Abstract
Abbott Molecular's m2000 system and RealTime HIV-1 assay (RealTime) were evaluated for sensitivity, reproducibility, linearity, ability to detect diverse HIV-1 subtypes/groups, and correlation to the Roche AMPLICOR HIV-1 MONITOR Test, Version 1.5 (Amplicor). The limit of detection was determined using the second International World Health Organization Standard and Viral Quality Assurance standard material. Serial dilutions of four patient samples were used to determine inter- and intra-assay reproducibility and linearity. Samples representing HIV-1 groups M, N, and O were evaluated in the RealTime, Amplicor, and Siemens Versant HIV-1 branched chain DNA 3.0 (Versant) assays. Archived Amplicor-tested samples were tested with the 1 ml, 0.5 ml, and 0.6 ml versions of the RealTime assay. Probit analysis predicts a limit of detection of 21.94 IU/ml using the World Health Organization Standard and 26.54 copies/ml using Viral Quality Assurance material with the 1 ml assay. Linearity and reproducibility were very good between ∼1.60 to 6.0 log10 copies/ml. All three assays produced similar measurements for all Group M subtypes tested; the RealTime assay was the only assay that detected all three Group O samples tested. Correlation with the Amplicor assay was good, although the RealTime assay measured between 0.342 and 0.716 log10 copies/ml lower on average, depending on the input volume. The automated RealTime assay exhibits excellent sensitivity, dynamic range, reproducibility, and group/subtype detection, albeit with consistently lower values than Amplicor.
HIV-1 infection continues to be a major cause of mortality in the world. According to the Joint United Nations Programme on HIV/AIDS (http://www.unaids.org), there were approximately 40 million people worldwide living with HIV in 2006, over 10% of which were newly infected. Nearly 3 million deaths in 2006 were attributed to AIDS. Clinical management of HIV patients requires establishing and monitoring the viral load before initiating and during the administration of anti-viral therapies. The response of viral loads to medication, typically in conjunction with drug resistance testing, determines treatment options.
Several commercial and laboratory-developed assays are available to monitor HIV-1 viral load, including assays based on end-point PCR, isothermal nucleic acid based amplification, branched chain DNA (bDNA) signal amplification, and real-time PCR, the characteristics and performance of which have been reviewed and compared in multiple studies.1,2,3,4,5,6,7,8,9 Each assay has its own advantages and disadvantages in terms of sensitivity, equipment requirements, throughput, dynamic range, subtype detection, and cost.
Abbott Molecular released the m2000 system (m2000sp for sample preparation and PCR setup; m2000rt for amplification, detection, and data analysis) and RealTime HIV-1 assay in the European Union with CE Mark certification. We used the m2000 system to evaluate the Abbott Molecular Investigational Use Only RealTime HIV-1 assay (RealTime) and compared some aspects of its performance to the Roche Amplicor HIV-1 Monitor Test, Version 1.5 (Amplicor) and Siemens Versant HIV-1 bDNA 3.0 assay (Versant). The RealTime HIV-1 assay recently obtained United States Food and Drug Administration approval.
Materials and Methods
Limit of Detection
To determine the limit of detection in the 1 ml RealTime assay, eleven dilutions of the World Health Organization reference material (HIV-1 RNA, second International Standard, 97/650, National Institute for Biological Standards and Control (NIBSC), Potters Bar, UK) or Viral Quality Assurance standard (VQA, Rush University Medical Center, Chicago, IL) were prepared in Basematrix Diluent (BBI Diagnostics, West Bridgewater, MA). At least six replicates at each concentration (139.2, 87.0, 69.6, 60.9, 55.7, 52.2, 43.5, 34.8, 17.4, 8.7, and 1.7 IU/ml for World Health Organization; 80, 50, 40, 35, 32, 30, 25, 20, 10, 5, and 1 copies/ml for VQA) were tested on a total of two runs. Probit analysis was performed to determine the limit of detection (95% detection rate) and 95% confidence interval.
Reproducibility and Linearity
Four patient samples with high viral titers (>6 log10 copies/ml) serially diluted in Basematrix were used to evaluate intra- and interassay reproducibility and linearity of the 1 ml RealTime assay. Sufficient volume for five aliquots of each dilution were prepared and stored at −70°C until use. For each sample, three of five replicates were tested on the same run to evaluate intra-assay reproducibility and linearity. Interassay reproducibility was evaluated by performing two additional runs containing one aliquot of each sample dilution on each run. The average values for the intra-assay replicates were combined with the values for the two additional runs to determine interassay reproducibility and linearity. Reproducibility was evaluated by determining the mean and SEM for each dilution. Linearity was evaluated by linear regression analysis, excluding the aliquots with the lowest coefficient of variation, because they were set at identity to establish the expected values.
