Abstract
The Komeda miniature rat Ishikawa (KMI) is a spontaneous animal model of dwarfism caused by a mutation in Prkg2, which encodes cGMP-dependent protein kinase type II (cGKII). This strain has been maintained as a segregating inbred strain for the mutated allele mri. In this study, we characterized the phenotype of the KMI strain, particularly growth traits, craniofacial measurements, and organ weights. The homozygous mutant (mri/mri) animals were approximately 70% to 80% of the size of normal, heterozygous (mri/+) animals in regard to body length, weight, and naso-occipital length of the calvarium, and the retroperitoneal fat of mri/mri rats was reduced greatly. In addition, among progeny of the (BN×KMI-mri/mri)F1×KMI-mri/mri backcross, animals with the KMI phenotype (mri/mri) were easily distinguished from those showing the wild-type phenotype (mri/+) by using growth traits such as body length and weight. Genetic analysis revealed that all of the backcrossed progeny exhibiting the KMI phenotype were homozygous for the KMI allele in the 1.2-cM region between D14Rat5 and D14Rat80 on chromosome 14, suggesting strongly that mri acts in a completely recessive manner. The KMI strain is the first and only rat model with a confirmed mutation in Prkg2 and is a valuable model for studying dwarfism and longitudinal growth traits in humans and for functional studies of cGKII.
Abbreviations: cGKII, cGMP-dependent protein kinase type II; CNP, C-type natriuretic peptide; KMI, Komeda miniature rat Ishikawa
Dwarfism is caused by both endocrinologic and nonendocrinologic defects. Most instances of dwarfism, including normal variants, are nonendocrinologic, and subjects retain growth hormone secretion. Although spontaneous rodent models of dwarfism with confirmed mutations have been reported—Snell dwarf mice with Pou1f1 (Pit1) mutation,14 Ames dwarf mice with Prop1 mutation,22 little mice with Ghrhr mutation,15 pygmy mice (also known as mini-mice) with Hmga2 (HMGI-C) mutation,26 spontaneous dwarf rats with Gh mutation,23 and rdw rats with Tg mutation9,11—most of these are models of endocrinologic dwarfism. A few models of nonendocrinologic dwarfism have been produced by gene manipulation techniques, such as transgenic and knockout strategies, and include Col2a1-transgenic mice,7,24 Col10a1-transgenic mice,10 and Fgfr3-knock-in mice.13
A novel spontaneous dwarf mutation, miniature rat Ishikawa (mri), was discovered in a closed colony of Wistar rats at Ishikawa Animal Laboratory (Saitama, Japan) and has been maintained on the genetic background of Wistar rats. This mutant strain, previously termed Miniature Rat Ishikawa (MRI), has recently been established as a segregating inbred strain on the Wistar genetic background, designated Komeda Miniature rat Ishikawa (KMI). The breeding record suggested that the mutation was inherited in an autosomal recessive mode. KMI rats show no abnormality in the basal amounts or distribution of several hormones, including growth hormone, luteinizing hormone, follicle-stimulating hormone, prolactin, thyroid-stimulating hormone, and adrenocorticotropic hormone, but growth hormone response to growth hormone releasing hormone is decreased.21
Using positional candidate cloning of mri, we recently identified a deletion mutation in Prkg2, which encodes cGMP-dependent protein kinase type II (cGKII), and clarified a role of cGKII as a molecular switch that couples cessation of proliferation and the start of hypertrophic differentiation of chondrocytes.2 Longitudinal skeletal growth is achieved by endochondral ossification in the growth plate, in which chondrocyte hypertrophic differentiation is an important step. Due to the impaired coupling of proliferation and hypertrophic differentiation in the growth plate chondrocytes, homozygous mutant (mri/mri) animals show longitudinal growth retardation.
In this study, we further characterize the phenotype of the KMI strain, including body length, body weight, organ weight, and craniofacial measurements. Furthermore, we describe phenotypic characteristics of the progeny produced from the (BN×KMI-mri/mri)F1×KMI-mri/mri backcross and provide updated genetic, physical, and comparative maps of the mri region.
Materials and Methods
Animals.
The KMI rat strain has been maintained at the Animal Research Center, Tokyo Medical University, since 1989. Although homozygous mutant (mri/mri) animals of both sexes have reproductive ability, the strain has been maintained by brother–sister mating between heterozygous (mri/+) females and homozygous (mri/mri) males, resulting in a segregating inbred strain. The KMI strain has been deposited into and is available from the National Bio-Resource Project of Rat in Japan (http://www.anim.med.kyoto-u.ac.jp/nbr/). Brown Norway (BN/Sea) rats were purchased from Seiwa Experimental Animals (Fukuoka, Japan).
Animal husbandry.
