Abstract
The retinoblastoma (RB) family of proteins, pRB, p107, and p130, have been postulated to be partially redundant in their ability to regulate progression through the G1 phase of the cell cycle. However, pRB appears to be unique in its capacity as a classical tumor suppressor, possibly because of a specialized role in maintaining the balance between proliferation and differentiation. A variety of studies have in fact revealed an apparent role for pRB in cellular differentiation and development. However, roles for p107 and p130 in differentiation have not yet been established, and knockout mouse studies have indicated that they may be functionally redundant during development, and possibly perform a role in differentiation distinct from that of pRB. Using adipogenesis as a model, we have indeed found distinct roles for the pRB family proteins in regulating differentiation. 3T3 fibroblasts deficient in p107 and p130 differentiate with high efficiency, whereas pRB−/− 3T3 cells exhibit defects in their differentiation potential. Moreover, over-expression of pRB in wild-type cells promotes differentiation, whereas over-expression of p107 antagonizes differentiation. The seemingly opposing roles of pRB family members in adipocyte differentiation can be explained, at least in part, by a requirement for pRB in maintaining cell cycle exit as well as potentiating the activity of the differentiation-associated transcription factor, C/EBPα. p107 does not affect C/EBPα-driven transcription and is not required for cell cycle exit, but instead, loss of p107 lowers the requirement for the differentiation factor PPARγ. These findings suggest contrasting biological roles for individual members of the pRB family of proteins that may explain why pRB, but not p107, is commonly mutated during human tumor development.
Abnormal regulation of cell proliferation and differentiation is a critical aspect of oncogenesis. During tumorigenesis, the balance between differentiation and proliferation is disrupted, in part, through the activation of oncogenes or the loss of tumor suppressor genes. Numerous tumor suppressor genes have been identified, and inheritance of germ-line mutations in these genes strongly predisposes to cancer development. The first tumor suppressor to be cloned was the retinoblastoma (Rb) gene (1, 2), which is frequently mutated in human tumors. In addition, mice heterozygous for Rb exhibit an increased predisposition for pituitary and thyroid tumors, which are associated with a loss of heterozygosity at the Rb locus (3, 4).
Mouse embryos lacking both alleles of Rb die between day 13.5 and 14.5 of gestation, and embryonic cells within the central nervous system as well as erythrocytes were found to lose their ability to commit to some aspects of terminal differentiation. Neural precursor cells lacking pRB continue to divide at inappropriate times, and erythrocytes remain in their nucleated stage before their final differentiation (for review see ref. 5). These results suggest that pRB is required in some cell types for terminal differentiation, a function that may be relevant for its role as a tumor suppressor.
Two proteins highly related to pRB, p107 and p130, have also been identified and disrupted in mice. There are, however, no reports of a predisposition for tumor formation in mice deficient in p107 or p130 (6), and germ-line mutations in the corresponding human genes have not been identified in cancer patients. Recently, it was reported that somatic loss of p130 occurs in a variety of tumors, suggesting that it may also play a role in tumorigenesis. Although p107 or p130 knockout mice do not display obvious developmental phenotypes in a C57BL6–SV129 mixed genetic background, p107−/−/p130−/− mice die shortly after birth and exhibit defects in bone development (7), indicating that they are partially redundant in function, and may also play a role in differentiation.
A role of pRB family members in cell cycle control has been studied largely in cultured cells. In over-expression experiments, pRB and p107 arrest some cell types in the G1 phase of the cell cycle (8–10). The growth suppressive functions of these proteins map to a domain shown to mediate interactions with several viral oncoproteins as well as members of the E2F family of cellular transcription factors (11–15). However, it is not clear that the effects of over-expression of these proteins reflect their normal physiological roles.
In addition to its ability to affect the activity of E2F, pRB has also been shown to bind to and regulate the activity of a number of other transcription factors, many of which bind to the same region of pRB that is required for E2F association. For example, pRB can cooperate with differentiation-specific transcription factors, such as MyoD and CCAAT enhancer binding protein (C/EBP), to activate genes involved in terminal differentiation (16–19). Whether modification of differentiation-specific transcriptional programs by pRB is direct or is a consequence of pRB's ability to repress E2F-regulated promoters and affect cell cycle position is unknown. Moreover, it is unclear to what extent differentiation-specific regulatory functions contribute to pRB's role as a tumor suppressor, or if p107 and p130 regulate transcriptional programs that define the differentiated state.
Our goal was to establish a genetically defined cellular system in which the role of pRB family members in the balance between proliferation and differentiation can be studied. We had generated 3T3 fibroblast cell lines from mouse embryos lacking pRB, p107, and/or p130 (20), and found that the duration of the cell cycle in each of these lines is similar. Subsequently, we examined the ability of these cells to differentiate, using adipogenesis as a model (21, 22). Dramatic differences were observed in the ability of the 3T3 cells lacking combinations of pRB family members to undergo adipogenesis, and we conclude that in this system, p107 has an opposing, nonoverlapping function relative to that of pRB. Furthermore, our experiments imply that pRB mediates cell cycle withdrawal and differentiation by integrating contributions from transcription factors important for proliferation with those involved in cell fate determination.
