Abstract
Previous studies indicate that two upstream CCAAT/enhancer-binding protein (C/EBP) sites and C/EBPβ are required for subtype B HIV-1 gene expression in cells of the monocyte–macrophage lineage. The mechanisms of C/EBP regulation of HIV-1 transcription and replication remain unclear. This review focuses on studies concerning the role of C/EBP factors in HIV-1, human T-cell leukemia virus type 1, and SIV transcription in various cell types and tissues cultured in vitro, animal models and during human infection. The structure and function of the C/EBPβ gene and the related protein isoforms are discussed along with the transcription factors, coactivators, viral proteins, cytokines and chemokines that affect C/EBP function.
Keywords: CCAAT/enhancer-binding protein, gene expression, HIV-1, human T -cell leukemia virus type-1, retrovirus, SIV, transcription
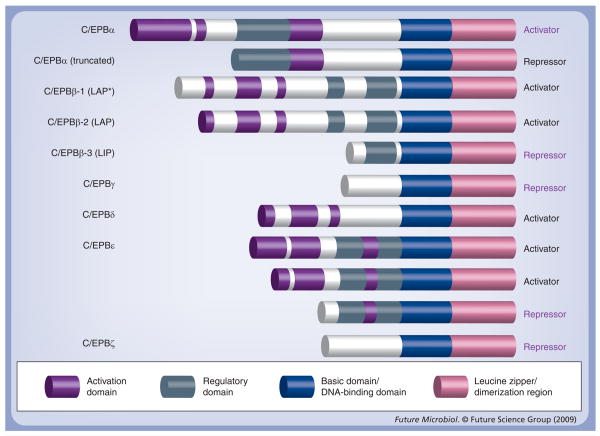
The CCAAT/enhancer-binding protein (C/EBP) family is one member of a large group of basic region leucine zipper (bZIP) transcription factors. Six C/EBP family members have been classified: C/EBPα, -β, -γ, -δ, -ε and -ζ (Figure 1) [1–4]. All members of this family share a highly conserved C-terminal region (bZIP), consisting of a basic amino acid-rich DNA-binding domain followed by a leucine zipper dimerization motif. However, the N-termini of C/EBP factors are diverse. Some members (C/EBPα, -β, -δ and -ε) contain both activation and regulatory domains, allowing them to function as activators, whereas C/EBPγ and -ζ lack activation domains and, as a result, function as inhibitors [1,5–11]. In addition, multiple isoforms of C/EBPα, -β and -ε have been identified. Different isoforms may function as either activators or inhibitors, depending largely on the number of activation domains contained within their N-terminal region (Figure 1) [10,12,13]. The functional properties of C/EBP family members have been well characterized. These properties include their ability to regulate cellular growth and differentiation, immune and inflammatory responses, as well as their specific role within the context of various disease processes [14]. For example, C/EBPs play an important role in the regulation of gene expression in myelomonocytic cells and cells of the monocyte–macrophage lineage [11], Additionally, C/EBPα is involved in the regulation of granulopoiesis and hepatic gene expression [15] and C/EBPβ modulates gluconeogenesis [16] and adipogenesis gene expression [17]. In addition, C/EBPs are able to regulate the expression of a number of cytokines and chemokines, including IL-6, IL-8 and monocyte chemoattractant protein-1 (MCP-1) [18–21].
Figure 1. Structure of the CCAAT/enhancer-binding protein transcription factor family.
Two isoforms of C/EBPα, three isoforms of C/EBPβ, and C/EBPε, C/EBPγ, C/EBPδ, and C/EBPζ are shown. They all share a high degree of sequence similarity at the C-terminus, including the leucine zipper domain and basic region. At the N-terminus the structures vary among different family members. LAP* indicates full-length liver activator protein; LAP is translated from the second ATG; LIP is translated from the third ATG.
C/EBP: CCAAT/enhancer-binding protein; LAP: Liver activator protein; LIP: Liver inhibitory protein.
The expression patterns of different C/EBP family members vary widely. The level of C/EBPα expression increases during the course of adipocyte differentiation and reaches its highest level when cells differentiate into nondividing adipocytes. In contrast, C/EBPβ and -δ are expressed at their highest levels during the early stages of adipocyte development [1,22]. However, during granulocyte differentiation, the levels of C/EBPα expression increase first and are expressed maximally in cells that retain the ability to divide; eventually, the level of expression is reduced. The levels of C/EBPδ expression increase markedly early during induction of granulocyte differentiation. Expression then remains fairly constant, with small reductions in expression eventually observed in more differentiated granulocyte populations. In contrast to the expression levels of C/EBPα and C/EBPδ, those of C/EBPβ increase steadily throughout the progressive stages of granulocyte differentiation [22]. In the monocyte–macrophage cell lineage, C/EBPβ expression levels increase during the differentiation of progenitor cells. Additionally, it is the only family member whose expression level is increased upon activation of the U-937 promonocytic cell line [23]. Exposure of macrophages to lipopolysaccharide (LPS), IL-1, IFN-γ and TNF-α, all of which are present at elevated levels during the course of HIV-1 infection, leads to a reduction in the level of C/EBPα mRNA, whereas the levels of C/EBPβ and C/EBPδ expression increase [24]. Decreased C/EBPα levels have been observed in HIV-1-infected patients receiving HAART. These patients have serious metabolic difficulties such as combined insulin-resistance, dyslipidemia, central adiposity and peripheral lipotrophy. The inhibitory effects of a protease inhibitor on adipogenesis have been related to decreased expression of C/EBPα, and overexpression of C/EBPα was able to rescue some of the anti-adipogenic effects of the protease inhibitor [25–27]. However, the expression levels of C/EBPβ were not affected by HIV-1 protease-inhibitor treatment [26,27]. The phenomenon may be caused by protease inhibitors acting on specific stages of adipocyte differentiation [27] or the molecular targets affected by the anti-adipogenic properties of the antiretroviral drug, likely to be located downstream from C/EBPβ [26]. For example, nelfinavir affects steps subsequent to critical early events in preadipoctye differentiation [27], and efavirenz may affect transcription factors located downstream of C/EBPβ [26]. C/EBPδ is also involved in adipogenesis. Because of this, it would be interesting to determine whether C/EBPδ expression is affected during treatment with antiretroviral drugs.
Under some physiologic circumstances during the course of HIV disease, cells of the monocyte–macrophage lineage appear to be resistant to the cytopathic effects of HIV-1 and may serve as a viral reservoir during persistent viral infection. The capacity of these cells to migrate into various organs and tissues makes them potential conveyors of HIV-1 to multiple organ systems including the brain and kidney [28,29]. Considering the important role that the monocyte–macrophage lineage likely plays in HIV infection, C/EBPβ has received more attention than other members of the C/EBP family of proteins.
This review examines the C/EBP family of proteins, especially C/EBPβ, and its roles in HIV-1 infection and associated diseases as well as other retroviral infections. Five aspects to be considered are:
The C/EBPβ gene and corresponding protein isoforms;
The effects of C/EBP on HIV-1 long terminal repeat (LTR)-directed transcription within different cell types and tissues;
The effects of C/EBPβ on the expression of the CCR5 coreceptor in cells susceptible to HIV-1 infection;
Specific proteins that interact with C/EBPβ to regulate HIV-1 gene expression;
The role that C/EBP plays within the context of other retrovirus-associated diseases.
C/EBPβ gene & corresponding proteins
Studies from a rodent system indicate that three C/EBPβ activation domains (AD) – AD1, AD2 and AD3 – are localized within the first 131 amino acids of the protein. Although each AD exhibits low intrinsic activity, collectively the three intact ADs result in full protein activity [4,12,30]. Two regulatory domains (RDs) – RD1 and RD2 – are located between the DNA-binding domain and ADs (Figure 2A) [4,12,30]. Mutations within either of the RDs lead to an increase in C/EBPβ activity. RD1 consists of residues within the internal region of the protein (amino acids 139–142) and has been shown to weakly inhibit C/EBPβ transactivation function. RD2 is localized within amino acid residues 163–191 and has been shown to be responsible for inhibiting C/EBPβ DNA-binding affinity in a cell-type specific manner. Several studies have demonstrated that phosphorylation within an inhibitory domain will likely result in liberation of transactivation and DNA-binding activities from an inhibitory state [2,6,12,30]. C/EBPβ can be phosphorylated in a number of cells by multiple stimuli and through various protein kinase pathways, and protein phosphorylation has been shown to be a major regulator of C/EBPβ function [31–34]. For example, phosphorylation of threonine 235 in the inhibitory domain of human C/EBPβ by a ras-dependent MAPK increased transcriptional activation [34], and phosphorylation of serine 299 of rat C/EBPβ (or Ser288 of human C/EBPβ) by cAMP-dependent protein kinase A resulted in nuclear translocation and subsequent transactivation [33]. Furthermore, a higher level of phosphorylated C/EBPβ (at Thr235) has been found in human macrophages compared within monocytes, indicating that C/EBPβ phosphorylation may be important in lineage commitment [35]. Recent studies have demonstrated that cyclin-dependent kinase 9 is able to phosphorylate C/EBPβ and leads to an increase in HIV-1 gene expression [36]. At the C-terminus, the leucine zipper region has been shown to be responsible for C/EBPβ homo- and heterodimerization with other C/EBP or bZIP protein family members. Dimerization has been shown to be a prerequisite for the binding of C/EBP factors to the corresponding DNA recognition element [4,12,37]. Previous studies indicate that the DNA-binding basic region is approximately 20 amino acids long, lies upstream of the leucine zipper region and interacts with the major groove of DNA in a sequence-specific manner [38].
Figure 2. Structure, isoforms and amino acids of CCAAT/enhanced-binding protein β.
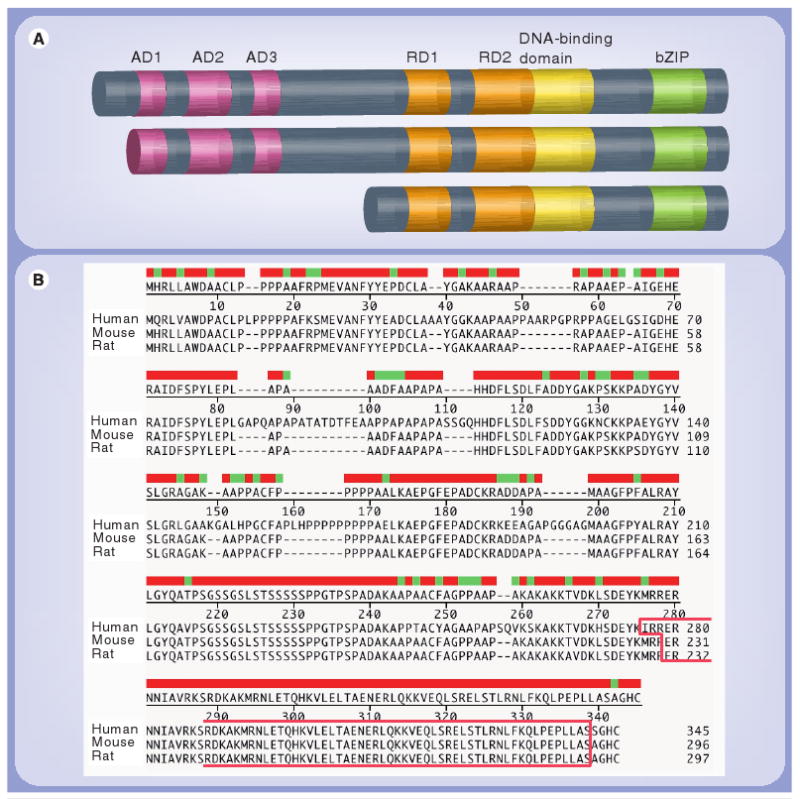
(A) Rodent CCAAT/enhanced-binding protein (C/EBP)β is presented as an example, and the different functional domains are shown. Pink boxes represent activation domains; orange boxes represent regulatory domains; yellow boxes represent DNA-binding domains; green boxes represent bZIPs.
(B) Amino acid-sequence comparison between rodent and human C/EBPβ. Full-length consensus sequences of human, mouse and rat C/EBPβ were compared by Clustal W analysis. Red shows conserved residues, green shows sequence heterogeneity. Gaps indicate the absence of specific amino acid residues. The bZIP region is highlighted.
AD: Activation domain; bZIP: Basic region leucine zipper; RD: Regulatory domain.
C/EBPβ (also referred to as nuclear factor IL-6 in human and liver-enriched activating protein [LAP], in rodents) has been shown to be located on human chromosome 20q13.13 [39] and on mouse chromosome 2 [40], respectively. It was first characterized as a protein that binds to the IL-1-responsive element within the promoter region of the IL-6 gene [2]. To date, the C/EBPβ gene has been described in human, mouse, rat, chicken, Xenopus laevis, fish and bovine species [32]. Among these, the human, mouse and rat C/EBPβ genes and gene products have been used in HIV-1 studies. C/EBPβ is encoded by a gene containing a single exon without introns that is transcribed into a 1.4 kB mRNA transcript [39–41]. The single C/EBPβ mRNA can be translated into three distinct protein products as a result of a leaky ribosome scanning mechanism that uses alternative in-frame translational initiation sites [12,13]. In the rodent system, three isoforms of C/EBPβ are produced: LAP*, LAP and liver-enriched inhibitory protein (LIP) (Figure 2A). LAP* is translated from the first AUG sequence and is 297 amino acids in length, resulting in a 38 kDa C/EBPβ isoform. LAP is translated from the second AUG sequence, generating a 35 kDa isoform, which lacks the first 21 N-terminal amino acids but leaves the transactivation domains intact. LIP is a 16–20 kDa isoform generated from the third AUG sequence and lacks the N-terminal trans-activation domain. LIP contains the C-terminal DNA-binding and dimerization domains and functions mainly as a dominant–negative C/EBPβ isoform [12,13]. Previous studies have shown that LIP predominates when the ratio of LIP:LAP is greater than 0.2 [12]. Other studies have shown that LIP may be generated by proteolytic cleavage during the preparation of cell extracts [42–44], and the expression level of LIP is mainly determined by the procedures used to prepare cell lysates [42]. The mechanisms regulating C/EBPβ have not been well characterized. Recent studies have suggested that C/EBPβ could be degraded by a calpain-dependent mechanism in skeletal muscle cells [45]. Similarly, three isoforms of C/EBPβ have also been reported in humans: C/EBPβ-1, -2 and -3. The full-length human C/EBPβ (C/EBPβ-1) is 346 amino acids in length. C/EBPβ-2 is translated from the second start codon, resulting in a 323 amino acid protein. C/EBPβ-3 is 146 amino acids in length. Experimentally, the three isoforms were identified as p55, a p45/p42 doublet and p20, respectively [46]. Amino acid sequence analyses have shown that human and mouse/rat C/EBPβ share a highly conserved sequence identity within the C-terminus, whereas N-terminal and internal region sequences show greater variability (Figure 2B), which may suggest that human and rodent C/EBPβ could exhibit distinct activation pathways, thus their biological function might be different. This observation is of interest because many studies have been performed in heterologous experimental systems including studies using a human virus and mouse C/EBPβ.