All animals were maintained at the Animal Research Center, Tokyo Medical University under specific-pathogen-free conditions, as described previously.18 Briefly, the animals were monitored and were maintained free of the following microorganisms: Salmonella typhimurium, Corynebacterium kutscheri, Clostridium piliforme, Mycoplasma pulmonis, Hantaan virus, Kilham rat virus, lymphocytic choriomeningitis virus, mouse adenovirus FL/K87, mouse pneumonia virus, mouse polio virus, Sendai virus, and Toolan H1 virus. The health status of the rats was evaluated by a commercial laboratory (Diagnostics Services, Charles River Laboratories, Wilmington, MA) according to their protocol. All rats were housed in a room maintained at 23 ± 2 °C and 55% ± 10% relative humidity, with a 12:12-h light:dark cycle, and were provided a commercial diet (FR1, Funabashi Farm, Chiba, Japan), and water ad libitum. All animal care and procedures were approved by the Steering Committee of Research-Related Laboratory Animals of Tokyo Medical University.
Phenotypic characterization.
From 3 to 8 wk of age, total lengths (from head to tail) and body weights of homozygous (mri/mri) and heterozygous (mri/+) animals were recorded weekly. In addition, craniofacial measurements of 8-wk-old rats (n = 5 for each group) were recorded. Total lengths, head-to-anus lengths, tail lengths, body weights, and organ weights of homozygous (mri/mri) and heterozygous (mri/+) rats were recorded at 10 wk of age (n = 10 for each group).
Genetic linkage mapping.
For genetic analysis of mri, female BN rats were crossed with homozygous mutant (mri/mri) male KMI to produce F1 animals. Female F1 animals (mri/+) were crossed with homozygous mutant (mri/mri) male rats to obtain backcrossed progeny: (BN×KMI-mri/mri)F1×KMI-mri/mri. Total length (from head to tail), head-to-anus length, tail length, body weight, and retroperitoneal fat weight of the backcrossed progeny were recorded at 50 d of age, and animals showing small body size were defined as those with the KMI phenotype (mri/mri). Genomic DNA was extracted from the spleen, and the DNA pooling method1 was used to detect initial linkage of SSLP markers with the mutation. Fifty SSLP markers spanning all the autosomes were genotyped in 2 pooled DNA samples, each consisting of 25 animals of the KMI (mri/mri) or wild-type (mri/+) phenotype. Fine mapping then was performed by genotyping of 241 animals showing the KMI phenotype (mri/mri) on 31 rat chromosome 14-specific SSLP markers. SSLP markers used in this study have been described elsewhere (RATMAP, available at http://ratmap.gen.gu.se/; Rat Genome Database, available at http://rgd.mcw.edu/). PCR and electrophoresis techniques have been described previously,18,27 except for D14Wox27 (Bmp3). PCR products of D14Wox27 (Bmp3) were resolved by automated sequencing (ABI PRISM 3100 Genetic Analyzer with GeneScan software; Applied Biosystems, Foster City, CA). The segregation patterns of the markers in the backcrossed progeny were analyzed with the Map Manager program.16
Physical and comparative mapping.
The physical map of rat chromosome 14 and rat–mouse–human comparative map was constructed based on data obtained in this study and information from the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/) and Rat Genome Database (http://rgd.mcw.edu/).
Statistical analysis.
Data are shown as mean ± SD. Two-tailed Student t tests were used to compare means between homozygous (mri/mri) and heterozygous (mri/+) animals and between animals of the KMI phenotype (mri/mri) and those of wild-type phenotype (mri/+) in the backcrossed progeny. Differences for which the P value was less than 0.05 were regarded as significant.
Results
Phenotypic characterization of the mri mutation.
Because the KMI strain has been maintained by brother–sister mating between heterozygous (mri/+) females and homozygous mutant (mri/mri) males, we obtained homozygous mutant (mri/mri) and heterozygous (mri/+) animals at each generation. Growth of homozygous mutant (mri/mri) animals was retarded to 70% to 80% of that of heterozygous (mri/+) animals (Figures 1 and 2). There was no obvious difference in growth between heterozygous (mri/+) and wild-type (+/+) animals (data not shown).
Figure 1.
The KMI rat strain at 8 wk of age. The homozygous mutant (mri/mri) animals and heterozygous (mri/+) littermates of both sexes are shown.
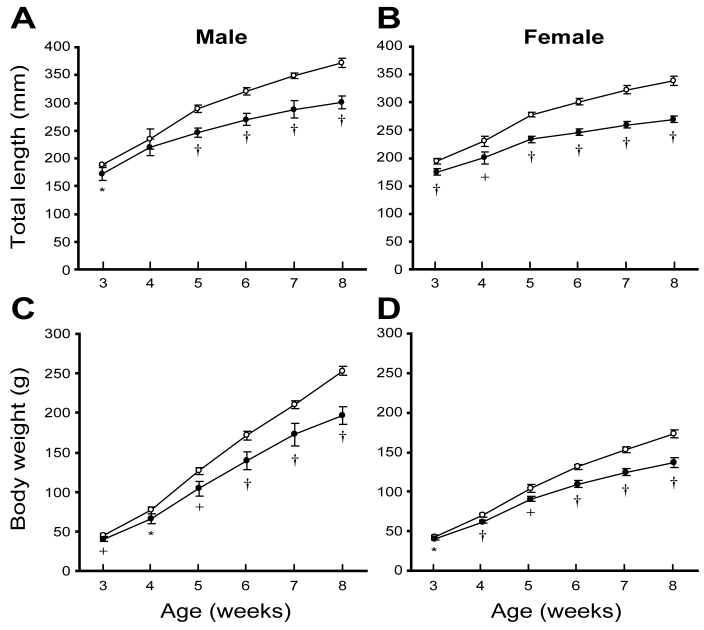
Figure 2.
