Abstract
Stem-cell transplant recipients are at risk of developing ganciclovir-resistant human cytomegalovirus (HCMV) infection caused by mutations in the viral UL97 gene. Knowledge of the relative proportions of coexisting HCMV wild-type and mutant strains may contribute to a better understanding of the dynamics of in vivo mutant strain selection under ganciclovir. Currently, genotype resistance screening for UL97 is routinely performed by restriction fragment length polymorphism detection and sequencing. We present here the longitudinal course of a pediatric recipient of an allogeneic stem-cell transplant infected with a ganciclovir-resistant HCMV strain. EDTA-treated blood samples were analyzed longitudinally. The patient acquired a primary HCMV infection shortly before transplantation and reactivated the virus following allogeneic hematopoietic stem cell transplantation, thus receiving an intensive antiviral treatment schedule. Three different methods for UL97 mutation analysis, restriction fragment length polymorphism detection, sequencing, and a new, real-time PCR approach were performed. In conclusion, for our pediatric patient, during peak viral load, the UL97 wild-type strain predominates, while during clinical deterioration with low viral load, the predominant mutant strain persists.
The emergence of an human cytomegalovirus (HCMV) infection needing ganciclovir (GCV) therapy in stem-cell transplant recipients is a severe complication. The incidence of resistance to GCV in pediatric stem-cell transplant recipients was roughly estimated at 3.8%.1 Early and rapid detection of GCV resistance in human pediatric and adult hematopoietic stem-cell transplantation (HSCT) as well as in solid organ-transplantation is very important for determining therapeutic course.2,3,4
GCV resistance is caused by mutations in the UL97 and the UL54 genes.2 Most clinical GCV-resistant isolates have clustered mutations in the UL97 gene, especially in the region between codons 400 and 665.5,6 The determination of relative proportions of wild-type and mutant strains out of mixed viral populations provides important information with regard to therapeutic options.1,7 Phenotypic and genotypic assays are used for the diagnosis of GCV-resistant HCMV infections. For a culture-based phenotypic assay a viral isolate is necessary. Even rapid modifications using cell-associated virus strains in an HCMV immediate early plaque reduction assay8 are very time-consuming and laborious, as compared with genotypic methods. In contrast, both sequencing and PCR-based restriction fragment length polymorphism (RFLP) assays are suitable to detect mutations in the HCMV phosphotransferase gene (UL97) associated with GCV resistance. Mutations in the polymerase-gene (UL54), which contains no clustered regions for mutations involved in drug resistance for cidofovir and foscarnet, are detectable only by sequencing.
Here, we present a case of an HCMV-infected infant after allogeneic stem cell-transplantation developing the UL97-mutation C603W, which conferred GCV resistance with fatal outcome. To screen the dynamics of the development of GCV resistance, three genotypic screening methods for the detection of this mutation were compared: RFLP assay, sequencing analysis, and our newly established LightCycler real-time PCR approach.9,10
Case Report
We report a 4-month-old girl with hemophagocytic lymphohistiocytosis who received a HSCT from a 10-out-of-10 human leukocyte antigen-matched unrelated female donor. The girl was in complete remission before HSCT and received a myeloablative conditioning regimen with busulfan, etoposide phosphate (VP16), cyclophosphamide, and anti-thymocyte globulin. Bone marrow was infused at day 0, containing 6.52 × 106 CD34+ stem cells per kg body weight and 3.1× 106 CD3+ T-cells per kg body weight. Both the donor and the recipient were initially HCMV IgG negative before HSCT. The patient acquired an HCMV primary infection 3 months before HSCT with HCMV-DNA present in throat swabs, blood, and urine. The HCMV infection was treated with GCV and valganciclovir and had a course over more than 2 months. During the weeks before HSCT, HCMV-PCR became negative and anti-HCMV IgM re-seroconversion was detectable at the beginning of the conditioning regimen.
Antiviral prophylaxis (Figure 1) was given as aciclovir day −9 (before HSCT) to day 2 (post HSCT), ganciclovir day −8 to −1, ribavirin day −8 to day 31 and forcarnet from day 2 to day 26. HCMV reactivation was detectable in peripheral blood from day 11. Reactivation of HCMV was initially treated with foscarnet and ganciclovir until PCR in peripheral blood became negative on day 26 and than continued with ganciclovir alone. On the increase of the viral load after day 60, treatment was switched to an outpatient regimen with foscarnet once daily and valganciclovir. After detection of GCV resistance, treatment was continued with foscarnet three times per day (Figure 1).