Although C/EBPβ-1 and C/EBPβ-2 differ by only 21 amino acids in mouse, rat and chicken proteins and by 23 amino acids in the human protein, functional differences between the two isoforms have been reported [47,48]. Three factors contribute to the differences observed between LAP* and LAP or C/EBPβ-1 and C/EBPβ-2. First, differences within the first 21 amino acids of the N-terminus of LAP*, a region responsible for functional interaction with the switch mating type/sucrose nonfermenting (SWI/SNF) complex, have been shown. This complex is a histone remodeling enzyme that uses energy derived from ATP hydrolysis to model chromatin, thus allowing transcriptional machinery to access DNA in the presence of LAP* but not LAP, suggesting that differences may exist between LAP* and LAP when interacting with chromosome-embedded genes [49]. Second, six cysteine residues exist within mouse LAP* (i.e., Cys11, Cys33, Cys123, Cys143, Cys201 and Cys296), five in LAP (Cys33, Cys123, Cys143, Cys201 and Cys296), and two in LIP (Cys201 and Cys296). The presence of additional disulfide bonds between Cys11 and Cys33 in LAP* was shown to be related to the ability of LAP* to regulate LPS-induced IL-6 gene expression under a sudden surge of reducing potential [50]. Third, studies from human C/EBPβ have demonstrated that C/EBPβ-1, but not C/EBPβ-2, was able to conjugate to the small ubiquitin-like modifier (SUMO) family members, SUMO-2 and SUMO-3. This incorporation required the integrity of the extreme N-terminus of C/EBPβ-1 and lysine 173 [51]. Since sumolyation affects protein–protein interactions, the difference in linkage to SUMO by C/EBPβ-1 and C/EBPβ-2 may also contribute to differential interactions with coactivator proteins and chromatin remodeling proteins, thus contributing to functional differences observed between C/EBPβ-1 and C/EBPβ-2 [51]. However, differences between the roles of C/EBPβ-1 and C/EBPβ-2 in mediating HIV-1 transcription have not been reported. Based on the demonstrated functional differences between C/EBPβ-1 and C/EBPβ-2, it is possible that functional differences exist between the two isoforms with respect to regulating HIV-1 gene expression. For example, it has been reported that IL-6 stimulation may increase C/EBPβ-2 but not C/EBPβ-1 or -3 expression [52]. Thus, HIV-1 infection resulting in the upregulation of specific cytokines such as IL-6, IL-1 and TNF-α, may also alter the expression of different C/EBPβ isoforms, further modulating HIV-1 gene expression by interacting with different co-activators and/or chromatin remodeling proteins. IFN-β or IL-10 is able to induce C/EBPβ-3 production in macrophages, which may be important for HIV-1 latent infection and may provide a selective advantage by facilitating the establishment of a viral reservoir [53]. Subsequently, the different C/EBPβ isoforms may have different roles in determining human macrophage susceptibility to productive HIV-1 infection and therefore require more investigation.
Role of C/EBPβ & C/EBP-binding sites in HIV-1 transcription in cells of the monocyte–macrophage lineage
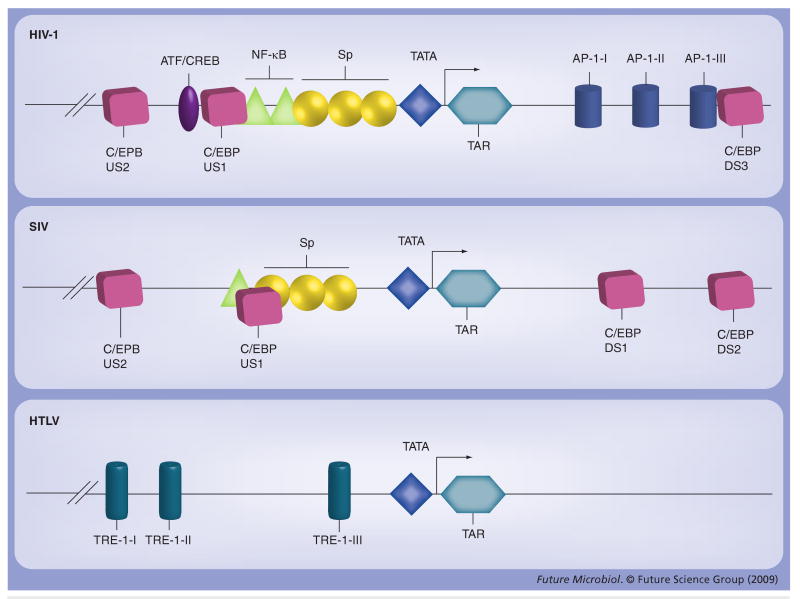
It is not totally understood how C/EBPβ factors regulate HIV-1 transcription. Three C/EBP-binding sites have been identified upstream of the transcriptional start site within the HIV-1 promoter or LTR [54]. One site is located immediately 5′ of the distal nuclear factor (NF)-κB binding site (upstream site [US] 1, -105 to -116, relative to the transcription start site); another is located upstream (US2 -164 to -173); and a third (US3 -245 to -253) is located further upstream of US2 (Figure 3). US1 and 2 include a consensus C/EBPβ-binding site, though this consensus sequence is not apparent within US3 [55]. C/EBPβ protein and either HIV-1 LTR C/EBP-binding sites US1 or US2 are required for HIV-1 replication in the U-937 monocytic cell line as well as in primary cells of the monocyte–macrophage linage. The two C/EBPβ-binding sites appear to be functionally equivalent [23,56]. A number of studies have demonstrated that specific LTR C/EBP-binding site configurations exhibit different C/EBPβ DNA-binding affinities, and the prevalence of HIV-1 LTRs containing these configurations correlates with specific stages of HIV-1 disease [57–60]. Specifically, the 6G configuration of C/EBP US1 (T-to-G change at nucleotide position 6 of consensus subtype B US1) exhibiting a moderate affinity for C/EBP factors was commonly found in brain-derived LTRs, but was infrequently encountered in peripheral blood-derived LTRs [60]. In contrast, the 3T C/EBP US1 (C-to-T change at nucleotide position 3 of consensus subtype B US1) and consensus US2 sequence were found predominantly in LTRs derived from peripheral blood mononuclear cells in late stage HIV disease, and brain tissues of HIV-associated dementia (HAD) patients. Interestingly, the 3T configuration of the C/EBP US1 has been shown to be preferentially encountered in LTRs from late-stage disease with a 5T configuration of Sp site III (a C-to-T change at nucleotide position 5 of the subtype B consensus sequence of Sp site III).
Figure 3. Structural comparison of HIV-1, SIV and HTLV-1 long terminal repeats.
Locations of the binding sites for C/EBP, TATA binding protein, Sp factors, NF-κB and ATF/CREB factors are shown (not to scale). Although no canonical C/EBP-binding sites have been identified within the HTLV-1 long terminal repeat, C/EBP has been shown to be able to bind to TRE-1 repeat III. The three 21 bp repeats of TRE-1 are illustrated on the HTLV-1 long terminal repeat.
ATF: Activating transcription factor; C/EBP: CCAAT/enhancer protein; CREB: cAMP response element binding protein; HTLV: Human T-cell leukemia virus type 1; NF-κB: Nuclear factor-κB; TRE: Tax-responsive element.
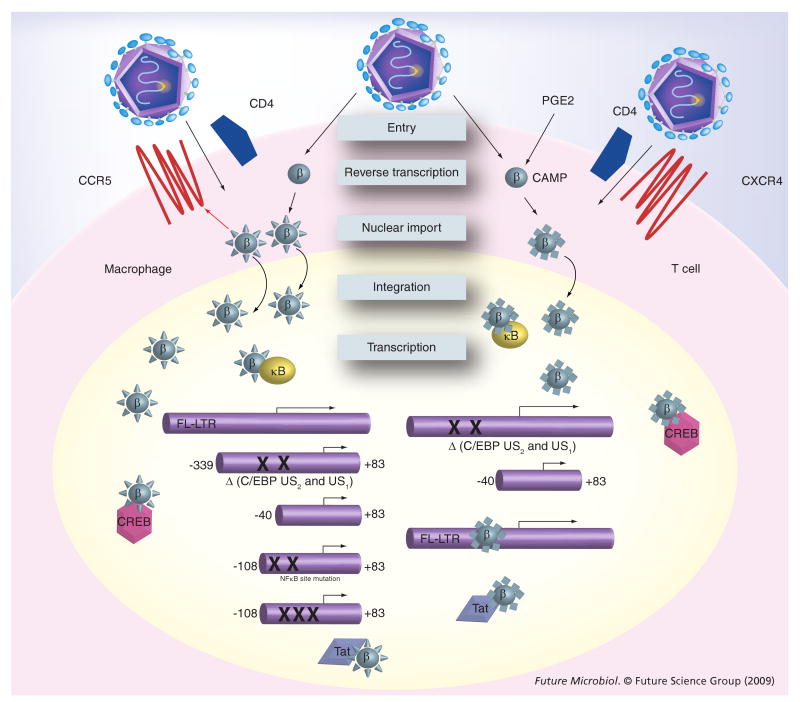
In addition to activating HIV-1 transcription by binding to US1 and US2, C/EBPβ has the ability to activate the HIV-1 LTR in the absence of these binding sites in U-937 monocytic cells (Figure 4) [54,61]. For example, C/EBPβ transactivated a mutant HIV-1 LTR, which contains the sequence from -339 to +83 (relative to the transcription start site) with a deletion of both US1 and US2 binding sites. C/EBPβ also elicited a relatively low level of gene activation from the basal LTR element (-40 to +83) or from a partial LTR region (-80 to +83), although the maximal levels of LTR response to C/EBPβ still required the upstream elements [54,61]. Additionally, exogenous C/EBPβ activated mutant LTRs that contain only a partial LTR element (-108 to +83), including NF-κB binding sites without the three Sp binding sites or intact Sp binding sites with mutant NF-κB binding sites in U-937 monocytic cells, in the absence or presence of combined LPS and phorbol-12-myristate-13-acetate (PMA) stimulation, suggesting the critical role of C/EBPβ in the context of HIV-1 LTR variants containing multiple mutated and/or deleted transcription factor binding sites [54,61]. However, the expression of C/EBPζ (C/EBP-homologous protein), which is a negative regulator of C/EBPs, inhibits C/EBPβ transcriptional ability and leads to a delay of HIV-1 replication in U-937 cells [36]. These studies suggest that either some unidentified C/EBP-binding sites exist within the HIV-1 LTR or that C/EBP factors could be recruited to the LTR via direct or indirect interaction with other transcription factors and related signaling pathways and/or other transcription factor-binding sites. For example, a highly conserved C/EBP-binding site (DS3) which is located at position +158 to +175 relative to the transcriptional start site in the HIV-1 LTR, has been recently identified within the HIV-1 subtype B LTR. This C/EBP-binding site affects the basal and transactivator of transcription (Tat)-mediated HIV-1 subtype B transcription and replication in U-937 cell lines [Liu Y, Wigdahl B, Unpublished Data] (Figure 3).
Figure 4. CCAAT/enhancer-binding protein-β regulates HIV-1 gene expression in macrophages and T lymphocytes.
Different LTR forms activated by C/EBPβ, the different active forms of C/EBPβ and specific protein–C/EBPβ complexes in macrophages and T lymphocytes are shown. See the text for complete description.
C/EBP: CCAAT/enhancer-binding protein; CREB: cAMP response element binding protein; LTR: Long terminal repeat; NF-κB: Nuclear factor-κB; Tat: Transactivator of transcription; US: Upstream site. Adapted from [194].
Role of C/EBPβ in HIV-1 transcription in T cells
CD4+ T cells are the major targets of HIV-1 infection; however, the role of C/EBPβ in HIV-1 infection of T cells has not been well characterized. The expression levels of C/EBPβ mRNA and protein have been characterized in the Jurkat T-cell line [62–64] and are increased in T cells isolated from peripheral blood mononuclear cells of HIV-1-infected patients [65]. However, C/EBP US2 is unable to form specific DNA–protein complexes with nuclear extracts from Jurkat T cells with or without LPS and PMA stimulation [62], indicating that either C/EBPβ expressed in Jurkat T cells is different from that expressed in cells of the monocyte–macrophage lineage, or that combined LPS and PMA treatment could not release the suppression of C/EBPβ, by RD2, which is predominately responsible for inhibiting C/EBPβ DNA binding affinity [12]. It has also been demonstrated that C/EBP US1 and US2 are dispensable for HIV-1 replication in the Jurkat T-cell lines as well as in primary T cells [62]. Interestingly, in Jurkat T cells stimulated with LPS and PMA, the transcriptional level of the HIV-1 LTR is increased by exogenous C/EBPβ. Both the basal region (-40 to +83) and the TATAA element (-35 to -15) of the HIV-1 LTR respond to C/EBPβ-mediated upregulation of HIV-1 transcription, indicating that the basal region of the HIV-1 LTR may contain unidentified C/EBP-binding sites or that specific interactions may occur between C/EBPβ and TATAA-binding proteins, which may promote HIV-1 LTR activation (Figure 4) [66]. Consistent with this concept, interaction between C/EBPα and TATAA-binding proteins has been observed [67,68]. Prostaglandin E2 and forskolin, both of which increase the level of intracellular cAMP, are able to potently activate C/EBPβ in human T cells, and the prostaglandin E2-mediated increase in HIV-1 LTR activity is dependent on the C/EBP-binding sites (Figure 4) [69]. By contrast, C/EBP-binding sites do not appear to be involved in the induction of LTR activity in response to phytohemagglutinin combined with either PMA or TNF-α, none of which can increase cAMP levels [69,70]. Therefore, it is possible that intracellular cAMP levels have the potential to stimulate C/EBPβ expression and affect HIV-1 transcription in T cells. Therefore, these studies provide further evidence of the ability of C/EBPβ to regulate HIV-1 transcription in a cell-type specific manner.