Growth curves for (A and B) total lengths and (C and D) body weights of the KMI strain. Values shown are mean ± SD (n = 5). Open circles, heterozygous (mri/+) animals; filled circles, homozygous mutant (mri/mri) animals. *, P < 0.05; +, P < 0.01; †P < 0.001 versus values for heterozygous animals.
The growth retardation of homozygous mutant (mri/mri) animals became obvious at about 5 wk of age (Figure 2). At this age, the total length (mean ± SD) of homozygous mutant (mri/mri) animals (male [n = 5], 246.7 ± 8.6 mm; female [n = 5], 233.5 ± 5.6 mm) was significantly less than that of heterozygous (mri/+) animals (male [n = 5], 288.8 ± 6.9 mm; female [n = 5], 277.5 ± 3.9 mm; P < 0.001 for both comparisons). In addition, the body weight of homozygous mutant (mri/mri) animals (male [n = 5], 104.0 ± 8.9 g; female [n = 5], 90.8 ± 3.2 g) was significantly lower than that of heterozygous (mri/+) animals (male [n = 5], 126.4 ± 4.9 g; female [n = 5], 103.6 ± 5.0 g; P < 0.01 for both comparisons). The differences in body size became more pronounced with age (Figure 2). At 8 wk, the total length of homozygous mutant (mri/mri) animals (male [n = 5], 300.8 ± 11.6 mm; female [n = 5], 269.3 ± 5.9 mm) was markedly smaller than that of heterozygous (mri/+) animals (male [n = 5], 371.7 ± 8.1 mm; female [n = 5], 338.6 ± 8.3 mm; P < 0.001 for both comparisons). Body weight at 8 wk of age of homozygous mutant (mri/mri) animals (male [n = 5], 196.8 ± 11.2 g; female [n = 5], 136.6 ± 6.0 g) was much lower than that of heterozygous (mri/+) animals (male [n = 5], 252.8 ± 5.6 g; female [n=5], 173.0 ± 5.2 g; P < 0.001 for both comparisons).
Previous histopathologic examinations of pituitary, thyroid, adrenal, pancreas, cerebrum, cerebellum, thymus, heart, spleen, kidney, ovary, uterus, testis, and prostate revealed no significant differences between homozygous mutant (mri/mri) and heterozygous (mri/+) animals in KMI strain.21 In addition, there were no significant differences between homozygous mutant (mri/mri) and heterozygous (mri/+) animals in blood biochemistry, hematology (including blood coagulation analysis), and urinalysis (data not shown). To investigate the effects of the mri mutation on organ and tissue weights, we compared homozygous mutant (mri/mri) and heterozygous (mri/+) animals at 10 wk of age. Overall weights of most organs and tissues evaluated were markedly lower in homozygous mutant (mri/mri) animals than heterozygous (mri/+) rats (data not shown), suggesting a proportional decrease in homozygous mutant (mri/mri) animals. To further clarify this hypothesis, we compared relative organ and tissue weights (mg/100 g body weight; Table 1). Relative weights of lung, pancreas, and uterus did not differ between genotypes. Relative weights of brain, heart, kidney, spleen, adrenal, thymus, salivary gland, thyroid gland, pituitary, ovary, testis, seminal vesicle, and prostate in homozygous mutant (mri/mri) animals were somewhat higher than those in heterozygous (mri/+) animals. In contrast, the relative weight of retroperitoneal fat of homozygous mutant (mri/mri) animals (male [n = 7], 231.7 ± 64.6 mg/100 g body weight; female [n = 7], 246.9 ± 52.4 mg/100 g body weight) was greatly reduced compared with that of heterozygous (mri/+) animals (male [n = 7], 526.9 ± 73.0 mg/100 g body weight; female, 547.2 ± 58.1 mg/100 g body weight; P < 0.001 for both comparisons).
Table 1.