Figure 1.
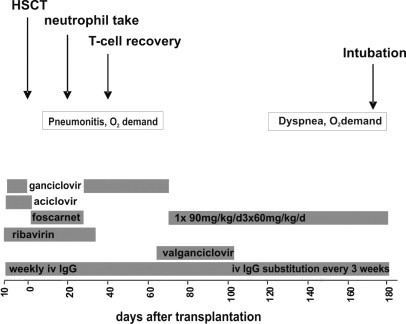
Clinical course and antiviral treatment of a 4-month old girl with hemophagocytic lymphohistiocytosis after allogeneic stem-cell transplantation from a HLA matched unrelated donor. The patient acquired a HCMV primary infection shortly before and reactivated 60 days after HSCT, thus receiving an intensive antiviral treatment schedule. The gray bars represent the period of antiviral treatment.
From day 10 after HSCT the girl had continuous O2 demand over nasal cannula. Cardiac function was normal and pulmonary functions test revealed an impaired diffusion capacity with no airway obstruction. CT scans on the lungs showed a slight infiltrate. Therefore differential diagnosis of the ongoing O2 demand was the HCMV infection and a microangiopathy of the lungs due to the underlying hemophagocytic lymphohistiocytosis. After day 120 post-HSCT, hospital admission was necessary because HCMV load increased and pulmonary condition deteriorated, resulting in tracheal intubation and mechanical assisted ventilation in the intensive care unit. Finally the patient died with delayed pulmonary toxicity syndrome at day 247 post HSCT.
Materials and Methods
Quantitative HCMV-PCR
For detection of HCMV viral load we performed the COBAS Amplicor CMV Test (Roche Diagnostics, Mannheim, Germany) from longitudinally collected plasma of the patient. For confirmation of peak viral load data, we performed three replicates of each time point, if enough plasma was available.
UL97-RFLP
The RFLP analysis for codon 603 was described previously.10 The AvaII (G′GWCC) restriction pattern for the wild-type C603 revealed fragments of 173 bp and 16 bp while the mutant strain C603W was digested into three fragments of 104 bp, 69 bp, and 16 bp.
Sequencing Analysis
Sequencing analysis was performed by PCR with primers 595F/R, cleaned up with the Qiagen PCR Purification Kit (Qiagen, Hilden, Germany) as described previously.3
UL97 Real-Time PCR
The LightCycler PCR-assay for detection of mutations in codon 603 was described recently.10 The real-time-PCR-assay for characterization of mutations in codons 603 and 607 was established using a dual color format with two pairs of hybridization-probes labeled each with two different fluorescent dyes.10 The detection of UL97-mutations was achieved by melting-curve analysis following PCR. For the detection of mutations by 1. RFLP analysis in codon 603 the amplicon size was 201 bp using primers 603/607fw and 603/607rev.10
Longitudinal DNA specimens from the pediatric HSCT recipient with proven GCV resistance (days 61, 97, 111, 119, 130, and 137 post HSCT) containing the mutation C603W were analyzed using our LightCycler-PCR for codon 603/607 and compared with RFLP results and sequencing.
