email: siwtn@mahidol.ac.th.
Pa-thai Yenchitsomanus, Email: grpye@mahidol.ac.th.
References
- 1.Patrinos GP, Giardine B, Riemer C, Miller W, Chui DH, Anagnou NP, Wajcman H, Hardison RC. Improvements in the HbVar database of human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004;32(Database issue):D537–D541. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fucharoen S, Winichagoon P. Hemoglobinopathies in Southeast Asia. Hemoglobin. 1987;11:65–88. doi: 10.3109/03630268709036587. [DOI] [PubMed] [Google Scholar]
- 3.Levinson D. Ethnic groups worldwide: A ready reference handbook. Oryx Press; Phoenix: 1998. Thailand; pp. 286–289. [Google Scholar]
- 4.Chinchang W, Viprakasit V, Pung-Amritt P, Tanphaichitr VS, Yenchitsomanus PT. Molecular analysis of unknown beta-globin gene mutations using polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) technique and its application in Thai families with beta-thalassemias and beta-globin variants. Clin Biochem. 2005;38:987–996. doi: 10.1016/j.clinbiochem.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Old JM, Petrou M, Modell B, Weatherall DJ. Feasibility of antenatal diagnosis of beta thalassaemia by DNA polymorphisms in Asian Indian and Cypriot populations. Br J Haematol. 1984;57:255–263. [PubMed] [Google Scholar]
- 6.Maggio A, Giambona A, Cai SP, Wall J, Kan YW, Chehab FF. Rapid and simultaneous typing of hemoglobin S, hemoglobin C, and seven Mediterranean beta-thalassemia mutations by covalent reverse dot-blot analysis: application to prenatal diagnosis in Sicily. Blood. 1993;81:239–242. [PubMed] [Google Scholar]
- 7.Sutcharitchan P, Saiki R, Fucharoen S, Winichagoon P, Erlich H, Embury SH. Reverse dot-blot detection of Thai beta-thalassaemia mutations. Br J Haematol. 1995;90:809–816. doi: 10.1111/j.1365-2141.1995.tb05200.x. [DOI] [PubMed] [Google Scholar]
- 8.Winichagoon P, Saechan V, Sripanich R, Nopparatana C, Kanokpongsakdi S, Maggio A, Fucharoen S. Prenatal diagnosis of beta-thalassaemia by reverse dot-blot hybridization. Prenat Diagn. 1999;19:428–435. [PubMed] [Google Scholar]
- 9.Fortina P, Dotti G, Conant R, Monokian G, Parrella T, Hitchcock W, Rappaport E, Schwartz E, Surrey S. Detection of the most common mutations causing beta-thalassemia in Mediterraneans using a multiplex amplification refractory mutation system (MARMS) PCR Methods Appl. 1992;2:163–166. doi: 10.1101/gr.2.2.163. [DOI] [PubMed] [Google Scholar]
- 10.Lee HH, Chang JG, Chen RT, Yang ML, Choo KB. Prenatal diagnosis of beta-thalassemic mutations in Chinese by multiple restriction fragment-single strand conformation polymorphism analysis. Proc Natl Sci Counc Repub China B. 1994;18:112–117. [PubMed] [Google Scholar]
- 11.Ghanem N, Girodon E, Vidaud M, Martin J, Fanen P, Plassa F, Goossens M. A comprehensive scanning method for rapid detection of beta-globin gene mutations and polymorphisms. Hum Mutat. 1992;1:229–239. doi: 10.1002/humu.1380010310. [DOI] [PubMed] [Google Scholar]
- 12.Losekoot M, Fodde R, Harteveld CL, van Heeren H, Giordano PC, Bernini LF. Denaturing gradient gel electrophoresis and direct sequencing of PCR amplified genomic DNA: a rapid and reliable diagnostic approach to beta thalassaemia. Br J Haematol. 1990;76:269–274. doi: 10.1111/j.1365-2141.1990.tb07883.x. [DOI] [PubMed] [Google Scholar]
- 13.Wong C, Dowling CE, Saiki RK, Higuchi RG, Erlich HA, Kazazian HH., Jr Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single copy DNA. Nature. 1987;330:384–386. doi: 10.1038/330384a0. [DOI] [PubMed] [Google Scholar]
- 14.Chan K, Wong MS, Chan TK, Chan V. A thalassaemia array for Southeast Asia. Br J Haematol. 2004;124:232–239. doi: 10.1046/j.1365-2141.2003.04758.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Kham SK, Yeo GH, Quah TC, Chong SS. Multiplex minisequencing screen for common Southeast Asian and Indian beta-thalassemia mutations. Clin Chem. 2003;49:209–218. doi: 10.1373/49.2.209. [DOI] [PubMed] [Google Scholar]
- 16.Su YN, Lee CN, Hung CC, Chen CA, Cheng WF, Tsao PN, Yu CL, Hsieh FJ. Rapid detection of beta-globin gene (HBB) mutations coupling heteroduplex and primer-extension analysis by DHPLC. Hum Mutat. 2003;22:326–336. doi: 10.1002/humu.10265. [DOI] [PubMed] [Google Scholar]
- 17.Yip SP, Pun SF, Leung KH, Lee SY. Rapid, simultaneous genotyping of five common Southeast Asian beta-thalassemia mutations by multiplex minisequencing and denaturing HPLC. Clin Chem. 2003;49:1656–1659. doi: 10.1373/49.10.1656. [DOI] [PubMed] [Google Scholar]