Recent studies have shown that C/EBPβ is able to activate HIV-1 replication in primary CD4+ T cells by binding to the cytidine deaminase APOBEC3G and inhibiting APOBEC3G antiviral function [71]. Phosphorylation of serine at position of 288 of C/EBPβ is critical for binding to APOBEC3G [71]. APOBEC3G is a natural inhibitor of HIV-1 infection and the HIV-1 accessory protein Vif binds to APOBEC3G leading to proteosome-dependent degradation of APOBEC3G, thereby inhibiting its antiviral function (reviewed in [72]). Interestingly, C/EBPβ has been shown to be able to induce the replication of a Vif-deficient strain of HIV-1. Furthermore, C/EBPβ inhibition of APOBEC3G subsequently prevents G-to-A mutations in HIV-1 DNA usually caused by APOBEC3G and maintenance of HIV-1 genome stability, which is likely to be important in HIV-1 infectivity [71].
Role of C/EBP in HIV-1 transcription in the brain
HIV-1 infects the brain at an early stage of infection, and the brain serves as an important reservoir for HIV-1 [73]. In addition to the infected macrophages infiltrating the brain, microglial cells and astrocytes have been shown to be additional targets of HIV-1 infection in the brain [73,74]. C/EBPβ and C/EBPγ are expressed in glial and microglial cell lines, and C/EBPβ has been found in primary mouse astrocyte and microglial cell cultures [75,76]. C/EBPδ has been detected in rat cortical astrocytes but does not appear to be expressed in human microglial and glial cells [76]. Nuclear levels of C/EBPβ increase rapidly in astrocyte and microglial cells following LPS treatment, and studies show that MAPK and cAMP pathways are involved in this process [75,77]. Furthermore, systemic injection of LPS results in an increase of C/EBPβ nuclear levels in the mouse brain [75]. Like LPS, proinflammtory cytokines, such as IL-1β, TNF-α and IL-6 have the ability to enhance C/EBPβ expression in primary astrocytes [77]. Therefore, C/EBPβ could also be involved in the regulation of gene expression in astrocytes during inflammatory processes affecting the brain, including HIV-1-associated brain disease. Additionally, C/EBP-binding sites have been identified within the promoter regions of the IL-1β, TNF-α and IL-6 genes. Therefore, the induction of C/EBPβ in astrocytes could, in turn, promote expression of these genes, thus facilitating a positive-feedback loop and an increased inflammatory response [77].
Within microglial cells, the three known upstream C/EBP-binding sites have been shown to be nonessential for C/EBPβ-mediated transcription of the HIV-1 LTR [76]. Instead, C/EBPβ activation was mediated through the -68 to +80 LTR region. However, in oligodendroglioma cells (TC-620 cells), the upstream C/EBP sites were required for efficient C/EBPβ responsiveness, although C/EBPβ was still able to function through the truncated LTR from -40 to +80 in the absence of Sp binding sites [76]. These observations indicate that C/EBPβ can alter HIV-1 LTR-directed transcription in the absence or presence of known C/EBP-binding sites within the brain and that the effects are cell-type specific. Therefore, C/EBPβ could be important during the course of HIV-1-associated brain dysfunction. However, overexpression of C/EBPγ did not significantly change the HIV-1 basal activity but resulted in the inhibition of C/EBPβ-induced stimulation in microglial cells and TC-620 cells [76]. It will be important to understand the regulation of the expression of these two C/EBP factors in the brain and explore the relationship with HIV-1-associated brain disease.
Role of C/EBPβ in HIV-1 infection in the lung
Enhancement of HIV-1 replication occurs in primary blood monocytes cultured in vitro after infection with Mycobacterium tuberculosis; however, HIV-1 replication is repressed when infected monocytes are differentiated into macrophages. One explanation for this difference may be found in the observation that following infection with M. tuberculosis, only the stimulatory 37 kDa C/EBPβ (C/EBPβ-2 or LAP in mice) was induced in monocyte-like cells (THP-1). In contrast, M. tuberculosis infection of PMA-treated THP-1 cells or primary macrophages was shown to induce expression okf the inhibitory C/EBPβ (LIP or C/EBPβ-3) [78]. C/EBPβ-binding sites are required for repressing HIV-1 LTR transcription in macrophages infected with M. tuberculosis. Mutagenesis of the three C/EBP-binding sites abolished the repression of LTR activity caused by C/EBPβ-3 [78]. HIV-1 replication in vivo was inhibited in noninflammed lung macrophages but increased during co-infection with M. tuberculosis [79]. Specifically, resting alveolar macrophages isolated from patients with HIV/AIDS without detectable viral replication, expressed large quantities of C/EBPβ-3. However, the expression of C/EBPβ-3 was decreased in inflamed lung tissue with high levels of viral replication [78]. The ratio of inhibitory to stimulatory C/EBPβ is 1.0 in patients with HIV/AIDS without detectable viral replication. Thus, it has been proposed that pulmonary tuberculosis causes the abolition of inhibitory C/EBPβ expression in the lung and is responsible for enhanced HIV-1 replication among HIV-1-infected patients co-infected with M. tuberculosis. Therefore, PMA-treated THP-1 cells stimulated with M. tuberculosis are similar to alveolar macrophages in the noninflammed lung of HIV-1-infected patients, in whom HIV-1 replication was inhibited, whereas HIV-1 replication was increased in aleveloar macrophages infected with M. tuberculosis. The differences between the in vitro and in vivo observations might be caused by the host immune response to M. tuberculosis in vivo. For example, surfactant protein-A, which is part of the innate immune response in the lung [80], has the ability to reduce the expression of C/EBPβ-3 [81]. Furthermore, in the lung of HIV-1-infected patients with tuberculosis, the contact between CD4+ T lymphocytes and alveolar macrophages is important for maximal HIV-1 production in tissue macrophages, and this interaction may reduce the production of C/EBPβ-3 and activate NF-κB [82,83]. Z-100, which is an arabinomannan extracted from M. tuberculosis, has recently been shown to inhibit HIV-1 replication by induction of the inhibitory C/EBPβ isoform in human monocyte-derived macrophages [84] and acutely infected macrophages [85,86]. Thus, C/EBPβ may promote or inhibit HIV-1 transcription in the lung depending on the specific C/EBPβ isoforms being expressed in a given situation. More recent studies have demonstrated that the expression of tyrosine kinase hematopoietic cell kinase and relative amounts of C/EBPβ-2 and C/EBPβ-3 play important roles in determining the overall level of HIV-1 replication in macrophages. Decreasing the relative concentration of C/EBPβ-3, thereby enhancing the ratio of C/EBPβ-2 to C/EBPβ-3 by treatment with an antisense oligonucleotide probe for C/EBPβ-3 [87] or erythromycin derivatives [88], significantly stimulated HIV-1 replication in macrophages. Taken together, these studies suggest that enhancing the expression of C/EBPβ-3 may be a useful way to control HIV-1 replication in AIDS patients.
C/EBPβ alters coreceptor expression on HIV-1-infected cells
HIV-1 requires the presence of CD4 and either CCR5 or CXCR4 to successfully infect cells. Seven putative C/EBPβ-binding sites have been found within the CCR5 gene [65]. Nuclear extracts from U-937 cells transiently transfected with the LAP or LIP expression vectors formed specific DNA–C/EBP complexes with each of these seven C/EBP-binding sites. However, only two of seven, which are located in the 3′ portion of the intron, formed specific DNA–protein complexes with Jurkat T-cell nuclear extracts transiently transfected with the LAP or LIP expression vectors [65]. These observations further demonstrate that the interaction between C/EBPβ factors and their cognate binding sites occurs in a cell type-specific manner. Functionally, LAP or LIP was shown to alter CCR5 expression in U-937 cells and Jurkat T cells by different mechanisms. In U-937 cells, LAP or LIP was shown to activate or inhibit CCR5 expression through sites located either in the promoter region or in the intron of the CCR5 gene, whereas in Jurkat T cells, the presence of the intronic cis-regulatory regions was required for LAP- or LIP-mediated activation or repression [65]. Compared with that observed in healthy donors, a significant increase in LAP expression in primary peripheral blood mononuclear cells has been observed in HIV-1-infected individuals [65]. Thus, it has been suggested that the enhanced LAP activity may correlate with the higher expression of CCR5 on circulating lymphocytes in AIDS patients. Interestingly, the binding of HIV-1 envelope glycoprotein gp120 to the CD4 receptor on T cells results in the rapid accumulation of C/EBPβ and enhanced DNA binding ability with respect to the C/EBP consensus binding site [89]. These studies indicate that LAP induction by HIV-1 could increase the number of cells susceptible to infection by stimulation of CCR5 expression in CD4+ cells [65]. However, overexpression of LIP did not affect CD4 or CXCR4 expression on the U-937 cell surface, and LIP was unable to block HIV-1 entry (CCR5 was not detectable in their studies) [90]. Thus, in addition to regulating HIV-1 LTR function, C/EBPβ has the ability to affect HIV-1 coreceptor expression and influence HIV-1 infection during the course of viral disease.
Proteins that interact with C/EBPβ in HIV-1 infection
C/EBPβ can form dimers with other transcription factors through the leucine-zipper domain or interact with co-activators via the trans-activation domain, which has been shown to enhance the capability of C/EBPβ to influence HIV-1 gene expression and overall viral replication (Figure 5). These protein partners include chromatin remodeling proteins, transcription factors and viral proteins.
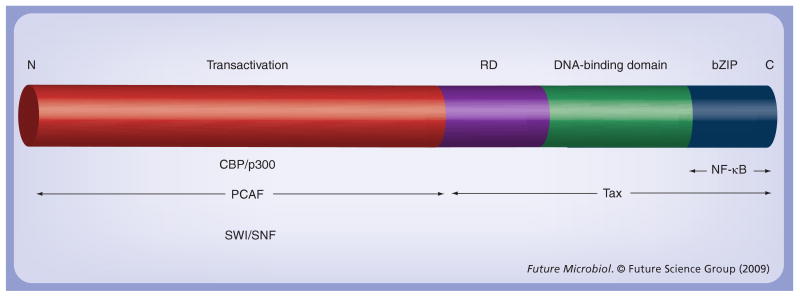
Figure 5. Cellular factors associated with CCAAT/enhancer-binding protein-β.
Several examples of cellular factors and viral proteins associated with different regions of CCAAT/enhancer-binding protein β are presented.
bZIP: Basic region leucine zipper; CBP: cAMP response element binding protein-binding protein; NF-κB: Nuclear factor-κB; PCAF: p300/CBP-associated factor; RD: Regulatory domain; SWI/SNF: Switch mating type/sucrose nonfermenting complex.
Chromatin remodeling proteins
C/EBPβ-2 or LAP can activate HIV-1 transcription by recruiting histone acetyltransferase, including cAMP response element binding protein (CREB) binding protein (CBP)/p300, p300/CBP-associated factor (PCAF) which can modify chromatin structure and make DNA more accessible to transcription factors [91]. Therefore, the interaction between C/EBPβ and histone acetyltransferase may facilitate HIV-1 transcription by promoting the remodeling of chromatin structure. CBP/p300 does not interact with specific DNA binding sites directly; rather, they link specific transcription factors to the basal transcriptional machinery via protein–protein interactions. The N-terminus of C/EBPβ (Figure 5) and the C-terminal half of p300 (amino acids 1752–1859), which partially overlaps with the E1A binding domain of p300, are responsible for this specific interaction [92]. E1A, which binds directly to CBP/p300, suppressed chicken C/EBPβ-mediated transactivation. This inhibition was restored by overexpression of p300 [92–94]. Co-transfection of C/EBPβ and CBP expression constructs results in synergistic activation of the HIV-1 LTR, whereas co-expression of E1A and CBP/p300 inhibits C/EBPβ-mediated HIV-1 LTR transactivation [90]. Additionally, full-length C/EBPβ induced phosphorylation of p300 at multiple sites within the C-terminus of p300 and changed the subnuclear localization of p300. Specifically, p300 has a heterogenous cellular distribution in the absence of C/EBPβ, whereas in the presence of C/EBPβ it was evenly distributed. Furthermore, C/EBPβ-induced phosphorylation of p300 was required for full activity of p300 as a coactivator of C/EBPβ function [95]. Synergistic activation of the endogenous C/EBPβ-dependent gene by p300 and C/EBPβ has been reported, and the synergistic interaction between Myb and C/EBPβ is increased by p300 [92]. C/EBPβ-2 or LAP interacts physically with PCAF through its transactivation domain (Figure 5), thereby activating the HIV-1 LTR in a dose-dependent manner. However, C/EBPβ-3, which lacks the transactivation domain, does not have the ability to interact with PCAF and is unable to mediate the displacement of nucleosomes that repress HIV-1 transcriptional initiation and fails to activate HIV-1 transcription [90]. As discussed previously, the N-terminal domain of LAP* is able to interact with SWI/SNF [49]. Since SWI/SNF has the ability to alter the histone structure, it is reasonable to suppose that the LAP*–SWI/SNF interactions could also affect HIV-1 gene expression. In addition, the physical and functional interactions between C/EBPα and histone acetyltransferase Tat-interactive protein (TIP) 60 have been shown to be important for leukemiogenesis, and TIP 60 was able to acetylate the histones at C/EBPα-dependent genes [96,97]. Thus, if the interaction between C/EBPα or other C/EBP factors and TIP 60, or between C/EBPβ and other histone acetyltransferases could influence HIV-1 transcription, it will be important to understand the detailed signaling and transcriptional control pathways involved in the regulation of HIV-1 gene expression by members of the C/EBP family.
In contrast, histone deacetylase (HDAC) has been shown to mediate deacetylation of histone tails leading to the inhibition of binding of basal transcription factors to their DNA targets. HDAC inhibitors have also been shown to induce expression of latent viruses in resting CD4+ T cells without fully activating cells or enhancing de novo viral replication, suggesting that HDAC inhibitors may be a new strategy to prevent the activation of latent HIV proviral DNA [98–100]. Specifically, the NF-κB–HDAC1 complex has been shown to constitutively bind the latent HIV-1 LTR and cause histone deacetylation and the formation of a repressive chromatin structure, thereby affecting RNA polymerase II recruitment to the LTR and inhibiting transcription initiation [98]. In addition to directly interacting with HDAC, the transcription factor Sp1 has been shown to be able to recruit HDAC1, first by interacting with c-myc with the complex Sp1–c-myc, in turn recruiting HDAC1 and repressing HIV-1 LTR expression [100]. Interestingly, C/EBPβ has the ability to inhibit peroxisome proliferator-activated receptor-β gene expression through recruiting the transcription repressor complex containing HDAC1 to a specific C/EBP-binding site on the peroxisome proliferator-activated receptor-β promoter [101]. Dissociation of C/EBPβ from the corepressor complex containing HDAC1 has been shown to be involved in C/EBPα promoter activation and is important for preadipocyte differentiation in the presence of glucocorticoid [102–104]. Since C/EBPβ interacts with HDAC1 and HDAC1 has been shown to be involved in inhibiting HIV-1 repression, it could be possible that C/EBPβ may also repress HIV-1 transcription by forming a repressor complex with HDACs, especially HDAC1. Myc has been shown to mediate transcriptional repression through the recruitment of HDACs [105]. Therefore, it is possible that Myc is also involved in HIV-1 gene repression by forming repressor complexes such as C/EBPβ–Myc, or C/EBPβ–CBP/p300–Myc complex whose interaction (CBP/p300) has been observed [106,107].