Results and Discussion
At day 61 post HSCT (P9648), an exclusively wild-type sequence was present. RFLP and LightCycler analysis showed the expected restriction patterns and the melting-point corresponded to the wild-type (Figure 2C and F). The viral load at this time point of infection was 3210 copies/ml (Figure 3A). On day 97 post HSCT (+97) the RFLP results showed the expected restriction pattern of the AvaII digest for codon 603 during increase of plasma viral load. The wild-type control was cut into two fragments with 173 bp and 16 bp. The sample containing the mixture of wild-type and C603W mutation showed the specific restriction pattern for both strains with fragments of 173 bp, 104 bp, 69 bp, and 16 bp, and a proportion of 90% wild-type versus 10% mutant strain (Figure 2F). These results were in very good concordance to the results of the LightCycler assay. The specific melting point of the wild-type was 66.2°C while a mutation in codon 603 reduced the melting-point to 55.3°C (Figure 2C). The viral load on day +97 increased to 25,800 copies/ml or about 23,220 copies/ml for wild-type, and 2,580 copies/ml for the mutant strain, respectively (Figure 3A). On day +111 the ratio of wild-type to mutant strain changed to 60% wild-type versus 40% mutant strain in RFLP as well as in LightCycler analysis (Figure 2, C and F). At this time the total viral load had a peak value of 95,800 copies/ml (Figure 3A). Regarding the proportion of 60% wild-type versus 40% mutant strain resulting from real-time PCR the viral load for the individual strains was splitting to 57,480 copies/ml for wild-type, and 38,320 copies/ml for the mutant strain (Figure 3A). One week later, at day +119 the ratio remained unaltered with 60% wild-type versus 40% mutant strain (Figure 2, A and D), but with a decreasing total viral load of 59,800 copies/ml (Figure 3A). A viral isolate from urine on day +119 showed also phenotypic GCV resistance with an IC50 value of 17.0 μmol/L (cut of for GCV resistance: >6 μmol/L) (Figure 3A). On day +120 the pulmonary situation worsened and the patient was intubated. On day +130 plasma and leukocytes were analyzed again (P697). The LightCycler showed a ratio of 10% wild-type versus 90% of predominant mutant strain, while RFLP and sequencing detected only mutant strain with 100% (Figure 2, B and E). The viral load at this time point of infection was 14,000 copies/ml with a predominant viral load of the mutant strain with 12,600 copies/ml (Figure 3A). On day +137 EDTA blood sample (H47) showed again with all three methods a mixture of wild-type and mutant C603W strains. In the LightCycler-assay the ratio could be given with 55% wild-type versus 45% mutant strain confirmed by RFLP analysis (Figure 2, B and E). The viral load decreased to 3,440 copies/ml whereas the viral load of the wild-type strain could be given with 1,548 copies/ml while the mutant-strain viral load was 1,892 copies/ml (Figure 3A). On day +189 only the mutant strain could be detected in RFLP and LightCycler analysis. The viral load at this time point of infection was 3,920 copies/ml (Figure 3A). The corresponding UL97-sequences for the wild-type strain AD169 and the patient strains H12, P697, and H47 showed a TGC sequence for the wild-type 603 codon and TGG sequence for the recombinant plasmid pC603W10 with different ratios at the different time points of infection (Figure 3B).
Figure 2.
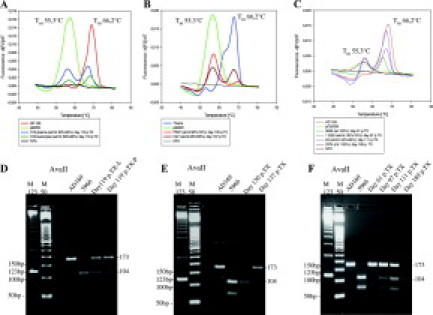
Comparison of UL97 mutation analysis by LightCycler PCR (A-C) and RFLP assay (D-F) of sequential DNA-specimens from blood of a pediatric HSCT recipient with GCV-resistance based on UL97 mutation C603W. Specific melting point analysis and restriction patterns of the investigated patient DNA in comparison with the laboratory wild-type strain AD 169 and a clinical reference strain (5966) or plasmid (pC603W) containing the mutation C603W are shown.
Figure 3.
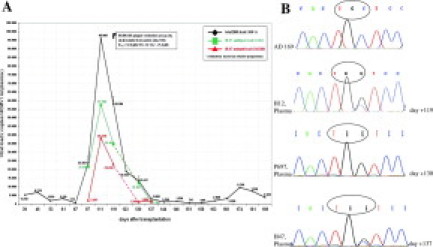
A: Estimations of sequential DNA viral load data from a pediatric HSCT recipient. Total viral loads are given as well as the deduced viral load ratios of wild-type and mutant strain resulting from a semiquantitative real-time mutation analysis. B: Sequencing analysis of longitudinal blood samples (H12, P697, and H47) with the UL97 mutation C603W are shown in comparison with the laboratory wild-type strain AD169.