Subtype Panels
To assess the ability of the 1 ml RealTime assay to measure viral loads in samples containing diverse HIV subtypes and groups, two subtype panels were tested, including group M subtypes A, B, C, D, F, G, circulating recombinant forms AE, AG, AG-GH, and Group N and Group O samples. Abbott Molecular provided eighteen blinded samples. Ten samples in the HIV-1 RNA Genotype Panel for NAT Assays (01/466) (NIBSC) were diluted approximately 1.7-fold in Basematrix. Each sample was stored at −70°C, and tested by the Amplicor, RealTime, and Versant assays using standard procedures.
Correlation Samples
Archived frozen plasma samples previously submitted to ARUP for HIV-1 viral load testing by the Roche Amplicor HIV-1 Monitor Test, version 1.5 were retrieved from storage, de-identified, thawed at room temperature, transferred to barcoded 4 ml Simport tubes (Beloeil, Quebec, Canada), and stored at −70°C before testing in the RealTime assay. Patient samples with viral loads spanning the range of both assays were selected to evaluate assay correlation using the 1 ml (n = 171 samples), 0.5 ml (n = 135 samples), and newly approved 0.6 ml (n = 180 samples) RealTime protocol. Forty-four (44) samples with viral loads below the limit of detection of the UltraSensitive Amplicor assay (<50 copies/ml) were selected for retesting in the 1 ml RealTime assay.
Roche Amplicor Assay
The Roche Amplicor HIV-1 Monitor Test, version 1.5 (Roche Molecular Systems, Inc., Branchburg, NJ) was performed as prescribed by the manufacturer using manual extraction and PCR setup. The Standard version of the assay isolates HIV-1 RNA from 200 μl of plasma by lysis and alcohol precipitation. In the UltraSensitive version of the assay, virus is concentrated by centrifugation of 500 μl of plasma before extraction. The amplifications were performed on Applied Biosystems GeneAmp PCR System 9600 thermal cyclers. The microplates were washed using a Bio-Tek Instruments ELP-40 and read using a Bio-Tek Instruments ELx800.
Abbott RealTime Assay
Nucleic acid extraction, PCR reaction preparation, and amplification for the Abbott assay were performed on the m2000 system using manufacturer provided Investigational Use Only protocols, reagents, and software. Investigational Use Only reagents are used for clinical trials and represent the released product. They are manufactured under Good Manufacturing Practice and with the same quality control as for in vitro diagnostic product (personal communication, Klara Abravaya). The m2000 system extracts nucleic acid from 1.0, 0.5, or 0.6 ml of plasma using magnetic particles as previously described.6,7,8,10 The primers and probe system targeting the integrase region of the HIV-1 genome were recently described.11
Versant HIV-1 RNA 3.0 Assay (bDNA)
The assay was performed as prescribed by the manufacturer, using the System 340 bDNA Analyzer (Siemens Health care Diagnostics, Deerfield, IL).
Workflow Comparisons
Detailed comparisons of assay workflow for Amplicor, RealTime, and Versant have been described previously.12,13
Statistical Analysis
Probit was performed using R software (The R Project for Statistical Computing – http://www.r-project.org). Deming regression14 was calculated using a Microsoft Excel macro written at ARUP.
Results
The limit of detection of the 1 ml RealTime assay was determined using replicates of each of eleven dilutions of second International Standard World Health Organization material ranging between 1.7 to 139.2 IU/ml and VQA standard material ranging between 1 and 80 copies/ml (Table 1). Probit analysis predicts a 95% limit of detection of 21.94 IU/ml (95% confidence interval: 13.28 to 36.26) for World Health Organization material and 26.54 copies/ml (95% confidence interval: 16.20 to 43.47) for VQA material.
Table 1.
Limit of Detection and Probit Analysis
| 2nd International WHO standard |
Viral quality assurance standard |
||||||
|---|---|---|---|---|---|---|---|
| IU/ml | Number tested | Number detected | Percent detected | Copies/ml | Number tested | Number detected | Percent detected |
| 139.2 | 6 | 6 | 100% | 80 | 6 | 6 | 100% |
| 87.0 | 7 | 7 | 100% | 50 | 7 | 7 | 100% |
| 69.6 | 6 | 6 | 100% | 40 | 7 | 7 | 100% |
| 60.9 | 7 | 7 | 100% | 35 | 7 | 7 | 100% |
| 55.7 | 7 | 7 | 100% | 32 | 7 | 7 | 100% |
| 52.2 | 7 | 7 | 100% | 30 | 7 | 6 | 86% |
| 43.5 | 7 | 7 | 100% | 25 | 7 | 7 | 100% |
| 34.8 | 7 | 7 | 100% | 20 | 7 | 7 | 100% |
| 17.4 | 7 | 6 | 86% | 10 | 7 | 4 | 57% |
| 8.7 | 7 | 3 | 43% | 5 | 7 | 3 | 43% |
| 1.7 | 7 | 0 | 0% | 1 | 7 | 1 | 14% |
| Probit Results | Probit Results | ||||||
| 21.94 IU/ml (95% CI: 13.28 to 36.26) | 26.54 copies/ml (95% CI: 16.20 to 43.47) | ||||||
Eleven dilutions of the 2nd International Standard or Viral Quality Assurance standard were prepared in Basematrix Diluent. Six or seven replicates at each concentration were tested on a total of two runs. Probit analysis was performed to determine the limit of detection (95% detection rate) and 95% confidence interval.