Transcription factors
NF-κB (p50 & p65)
NF-κB and the DNA recognition elements that mediate its interaction with the integrated proviral DNA play a critical role in the regulation of HIV-1 gene expression [108–111]. The Rel homology domain of NF-κB and the bZIP region of C/EBPβ mediate the physical interaction between NF-κB and C/EBPβ (Figure 5). Functionally, coexpression of C/EBPβ and NF-κB inhibited NF-κB-induced expression of promoters containing NF-κB binding sites but synergistically increased the expression of promoters containing C/EBP-binding sites [112]. The transcriptional activation of the HIV-1 LTR by p50–C/EBPβ complexes requires the DNA-binding domain of p50 and the transcription activation domain of C/EBPβ in NTera-2 cells, a human teratocarcinoma cell line [65,113]. As discussed previously, overexpression of C/EBPβ in U-937 cells results in activation of a deletion mutation within the HIV-1 LTR that contains bases -108 to +83 of the LTR, including both NF-κB binding sites but without the three adjacent Sp binding sites [61]. Recent studies have indicated that ectopic expression of C/EBPβ and p65 is able to significantly increase the ability of p65 to bind to the HIV-1 LTR while slightly decreasing the ability of C/EBPβ to bind the HIV-1 LTR. However, in the presence of Tat, co-transfection of C/EBPβ and p65 decreased p65 DNA binding ability [114]. This observation could be explained by the formation of a Tat–C/EBPβ complex, which could result in the inhibition of p65–DNA complex formation [115]. Functionally, C/EBPβ decreases the cooperation of HIV-1 LTR-directed transcription by p65 and Tat, whereas p65 does not affect Tat and C/EBPβ synergistically transactivating the HIV-1 LTR [114].
Activating transcription factor/CREB
The C/EBP US1 binding site within the HIV-1 LTR is located immediately downstream of the activating transcription factor (ATF)/CREB binding site and immediately upstream of the distal NF-κB binding site in the HIV-1 LTR [116]. Consequently, it is possible that the nucleotide sequence variation observed in the ATF/CREB or NF-κB binding site could affect C/EBP US1 DNA-binding ability. Electrophoretic mobility shift assays have shown that ATF/CREB binding sites in the HIV-1 LTR with high affinity for CREB-1 are able to enhance C/EBPβ-binding to relatively weak C/EBP US1 binding sites [117]. Furthermore, different ATF/CREB binding site configurations have been shown to dictate the type of cognate factor recruited to the ATF/CREB site. The differential recruitment of factors to this site may play an important role in the recruitment of cognate factors to the immediately adjacent C/EBP site. Consistent with these observations, low-affinity ATF/CREB binding sites do not exhibit nearly the same level of enhanced C/EBPβ DNA binding capacity [117]. In addition, a high affinity C/EBP US1 binding site results in enhanced CREB-1 recruitment to a low affinity ATF/CREB site. Functional studies have shown that ATF/CREB binding sites affect the activity of the HIV-1 LTR with low affinity C/EBP US1 after IL-6 stimulation [117]. These studies support the hypothesis that protein–protein interactions of C/EBP factors with members of the ATF/CREB family may be important for HIV-1 transcription and that C/EBP–DNA interactions could also be affected by other DNA–protein interactions that occur at other neighboring or possibly distant transcription factor binding sites.
Rad51
Rad51 is a major component of the DNA repair system, playing an important role in homologous recombination by pairing and transferring single strands of DNA to a homologous duplex DNA [118]. DNA repair processes are important for sustaining chromosomal integrity and genomic stability. HIV-1 infection can cause significant changes in host-gene expression, including genes encoding DNA repair proteins. Previous studies have indicated that HIV-1 infection was able to increase Rad51 gene expression in astrocyte and neuronal cell cultures [119]. A physical interaction between Rad51 and C/EBPβ has been observed, and Rad51 has been shown to slightly increase C/EBPβ DNA binding ability to US1 in vivo and in vitro, which may be caused by Rad51-induced changes in DNA structure leading to a more functional interaction between C/EBPβ and its binding sites, thus promoting HIV-1 LTR-directed transcription [120]. Moreover, overexpression of Rad51 in astrocytes is able to synergistically transactivate the HIV-1 LTR either with C/EBPβ alone or with combined C/EBPβ and Tat together [120]. Additionally, C/EBPζ, which can act as an inhibitor of C/EBPβ or as activator of selected C/EBPβ target genes, also synergizes Rad51 activation and Tat-induced transcription of the HIV-1 LTR in cultured astrocytes [120].
Viral proteins
Tat
Tat has been shown to be critically important to achieve high levels of HIV-1 gene expression [121–123] and physically interacts with C/EBPβ in vitro and in vivo [124]. The amino acid residues 47–67 of Tat are critical for interaction with C/EBPβ. Functionally, Tat enhances C/EBPβ DNA binding to the HIV-1 LTR and IL-6 promoter [120,124,125] and activates C/EBPβ expression in human U-373MG astroglial cells in a dose-dependent manner [126]. Furthermore, Tat specifically increases the nuclear levels of C/EBPβ. However, the expression of active C/EBPβ has been shown to stimulate a Tat-induced production of nitric oxide, which may be related to neuronal abnormalities in HAD, whereas the expression of inhibitory C/EBPβ represses the production of nitric oxide [2,127,128].
In addition, the cooperative interaction of C/EBPβ and Tat modulates MCP-1 gene transcription in astrocytes [115]. C/EBPβ has the ability to stimulate basal and Tat-mediated MCP-1 transcription, and Tat has been shown to increase binding of C/EBPβ to the MCP-1 promoter. MCP-1 plays a critical role in monocytic infiltration to the site of injury or inflammation, and HIV-1-infected patients with HAD express higher levels of MCP-1 in the brain, cerebrospinal fluid and macrophages [129–131]. The cooperation between Tat and C/EBPβ is important in HIV-1 infection, especially in HIV-1-associated neurologic dysfunction, since:
Brain macrophages and microglia are the primany productively infected cell types within the CNS during all stages of HIV-1 infection [132];
C/EBPβ plays an important role in HIV-1 infection in macrophages that can migrate to the brain;
Tat itself has been shown to induce neuronal dysfunction/toxicity [133–135].
Additionally, physical and functional interactions between C/EBPβ and Tat's partners, cyclin T1 and CDK9, have been observed [36]. Activation of the HIV-1 LTR by C/EBPβ was further increased in the presence of CDK9 or CDK9 and Tat, which may be related to the phosporylation of C/EBPβ by CDK9 [36]. However, these effects are inhibited in the presence of C/EBPζ [36].
Viral protein R
Viral protein R (Vpr) is a multifunctional protein encoded by the HIV-1 genome expressed in the early stages of infection and is present in the intact infectious virion [136–138]. Previous studies have demonstrated that Vpr activates HIV-1 transactivation by interacting with transcription factors bound to the LTR or by directly interacting with the LTR [139–142]. Specifically, Vpr has been shown to associate with Sp1 in the context of the G/C box array but with neither Sp1 nor the Sp binding sites alone [141]. Recent studies have demonstrated that Vpr binds with a relatively higher affinity to an oligonucleotide probe spanning 37 base pairs compared with an oligonucleotide probe spanning 18 base pairs, both of the oliognucleotide probes being centered on C/EBP US1 [143]; the sequence variations at C/EBP US1 results in significant alterations in the affinity of this region for the viral promoter of Vpr [143]. Additional studies demonstrate that the LTR region required for Vpr binding physically maps to sequences contained within the C/EBP US1 and NF-κB binding site II. In particular, the studies further demonstrate a correlation between the diagnosis of HAD and the increased prevalence of HIV-1 LTRs containing a US1–3T variant (C-to-T change at position 3 compared with consensus subtype B US1). Interestingly, the US1–3T variation exhibited a relatively low DNA binding affinity for C/EBP but a relatively high affinity for Vpr, suggesting that sequence-specific interactions between cis-acting elements in the LTR, C/EBP factors and Vpr may play important roles in the pathogenesis of HAD or possibly other forms of HIV-1-associated neurologic dysfunction [144]. In addition, Vpr has been shown to enhance the DNA binding affinity of C/EBPβ during Vpr induction of IL-6, IL-8 and IL-10 expression [145].
Interactions of C/EBPβ with cytokine & chemokines involved in HIV-1 infection
HIV-1 infection results in the abnormal secretion of a number of different cytokines, which is likely to be integrally involved in the pathogenesis of HIV-1 infection. It has been well documented that the expression levels of IL-1, IL-6 and TNF-α are elevated during HIV-1 infection, which can stimulate virus replication, whereas the expression levels of IFN-α, IFN-β and IL-16, which inhibit HIV-1 replication, are decreased during HIV-1 infection [146,147]. Many genes that encode cytokines and chemokines have C/EBP-binding sites within their promoter. Deletion of the C/EBPβ gene in mice causes elevated serum IL-6 levels and lowered IL-12 levels [148]. Furthermore, the expression of IL-6, IL-1β, IL-12, p35 and TNF-α mRNAs are impaired in C/EBP-/- macrophages, while the level of IL-12 p40, RANTES, and MIP-1β is upregulated [149]. C/EBPβ uses two major mechanisms to regulate the production of cytokines and chemokines: C/EBPβ binds to C/EBP-binding sites located within the promoter of a number of different genes that include IL-6, IL-1β, TNF-α and MCP-1 [150–153], and C/EBPβ interacts with signal pathway factors (Smad-3 and -4) of the cytokine TGF-β pathway [154]. Specifically, C/EBPβ-2 has been shown to activate human TNF-α gene transcription in cells of the monocyte–macrophage lineage via direct binding to the region between -95 and -36, relative to the transcription start site. However, C/EBPβ-3 is unable to activate the TNF-α promoter, and expression of C/EBPβ-3 inhibits the activation by C/EBPβ-2 in cells of the monocyte–macrophage lineage [155–157]. Although a number of studies have demonstrated the regulation of IL-6 by C/EBPβ in various cell types including macrophages, fibrosarcoma cells, embryonic kidney cells, vascular smooth muscle cells and enterocytes [150,151], the mechanism whereby C/EBPβ regulates IL-6 secretion is unclear. Phosphorylation of C/EBPβ at threonine 189 has been shown to increase its ability to activate the IL-6 promoter [34]. However, another study reported that the C/EBPβ bZIP domain was sufficient to support LPS-induced activation of IL-6 expression [20] and the derepression of the regulatory region was unable to impact IL-6 gene expression [158]. IL-1β is a potent inflammatory cytokine that is primarily expressed within activated cells of the monocyte–macrophage lineage. C/EBPβ has been shown to activate LPS-induced IL-1β gene expression in THP-1 cells [159]. Two C/EBP-binding sites have been identified at -2579 to -2591 and -3107 to -3118 base pairs relative to the transcription start site within the promoter of the MCP-1 gene [160]. C/EBPβ is able to activate LPS-inducible expression of MCP-1 in the B lymphoblastic P388 cell line [161]. The region from amino acids 64 to 101 of C/EBPβ, which contains parts of AD2 and AD3 [162] as well as the bZIP domain [20], has been shown to be responsible for activation of LPS-inducible MCP-1 expression, whereas the regulatory regions suppress C/EBPβ activity with respect to MCP-1 expression [162].
The expression level of TGF-β has been shown to be elevated in the CNS of patients infected with HIV-1 or cells treated with Tat protein [163–165]. Members of the TGF-β family mediate their functions from membrane to nucleus through their downstream effectors, Smad-3 and -4 [166,167]. Smad-3 and -4 interact functionally and physically with C/EBPβ [154]. Specifically, C/EBPβ binds to Smad-4 alone or to Smad-3 and Smad-4 together but not to Smad-3 alone. Functionally, Smad-3 does not affect C/EBPβ transactivation, whereas Smad-3 and Smad-4 together significantly decrease C/EBPβ transactivation. Smad-3 and -4, either alone or together, inhibit the interaction of C/EBPβ with DNA binding sites through protein–protein interactions. Furthermore, the interaction of C/EBPβ and Tat decreased in the presence of Smad-3 [115,154]. Therefore, C/EBPβ may be involved in the way that TGF-β signaling factors regulate HIV-1 LTR transcription through interaction with Smad-3 and Smad-4.
C/EBPβ & human T-cell leukemia virus type-1
Human T-cell leukemia virus type-1 (HTLV-1) causes adult T-cell leukemia (ATL) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP); however, approximately only 5% of individuals infected with HTLV-1 develop ATL or HAM/TSP, and approximately 95% remain healthy carriers [168–170]. The virus-encoded Tax protein plays an important role in HTLV-1 transcription and replication [171,172]. Tax itself is unable to bind DNA directly but activates transcription by interacting with, or modifying, other cellular transcription factors, such as ATF/CREB and CBP/p300 [173–176]. The interaction between Tax and ATF/CREB is important for Tax-mediated HTLV-1 LTR transactivation [177,178]. Tax directly interacts with members of the ATF/CREB family and promotes the dimerization of ATF/CREB factors, which has been shown to increase the DNA-binding ability of the ATF/CREB–Tax complex to the viral promoter [173,179].