Phenotypic characteristics of homozygous mutant (mri/mri) and heterozygous (mri/+) KMI rats at 10 wk of age
| Male |
Female |
|||||
| mri/+ | mri/mri | mri/+ | mri/mri | |||
| Total length (mm) | 388.4 ± 2.9 | >>> | 295.5 ± 5.2c | 353.2 ± 6.4 | >>> | 273.0 ± 4.0c |
| Head-to-anus length (mm) | 218.8 ± 3.1 | >>> | 175.9 ± 3.6c | 193.5 ± 4.4 | >>> | 156.4 ± 2.9c |
| Tail length (mm) | 169.6 ± 2.5 | >>> | 119.6 ± 2.8c | 159.7 ± 2.8 | >>> | 116.6 ± 2.2c |
| Body weight (g) | 313.4 ± 10.8 | >>> | 232.2 ± 18.7c | 204.1 ± 11.1 | >>> | 152.2 ± 9.4c |
| Retroperitoneal fat (mg/100 g BW) | 526.9 ± 73.0 | >>> | 231.7 ± 64.6c | 547.2 ± 58.1 | >>> | 246.9 ± 52.4c |
| Brain (mg/100 g BW) | 616.4 ± 16.6 | <<< | 804.3 ± 64.7c | 877.6 ± 30.9 | <<< | 1127.4 ± 49.4c |
| Heart (mg/100 g BW) | 279.6 ± 6.0 | <<< | 303.6 ± 12.7c | 320.6 ± 6.5 | <<< | 342.8 ± 15.7c |
| Lung (mg/100 g BW) | 365.5 ± 15.0 | 372.5 ± 42.7 | 450.4 ± 20.6 | 464.6 ± 41.2 | ||
| Liver (mg/100 g BW) | 2912.7 ± 121.2 | > | 2759.9 ± 119.8a | 2671.8 ± 94.5 | 2767.4 ± 145.4 | |
| Kidney (mg/100 g BW) | 708.6 ± 37.9 | <<< | 767.4 ± 14.7c | 719.2 ± 36.1 | << | 777.0 ± 34.6b |
| Pancreas (mg/100 g BW) | 347.8 ± 47.9 | 400.5 ± 77.6 | 399.3 ± 38.1 | 432.6 ± 52.0 | ||
| Spleen (mg/100 g BW) | 214.7 ± 14.8 | < | 228.5 ± 11.0a | 246.0 ± 8.2 | <<< | 268.1 ± 15.6c |
| Adrenal (mg/100 g BW) | 14.5 ± 1.1 | <<< | 18.6 ± 1.9c | 30.3 ± 2.5 | <<< | 36.1 ± 3.6c |
| Thymus (mg/100 g BW) | 145.8 ± 14.2 | 160.9 ± 23.2 | 221.3 ± 12.0 | < | 241.4 ± 20.6a | |
| Salivary gland (mg/100 g BW) | 152.5 ± 6.0 | <<< | 183.4 ± 14.6c | 174.5 ± 3.9 | <<< | 208.5 ± 9.2c |
| Thyroid gland (mg/100 g BW) | 6.68 ± 1.05 | << | 8.16 ± 1.02b | 8.63 ± 0.78 | < | 9.94 ± 1.34a |
| Pituitary (mg/100 g BW) | 2.5 ± 0.3 | <<< | 3.2 ± 0.2c | 4.3 ± 0.8 | < | 5.0 ± 0.6a |
| Ovary (mg/100 g BW) | NA | NA | 33.1 ± 1.4 | < | 38.4 ± 6.9a | |
| Uterus (mg/100 g BW) | NA | NA | 397.8 ± 164.0 | 382.6 ± 143.9 | ||
| Testis (mg/100 g BW) | 995.4 ± 31.8 | <<< | 1256.4 ± 48.6c | NA | NA | |
| Seminal vesicle (mg/100 g BW) | 99.7 ± 12.9 | << | 121.9 ± 12.2b | NA | NA | |
| Prostate (mg/100 g BW) | 82.9 ± 8.3 | < | 96.6 ± 13.5a | NA | NA | |
BW, body weight; NA, not applicable.
Values shown are mean ± SD (n = 10) except for retroperitoneal fat (n = 7).
aP < 0.05, bP < 0.01, cP < 0.001 versus value for heterozygous animals.