The reproducibility and linearity of the RealTime assay were determined by replicate measurements of a dilution series derived from four high-titer patient samples and analyzed by linear regression (Figure 1 and Table 2). The maximum coefficient of variation was 6.7% for intra-assay data and 5.8% for interassay data, both occurring at approximately 2 log10 copies/ml, indicating high precision. Intra-assay slopes ranged from 0.944 ± 0.013 to 1.015 ± 0.007 and interassay slopes ranged from 0.895 ± 0.019 to 1.007 ± 0.013. The intercept ranges were −0.125 ± 0.028 to 0.175 ± 0.051 and −0.071 ± 0.051 to 0.369 ± 0.072 for the intra- and interassay regressions, respectively.
Figure 1.
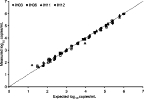
RealTime linearity and reproducibility. Serial dilutions of four high-titer clinical samples (IH03, IH06, IH11, and IH12) were prepared, five replicates each, and tested in the 1 ml RealTime assay. Three replicates were tested on a single run to evaluate intra-assay reproducibility, and the remaining two replicates were tested on a second and third run (and compared with the mean of the intra-assay data) to evaluate interassay reproducibility. The expected values were calculated by setting the concentration with the lowest coefficient of variation at identity, and using the dilution factor to calculate the remaining expected concentrations. Replicates that were below the assay's quantification limit are not shown.
Table 2.
Intra- and Inter-Assay Precision and Linearity
| A | Intra-assay mean ± SEM log10 copies/ml (n = 3, except*) |
|||
|---|---|---|---|---|
| Dilution No. | IH03 | IH06 | IH11 | IH12 |
| 1 | 5.238 ± 0.022 | 5.025 ± 0.024 | 5.832 ± 0.025 | 5.984 ± 0.012 |
| 2 | 4.791 ± 0.014 | 4.467 ± 0.013† | 5.179 ± 0.013 | 5.414 ± 0.009 |
| 3 | 4.153 ± 0.033 | 3.895 ± 0.011 | 4.573 ± 0.014 | 4.819 ± 0.006 |
| 4 | 3.619 ± 0.009 | 3.258 ± 0.085 | 3.840 ± 0.131 | 4.116 ± 0.036 |
| 5 | 3.034 ± 0.040 | 2.748 ± 0.021 | NT‡ | 3.526 ± 0.013 |
| 6 | 2.469 ± 0.048 | 2.057 ± 0.079 | 2.763 ± 0.012 | 2.969 ± 0.022 |
| 7 | 1.841 ± 0.056 (2)* | (0) | 2.142 ± 0.035 | 2.311 ± 0.026 |
| 8 | (0) | NT | 1.602 (1) | 1.724 ± 0.016 (2) |
| B | Inter-assay mean ± SEM log10 copies/ml (n = 3, except*) |
|||
|---|---|---|---|---|
| Dilution No. | IH03 | IH06 | IH11 | IH12 |
| 1 | 5.255 ± 0.051 | 4.927 ± 0.049 | 5.768 ± 0.047 | 5.972 ± 0.009 |
| 2 | 4.684 ± 0.054 | 4.433 ± 0.034 (2) | 5.161 ± 0.023 | 5.369 ± 0.053 |
| 3 | 4.152 ± 0.020 | 3.894 ± 0.008 | 4.576 ± 0.002 | 4.736 ± 0.053 |
| 4 | 3.616 ± 0.006 | 3.272 ± 0.030 | 3.880 ± 0.023 | 4.048 ± 0.036 |
| 5 | 3.017 ± 0.026 | 2.675 ± 0.059 | NT | 3.537 ± 0.010 |
| 6 | 2.471 ± 0.017 | 2.152 ± 0.071 | 2.777 ± 0.014 | 2.934 ± 0.021 |
| 7 | 1.914 ± 0.073 (2) | 1.688 ± 0.045 (2) | 2.200 ± 0.036 | 2.292 ± 0.054 |
| 8 | 1.771 (1) | NT | 1.602 (1) | 1.716 ± 0.008 (2) |
| C | Intra-assay regression analysis |
Inter-assay regression analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Slope | Slope standard error | Intercept | Intercept standard error | Slope | Slope standard error | Intercept | Intercept standard error |
| IH03 | 0.944 | 0.013 | 0.175 | 0.051 | 0.895 | 0.019 | 0.369 | 0.072 |
| IH06 | 0.978 | 0.023 | 0.088 | 0.081 | 0.916 | 0.017 | 0.260 | 0.058 |
| IH11 | 1.009 | 0.020 | −0.054 | 0.080 | 0.987 | 0.011 | 0.037 | 0.043 |
| IH12 | 1.015 | 0.007 | −0.125 | 0.028 | 1.007 | 0.013 | −0.071 | 0.051 |
The (A) intra- and (B) inter-assay precision and (C) linearity were evaluated by fourfold serial dilutions of four high-titer patient samples (IH03, IH06, IH11, and IH12). Five replicates were prepared and tested in the 1 ml RealTime assay. Three replicates were tested on a single run to evaluate intra-assay reproducibility, and the remaining two replicates were tested on a second and third run (and compared to the mean of the intra-assay data) to evaluate inter-assay reproducibility. The expected values were calculated by setting the concentration with the lowest coefficient of variation at identity (values in bold), and using the dilution factor to calculate the remaining expected concentrations. The data assigned to identity in this manner were excluded from the regression analysis.