Expression of C/EBPβ-1 or LAP* in Jurkat T cells and U-937 promyelocytic cell lines causes a modest upregulation of basal HTLV-1 LTR activation. However, the expression of LAP*, LAP or LIP results in the repression of Tax-mediated LTR transactivation in both Jurkat T cells and U-937 cells [180,181]. Studies with human C/EBPβ indicate that this repression is the result of a direct interaction between the central region of C/EBPβ (amino acids 112–271) and the Tax protein [181]. In particular, human C/EBPβ mutants (from amino acids 1 to 315), which lack the bZIP domain, were still able to inhibit Tax-dependent transcription, and the inhibitory activity was similar to that of full-length C/EBPβ (amino acids 1–345). In these studies, a C/EBPβ mutant containing only the bZIP region (from amino acid 271 to 345) was unable to impact Tax-mediated transactivation [181]. However, another study showed that both rodent LAP and LIP were able to inhibit Tax-mediated LTR transactivation. Rodent C/EBPβ mutants that lack sequences within the basic region or the leucine-zipper region were unable to inhibit Tax-mediated transactivation of the HTLV-1 LTR, indicating that the C-terminal bZIP region was required for C/EBPβ inhibition of Tax-mediated HTLV-1 LTR transcription [180]. The difference between these results may be related to the small degree of structural disparity between rodent and human C/EBPβ. However, based on the two studies, it has been suggested that the center and C-terminus of C/EBPβ, rather than the AD located at the N-terminus, are required for repressing Tax-mediated transcription of the HTLV-1 LTR (Figure 5). Alternatively, the ability of C/EBPβ to inhibit Tax-dependent trans-activation of the HTLV-1 LTR may involve the formation of heterodimers with CREB-2, and C/EBPβ–CREB heterodimerization may prevent ATF/CREB-mediated Tax binding to the HTLV-1 promoter. The deletion of the leucine-zipper region disrupts C/EBPβ interactions with CREB-2, preventing C/EBPβ from inhibiting Tax-mediated HTLV-1 LTR transcription. Thus, C/EBPβ may inhibit Tax function by competing for Tax binding to CREB-2 [180].
Although C/EBPβ forms DNA-protein complexes with Tax-responsive element 1 repeat III (Figure 3) in the HTLV-1 LTR, binding to the LTR is not required in order for C/EBPβ to repress Tax-dependent LTR activity. C/EBPβ is still able to inhibit Tax-mediated transactivation of the HTLV-1 LTR in the presence of mutated Tax-responsive element-1 repeat III [180]. In addition to repressing Tax-mediated transactivation, C/EBPβ is able to efficiently decrease Tax synthesis from an infectious HTLV-1 molecular clone (ACH) [181]. C/EBPβ repression of Tax-dependent HTLV-1 LTR transactivation may partly explain why only approximately 5% of individuals infected with HTLV-1 develop ATL or HAM/TSP. C/EBPβ may help infected T cells to escape immune surveillance by inducing a low level of Tax production in infected T cells and/or inhibiting Tax-mediating transactivation. Interestingly, some studies have also demonstrated that Tax can enhance the DNA binding activity of C/EBPβ, thereby mildly stimulating transcription [182], whereas other studies have shown that Tax can repress C/EBPβ-mediated transactivation [181]. These studies clearly suggest that the interaction between Tax and C/EBPβ requires additional investigation to clarify the intracellular mechanisms whereby these proteins control cellular and/or viral gene expression.
C/EBPβ & SIV
SIV infection of macaques has provided an excellent model for studying HIV-1 infection. SIV causes clinical and pathological manifestations that are similar to HIV infection. SIV-infected macaques develop diseases that include AIDS and associated neuropathologic conditions [183]. Some degree of structural similarity has also been observed between the HIV-1 and SIV LTRs. Electrophoretic mobility shift analyses using U-937 nuclear extracts demonstrate the existence of four C/EBP-binding sites within the SIV LTR, designated US2, US1, DS1 and DS2 (US2: nt -385 to -372, US1: nt -100 to -87, DS1: nt +133 to +146 and DS2: nt +267 to +281, relative to the transcription start site) (Figure 3) [184]. LAP or LIP form specific DNA–protein complexes with the four elements [184,185]. Furthermore, each C/EBP-binding site is involved in SIV LTR basal transcription. Specifically, US2, DS1 and DS2 were shown to be negative regulators and US1 was shown to be a positive regulator [Nonnemacher and Wigdahl, Unpublished Data]. LAP activated basal SIV LTR-directed transcription, whereas an increase in the ratio of LIP to LAP inhibited LAP-mediated SIV LTR-directed transcription, indicating that, like the HIV-1 LTR, SIV LTR activation can also be differentially regulated by the ratio of LAP to LIP [186]. The mechanism whereby LIP inhibits LAP activity is related to LIP suppression of LAP-related acetylation of histone H4 at the SIV LTR [186].
An accelerated and consistent model of SIV CNS disease was recently developed. All infected macaques developed AIDS, and more than 90% developed encephalitis within 3 months post inoculation [185,187–189]. These animals exhibited the classical stages of HIV/SIV disease: acute virus replication, an asymptomatic stage and resurgence of virus replication in the brain, concomitant with the development of AIDS [185]. During the most advanced stage of the disease, the CNS of infected macaques exhibited characteristics similar to those of HAD [185]. Reports indicate that IFN-β or LPS inhibited HIV replication in macrophages in vitro by a mechanism involving the increased expression of LIP and a resultant change in the ratio of LIP to LAP in promonocytic U-937 cells [78,131,186]. Similarly, IFN-β inhibited SIV replication in macrophages isolated from the infected macaques and cultured in vitro. The levels of IFN-β in the brains of SIV-infected macaques increased from 7 to 21 days after infection with a consequential increase in the expression of LIP [185]. Similarly to what happens in the brain, IFN-β and LIP also contribute to the suppression of acute SIV replication in the lung and result in low levels of viral replication during the asymptomatic stage [186]. Specifically, the levels of LIP expression in SIV-infected lungs increased between 10 and 21 days after inoculation [186]. As a result, it was proposed that SIV/HIV may invade the CNS during acute infection when higher levels of LAP as opposed to LIP, and chromatin remodeling events, facilitate SIV/HIV LTR-dependent transcription. In this regard, acute infection leads to IFN-β production in the brain that eventually leads to an increase in the expression of LIP expression, resulting in a decrease in the ratio of LAP to LIP, thus repressing LAP involvement in LTR transcription toward the end of the acute infection stage. Therefore, LAP/LIP and IFN-β may play critical roles in SIV/HIV latency and might be important for the control of acute SIV replication in tissue in which macrophages are the primarily infected cells [186]. Recent studies from SIV-infected rhesus macaques with chronic diarrhea have shown that C/EBPβ is the mediator of the observed inflammation. The significantly increased expression of C/EBPβ is associated with persistent localized immune reaction, damage to the intestinal mucosal barrier and function and facilitates viral replication [190].
Conclusion & future perspective
C/EBP family members, especially C/EBPβ, are known to regulate the gene expression of HIV-1 and other retroviruses (HTLV and SIV) through a variety of mechanisms, including affecting CCR5 expression, virus promoter activities, cytokine and chemokine production and virus protein function. The regulation of retrovirus transcription is highly complex and requires a detailed understanding of how these transcriptional factors and signaling pathways function. Many key questions remain to be answered. As C/EBPβ has been implicated in interacting with chromatin remodeling proteins, it is likely that C/EBPβ has the ability to modify nucleosome position, alter the promoter accessibility and affect the accessibility of other transcription factors to the cellular or viral promoter. Thus, it is essential to understand how C/EBPβ affects promoter activities by remodeling HIV-1 chromosome structure. Further characterization of the relationship between the C/EBP and histone acetyltransferase, HDAC and HDAC inhibitors would contribute to this study. Nucleotide polymorphisms within HIV-1 LTR C/EBP-binding sites have been observed within integrated proviruses in the peripheral blood or brain of HIV-1-infected patients and may be useful as markers with respect to predicting HIV disease progression. Relevant to the control of cellular and viral genes by members of the C/EBP family, a number of studies have demonstrated that mutations within the genes encoding C/EBP factors are linked to the occurrence of certain cancers [191–193]. However, it is not known whether C/EBP factors containing amino acid polymorphisms may impact HIV-1-associated disease processes or whether the interaction of C/EBP factors containing polymorphic alterations in amino acid sequences would exhibit altered affinity for different C/EBP-binding site configurations, thereby altering regulatory patterns observed in cellular and/or viral promoters containing these elements. Overall, C/EBPβ has three isoforms that act as activators or suppressors in a given environment, depending on which isoform predominates. Therefore, exploring how each isoform is regulated and involved in determining positive or negative regulation with regard to retrovirus gene expression and what co-activators or co-repressors are involved in these activities are also especially important. In addition, it will be important to characterize how each C/EBP family member regulates retrovirus gene expression in different cell types and physiological states. An improved understanding of the network of interactions involving C/EBPs with other proteins, transcription binding sites and regulatory pathways could enhance our ability to control retrovirus pathogenesis and disease.
Executive summary.
Overview of CCAAT/enhancer-binding protein family
The CCAAT/enhancer-binding protein (C/EBP) family includes six members that share a highly conserved C-terminal region.
The expression patterns of C/EBP factors are diverse in different cell types.
C/EBPβ gene & corresponding proteins
C/EBPβ functional domains include three activation domains, two regulatory domains, and a basic region leucine zipper DNA binding region.
C/EBPβ proteins have three isoforms (C/EBPβ-1, 2 and 3 in humans, and full-length liver activator protein, liver activator protein and liver inhibitory protein in rodents).
C/EBP & HIV-1 transcription in cells of the monocyte–macrophage lineage
Three C/EBP binding sites (US1, US2, and US3) are located upstream and one binding site is located downstream (DS3) of the transcriptional start site within the HIV-1 long terminal repeat (LTR).
C/EBPβ is able to regulate HIV-1 transcription independent of C/EBP-binding sites.
C/EBPβ & HIV-1 transcription in T cells
The expression of C/EBPβ has been observed in T-cell lines and primary T cells.
C/EBP-binding sites are dispensable for HIV-1 replication in T cells.
The role of C/EBP in HIV-1 transcription in the brain
C/EBPβ, C/EBPδ, and C/EBPγ are expressed in astrocytes and in microglial and glial cells.
Proinflammatory cytokines are able to enhance C/EBPβ expression in primary astrocytes, and C/EBPβ may affect the production of these cytokines.
The role of C/EBPβ in HIV-1 infection in the lung
HIV-1 replication in the lung is dependent on the ratio of inhibitory (C/EBPβ-3) to stimulatory (C/EBPβ-2) protein expressed within patient-derived alveolar macrophages.
C/EBPβ alters CCR5 expression on HIV-1-infected cells
Seven putative C/EBP binding sites have been found within the CCR5 gene.
C/EBP affects CCR5 expression in U-937 cells and Jurkat T cells in cell type-specific ways.
Increased C/EBP levels may be related to higher expression of CCR5 on lymphocytes in patients with AIDS.
Proteins that interact with C/EBPβ during HIV-1 infection
Chromatin remodeling proteins: cAMP response element binding protein (CREB)-binding protein(CBP)/p300, p300/CBP-associated factor and switch mating type/sucrose nonfermenting complex.
Transcription factors: nuclear factor-κB, Sp1 and activating transcription factor/CREB.
DNA repair proteins: Rad51.
Viral proteins: transactivator of transcription and viral protein R.
Interaction of C/EBPβ with cytokines or chemokines involved in HIV-1 infection
Two mechanisms are involved: C/EBPβ binding to the C/EBP binding sites on the gene promoter (TNF-α, IL-6, and MCP-1) and/or C/EBPβ interacting with signal pathway factors (TGF-β).
C/EBPβ & human T-cell leukemia virus type-1
Center and C-terminus of C/EBPβ are necessary for repressing Tax-mediated transcription of the human T-cell leukemia virus type-1 LTR.
C/EBPβ–CREB heterodimerization may compete with the Tax–CREB interaction, resulting in inhibition of Tax transactivation of the human T-cell leukemia virus type-1 LTR.
C/EBPβ binding to the HTLV-1 LTR is not required for C/EBPβ to repress Tax-dependent LTR activity.
C/EBPβ & SIV
Four C/EBP-binding sites have been identified within the SIV LTR.
Acute infection leads to IFN-β production and subsequently to an increase in inhibitory C/EBPβ expression (LIP), which may contribute to viral latency in the lung, brain and potentially other tissues.
Conclusion & future perspective
C/EBP factors regulate retrovirus gene expression through a variety of mechanisms.
Investigations concerning the mechanism of C/EBPβ regulation of HIV-1 promoter that involve remodeling chromatin structure will be important.
The relationship between amino acid polymorphisms of C/EBPβ and HIV-1 disease progression remains unexplored.
Footnotes
Financial & competing interests disclosure: The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Yujie Liu, Department of Microbiology & Immunology, Center for Molecular Virology & Neuroimmunology, Center for Cancer Biology, Philadelphia, PA 19129, USA.
Michael R Nonnemacher, Department of Microbiology & Immunology, Center for Molecular Virology & Neuroimmunology, Center for Cancer Biology, Philadelphia, PA 19129, USA.
Brian Wigdahl, Department of Microbiology & Immunology, Center for Molecular Virology & Neuroimmunology, Center for Cancer Biology, Philadelphia, PA 19129, USA, Tel.: +1 215 762 8399, Fax: +1 215 762 1955, bwigdahl@drexelmed.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Isshiki H, Sugita T, et al. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ First report to identify CCAAT/enhancer-binding protein (C/EBP)β.
- 3.Chang CJ, Chen TT, Lei HY, Chen DS, Lee SC. Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Mol Cell Biol. 1990;10:6642–6653. doi: 10.1128/mcb.10.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poli V, Mancini FP, Cortese R. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell. 1990;63:643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- 5.Pei DQ, Shih CH. An “attenuator domain” is sandwiched by two distinct transactivation domains in the transcription factor C/EBP. Mol Cell Biol. 1991;11:1480–1487. doi: 10.1128/mcb.11.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBPβ (NF-M) transcriptional control: activation through derepression. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 7.Antonson P, Stellan B, Yamanaka R, Xanthopoulos KG. A novel human CCAAT/enhancer binding protein gene, C/EBPε, is expressed in cells of lymphoid and myeloid lineages and is localized on chromosome 14q11.2 close to the T-cell receptor α/δ locus. Genomics. 1996;35:30–38. doi: 10.1006/geno.1996.0319. [DOI] [PubMed] [Google Scholar]
- 8.Wang XZ, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 9.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant–negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka R, Kim GD, Radomska HS, et al. CCAAT/enhancer binding protein ε is preferentially up-regulated during granulocytic differentiation and its functional versatility is determined by alternative use of promoters and differential splicing. Proc Natl Acad Sci USA. 1997;94:6462–6467. doi: 10.1073/pnas.94.12.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper C, Henderson A, Artandi S, Avitahl N, Calame K. Ig/EBP (C/EBPγ) is a transdominant negative inhibitor of C/EBP family transcriptional activators. Nucleic Acids Res. 1995;23:4371–4377. doi: 10.1093/nar/23.21.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]; ▪▪ Demonstrates the isoform of C/EBPβ.