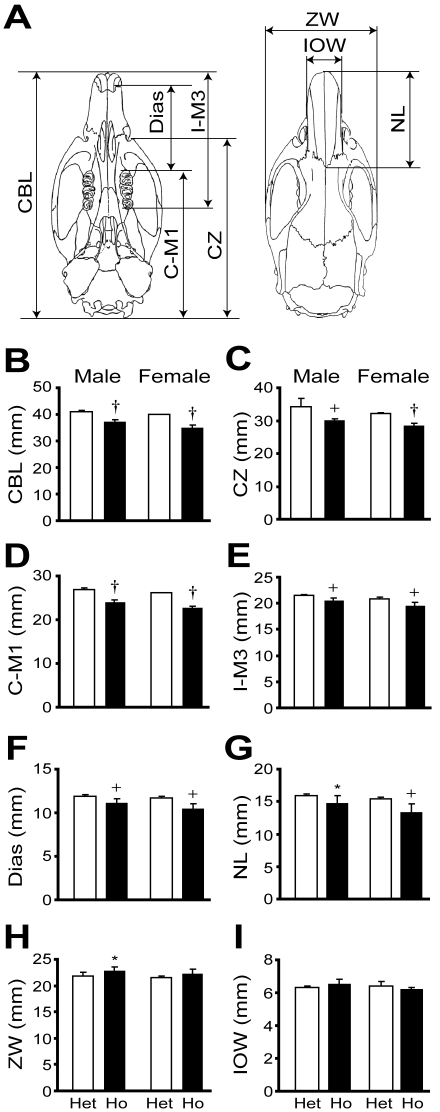
We previously reported that the longitudinal lengths of femora, tibiae, and vertebrae were significantly shorter in homozygous mutant (mri/mri) animals than in heterozygous (mri/+) animals.2 We here have characterized craniofacial morphology at 8 wk of age (Figure 3). Interparietal width measurements of the calvarium (Figure 3 H, I), which depend on membranous ossification, did not differ between groups. In contrast, naso-occipital measurements of the calvarium (for example, Figure 3B), which depend on endochondral ossification, were significantly smaller in homozygous mutant (mri/mri) animals than in heterozygous (mri/+) animals (mri/mri male, 37.2 ± 0.8 mm; mri/+ male, 41.1 ± 0.5 mm; mri/mri female, 35.1 ± 0.9 mm; mri/+ female, 40.0 ± 0.1 mm; P < 0.001 for both comparisons; n = 5 for all groups). These findings indicate that endochondral ossification, but not membranous ossification, is impaired in homozygous mutant (mri/mri) animals.
Figure 3.
Craniofacial morphology of the KMI strain at 8 wk of age. (A) Illustration of craniofacial measurements. CBL, condylobasal length; CZ, condylozygomatic length; C–M1, condylomolar (molar 1) length; I–M3, incisor–molar 3 length; Dias, length of diastema; NL, nasal length; ZW, zygomatic width; and IOW, interorbital width. (B through I) Comparisons of craniofacial measurements between heterozygous (mri/+, Het) and homozygous mutant (mri/mri, Ho) animals. Values shown are mean ± SD (n = 5). *, P < 0.05; +, P < 0.01; †P < 0.001 versus values for heterozygous animals.
Phenotypic characteristics of (BN×KMI)F1×KMI progeny and detailed genetic, physical, and comparative maps of the mri region.
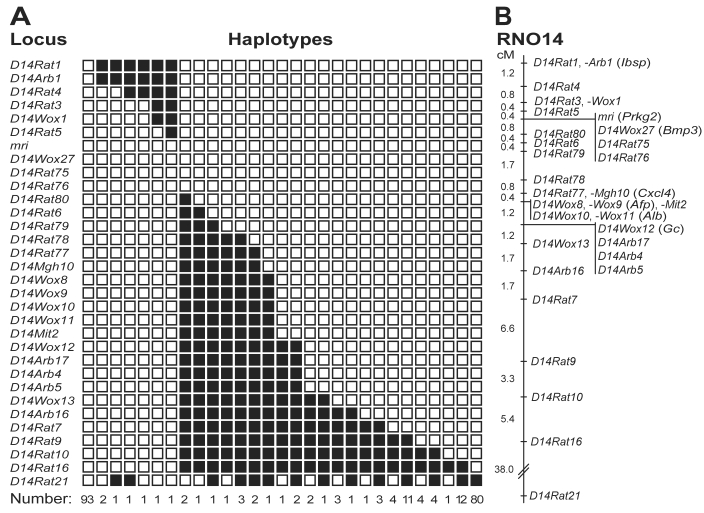
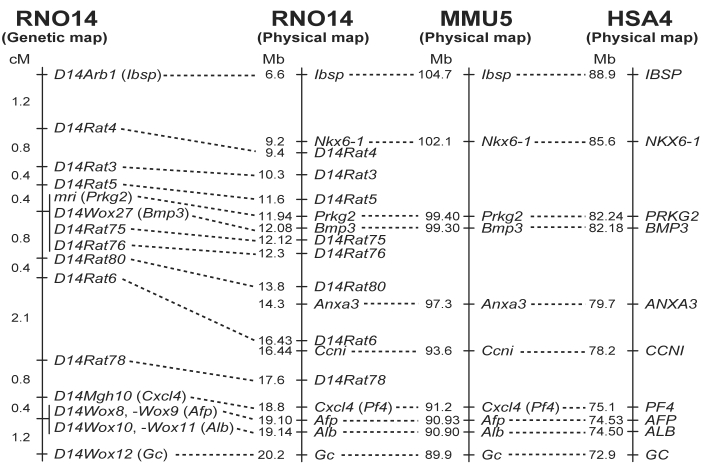
We recently reported a genetic linkage map of rat chromosome 14 in the vicinity of mri.2 We here describe phenotypic characteristics of the progeny produced from the (BN×KMI-mri/mri)F1×KMI-mri/mri backcross and provide updated versions of the genetic, physical, and comparative maps of the mri region. Of 475 progeny produced from the backcross, we obtained 114 male and 127 female animals showing the KMI phenotype (that is, small body size; Table 2). At 50 d of age, the rats showing the KMI phenotype (mri/mri) were distinguished easily from those showing the wild-type phenotype (mri/+; for example, total length: mri/mri male, 273.8 ± 7.4 mm; mri/+ male, 342.4 ± 8.9 mm; mri/mri female, 257.1 ± 6.3 mm; mri/+ female, 317.1 ± 10.9 mm; P < 0.001 for both comparisons). In addition, the body weight and retroperitoneal fat weight of animals showing the KMI phenotype (mri/mri) were greatly lower than those of rats showing the wild-type phenotype. The proportion of animals showing the KMI phenotype (50.7%, 241 of 475 rats) indicates that the mri mutation acts in an autosomal recessive manner. Genotyping of chromosome 14-specific SSLP markers and a haplotype analysis of the homozygous mutant (mri/mri) animals placed the mri locus in a genomic segment of 1.2 cM between D14Rat5 and D14Rat80, in the vicinity of the Bmp3 locus4 (Figure 4). All of the animals exhibiting the KMI phenotype were homozygous for the KMI allele at the mri region (Figure 4 A), suggesting strongly that mri acts in a completely recessive manner. Information from the physical genomic map of the rat (NCBI Map Viewer, available at http://www.ncbi.nlm.nih.gov/) clarified the position of the mri locus in a genomic segment of 2.2 Mb harboring Prkg2 (Figure 5). Cosegregating markers—D14Wox27 (Bmp3), D14Rat75, and D14Rat76— also were located in this segment, further supporting the results of genetic analysis. A comparative map of the mri regions on the rat, mouse, and human chromosomes indicates that more than 10 Mb of the genomic segment including mri (Prkg2) is highly conserved among these species (Figure 5).