If the number of replicates that produced quantitative results was less than the number of samples tested, that number is indicated in parentheses.
Bold text indicates values that were set at identity.
Not tested.
Interassay reproducibility was also evaluated by examining the RealTime assay control values. A total of seventeen runs were performed using two lots of reagents, including both the 1 ml and 0.5 ml protocols. Box and whisker plots for the controls are presented in Figure 2A and B. The controls from one run were determined to be “minor outliers” (between 1.5 and 3 times the interquartile range lower than the first quartile).
Figure 2.

RealTime control values. Box-and-whisker plots for the RealTime controls are shown for each run (n = 17) versus their expected values (diamond markers). A: Low positive control = 2.85 log10 copies/ml; and B: High positive control = 4.90 log10 copies/ml). Open circles indicate “mild outliers” (between 1.5 and 3 times the interquartile range lower than the first quartile). The negative control was “Not Detected” in all seventeen runs. The control values from both the 1 ml and 0.5 ml assays are included.
Two sets of subtype samples (one blinded set provided by Abbott, and a panel from NIBSC) were evaluated (Figure 3, A and B) in the 1 ml RealTime, Amplicor, and Versant assays. These panels represent group M, N, and O samples, including some common circulating recombinant forms (CRFs). For most samples, the assays show good agreement, although the limited sample volume did not permit replicate measurements. However, two CRF02_AG samples (Figure 3A), two subtype C samples (Figure 3A), and one subtype B sample (Figure 3B) measured approximately 1.1, 0.8, and 0.6 log10 copies/ml higher in the RealTime assay than in the Amplicor assay, respectively. The RealTime assay was able to quantify all three group O samples, while only one was measured by the Versant assay (approximately 2 log10 lower than RealTime) and none were detected by the Amplicor assay. For the one Group N sample available, the Amplicor assay measured the viral load approximately one log10 higher than the other two assays.
Figure 3.
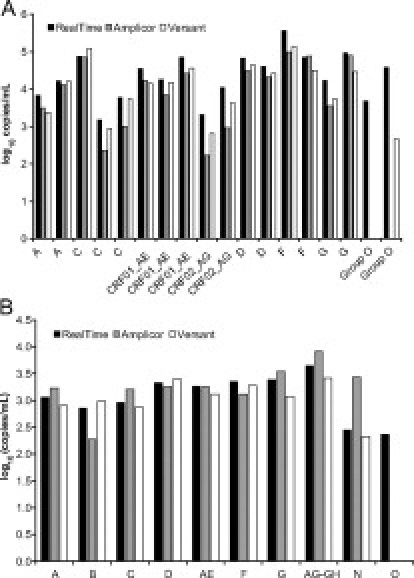
Subtype panels. A: Eighteen diverse HIV-1 plasma samples provided by Abbott were tested in a blinded evaluation of the 1 ml RealTime, Amplicor, and Versant assays. B: Dilutions (approximately 1.7-fold) of the 10 samples in the HIV-1 RNA Genotype Panel for NAT Assays (01/466) (NIBSC) were tested in the 1 ml RealTime, Amplicor, and Versant Assays.
The results of the correlation of the RealTime assay to the Amplicor assay using archived samples are shown in Table 3 and Figure 4, A–C. One hundred seventy-one samples were tested in the 1 ml RealTime assay. One hundred thirty-three 1 ml samples produced valid quantitative results with a Deming regression of RealTime = 0.971 (Amplicor) − 0.601, R2 = 0.849, Sy/x|D = 0.361 (SE of the estimates of y given x). One hundred thirty-five samples were tested in the 0.5 ml RealTime assay. Ninety-eight 0.5 ml samples produced valid quantitative results with a Deming regression of RealTime = 1.021 (Amplicor) − 0.659, R2 = 0.793, Sy/x|D = 0.390. On average, the Amplicor assay results were 0.716 log10 copies/ml higher than the 1 ml RealTime assay and 0.570 log10 copies/ml higher than the 0.5 ml RealTime assay. Thirty percent of the 1 ml samples and 40% of the 0.5 ml samples differed by less than 0.5 log10 copies/ml. Seventy-seven percent of the 1 ml samples and 85% of the 0.5 ml samples differed by less than 1.0 log10 copies/ml.