- 13.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross SE, Radomska HS, Wu B, et al. Phosphorylation of C/EBPα inhibits granulopoiesis. Mol Cell Biol. 2004;24:675–686. doi: 10.1128/MCB.24.2.675-686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arizmendi C, Liu S, Croniger C, Poli V, Friedman JE. The transcription factor CCAAT/enhancer-binding protein β regulates gluconeogenesis and phosphoenolpyruvate carboxykinase (GTP) gene transcription during diabetes. J Biol Chem. 1999;274:13033–13040. doi: 10.1074/jbc.274.19.13033. [DOI] [PubMed] [Google Scholar]
- 17.Darlington GJ, Ross SE, MacDougald OA. The role of C/EBP genes in adipocyte differentiation. J Biol Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 18.Hu HM, Baer M, Williams SC, Johnson PF, Schwartz RC. Redundancy of C/EBP-α, -β, and -δ in supporting the lipopolysaccharide-induced transcription of IL-6 and monocyte chemoattractant protein-1. J Immunol. 1998;160:2334–2342. [PubMed] [Google Scholar]
- 19.Cloutier A, Guindi C, Larivee P, et al. Inflammatory cytokine production by human neutrophils involves C/EBP transcription factors. J Immunol. 2009;182:563–571. doi: 10.4049/jimmunol.182.1.563. [DOI] [PubMed] [Google Scholar]
- 20.Hu HM, Tian Q, Baer M, et al. The C/EBP bZIP domain can mediate lipopolysaccharide induction of the proinflammatory cytokines interleukin-6 and monocyte chemoattractant protein-1. J Biol Chem. 2000;275:16373–16381. doi: 10.1074/jbc.M910269199. [DOI] [PubMed] [Google Scholar]
- 21.Gao H, Parkin S, Johnson PF, Schwartz RC. C/EBPγ has a stimulatory role on the IL-6 and IL-8 promoters. J Biol Chem. 2002;277:38827–38837. doi: 10.1074/jbc.M206224200. [DOI] [PubMed] [Google Scholar]
- 22.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 23.Henderson AJ, Zou X, Calame KL. C/EBP proteins activate transcription from the human immunodeficiency virus type-1 long terminal repeat in macrophages/monocytes. J Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ First report to demonstrate that C/EBP is required for HIV-1 transcription in cells of the monocyte–macrophage lineage.
- 24.Tengku-Muhammad TS, Hughes TR, Ranki H, Cryer A, Ramji DP. Differential regulation of macrophage CCAAT-enhancer binding protein isoforms by lipopolysaccharide and cytokines. Cytokine. 2000;12:1430–1436. doi: 10.1006/cyto.2000.0711. [DOI] [PubMed] [Google Scholar]
- 25.Dowell P, Lane MD. C/EBPα reverses the anti-adipogenic effects of the HIV protease inhibitor nelfinavir. Biochem Biophys Res Commun. 2005;327:571–574. doi: 10.1016/j.bbrc.2004.11.169. [DOI] [PubMed] [Google Scholar]
- 26.El Hadri K, Glorian M, Monsempes C, et al. In vitro suppression of the lipogenic pathway by the nonnucleoside reverse transcriptase inhibitor efavirenz in 3T3 and human preadipocytes or adipocytes. J Biol Chem. 2004;279:15130–15141. doi: 10.1074/jbc.M312875200. [DOI] [PubMed] [Google Scholar]
- 27.Dowell P, Flexner C, Kwiterovich PO, Lane MD. Suppression of preadipocyte differentiation and promotion of adipocyte death by HIV protease inhibitors. J Biol Chem. 2000;275:41325–41332. doi: 10.1074/jbc.M006474200. [DOI] [PubMed] [Google Scholar]
- 28.Bradley MN, Zhou L, Smale ST. C/EBPβ regulation in lipopolysaccharide-stimulated macrophages. Mol Cell Biol. 2003;23:4841–4858. doi: 10.1128/MCB.23.14.4841-4858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natsuka S, Akira S, Nishio Y, et al. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 30.Williams SC, Baer M, Dillner AJ, Johnson PF. CRP2 (C/EBPβ) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪▪ Describes the functional domains of C/EBPβ.
- 31.Takiguchi M. The C/EBP family of transcription factors in the liver and other organs. Int J Exp Pathol. 1998;79:369–391. doi: 10.1046/j.1365-2613.1998.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm SL, Rosen JM. The role of C/EBPβ in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:191–204. doi: 10.1023/a:1025900908026. [DOI] [PubMed] [Google Scholar]
- 33.Chinery R, Brockman JA, Dransfield DT, Coffey RJ. Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein β. A critical role for protein kinase A-mediated phosphorylation of Ser299. J Biol Chem. 1997;272:30356–30361. doi: 10.1074/jbc.272.48.30356. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima T, Kinoshita S, Sasagawa T, et al. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham TH, Langmann S, Schwarzfischer L, et al. CCAAT enhancer-binding protein β regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J Biol Chem. 2007;282:21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 36.Mameli G, Deshmane SL, Ghafouri M, et al. C/EBPβ regulates human immunodeficiency virus 1 gene expression through its association with cdk9. J Gen Virol. 2007;88:631–640. doi: 10.1099/vir.0.82487-0. [DOI] [PubMed] [Google Scholar]
- 37.Williams SC, Cantwell CA, Johnson PF. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 38.Nye JA, Graves BJ. Alkylation interference identifies essential DNA contacts for sequence-specific binding of the eukaryotic transcription factor C/EBP. Proc Natl Acad Sci USA. 1990;87:3992–3996. doi: 10.1073/pnas.87.10.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendricks-Taylor LR, Bachinski LL, Siciliano MJ, et al. The CCAAT/enhancer binding protein (C/EBPα) gene (CEBPA) maps to human chromosome 19q13.1 and the related nuclear factor NF-IL6 (C/EBPβ) gene (CEBPB) maps to human chromosome 20q13.1. Genomics. 1992;14:12–17. doi: 10.1016/s0888-7543(05)80276-9. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins NA, Gilbert DJ, Cho BC, et al. Mouse chromosomal location of the CCAAT/enhancer binding proteins C/EBPβ (Cebpb), C/EBPδ (Cebpd), and CRP1 (Cebpe) Genomics. 1995;28:333–336. doi: 10.1006/geno.1995.1150. [DOI] [PubMed] [Google Scholar]
- 41.Descombes P, Chojkier M, Lichtsteiner S, Falvey E, Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990;4:1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- 42.Baer M, Johnson PF. Generation of truncated C/EBPβ isoforms by in vitro proteolysis. J Biol Chem. 2000;275:26582–26590. doi: 10.1074/jbc.M004268200. [DOI] [PubMed] [Google Scholar]
- 43.Baer M, Williams SC, Dillner A, Schwartz RC, Johnson PF. Autocrine signals control CCAAT/enhancer binding protein β expression, localization, and activity in macrophages. Blood. 1998;92:4353–4365. [PubMed] [Google Scholar]
- 44.Welm AL, Timchenko NA, Darlington GJ. C/EBPα regulates generation of C/EBPβ isoforms through activation of specific proteolytic cleavage. Mol Cell Biol. 1999;19:1695–1704. doi: 10.1128/mcb.19.3.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei W, Yang H, Cao P, et al. Degradation of C/EBPβ in cultured myotubes is calpain-dependent. J Cell Physiol. 2006;208:386–398. doi: 10.1002/jcp.20684. [DOI] [PubMed] [Google Scholar]
- 46.Eaton EM, Hanlon M, Bundy L, Sealy L. Characterization of C/EBPβ isoforms in normal versus neoplastic mammary epithelial cells. J Cell Physiol. 2001;189:91–105. doi: 10.1002/jcp.1139. [DOI] [PubMed] [Google Scholar]; ▪▪ Characterizes the three human C/EBPβ isoforms.
- 47.Bundy LM, Sealy L. CCAAT/enhancer binding protein β (C/EBPβ)-2 transforms normal mammary epithelial cells and induces epithelial to mesenchymal transition in culture. Oncogene. 2003;22:869–883. doi: 10.1038/sj.onc.1206216. [DOI] [PubMed] [Google Scholar]
- 48.Lee YM, Miau LH, Chang CJ, Lee SC. Transcriptional induction of the α-1 acid glycoprotein (AGP) gene by synergistic interaction of two alternative activator forms of AGP/enhancer-binding protein (C/EBPβ) and NF-κB or Nopp140. Mol Cell Biol. 1996;16:4257–4263. doi: 10.1128/mcb.16.8.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kowenz-Leutz E, Leutz A. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 50.Su WC, Chou HY, Chang CJ, et al. Differential activation of a C/EBPβ isoform by a novel redox switch may confer the lipopolysaccharide-inducible expression of interleukin-6 gene. J Biol Chem. 2003;278:51150–51158. doi: 10.1074/jbc.M305501200. [DOI] [PubMed] [Google Scholar]
- 51.Eaton EM, Sealy L. Modification of CCAAT/enhancer-binding protein-β by the small ubiquitin-like modifier (SUMO) family members, SUMO-2 and SUMO-3. J Biol Chem. 2003;278:33416–33421. doi: 10.1074/jbc.M305680200. [DOI] [PubMed] [Google Scholar]
- 52.Hsu W, Chen-Kiang S. Convergent regulation of NF-IL6 and Oct-1 synthesis by interleukin-6 and retinoic acid signaling in embryonal carcinoma cells. Mol Cell Biol. 1993;13:2515–2523. doi: 10.1128/mcb.13.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka N, Hoshino Y, Gold J, et al. Interleukin-10 induces inhibitory C/EBPβ through STAT-3 and represses HIV-1 transcription in macrophages. Am J Respir Cell Mol Biol. 2005;33:406–411. doi: 10.1165/rcmb.2005-0140OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tesmer VM, Bina M. Regulation of HIV-1 gene expression by NF-IL6. J Mol Biol. 1996;262:327–335. doi: 10.1006/jmbi.1996.0516. [DOI] [PubMed] [Google Scholar]
- 55.Tesmer VM, Rajadhyaksha A, Babin J, Bina M. NF-IL6-mediated transcriptional activation of the long terminal repeat of the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:7298–7302. doi: 10.1073/pnas.90.15.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ First report to identify three C/EBP binding sites within the HIV-1 long terminal repeat (LTR).
- 56.Henderson AJ, Connor RI, Calame KL. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates the important function of C/EBP in HIV-1 gene expression in monocytes.