Table 2.
Phenotypic characteristics at 50 d of age of progeny obtained from the (BN×KMI)F1×KMI backcross
| Male |
Female |
|||
| Wild-type | KMI-type | Wild-type | KMI-type | |
| Total length (mm) | 342.4 ± 8.9 | 273.8 ± 7.4a | 317.1 ± 10.9 | 257.1 ± 6.3a |
| Head-to-anus length (mm) | 194.6 ± 4.7 | 162.7 ± 4.9a | 175.9 ± 5.0 | 149.8 ± 3.6a |
| Tail length (mm) | 147.8 ± 6.3 | 111.1 ± 5.0a | 141.2 ± 7.0 | 107.2 ± 4.2a |
| Body weight (g) | 212.9 ± 16.9 | 172.2 ± 12.1a | 151.6 ± 9.2 | 125.5 ± 8.7a |
| Retroperitoneal fat (mg/100 g body weight) | 379.9 ± 70.1 | 243.3 ± 69.7a | 328.8 ± 74.7 | 192.9 ± 43.1a |
Values shown are mean ± SD of 130 wild-type male, 114 KMI-type male, 104 wild-type female, and 127 KMI-type female rats, except for retroperitoneal fat (47, 42, 41, and 42, respectively).
P < 0.001 versus wild type.
Figure 4.
Genetic linkage mapping of the mri mutation. (A) Distribution of haplotypes for rat chromosome 14 in 241 (BN×KMI)F1×KMI backcrossed progeny with the KMI phenotype. Black boxes, heterozygosity for the BN allele; white, homozygosity for the KMI allele. (B) Genetic linkage map of rat chromosome 14 including the mri (Prkg2) locus. Distances between loci (cM) are shown on the left.
Figure 5.
Comparative map of the mri region in the rat, mouse, and human. The comparative map of rat chromosome 14 (RNO14), mouse chromosome 5 (MMU5), and human chromosome 4 (HSA4) was constructed based on data obtained in this study and information from several databases (see text for details). Distances between loci (cM) are shown to the left of the genetic map. Physical map positions (Mb) of loci are shown to the left of each physical map.
Discussion
In this study, we phenotypically characterized a novel spontaneous dwarf mutant rat strain, KMI, and found that the homozygous mutant (mri/mri) animals showed growth retardation in body length, body weight, and naso-occipital lengths of the calvarium and greatly reduced retroperitoneal fat weight. We also describe phenotypic characteristics of the (BN×KMI-mri/mri)F1×KMI-mri/mri backcrossed progeny and provide detailed genetic, physical, and comparative maps of the mri region. These data support previous findings2 and further clarify the phenotype of the KMI strain.
The reductions in all of the naso-occipital length measurements of the calvarium in homozygous mutant (mri/mri) animals supports the previous finding that endochondral ossification is impaired in these animals.2 We here described organ and tissue weights of homozygous mutant (mri/mri) animals. Although relative weights (mg/100 g body weight) of most organs and tissues in homozygous mutant (mri/mri) animals were comparable with or somewhat higher than those in heterozygous (mri/+) animals, the relative weight of retroperitoneal fat was greatly reduced in homozygous mutant (mri/mri) animals. The mechanism underlying this reduction in the fat mass of the homozygous mutant (mri/mri) animals remains unclear at present, but the finding suggests the involvement of cGKII in fat metabolism and deposition.