Table 3.
Correlation Sample RealTime Results Summary
| Assay input volume |
|||
|---|---|---|---|
| 1 ml | 0.5 ml | 0.6 ml | |
| Tested | 171 | 135 | 180 |
| Delayed IC cycle number | 14 | 2 | 2 |
| Repeated | 3 | 0 | 0 |
| Detected but not quantified | 19 | 25 | 13 |
| Not detected | 8 | 10 | 10 |
| Total for Deming regression | 133 | 98 | 155 |
The results of testing samples previously assayed using Amplicor are shown. One hundred seventy-one (171), 135, and 180 samples were tested in the 1.0 ml, 0.5 ml, and 0.6 ml RealTime assays, respectively. Samples with delayed IC cycle numbers produced errors that prevented results from being calculated; a small number of samples had sufficient volume to permit repeat testing. Samples that were “Detected but not quantified” or “Not detected” were not retested due to insufficient volume.
Figure 4.

Correlation samples. Samples previously tested in the Amplicor assay were tested in the RealTime assay. A: 171 samples were tested in the 1 ml RealTime assay; B: 135 samples were tested in the 0.5 ml RealTime assay; and C: 180 samples were tested in the 0.6 ml assay (see Discussion). Left panels: The data were analyzed by Deming regression, shown by the solid line. The dashed line represents the line of identity. Right panels: The data were analyzed by the method of Bland and Altman. The bold lines indicate the average difference and average ± 2 standard deviations. A: RealTime = 0.971 (Amplicor) − 0.601, R2 = 0.849, Sy/x|D = 0.361, Average Amplicor − RealTime log10 copies/ml = 0.716, n = 133. B: RealTime = 1.021 (Amplicor) − 0.659, R2 = 0.793, Sy/x | D = 0.390, Average Amplicor − RealTime log10 copies/ml = 0.570, n = 98. C: RealTime = 1.061 (Amplicor) − 0.589, R2 = 0.910, Sy/x | D = 0.329, Average Amplicor − RealTime log10 copies/ml = 0.342, n = 155.
Due to the larger than expected differences observed in the 1 ml and 0.5 ml assays, we performed follow-up testing using the more recently approved 0.6 ml assay (see Discussion). In this version of the assay 180 samples were tested, 155 of which produced valid quantitative results with a Deming regression of RealTime = 1.061 (Amplicor) − 0.589, R2 = 0.910, Sy/x|D = 0.329. On average, the Amplicor results were 0.342 log10 copies/ml higher than the 0.6 ml RealTime assay. Seventy-one percent of the 0.6 ml samples differed by less than 0.5 log10 copies/ml; 95% differed by less than 1.0 log10 copies/ml.
Forty-four samples that had previously given results below the limit of detection in the UltraSensitive Amplicor assay (<50 copies/ml) were retested with the RealTime assay (Table 4). One sample was flagged with an error indicating it had an unacceptably high internal control cycle number. Twenty-nine samples had no detectable HIV-1 RNA. Twelve samples were detected, but were below the quantification limit of the assay (40 copies/ml). Two samples were quantified, one of which measured 47 copies/ml; the other sample measured 83 copies/ml. The first sample had insufficient volume for retesting; the second sample was retested using Amplicor, which also produced a result of 83 copies/ml.
Table 4.
RealTime Results for Samples Reported as “<50 copies/ml” Using Amplicor
| RealTime result | Samples |
|---|---|
| Delayed IC cycle number | 1 |
| Not detected | 29 |
| Detected but not quantified | 12 |
| Quantified | 2 |
| Total tested | 44 |
The 1 ml RealTime results for 44 samples previously assayed below the limit of detection (<50 copies/ml) with UltraSensitive Amplicor are shown. Of the “Quantified” samples, one sample produced a result of 47 copies/ml; the other produced a result of 83 copies/ml. When this latter sample was retested in Amplicor, the Amplicor assay also measured a viral load of 83 copies/ml.
Discussion
Viral load testing has long been the standard of care for HIV-infected patients. Accurate measurements are critical for determining disease prognosis and guiding the course of treatment. Failure to accurately quantify HIV viral load can lead to inappropriate clinical management of patients and unnecessary virus transmission.15
We evaluated several key performance characteristics of the newly Food and Drug Administration-approved RealTime assay. Overall, the RealTime assay's performance was very good.