- 57.Hogan TH, Krebs FC, Wigdahl B. Regulation of human immunodeficiency virus type 1 gene expression and pathogenesis by CCAAT/enhancer binding proteins in cells of the monocyte/macrophage lineage. J Neurovirol. 2002;8(Suppl 2):21–26. doi: 10.1080/13550280290167911. [DOI] [PubMed] [Google Scholar]
- 58.Burdo TH, Gartner S, Mauger D, Wigdahl B. Region-specific distribution of human immunodeficiency virus type 1 long terminal repeats containing specific configurations of CCAAT/enhancer-binding protein site II in brains derived from demented and nondemented patients. J Neurovirol. 2004;10(Suppl 1):7–14. doi: 10.1080/753312746. [DOI] [PubMed] [Google Scholar]
- 59.Krebs FC, Hogan TH, Quiterio S, Gartner S, Wigdahl B. Lentiviral LTR-directed expression, sequence variation, and disease pathogenesis. In: Kuiken C, Foley B, Hahn B, et al., editors. HIV Sequence Compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM, USA: 2001. pp. 29–70. LA-UR 02-2877. [Google Scholar]
- 60.Ross HL, Gartner S, McArthur JC, et al. HIV-1 LTR C/EBP-binding site sequence configurations preferentially encountered in brain lead to enhanced C/EBP factor binding and increased LTR-specific activity. J Neurovirol. 2001;7:235–249. doi: 10.1080/13550280152403281. [DOI] [PubMed] [Google Scholar]
- 61.Yang Y, Tesmer VM, Bina M. Regulation of HIV-1 transcription in activated monocyte macrophages. Virology. 2002;299:256–265. doi: 10.1006/viro.2001.1530. [DOI] [PubMed] [Google Scholar]
- 62.Henderson AJ, Calame KL. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4+ T cells. Proc Natl Acad Sci USA. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davydov IV, Krammer PH, Li-Weber M. Nuclear factor-IL6 activates the human IL-4 promoter in T cells. J Immunol. 1995;155:5273–5279. [PubMed] [Google Scholar]
- 64.Berberich-Siebelt F, Berberich I, Andrulis M, et al. SUMOylation interferes with CCAAT/enhancer-binding protein-β-mediated c-myc repression, but not IL-4 activation in T cells. J Immunol. 2006;176:4843–4851. doi: 10.4049/jimmunol.176.8.4843. [DOI] [PubMed] [Google Scholar]
- 65.Rosati M, Valentin A, Patenaude DJ, Pavlakis GN. CCAAT-enhancer-binding protein-β (C/EBPβ) activates CCR5 promoter: increased C/EBPβ and CCR5 in T lymphocytes from HIV-1-infected individuals. J Immunol. 2001;167:1654–1662. doi: 10.4049/jimmunol.167.3.1654. [DOI] [PubMed] [Google Scholar]
- 66.Buckner AE, Tesmer VM, Bina M. Regulation of HIV-1 transcription by NF-IL6 in activated Jurkat T cells. Virus Res. 2002;89:53–63. doi: 10.1016/s0168-1702(02)00110-7. [DOI] [PubMed] [Google Scholar]
- 67.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dumais N, Bounou S, Olivier M, Tremblay MJ. Prostaglandin E(2)-mediated activation of HIV-1 long terminal repeat transcription in human T cells necessitates CCAAT/enhancer binding protein (C/EBP) binding sites in addition to cooperative interactions between C/EBPβ and cyclic adenosine 5′-monophosphate response element binding protein. J Immunol. 2002;168:274–282. doi: 10.4049/jimmunol.168.1.274. [DOI] [PubMed] [Google Scholar]
- 70.Park EA, Gurney AL, Nizielski SE, et al. Relative roles of CCAAT/enhancer-binding protein β and cAMP regulatory element-binding protein in controlling transcription of the gene for phosphoenolpyruvate carboxykinase (GTP) J Biol Chem. 1993;268:613–619. [PubMed] [Google Scholar]
- 71.Kinoshita SM, Taguchi S. NF-IL6 (C/EBPβ) induces HIV-1 replication by inhibiting cytidine deaminase APOBEC3G. Proc Natl Acad Sci USA. 2008;105:15022–15027. doi: 10.1073/pnas.0807269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 73.Kramer-Hammerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111:194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 74.Ghafouri M, Amini S, Khalili K, Sawaya BE. HIV-1 associated dementia: symptoms and causes. Retrovirology. 2006;3:28. doi: 10.1186/1742-4690-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ejarque-Ortiz A, Medina MG, Tusell JM, et al. Upregulation of CCAAT/enhancer binding protein β in activated astrocytes and microglia. Glia. 2007;55:178–188. doi: 10.1002/glia.20446. [DOI] [PubMed] [Google Scholar]
- 76.Schwartz C, Catez P, Rohr O, et al. Functional interactions between C/EBP, Sp1, and COUP-TF regulate human immunodeficiency virus type 1 gene transcription in human brain cells. J Virol. 2000;74:65–73. doi: 10.1128/jvi.74.1.65-73.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cardinaux JR, Allaman I, Magistretti PJ. Proinflammatory cytokines induce the transcription factors C/EBPβ and C/EBPδ in astrocytes. Glia. 2000;29:91–97. [PubMed] [Google Scholar]
- 78.Honda Y, Rogers L, Nakata K, et al. Type I interferon induces inhibitory 16 kD CCAAT/enhancer binding protein (C/EBP)β, repressing the HIV-1 long terminal repeat in macrophages: pulmonary tuberculosis alters C/EBP expression, enhancing HIV-1 replication. J Exp Med. 1998;188:1255–1265. doi: 10.1084/jem.188.7.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakata K, Rom WN, Honda Y, et al. Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication in the lung. Am J Respir Crit Care Med. 1997;155:996–1003. doi: 10.1164/ajrccm.155.3.9117038. [DOI] [PubMed] [Google Scholar]
- 80.Wright JR. Immunomodulatory functions of surfactant. Physiol Rev. 1997;77:931–962. doi: 10.1152/physrev.1997.77.4.931. [DOI] [PubMed] [Google Scholar]
- 81.Gold JA, Hoshino Y, Tanaka N, et al. Surfactant protein A modulates the inflammatory response in macrophages during tuberculosis. Infect Immun. 2004;72:645–650. doi: 10.1128/IAI.72.2.645-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoshino Y, Nakata K, Hoshino S, et al. Maximal HIV-1 replication in alveolar macrophages during tuberculosis requires both lymphocyte contact and cytokines. J Exp Med. 2002;195:495–505. doi: 10.1084/jem.20011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakata K, Keicho N, Honda Y, et al. The mechanism of HIV replication at the site of inflammation coinfected with HIV and M. tuberculosis. Kekkaku. 2002;77:687–692. [PubMed] [Google Scholar]
- 84.Emori Y, Ikeda T, Ohashi T, et al. Inhibition of human immunodeficiency virus type 1 replication by Z-100, an immunomodulator extracted from human-type tubercle bacilli, in macrophages. J Gen Virol. 2004;85:2603–2613. doi: 10.1099/vir.0.80046-0. [DOI] [PubMed] [Google Scholar]
- 85.Darcissac EC, Truong MJ, Dewulf J, et al. The synthetic immunomodulator murabutide controls human immunodeficiency virus type-1 replication at multiple levels in macrophages and dendritic cells. J Virol. 2000;74:7794–7802. doi: 10.1128/jvi.74.17.7794-7802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vidal VF, Casteran N, Riendeau CJ, et al. Macrophage stimulation with murabutide, an HIV-suppressive muramyl peptide derivative, selectively activates extracellular signal-regulated kinases 1 and 2, C/EBPβ and STAT1: role of CD14 and Toll-like receptors 2 and 4. Eur J Immunol. 2001;31:1962–1971. doi: 10.1002/1521-4141(200107)31:7<1962::aid-immu1962>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 87.Komuro I, Yokota Y, Yasuda S, Iwamoto A, Kagawa KS. CSF-induced and HIV-1-mediated distinct regulation of Hck and C/EBPβ represent a heterogeneous susceptibility of monocyte-derived macrophages to M-tropic HIV-1 infection. J Exp Med. 2003;198:443–453. doi: 10.1084/jem.20022018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Komuro I, Sunazuka T, Akagawa KS, et al. Erythromycin derivatives inhibit HIV-1 replication in macrophages through modulation of MAPK activity to induce small isoforms of C/EBPβ. Proc Natl Acad Sci USA. 2008;105:12509–12514. doi: 10.1073/pnas.0805504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Popik W, Hesselgesser JE, Pitha PM. Binding of human immunodeficiency virus type 1 to CD4 and CXCR4 receptors differentially regulates expression of inflammatory genes and activates the MEK/ERK signaling pathway. J Virol. 1998;72:6406–6413. doi: 10.1128/jvi.72.8.6406-6413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee ES, Sarma D, Zhou H, Henderson AJ. CCAAT/enhancer binding proteins are not required for HIV-1 entry but regulate proviral transcription by recruiting coactivators to the long-terminal repeat in monocytic cells. Virology. 2002;299:20–31. doi: 10.1006/viro.2002.1500. [DOI] [PubMed] [Google Scholar]
- 91.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 92.Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chakravarti D, Ogryzko V, Kao HY, et al. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 94.Hamamori Y, Sartorelli V, Ogryzko V, et al. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 95.Schwartz C, Beck K, Mink S, et al. Recruitment of p300 by C/EBPβ triggers phosphorylation of p300 and modulates coactivator activity. EMBO J. 2003;22:882–892. doi: 10.1093/emboj/cdg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.So CW, van der Reijden BA. C/EBPα, do not forget your TIP60. Leukemia. 2008;22:676–677. doi: 10.1038/leu.2008.16. [DOI] [PubMed] [Google Scholar]
- 97.Bararia D, Trivedi AK, Zada AA, et al. Proteomic identification of the MYST domain histone acetyltransferase TIP60 (HTATIP) as a co-activator of the myeloid transcription factor C/EBPα. Leukemia. 2008;22:800–807. doi: 10.1038/sj.leu.2405101. [DOI] [PubMed] [Google Scholar]
- 98.Williams SA, Chen LF, Kwon H, et al. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18:1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 100.Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol. 2007;81:10914–10923. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Di-Poi N, Desvergne B, Michalik L, Wahli W. Transcriptional repression of peroxisome proliferator-activated receptor β/δ in murine keratinocytes by CCAAT/enhancer-binding proteins. J Biol Chem. 2005;280:38700–38710. doi: 10.1074/jbc.M507782200. [DOI] [PubMed] [Google Scholar]
- 102.Wiper-Bergeron N, Wu D, Pope L, Schild-Poulter C, Hache RJ. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J. 2003;22:2135–2145. doi: 10.1093/emboj/cdg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zuo Y, Qiang L, Farmer SR. Activation of CCAAT/enhancer-binding protein (C/EBP) α expression by C/EBPβ during adipogenesis requires a peroxisome proliferator-activated receptor-γ-associated repression of HDAC1 at the C/EBP α gene promoter. J Biol Chem. 2006;281:7960–7967. doi: 10.1074/jbc.M510682200. [DOI] [PubMed] [Google Scholar]
- 104.Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Hache RJ. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPβ by GCN5. Proc Natl Acad Sci USA. 2007;104:2703–2708. doi: 10.1073/pnas.0607378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 106.Faiola F, Liu X, Lo S, et al. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vervoorts J, Luscher-Firzlaff JM, Rottmann S, et al. Stimulation of c-Myc transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bachelerie F, Alcami J, Arenzana-Seisdedos F, Virelizier JL. HIV enhancer activity perpetuated by NF-κB induction on infection of monocytes. Nature. 1991;350:709–712. doi: 10.1038/350709a0. [DOI] [PubMed] [Google Scholar]
- 109.Alcami J, Lain de Lera T, Folgueira L, et al. Absolute dependence on κB responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 111.Asin S, Bren GD, Carmona EM, Solan NJ, Paya CV. NF-κB cis-acting motifs of the human immunodeficiency virus (HIV) long terminal repeat regulate HIV transcription in human macrophages. J Virol. 2001;75:11408–11416. doi: 10.1128/JVI.75.23.11408-11416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stein B, Cogswell PC, Baldwin AS., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ruocco MR, Chen X, Ambrosino C, et al. Regulation of HIV-1 long terminal repeats by interaction of C/EBP (NF-IL6) and NF-κB/Rel transcription factors. J Biol Chem. 1996;271:22479–22486. doi: 10.1074/jbc.271.37.22479. [DOI] [PubMed] [Google Scholar]
- 114.Mukerjee R, Sawaya BE, Khalili K, Amini S. Association of p65 and C/EBPβ with HIV-1 LTR modulates transcription of the viral promoter. J Cell Biochem. 2007;100:1210–1216. doi: 10.1002/jcb.21109. [DOI] [PubMed] [Google Scholar]
- 115.Abraham S, Sweet T, Sawaya BE, et al. Cooperative interaction of C/EBPβ and Tat modulates MCP-1 gene transcription in astrocytes. J Neuroimmunol. 2005;160:219–227. doi: 10.1016/j.jneuroim.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 116.Krebs FC, Goodenow MM, Wigdahl B. Neuroglial ATF/CREB factors interact with the human immunodeficiency virus type 1 long terminal repeat. J Neurovirol. 1997;3(Suppl 1):S28–S32. [PubMed] [Google Scholar]
- 117.Ross HL, Nonnemacher MR, Hogan TH, et al. Interaction between CCAAT/enhancer binding protein and cyclic AMP response element binding protein 1 regulates human immunodeficiency virus type 1 transcription in cells of the monocyte/macrophage lineage. J Virol. 2001;75:1842–1856. doi: 10.1128/JVI.75.4.1842-1856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aten JA, Stap J, Krawczyk PM, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 119.Chipitsyna G, Slonina D, Siddiqui K, et al. HIV-1 Tat increases cell survival in response to cisplatin by stimulating Rad51 gene expression. Oncogene. 2004;23:2664–2671. doi: 10.1038/sj.onc.1207417. [DOI] [PubMed] [Google Scholar]
- 120.Chipitsyna G, Sawaya BE, Khalili K, Amini S. Cooperativity between Rad51 and C/EBP family transcription factors modulates basal and Tat-induced activation of the HIV-1 LTR in astrocytes. J Cell Physiol. 2006;207:605–613. doi: 10.1002/jcp.20612. [DOI] [PubMed] [Google Scholar]
- 121.Liou LY, Herrmann CH, Rice AP. HIV-1 infection and regulation of Tat function in macrophages. Int J Biochem Cell Biol. 2004;36:1767–1775. doi: 10.1016/j.biocel.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 122.Brady J, Kashanchi F. Tat gets the “green” light on transcription initiation. Retrovirology. 2005;2:69. doi: 10.1186/1742-4690-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huigen MC, Kamp W, Nottet HS. Multiple effects of HIV-1 trans-activator protein on the pathogenesis of HIV-1 infection. Eur J Clin Invest. 2004;34:57–66. doi: 10.1111/j.1365-2362.2004.01282.x. [DOI] [PubMed] [Google Scholar]
- 124.Ambrosino C, Ruocco MR, Chen X, et al. HIV-1 Tat induces the expression of the interleukin-6 (IL6) gene by binding to the IL6 leader RNA and by interacting with CAAT enhancer-binding protein β (NF-IL6) transcription factors. J Biol Chem. 1997;272:14883–14892. doi: 10.1074/jbc.272.23.14883. [DOI] [PubMed] [Google Scholar]
- 125.Mukerjee R, Sawaya BE, Khalili K, Amini S. Association of p65 and C/EBPβ with HIV-1 LTR modulates transcription of the viral promoter. J Cell Biochem. 2006;100(5):1210–1216. doi: 10.1002/jcb.21109. [DOI] [PubMed] [Google Scholar]
- 126.Liu X, Jana M, Dasgupta S, et al. Human immunodeficiency virus type 1 (HIV-1) tat induces nitric-oxide synthase in human astroglia. J Biol Chem. 2002;277:39312–39319. doi: 10.1074/jbc.M205107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hori K, Hatfield D, Maldarelli F, Lee BJ, Clouse KA. Selenium supplementation suppresses tumor necrosis factor α-induced human immunodeficiency virus type 1 replication in vitro. AIDS Res Hum Retroviruses. 1997;13:1325–1332. doi: 10.1089/aid.1997.13.1325. [DOI] [PubMed] [Google Scholar]
- 128.Adamson DC, Wildemann B, Sasaki M, et al. Immunologic NO synthase: elevation in severe AIDS dementia and induction by HIV-1 gp41. Science. 1996;274:1917–1921. doi: 10.1126/science.274.5294.1917. [DOI] [PubMed] [Google Scholar]