Similar to homozygous mutant (mri/mri) animals in the KMI strain, Prkg2-deficient mice are fertile with no impairment in embryonic and fetal development and exhibit dwarfism with a defect in endochondral ossification at the growth plates.20 cGKII also is expressed in nonosseous tissues including intestinal mucosa, juxtaglomerular cells of kidney, and lung, and exerts diverse physiologic functions.6,25 For example, cGKII is highly concentrated in intestinal mucosa and mediates the intestinal secretion of water and electrolytes induced by E. coli toxin STa, an enterotoxin that increases cellular cGMP concentrations and intestinal fluid secretion, resulting in diarrhea.5,17 Consistent with this role of cGKII, Prkg2-deficient mice showed no accumulation of fluid in the intestine after treatment with STa20 and thus are resistant to this toxin.
Mice deficient in C-type natriuretic peptide (CNP; Nppc−/−) resemble Prkg2-deficient mice but show more severe dwarfism and early death.3 Furthermore, the defect in skeletal growth of Prkg2-deficient mice was not rescued by CNP expression in the growth-plate chondrocytes, indicating that CNP acts as a positive regulator of endochondral ossification upstream of cGKII.19 Together, these findings suggest the involvement of noncGKII signaling pathways in CNP-mediated endochondral ossification and explain the more severe phenotype of CNP-deficient (Nppc−/−) mice compared with that of Prkg2-deficient mice. Prkg2 deficiency results in live rodents that are fertile but with shortened trunks.
Accordingly, variants or mutations of PRKG2 might not be involved in severe growth defects (for example, skeletal dysplasia) but rather might contribute to normal variations in longitudinal growth in humans. Associations between common variants in PRKG2 and height in humans have been reported recently.8,12 The KMI strain is the first rat model with a confirmed mutation in Prkg2 and likely is a valuable model for studying dwarfism and longitudinal growth traits in humans and for physiologic studies of cGKII function.
Acknowledgments
This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by Scientific Research Grants from the Ministry of Health, Labour, and Welfare of Japan. The main part of this study was conducted at the Division of Laboratory Animal Science (Animal Research Center, Tokyo Medical University) with which MN and MF were affiliated formerly.
References
- 1.Asada Y, Varnum DS, Frankel WN, Nadeau JH. 1994. A mutation in the Ter gene causing increased susceptibility to testicular teratomas maps to mouse chromosome 18. Nat Genet 6:363–368 [DOI] [PubMed] [Google Scholar]
- 2.Chikuda H, Kugimiya F, Hoshi K, Ikeda T, Ogasawara T, Shimoaka T, Kawano H, Kamekura S, Tsuchida A, Yokoi N, Nakamura K, Komeda K, Chung UI, Kawaguchi H. 2004. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev 18:2418–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chusho H, Tamura N, Ogawa Y, Yasoda A, Suda M, Miyazawa T, Nakamura K, Nakao K, Kurihara T, Komatsu Y, Itoh H, Tanaka K, Saito Y, Katsuki M, Nakao K. 2001. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc Natl Acad Sci USA 98:4016–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM. 2001. Bone morphogenetic protein 3 is a negative regulator of bone density. Nat Genet 27:84–88 [DOI] [PubMed] [Google Scholar]
- 5.French PJ, Bijman J, Edixhoven M, Vaandrager AB, Scholte BJ, Lohmann SM, Nairn AC, de Jonge HR. 1995. Isotype-specific activation of cystic fibrosis transmembrane conductance regulator chloride channels by cGMP-dependent protein kinase II. J Biol Chem 270:26626–26631 [DOI] [PubMed] [Google Scholar]
- 6.Gambaryan S, Häusler C, Markert T, Pöhler D, Jarchau T, Walter U, Haase W, Kurtz A, Lohmann SM. 1996. Expression of type II cGMP-dependent protein kinase in rat kidney is regulated by dehydration and correlated with renin gene expression. J Clin Invest 98:662–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garofalo S, Vuorio E, Metsaranta M, Rosati R, Toman D, Vaughan J, Lozano G, Mayne R, Ellard J, Horton W, de Crombrugghe B. 1991. Reduced amounts of cartilage collagen fibrils and growth plate anomalies in transgenic mice harboring a glycine-to-cysteine mutation in the mouse type II procollagen α1 chain gene. Proc Natl Acad Sci USA 88:9648–9652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, den Heijer M, Franke B, Verbeek AL, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K. 2008. Many sequence variants affecting diversity of adult human height. Nat Genet 40:609–615 [DOI] [PubMed] [Google Scholar]