The limit of detection was determined by probit analysis of diluted VQA and World Health Organization standard replicate measurements. Using VQA standard, the limit of detection is 26.54 copies/ml (95% CI: 16.20 to 43.47), which is in agreement with prior reports.9 Using the second International Standard (World Health Organization), the limit of detection is 21.94 IU/ml (95% CI: 13.28 to 36.26). Since the RealTime assay is calibrated to the first International Standard (NIBSC 97/656), which is no longer available, the copies:IU conversion factor (1.74 copies/IU) stated by Abbott may not apply. Although the RealTime assay is not approved for diagnosis of HIV infection, its limit of detection is similar to the Aptima HIV-1 RNA Qualitative assay (Gen-probe, San Diego, CA), which was recently approved for this use.
The RealTime assay exhibited excellent reproducibility and linearity between ∼1.6 to 6.0 log10 copies/ml using dilutions of four clinical samples. Due to the lack of very high titer samples, we were unable to verify performance above approximately 6 log10 copies/ml, although the assay specifications indicate linearity up to 7 log10 copies/ml. The most variability (both intra- and interassay) was present near the lower limit of the assay (approximately 2 log10 copies/ml). Because the initial dilution of four patient samples gave the expected results and further dilutions remained linear, it is unlikely that the Basematrix diluent had a significant effect on the limit of detection, linearity, or reproducibility experiments. The assay-provided controls demonstrate high reproducibility, as variability was low, and no significant bias, as observed and expected values were similar. There was one run that produced mild outliers for both controls. This run also had the highest average internal control cycle numbers of all of the runs (data not shown), indicating mild inhibition. Since the internal control values for this run were within the range required by the assay and no errors were generated, we did not exclude these samples from analysis, since the typical user of the assay would not be aware that any anomaly had occurred.
With the exception of two CRF02_AG, two subtype C, and one subtype B sample, all three assays (Amplicor, RealTime, and Versant) quantified the group M subtypes equivalently, although due to the limited amount of sample available, we were unable to test replicates of the genotype sample specimens. Other investigators have reported discrepancies between assays for HIV subtypes and CRFs, especially CRF02_AG.4,16,17,18 At most, there was an approximate 1 log10 copy/ml difference between the Amplicor and RealTime assays in our evaluation of CRF02_AG samples. The RealTime assay is the only assay approved by the Food and Drug Administration with a claim to detect group O samples, which was verified on three samples in this study. The Versant assay detected one of the three group O samples, but the viral load was approximately 2 log10 copies/ml lower than RealTime. The Amplicor assay did not detect any group O samples.
Clinical samples tested in both assays had correlation coefficients of 0.793 for the 0.5 ml, 0.849 for the 1 ml, and 0.910 for the 0.6 ml RealTime assay. The most noticeable difference between the two assays was the high discrepancy between the average values obtained. The RealTime assay measured 0.716 log10 copies/ml, 0.570 log10 copies/ml, and 0.342 log10 copies/ml lower than Amplicor for the 1 ml, 0.5 ml, and 0.6 ml RealTime assay, respectively. This discrepancy was consistent across viral load levels.
Prior comparisons of the Amplicor and RealTime assays have shown average differences (Amplicor − RealTime) of −0.4042, −0.154, −0.148, 0.145, and 0.237. While our correlation results show a greater difference between the Amplicor and RealTime assays than in the published literature, our results with diverse genotype specimens do not show the same trend. The average difference (Amplicor − RealTime) for the 25 samples in the genotype panels that produced results for both assays was −0.25 log10 copies/ml. Despite analysis of operator, site, instrument, sample handling and storage, and calibration effects (data not shown), no compelling differences were observed between the assay configurations or between the correlation and genotype panel samples. Repeat Amplicor testing of nine samples (∼2.2 to 5.0 log10 copies/ml) after RealTime indicates that the additional freeze/thaw cycle is not the cause of the discrepancy (data not shown − maximum difference of 0.5 log10 copies/ml). Additionally, all Amplicor controls were within the parameters for clinical laboratory testing. However, the possibility of minor variation in the Amplicor assay cannot be ruled out. Although we did not make direct comparisons of different reagent lots, the positive control results (Figure 2) do not indicate appreciable lot to lot variability. In addition, any lot variability should be compensated for by reagent calibration. Although the subtypes of the correlation samples are unknown, they likely consist mainly of subtype B viruses, since they were derived from a pool of samples submitted from across the United States (unpublished observations at ARUP, and Osmanov et al19).
The results of testing samples that were below the limit of detection of the Amplicor assay (<50 copies/ml) are in agreement with the specifications of the RealTime assay and our probit analysis. However, these results disagree with the large negative bias observed in the correlation sample set. Assuming the average 0.716 log10 copies/ml lower results for RealTime in the correlation samples applies at levels below 50 copies/ml, fewer positive samples should have been detected in this experiment.