- 129.Johnson MD, Kim P, Tourtellotte W, Federspiel CF. Transforming growth factor-β and monocyte chemotactic protein-1 are elevated in cerebrospinal fluid of immunocompromised patients with HIV-1 infection. J NeuroAIDS. 2004;2:33–43. doi: 10.1300/J128v02n04_03. [DOI] [PubMed] [Google Scholar]
- 130.Conant K, Garzino-Demo A, Nath A, et al. Induction of monocyte chemoattractant protein-1 in HIV-1 Tat-stimulated astrocytes and elevation in AIDS dementia. Proc Natl Acad Sci USA. 1998;95:3117–3121. doi: 10.1073/pnas.95.6.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mengozzi M, De Filippi C, Transidico P, et al. Human immunodeficiency virus replication induces monocyte chemotactic protein-1 in human macrophages and U-937 promonocytic cells. Blood. 1999;93:1851–1857. [PubMed] [Google Scholar]
- 132.Wiley CA, Schrier RD, Nelson JA, Lampert PW, Oldstone MB. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc Natl Acad Sci USA. 1986;83:7089–7093. doi: 10.1073/pnas.83.18.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.King JE, Eugenin EA, Buckner CM, Berman JW. HIV tat and neurotoxicity. Microbes Infect. 2006;8:1347–1357. doi: 10.1016/j.micinf.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 134.Peruzzi F, Gordon J, Darbinian N, Amini S. Tat-induced deregulation of neuronal differentiation and survival by nerve growth factor pathway. J Neurovirol. 2002;8(Suppl 2):91–96. doi: 10.1080/13550280290167885. [DOI] [PubMed] [Google Scholar]
- 135.Haughey NJ, Mattson MP. Calcium dysregulation and neuronal apoptosis by the HIV-1 proteins Tat and gp120. J Acquir Immune Defic Syndr. 2002;31(Suppl 2):S55–S61. doi: 10.1097/00126334-200210012-00005. [DOI] [PubMed] [Google Scholar]
- 136.Li L, Li HS, Pauza CD, Bukrinsky M, Zhao RY. Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions. Cell Res. 2005;15:923–934. doi: 10.1038/sj.cr.7290370. [DOI] [PubMed] [Google Scholar]
- 137.Andersen JL, Planelles V. The role of Vpr in HIV-1 pathogenesis. Curr HIV Res. 2005;3:43–51. doi: 10.2174/1570162052772988. [DOI] [PubMed] [Google Scholar]
- 138.Kino T, Pavlakis GN. Partner molecules of accessory protein Vpr of the human immunodeficiency virus type 1. DNA Cell Biol. 2004;23:193–205. doi: 10.1089/104454904773819789. [DOI] [PubMed] [Google Scholar]
- 139.Philippon V, Matsuda Z, Essex M. Transactivation is a conserved function among primate lentivirus Vpr proteins but is not shared by Vpx. J Hum Virol. 1999;2:167–174. [PubMed] [Google Scholar]
- 140.Sawaya BE, Khalili K, Rappaport J, et al. Suppression of HIV-1 transcription and replication by a Vpr mutant. Gene Ther. 1999;6:947–950. doi: 10.1038/sj.gt.3300907. [DOI] [PubMed] [Google Scholar]
- 141.Wang L, Mukherjee S, Jia F, Narayan O, Zhao LJ. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 142.Zhang S, Pointer D, Singer G, et al. Direct binding to nucleic acids by Vpr of human immunodeficiency virus type 1. Gene. 1998;212:157–166. doi: 10.1016/s0378-1119(98)00178-4. [DOI] [PubMed] [Google Scholar]
- 143.Hogan TH, Nonnemacher MR, Krebs FC, Henderson A, Wigdahl B. HIV-1 Vpr binding to HIV-1 LTR C/EBP cis-acting elements and adjacent regions is sequence-specific. Biomed Pharmacother. 2003;57:41–48. doi: 10.1016/s0753-3322(02)00333-5. [DOI] [PubMed] [Google Scholar]
- 144.Burdo TH, Nonnemacher M, Irish BP, et al. High-affinity interaction between HIV-1 Vpr and specific sequences that span the C/EBP and adjacent NF-κB sites within the HIV-1 LTR correlate with HIV-1-associated dementia. DNA Cell Biol. 2004;23:261–269. doi: 10.1089/104454904773819842. [DOI] [PubMed] [Google Scholar]
- 145.Roux P, Alfieri C, Hrimech M, Cohen EA, Tanner JE. Activation of transcription factors NF-κB and NF-IL-6 by human immunodeficiency virus type 1 protein R (Vpr) induces interleukin-8 expression. J Virol. 2000;74:4658–4665. doi: 10.1128/jvi.74.10.4658-4665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tyor WR, Glass JD, Griffin JW, et al. Cytokine expression in the brain during the acquired immunodeficiency syndrome. Ann Neurol. 1992;31:349–360. doi: 10.1002/ana.410310402. [DOI] [PubMed] [Google Scholar]
- 147.Kedzierska K, Crowe SM. Cytokines and HIV-1: interactions and clinical implications. Antivir Chem Chemother. 2001;12:133–150. doi: 10.1177/095632020101200301. [DOI] [PubMed] [Google Scholar]
- 148.Screpanti I, Romani L, Musiani P, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBPβ-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gorgoni B, Maritano D, Marthyn P, Righi M, Poli V. C/EBPβ gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J Immunol. 2002;168:4055–4062. doi: 10.4049/jimmunol.168.8.4055. [DOI] [PubMed] [Google Scholar]
- 150.Akira S, Kishimoto T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol Rev. 1992;127:25–50. doi: 10.1111/j.1600-065x.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 151.Hungness ES, Luo GJ, Pritts TA, et al. Transcription factors C/EBP-β and -δ regulate IL-6 production in IL-1β-stimulated human enterocytes. J Cell Physiol. 2002;192:64–70. doi: 10.1002/jcp.10116. [DOI] [PubMed] [Google Scholar]
- 152.Shirakawa F, Saito K, Bonagura CA, et al. The human prointerleukin-1β gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993;13:1332–1344. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Y, Rom WN. Regulation of the interleukin-1β (IL-1β) gene by mycobacterial components and lipopolysaccharide is mediated by two nuclear factor-IL6 motifs. Mol Cell Biol. 1993;13:3831–3837. doi: 10.1128/mcb.13.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Coyle-Rink J, Sweet T, Abraham S, et al. Interaction between TGF-β signaling proteins and C/EBP controls basal and Tat-mediated transcription of HIV-1 LTR in astrocytes. Virology. 2002;299:240–247. doi: 10.1006/viro.2002.1439. [DOI] [PubMed] [Google Scholar]
- 155.Pope R, Mungre S, Liu H, Thimmapaya B. Regulation of TNF-α expression in normal macrophages: the role of C/EBPβ. Cytokine. 2000;12:1171–1181. doi: 10.1006/cyto.2000.0691. [DOI] [PubMed] [Google Scholar]
- 156.Pope RM, Leutz A, Ness SA. C/EBPβ regulation of the tumor necrosis factor-α gene. J Clin Invest. 1994;94:1449–1455. doi: 10.1172/JCI117482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zagariya A, Mungre S, Lovis R, et al. Tumor necrosis factor-α gene regulation: enhancement of C/EBPβ-induced activation by c-Jun. Mol Cell Biol. 1998;18:2815–2824. doi: 10.1128/mcb.18.5.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Piwien-Pilipuk G, MacDougald O, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem. 2002;277:44557–44565. doi: 10.1074/jbc.M206886200. [DOI] [PubMed] [Google Scholar]
- 159.Basak C, Pathak SK, Bhattacharyya A, et al. NF-κB- and C/EBPβ-driven interleukin-1β gene expression and PAK1-mediated caspase-1 activation play essential roles in interleukin-1β release from Helicobacter pylori lipopolysaccharide-stimulated macrophages. J Biol Chem. 2005;280:4279–4288. doi: 10.1074/jbc.M412820200. [DOI] [PubMed] [Google Scholar]
- 160.Sekine O, Nishio Y, Egawa K, et al. Insulin activates CCAAT/enhancer binding proteins and proinflammatory gene expression through the phosphatidylinositol 3-kinase pathway in vascular smooth muscle cells. J Biol Chem. 2002;277:36631–36639. doi: 10.1074/jbc.M206266200. [DOI] [PubMed] [Google Scholar]
- 161.Bretz JD, Williams SC, Baer M, Johnson PF, Schwartz RC. C/EBP-related protein 2 confers lipopolysaccharide-inducible expression of interleukin 6 and monocyte chemoattractant protein 1 to a lymphoblastic cell line. Proc Natl Acad Sci USA. 1994;91:7306–7310. doi: 10.1073/pnas.91.15.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Spooner CJ, Guo X, Johnson PF, Schwartz RC. Differential roles of C/EBPβ regulatory domains in specifying MCP-1 and IL-6 transcription. Mol Immunol. 2007;44:1384–1392. doi: 10.1016/j.molimm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 163.Cupp C, Taylor JP, Khalili K, Amini S. Evidence for stimulation of the transforming growth factor β1 promoter by HIV-1 Tat in cells derived from CNS. Oncogene. 1993;8:2231–2236. [PubMed] [Google Scholar]
- 164.Sawaya BE, Thatikunta P, Denisova L, et al. Regulation of TNF-α and TGFβ-1 gene transcription by HIV-1 Tat in CNS cells. J Neuroimmunol. 1998;87:33–42. doi: 10.1016/s0165-5728(98)00044-7. [DOI] [PubMed] [Google Scholar]
- 165.Thatikunta P, Sawaya BE, Denisova L, et al. Identification of a cellular protein that binds to Tat-responsive element of TGF β-1 promoter in glial cells. J Cell Biochem. 1997;67:466–477. [PubMed] [Google Scholar]
- 166.Heldin CH, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through Smad proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 167.Piek E, Heldin CH, ten Dijke P. Specificity, diversity, and regulation in TGF-β superfamily signaling. FASEB J. 1999;13:2105–2124. [PubMed] [Google Scholar]
- 168.Grant C, Barmak K, Alefantis T, et al. Human T-cell leukemia virus type I and neurologic disease: events in bone marrow, peripheral blood, and central nervous system during normal immune surveillance and neuroinflammation. J Cell Physiol. 2002;190:133–159. doi: 10.1002/jcp.10053. [DOI] [PubMed] [Google Scholar]
- 169.Hollsberg P, Hafler DA. Seminars in medicine of the Beth Israel Hospital, Boston. Pathogenesis of diseases induced by human lymphotropic virus type I infection. N Engl J Med. 1993;328:1173–1182. doi: 10.1056/NEJM199304223281608. [DOI] [PubMed] [Google Scholar]
- 170.Nakamura T. Immunopathogenesis of HTLV-I-associated myelopathy/tropical spastic paraparesis. Ann Med. 2000;32:600–607. doi: 10.3109/07853890009002030. [DOI] [PubMed] [Google Scholar]
- 171.Yoshida M. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annu Rev Immunol. 2001;19:475–496. doi: 10.1146/annurev.immunol.19.1.475. [DOI] [PubMed] [Google Scholar]
- 172.Kashanchi F, Brady JN. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene. 2005;24:5938–5951. doi: 10.1038/sj.onc.1208973. [DOI] [PubMed] [Google Scholar]
- 173.Kimzey AL, Dynan WS. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J Biol Chem. 1998;273:13768–13775. doi: 10.1074/jbc.273.22.13768. [DOI] [PubMed] [Google Scholar]
- 174.Lenzmeier BA, Giebler HA, Nyborg JK. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol Cell Biol. 1998;18:721–731. doi: 10.1128/mcb.18.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lemasson I, Polakowski NJ, Laybourn PJ, Nyborg JK. Transcription factor binding and histone modifications on the integrated proviral promoter in human T-cell leukemia virus-I-infected T-cells. J Biol Chem. 2002;277:49459–49465. doi: 10.1074/jbc.M209566200. [DOI] [PubMed] [Google Scholar]
- 176.Lu H, Pise-Masison CA, Fletcher TM, et al. Acetylation of nucleosomal histones by p300 facilitates transcription from tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol Cell Biol. 2002;22:4450–4462. doi: 10.1128/MCB.22.13.4450-4462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gachon F, Peleraux A, Thebault S, et al. CREB-2, a cellular CRE-dependent transcription repressor, functions in association with Tax as an activator of the human T-cell leukemia virus type 1 promoter. J Virol. 1998;72:8332–8337. doi: 10.1128/jvi.72.10.8332-8337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gachon F, Thebault S, Peleraux A, Devaux C, Mesnard JM. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4) Mol Cell Biol. 2000;20:3470–3481. doi: 10.1128/mcb.20.10.3470-3481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yin MJ, Paulssen E, Seeler J, Gaynor RB. Chimeric proteins composed of Jun and CREB define domains required for interaction with the human T-cell leukemia virus type 1 Tax protein. J Virol. 1995;69:6209–6218. doi: 10.1128/jvi.69.10.6209-6218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grant C, Nonnemacher M, Jain P, et al. CCAAT/enhancer-binding proteins modulate human T-cell leukemia virus type 1 long terminal repeat activation. Virology. 2006;348:354–369. doi: 10.1016/j.virol.2005.12.024. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates the effect of C/EBP on Tax-mediated human T-cell leukemia virus type-1 transcription utilizing rodent C/EBP.
- 181.Hivin P, Gaudray G, Devaux C, Mesnard JM. Interaction between C/EBPβ and Tax down-regulates human T-cell leukemia virus type I transcription. Virology. 2004;318:556–565. doi: 10.1016/j.virol.2003.10.027. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates the effect of C/EBP on Tax-mediated human T-cell leukemia virus type-1 transcription utilizing human C/EBP.
- 182.Li-Weber M, Giaisi M, Krammer PH. Roles of CCAAT/enhancer-binding protein in transcriptional regulation of the human IL-5 gene. Eur J Immunol. 2001;31:3694–3703. doi: 10.1002/1521-4141(200112)31:12<3694::aid-immu3694>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 183.Boche D, Khatissian E, Gray F, et al. Viral load and neuropathology in the SIV model. J Neurovirol. 1999;5:232–240. doi: 10.3109/13550289909015809. [DOI] [PubMed] [Google Scholar]
- 184.Nonnemacher MR, Hogan TH, Quiterio S, et al. Identification of binding sites for members of the CCAAT/enhancer binding protein transcription factor family in the Simian Immunodeficiency Virus long terminal repeat. Biomed Pharmacother. 2003;57:34–40. doi: 10.1016/s0753-3322(02)00334-7. [DOI] [PubMed] [Google Scholar]; ▪ First report to identify four C/EBP binding sites within the SIV LTR.
- 185.Barber SA, Herbst DS, Bullock BT, Gama L, Clements JE. Innate immune responses and control of acute simian immunodeficiency virus replication in the central nervous system. J Neurovirol. 2004;10(Suppl 1):15–20. doi: 10.1080/753312747. [DOI] [PubMed] [Google Scholar]
- 186.Barber SA, Gama L, Dudaronek JM, et al. Mechanism for the establishment of transcriptional HIV latency in the brain in a simian immunodeficiency virus-macaque model. J Infect Dis. 2006;193:963–970. doi: 10.1086/500983. [DOI] [PubMed] [Google Scholar]; ▪▪ Demonstrates that C/EBP could affect histone acetylation of SIV LTR.
- 187.Weed MR, Hienz RD, Brady JV, et al. Central nervous system correlates of behavioral deficits following simian immunodeficiency virus infection. J Neurovirol. 2003;9:452–464. doi: 10.1080/13550280390218751. [DOI] [PubMed] [Google Scholar]
- 188.Flaherty MT, Hauer DA, Mankowski JL, Zink MC, Clements JE. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J Virol. 1997;71:5790–5798. doi: 10.1128/jvi.71.8.5790-5798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mankowski JL, Flaherty MT, Spelman JP, et al. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055–6060. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. CCAAT/enhancer binding protein β is a major mediator of inflammation and viral replication in the gastrointestinal tract of simian immunodeficiency virus-infected rhesus macaques. Am J Pathol. 2008;173:106–118. doi: 10.2353/ajpath.2008.080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Leroy H, Roumier C, Huyghe P, et al. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 192.Fuchs O. Growth-inhibiting activity of transcription factor C/EBPα, its role in haematopoiesis and its tumour suppressor or oncogenic properties in leukaemias. Folia Biol (Praha) 2007;53:97–108. [PubMed] [Google Scholar]
- 193.Nerlov C. C/EBPα mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 194.Verani A, Gras G, Pancino G. Macrophages and HIV-1: dangerous liaisons. Mol Immunol. 2005;42:195–212. doi: 10.1016/j.molimm.2004.06.020. [DOI] [PubMed] [Google Scholar]