- 18.Ding C, Chiu RW, Lau TK, Leung TN, Chan LC, Chan AY, Charoenkwan P, Ng IS, Law HY, Ma ES, Xuk X, Wanapiraki C, Sanguansermsrii T, Liaol C, Aim MATJ, Chuin DKH, Cantora CR, Lo YMD. MS analysis of single-nucleotide differences in circulating nucleic acids: Application to noninvasive prenatal diagnosis. Proc Natl Acad Sci USA. 2004;101:10762–10767. doi: 10.1073/pnas.0403962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao HK, Su YN, Kao HY, Hung CC, Wang HT, Chen YJ. Parallel minisequencing followed by multiplex matrix-assisted laser desorption/ionization mass spectrometry assay for beta-thalassemia mutations. J Hum Genet. 2005;50:139–150. doi: 10.1007/s10038-005-0234-z. [DOI] [PubMed] [Google Scholar]
- 20.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nature Biotechnol. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, Ding H, Hung K, Guo B. A new MALDI-TOF based mini-sequencing assay for genotyping of SNPS. Nucleic Acids Res. 2000;28:E68. doi: 10.1093/nar/28.12.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blievernicht JK, Schaeffeler E, Klein K, Eichelbaum M, Schwab M, Zanger UM. MALDI-TOF mass spectrometry for multiplex genotyping of CYP2B6 single-nucleotide polymorphisms. Clin Chem. 2007;53:24–33. doi: 10.1373/clinchem.2006.074856. [DOI] [PubMed] [Google Scholar]
- 23.Humeny A, Bonk T, Berkholz A, Wildt L, Becker CM. Genotyping of thrombotic risk factors by MALDI-TOF mass spectrometry. Clin Biochem. 2001;34:531–536. doi: 10.1016/s0009-9120(01)00264-8. [DOI] [PubMed] [Google Scholar]
- 24.Nakai K, Habano W, Fujita T, Nakai K, Schnackenberg J, Kawazoe K, Suwabe A, Itoh C. Highly multiplexed genotyping of coronary artery disease-associated SNPs using MALDI-TOF mass spectrometry. Hum Mutat. 2002;20:133–138. doi: 10.1002/humu.10099. [DOI] [PubMed] [Google Scholar]
- 25.Tost J, Gut IG. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin Biochem. 2005;38:335–350. doi: 10.1016/j.clinbiochem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Bleicher K, Bayer E. Various factors influencing the signal intensity of oligonucleotides in electrospray mass spectrometry. Biol Mass Spectrom. 1994;23:320–322. doi: 10.1002/bms.1200230604. [DOI] [PubMed] [Google Scholar]
- 27.Kim S, Edwards JR, Deng L, Chung W, Ju J. Solid phase capturable dideoxynucleotides for multiplex genotyping using mass spectrometry. Nucleic Acids Res. 2002;30:e85. doi: 10.1093/nar/gnf084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haff LA, Smirnov IP. Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res. 1997;7:378–388. doi: 10.1101/gr.7.4.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho CM, Pena SD. Optimization of a multiplex minisequencing protocol for population studies and medical genetics. Genet Mol Res. 2005;4:115–125. [PubMed] [Google Scholar]
- 30.Roskey MT, Juhasz P, Smirnov IP, Takach EJ, Martin SA, Haff LA. DNA sequencing by delayed extraction-matrix-assisted laser desorption/ionization time of flight mass spectrometry. Proc Natl Acad Sci USA. 1996;93:4724–4729. doi: 10.1073/pnas.93.10.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer K, Fredriksen A, Ueland PM. High-level multiplex genotyping of polymorphisms involved in folate or homocysteine metabolism by matrix-assisted laser desorption/ionization mass spectrometry. Clin Chem. 2004;50:391–402. doi: 10.1373/clinchem.2003.026799. [DOI] [PubMed] [Google Scholar]
- 32.Bray MS, Boerwinkle E, Doris PA. High-throughput multiplex SNP genotyping with MALDI-TOF mass spectrometry: practice, problems and promise. Hum Mutat. 2001;17:296–304. doi: 10.1002/humu.27. [DOI] [PubMed] [Google Scholar]
- 33.Cotter RJ. Time-of-flight mass spectrometry for the structural analysis of biological molecules. Anal Chem. 1992;64:1027A–1039A. doi: 10.1021/ac00045a002. [DOI] [PubMed] [Google Scholar]
- 34.Paracchini S, Arredi B, Chalk R, Tyler-Smith C. Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res. 2002;30:e27. doi: 10.1093/nar/30.6.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Wang H, Wang J, Cai Y, Zhou G, He F, Qian X. Multiplex single-nucleotide polymorphism genotyping by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem. 2003;314:54–62. doi: 10.1016/s0003-2697(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 36.Mengel-Jorgensen J, Sanchez JJ, Borsting C, Kirpekar F, Morling N. MALDI-TOF mass spectrometric detection of multiplex single base extended primers. A study of 17 y-chromosome single-nucleotide polymorphisms. Anal Chem. 2004;76:6039–6045. doi: 10.1021/ac049264k. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Ulz ME, Nguyen T, Li CM, Sato T, Tycko B, Ju J. Thirty fold multiplex genotyping of the p53 gene using solid phase capturable dideoxynucleotides and mass spectrometry. Genomics. 2004;83:924–931. doi: 10.1016/j.ygeno.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Misra A, Hong JY, Kim S. Multiplex genotyping of cytochrome p450 single-nucleotide polymorphisms by use of MALDI-TOF mass spectrometry. Clin Chem. 2007;53:933–939. doi: 10.1373/clinchem.2006.080739. [DOI] [PubMed] [Google Scholar]