The RealTime assay targets the integrase region of HIV and uses a novel probe system and low detection temperature,9 which increase the ability to tolerate mismatches and therefore detect a wide variety of subtypes. Recently, the first integrase inhibitor therapy, raltegravir (Isentress, Merck & Co., Inc., Whitehouse Station, NJ), was Food and Drug Administration-approved for treatment-experienced patients. Analysis of integrase inhibitor-naïve patients by Lataillade et al 20 showed that 64% of the integrase amino acid positions were polymorphic, although mutations within the active sites and catalytic domain were infrequent. The analysis of 243 integrase sequences by Lataillade et al, revealed that the RealTime HIV-1 primer and probe sites are highly conserved. The frequency of observed polymorphisms was 0.2%, 0.6%, and 1.2% respectively, for the forward primer, probe, and reverse primer, respectively.11 One of the polymorphic positions in the forward primer (Q148) is associated with integrase inhibitor resistance. A systematic analysis using engineered transcripts containing all possible resistance-associated substitutions at codon 148 revealed no discernible impact on assay performance. Additionally, analysis of clinical specimens from patients with integrase inhibitor resistance mutations did not impact results with the Abbott RealTime assay (personal communication, J. Hackett, Jr.).
To summarize, the automated RealTime HIV-1 assay exhibits high sensitivity, wide dynamic range, high reproducibility, and the unique ability to reliably detect and quantify group O samples. With archived samples, the RealTime assay correlated well with Amplicor, but consistently yielded lower values. Despite additional testing, the reason for the discrepancy remains unknown. This difference may necessitate determining new baselines for patients when changing viral load assays.
The Centers for Disease Control and Prevention recently recommended that all patients 13 to 64 years of age in any health care setting be screened for infection by HIV.21 As screening measures, the genetic diversity in worldwide19 and United States22 infections, and global travel and immigration all increase, it is safe to assume that tools that can monitor diverse HIV-1 infections will be increasingly important.
Acknowledgements
This study was performed in compliance with regulations concerning human subject research and was approved by the University of Utah Institutional Review Board. We thank Derek Vanhille, Mamak Vedadi, and Denise Jones for preparing samples, the staff of the Molecular Hepatitis/Retrovirus Laboratory at ARUP for performing the Amplicor and Versant testing, and Andrew Wilson for assistance with statistics.
Footnotes
Supported by Abbott Molecular: all components for Abbott RealTime HIV-1 Assay testing, including the use of the m2000sp and m2000rt instruments, disposables, RealTime HIV-1 Assay reagents, genotype panels, WHO reference standards, and additional Amplicor and Versant testing were provided.
D.R.H. has received honoraria from Roche Diagnostics and Abbott Molecular. M.T.P. has received honoraria from Roche Diagnostics.
Current addresses of E.Q.K.: University of Utah School of Medicine, 30 N 1900 E, Salt Lake City, UT 84132; and A.P.: Cytel Inc., 675 Massachusetts Ave., Cambridge, MA 02139
References
- 1.Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, Respess R, Stevens W, the Forum for Collaborative HIV Research Alternative Viral Load Assay Working Group HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun P, Ehret R, Wiesmann F, Zabbai F, Knickmann M, Kühn R, Thamm S, Warnat G, Knechten H. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin Chem Lab Med. 2007;45:92–99. doi: 10.1515/CCLM.2007.008. [DOI] [PubMed] [Google Scholar]
- 3.Rouet F, Rouzioux C. The measurement of HIV-1 viral load in resource-limited settings: how and where? Clin Lab. 2007;53:135–148. [PubMed] [Google Scholar]
- 4.Gueudin M, Plantier JC, Lemée V, Schmitt MP, Chartier L, Bourlet T, Ruffault A, Damond F, Vray M, Simon F. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J Acquir Immune Defic Syndr. 2007;44:500–505. doi: 10.1097/QAI.0b013e31803260df. [DOI] [PubMed] [Google Scholar]