In conclusion, the results of our new LightCycler PCR assay and the RFLP analysis for the detection of the mutation C603W are in very good concordance. Using melting-point analysis we can also generate semiquantitative results of the wild-type/mutant ratios in mixed viral strains with both methods. In conjunction with the viral load, information of the relative amount of mutant virus can be given, which may be of interest for considerations of therapy switch. Using sequencing analysis alone, the detection of different ratios of wild-type/mutant strain was unreliable. In comparison with RFLP analysis and sequencing (Figures 2D-F and 3B), the real-time UL97-mutation analysis allowed a very rapid detection. With RFLP analysis and direct sequencing it was not always possible to detect small amounts of mutant strains in mixed viral populations, but the real-time PCR described here enabled a sensitive detection of nondominant ex vivo mutant strains.1,11,12 The manifestation of GCV resistance was associated with increasing plasma viral load and preceded pulmonary deterioration immediately before, leading to a fatal outcome. The relative proportion of C603 wild-type strain to C603W mutant strain was >1 during peak viral load. Surprisingly, the C603W mutant strain was selected after day +120 when the total viral load was already decreasing and was predominant only in low plasma viral loads suggesting an impaired CD8+ T-cell control of HCMV infection.
In conclusion, we present the in vivo dynamics of the emergence of a GCV-resistant HCMV mutant in a pediatric stem-cell transplant recipient using a new real-time PCR approach for UL97 mutation analysis.
Footnotes
K.G. and T.F. contributed equally to this work.
References
- 1.Eckle T, Jahn G, Hamprecht K. The influence of mixed HCMV UL97 wild-type and mutant strains on ganciclovir susceptibility in a cell associated plaque reduction assay. J Clin Virol. 2003;30:50–56. doi: 10.1016/j.jcv.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti F, Lurain N, Gerna G. Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs. Hum Immunol. 2004;65:403–409. doi: 10.1016/j.humimm.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Eckle T, Prix L, Jahn G, Klingebiel T, Hangretinger R, Selle B, Hamprecht K. Drug-resistant human cytomegalovirus infection in children after allogeneic stem cell transplantation may have different clinical outcomes. Blood. 2000;96:3286–3289. [PubMed] [Google Scholar]
- 4.Limaye AP. Antiviral resistance in cytomegalovirus: an emerging problem in organ transplant recipients. Sem Respir Infect. 2002;17:265–273. doi: 10.1053/srin.2002.36447. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert C, Bestman-Smith J, Boivin G. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanism. Drug Res Update. 2002;5:88–114. doi: 10.1016/s1368-7646(02)00021-3. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert C, Boivin G. Human cytomegalovirus resistance to antiviral drugs. Antimicrob Agents Chemother. 2005;49:873–883. doi: 10.1128/AAC.49.3.873-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voigt S, Michel D, Kershaw O, Kühl JS, Mertens T, Ebell W, Meisel H. Fatal reactivation of postnatal cytomegalovirus infection with rapid emergence of ganciclovir resistance in an infant after allogeneic stem cell transplantation. J Clin Microbiol. 2005;43:3551–3554. doi: 10.1128/JCM.43.7.3551-3554.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prix L, Maierl J, Jahn G, Hamprecht K. A simplified assay for screening of drug resistance of cell-associated cytomegalovirus strains. J Clin Virol. 1998;24:29–37. doi: 10.1016/s0928-0197(98)00043-9. [DOI] [PubMed] [Google Scholar]
- 9.Göhring K, Mikeler E, Jahn G, Hamprecht K. Rapid simultaneous detection by real-time PCR of cytomegalovirus UL97 mutations in codons 460 and 520 conferring ganciclovir resistance. J Clin Microbiol. 2006;44:4541–4544. doi: 10.1128/JCM.01141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Göhring K, Mikeler E, Jahn G, Hamprecht K. Rapid semiquantitative real-time PCR approach for the detection of HCMV UL97 mutations conferring GCV resistance. Antivir Ther. 2008;13:461–466. [PubMed] [Google Scholar]
- 11.Prix L, Hamprecht K, Holzhüter B, Hangretinger R, Klingebiel T, Jahn G. Comprehensive restriction analysis of UL97 region allows early detection of ganciclovir-resistant human cytomegalovirus in an immunocompromised child. J Infect Dis. 1999;180:491–495. doi: 10.1086/314877. [DOI] [PubMed] [Google Scholar]
- 12.Baldanti F, Underwood MR, Talarico CL, Simonici L, Sarasini A, Biron KK, Gerna G. The Cys 607→ Thyr change in the UL97 phosphotransferase confers ganciclovir resistance to two human cytomegalovirus strains recovered from two immunocompromised patients. Antimicrob Agents Chemother. 1998;42:444–446. doi: 10.1128/aac.42.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]