- 9.Hishinuma A, Furudate S, Oh-Ishi M, Nagakubo N, Namatame T, Ieiri T. 2000. A novel missense mutation (G2320R) in thyroglobulin causes hypothyroidism in rdw rats. Endocrinology 141:4050–4055 [DOI] [PubMed] [Google Scholar]
- 10.Jacenko O, LuValle PA, Olsen BR. 1993. Spondylometaphyseal dysplasia in mice carrying a dominant-negative mutation in a matrix protein specific for cartilage-to-bone transition. Nature 365:56–61 [DOI] [PubMed] [Google Scholar]
- 11.Kim PS, Ding M, Menon S, Jung CG, Cheng JM, Miyamoto T, Li B, Furudate S, Agui T. 2000. A missense mutation G2320R in the thyroglobulin gene causes nongoitrous congenital primary hypothyroidism in the WIC–rdw rat. Mol Endocrinol 14:1944–1953 [DOI] [PubMed] [Google Scholar]
- 12.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Diabetes Genetics Initiative; FUSION; KORA; Prostate, Lung Colorectal and Ovarian Cancer Screening Trial; Nurses' Health Study; SardiNIA. Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. 2008. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40:584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. 1999. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet 8:35–44 [DOI] [PubMed] [Google Scholar]
- 14.Li S, Crenshaw EB, 3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. 1990. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene Pit-1. Nature 347:528–533 [DOI] [PubMed] [Google Scholar]
- 15.Lin SC, Lin CR, Gukovsky I, Lusis AJ, Sawchenko PE, Rosenfeld MG. 1993. Molecular basis of the little mouse phenotype and implications for cell-type-specific growth. Nature 364:208–213 [DOI] [PubMed] [Google Scholar]
- 16.Manly KF. 1993. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm Genome 4:303–313 [DOI] [PubMed] [Google Scholar]
- 17.Markert T, Vaandrager AB, Gambaryan S, Pöhler D, Häusler C, Walter U, De Jonge HR, Jarchau T, Lohmann SM. 1995. Endogenous expression of type II cGMP-dependent protein kinase mRNA and protein in rat intestine. Implications for cystic fibrosis transmembrane conductance regulator. J Clin Invest 96:822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuyama T, Ishibiki J, Awata T, Noda M, Kanazawa Y, Sugawara M, Komeda K. 2000. An improved genetic linkage map of rat chromosome 20. Comp Med 50:369–373 [PubMed] [Google Scholar]
- 19.Miyazawa T, Ogawa Y, Chusho H, Yasoda A, Tamura N, Komatsu Y, Pfeifer A, Hofmann F, Nakao K. 2002. Cyclic GMP-dependent protein kinase II plays a critical role in C-type natriuretic peptide-mediated endochondral ossification. Endocrinology 143:3604–3610 [DOI] [PubMed] [Google Scholar]
- 20.Pfeifer A, Aszódi A, Seidler U, Ruth P, Hofmann F, Fässler R. 1996. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science 274:2082–2086 [DOI] [PubMed] [Google Scholar]
- 21.Serizawa N. 1993. [Initial characterization of a new miniature animal model in the rat: studies on anatomy, pituitary hormones and GH mRNA in miniature rat Ishikawa. In Japanese.] Nippon Naibunpi Gakkai Zasshi. 69:33–45 [DOI] [PubMed] [Google Scholar]
- 22.Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, O'Connell SM, Gukovsky I, Carrière C, Ryan AK, Miller AP, Zuo L, Gleiberman AS, Andersen B, Beamer WG, Rosenfeld MG. 1996. Pituitary lineage determination by the Prophet of Pit1 homeodomain factor defective in Ames dwarfism. Nature 384:327–333 [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi T, Suzuki H, Sakurai S, Nogami H, Okuma S, Ishikawa H. 1990. Molecular mechanism of growth hormone (GH) deficiency in the spontaneous dwarf rat: detection of abnormal splicing of GH messenger ribonucleic acid by the polymerase chain reaction. Endocrinology 126:31–38 [DOI] [PubMed] [Google Scholar]
- 24.Vandenberg P, Khillan JS, Prockop DJ, Helminen H, Kontusaari S, Ala-Kokko L. 1991. Expression of a partially deleted gene of a human type II procollagen (COL2A1) in transgenic mice produces a chondrodysplasia. Proc Natl Acad Sci USA 88:7640–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner C, Pfeifer A, Ruth P, Hofmann F, Kurtz A. 1998. Role of cGMP-kinase II in the control of renin secretion and renin expression. J Clin Invest 102:1576–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang X, Benson KF, Chada K. 1990. Mini-mouse: disruption of the pygmy locus in a transgenic insertional mutant. Science 247:967–969 [DOI] [PubMed] [Google Scholar]
- 27.Yokoi N, Kanazawa M, Kitada K, Tanaka A, Kanazawa Y, Suda S, Ito H, Serikawa T, Komeda K. 1997. A non-MHC locus essential for autoimmune type I diabetes in the Komeda Diabetes-Prone rat. J Clin Invest 100:2015–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]