- 5.Schutten M, Peters D, Back NKT, Beld M, Beuselinck K, Foulongne V, Geretti A-M, Pandiani L, Tiemann C, Niesters HGM. Multicenter evaluation of the new Abbott RealTime assays for quantitative detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J Clin Microbiol. 2007;45:1712–1717. doi: 10.1128/JCM.02385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson P, Huang S, Holzmayer V, Bodelle P, Yamaguchi J, Brennan C, Badaro R, Brites C, Abravaya K, Devare SG, Hackett J., Jr Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from Brazil. J Virol Meth. 2006;134:237–243. doi: 10.1016/j.jviromet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Swanson P, Holzmayer V, Huang S, Hay P, Adebiyi A, Rice P, Abravaya K, Thamm S, Devare SG, Hackett J., Jr Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from London: comparison to VERSANT HIV-1 RNA 3.0, AMPLICOR HIV-1 MONITOR v1.5, and LCx HIV RNA Quantitative assays. J Virol Meth. 2006;137:184–192. doi: 10.1016/j.jviromet.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Swanson P, Huang S, Abravaya K, de Mendoza C, Soriano V, Devare SG, Hackett J., Jr Evaluation of performance across the dynamic range of the Abbott RealTime HIV-1 assay as compared to VERSANT HIV-1 RNA 3.0 and AMPLICOR HIV-1 MONITOR v1.5 using serial dilutions of 39 group M and O viruses. J Virol Meth. 2007;141:49–57. doi: 10.1016/j.jviromet.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 9.Tang N, Huang S, Salituro J, Mak W-B, Cloherty G, Johanson J, Li YH, Schneider G, Robinson J, Hackett J, Jr, Swanson P, Abravaya K. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, froup O and group N samples. J Virol Meth. 2007;146:236–245. doi: 10.1016/j.jviromet.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Halfon P, Bourlière M, Pénaranda G, Khiri H, Ouzan D. Real-time PCR assays for hepatitis C virus (HCV) RNA quantitation are adequate for clinical management of patients with chronic HCV infection. J Clin Microbiol. 2006;44:2507–2511. doi: 10.1128/JCM.00163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S, Salituro J, Tang N, Luk K-C, Hackett J, Jr, Swanson P, Cloherty G, Mak W-B, Robinson J, Abravaya K. Thermodynamically modulated partially double-stranded linear DNA probe design for homogenous real-time PCR. Nucleic Acids Res. 2007;35:e101. doi: 10.1093/nar/gkm551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun P, Ehret R, Wiesmann F, Zabbai F, Knickmann M, Kühn R, Thamm S, Warnat G, Knechten H. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin Chem Lab Med. 2007;45:93–99. doi: 10.1515/CCLM.2007.008. [DOI] [PubMed] [Google Scholar]
- 13.Wolff D, Gerritzen A. Comparison of the Roche COBAS Amplicor™ Monitor. Roche COBAS Ampliprep™/COBAS Taqman™ and Abbott RealTime™ Test assays for quantification of hepatitis C virus and HIV RNA. Clin Chem Lab Med. 2007;45:917–922. doi: 10.1515/CCLM.2007.149. [DOI] [PubMed] [Google Scholar]
- 14.Martin RF. General Deming regression for estimating systematic bias and its confidence interval in method-comparison studies. Clin Chem. 2000;46:100–104. [PubMed] [Google Scholar]
- 15.Geelen S, Lange J, Borleffs J, Wolfs T, Weersink A, Schuurman R. Failure to detect a non-B HIV-1 subtype by the HIV-1 Amplicor Monitor test, version 1.5: a case of unexpected vertical transmission. AIDS. 2003;17:781–782. doi: 10.1097/00002030-200303280-00027. [DOI] [PubMed] [Google Scholar]
- 16.Colson P, Solas C, Moreau J, Motte A, Henry M, Tamalet C. Impaired quantification of plasma HIV-1 RNA with a commercialized real-time PCR assay in a couple of HIV-1-infected individuals. J Clin Virol. 2007;39:226–229. doi: 10.1016/j.jcv.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Amendola A, Bordi L, Angeletti C, Girardi E, Ippolito G, Capobianchi MR. Comparison of LCx with other current viral load assays for detecting and quantifying human immunodeficiency virus type 1 RNA in patients infected with the circulating recombinant form A/G (CRF02) J Clin Microbiol. 2004;42:811–815. doi: 10.1128/JCM.42.2.811-815.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foulongne V, Montes B, Didelot-Rousseau M-N, Segondy M. Comparison of the LCx human immunodeficiency virus (HIV) RNA quantitative, RealTime HIV, and COBAS Ampliprep-COBAS TaqMan assays for quantitation of HIV Type 1 RNA in plasma. J Clin Microbiol. 2006;44:2963–2966. doi: 10.1128/JCM.00341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osmanov S, Pattou C, Waler N, Schwardländer B, Esparza J, the WHO-UNAIDS Network for HIV Isolation and Characterization Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29:184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- 20.Lataillade M, Chiarella J, Kozal MJ. Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir Ther. 2007;12:563–570. [PubMed] [Google Scholar]
- 21.Armstrong WS, Taege AJ. HIV screening for all: the new standard of care. Cleve Clin J Med. 2007;74:297–301. doi: 10.3949/ccjm.74.4.297. [DOI] [PubMed] [Google Scholar]
- 22.Lin H-H, Gaschen BK, Collie M, El-Fishaway M, Chen Z, Korber BT, Beatrice ST, Zhang L. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquir Immune Defic Syndr. 2006;41:399–404. doi: 10.1097/01.qai.0000200663.47838.f1. [DOI] [PubMed] [Google Scholar]