Materials and Methods
Cell Culture, Differentiation, Stable Transfections, and Retroviral Infections.
The 3T3 cells were established as described in the accompanying paper (20). Differentiation was performed as described earlier (23). Briefly, when cells reached confluence, the media was replaced by a mixture consisting of 10% FBS, 10 μg/ml insulin (GIBCO/BRL), 0.5 μM 3-isobutyl-1-methylxanthine (MIX; Sigma), and 1 μM dexamethasone (DEX, Sigma). After 48 h, the medium was changed to DMEM containing 10% FBS and insulin (10 μg/ml). The medium was replenished at 2-day intervals, and the appearance of cytoplasmic triglycerides was monitored by microscopy and confirmed by staining with oil red O.
Transfection of the 3T3 cells was performed using calcium phosphate. Vectors expressing pRB, p107, and p130 were cotransfected with a puromycin-selectable marker (pBABE) and selected in DMEM/10% calf serum (CS) containing 2 μg/ml puromycin, pooled, and used for differentiation and expression studies. The retroviral constructs expressing peroxisome proliferator-activated receptor (PPAR)γ and C/EBPα were kindly provided by B. M. Spiegelman (Dana–Farber Cancer Institute). To generate retroviruses, the packaging line BOSC-23 (a gift from W. S. Pear, Department of Pathology, University of Pennsylvania) was seeded at 4 × 106 cells per 10-cm dish, and the cells were transfected with the various retroviral constructs for 24 h. Two days after transfection, culture medium containing virus was harvested and centrifuged at 1500 rpm for 5 min. The viral supernatants were used to infect 3T3 cells in the presence of 50 μg/ml Polybrene (Sigma) for 18 h followed by selection in puromycin (2 μg/ml) -containing medium. After reaching confluence, the infected cells were tested for their ability to differentiate using the protocol described above.
Transcriptional Assays.
Cells (at 75% density) were transfected using calcium phosphate precipitation at a total concentration of 5 μg of plasmid in a total volume of 500 μl added to 3 ml of medium in a six-well dish. After transfection, cells were kept in the differentiation mixture media for 2 days before harvesting. Luciferase assays (Promega), chloramphenicol acetyltransferase (CAT) assays (Boehringer Mannheim), and β-galactosidase assays (Promega) were performed according to the supplier. Results were obtained using an ELISA reader from Molecular Dynamics and a luminometer from EG & G Berthold (Wallac, Gaithersburg, MD).
Flow Cytometric Analysis.
Confluent cells, as well as cells maintained at confluence or in differentiation conditions for 6 days, were split to 7 × 105 cells per 10-cm dish (Fig. 3) and labeled with 10 μM BrdUrd cell-labeling reagent (Amersham Pharmacia) for 30 min, harvested, and analyzed according to the manufacturer (Becton Dickinson). In short, cells were fixed in 80% ethanol, the DNA was denatured by treatment with 2 M HCl/0.5% Triton X-100 at room temperature for 30 min followed by a neutralization step in 0.1 M Na2B4O7, pH 8.5. For immunofluorescence staining, anti-BrdUrd antibodies (Becton Dickinson) and FITC-conjugated horse anti-mouse antibodies (Vector Laboratories) were used. The immunofluorescence step is followed by an incubation in propidium iodide and RNase prior to two-dimensional FACS analysis. The number of gated cells incorporating BrdUrd at the time of the analysis is presented as percent cells in S phase.
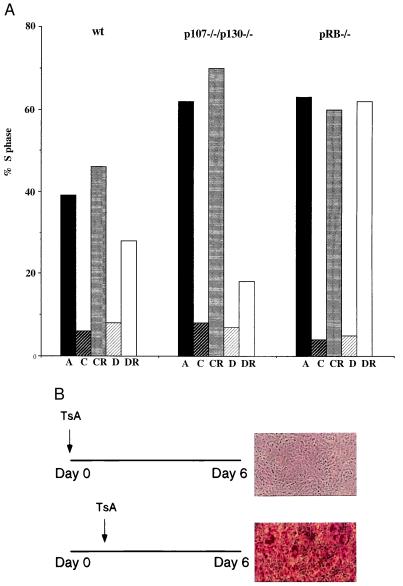
Figure 3.
pRB plays a role in permanent cell cycle exit. (A) wt, p107−/−/p130−/−, and pRB−/− cells were grown to confluence in DMEM containing 10% CS and thereafter maintained in either DMEM + 10% CS or differentiation mixture before they were split into subconfluent conditions in DMEM + 10% CS for 24 h. The percentage of cells in S phase was judged by BrdUrd incorporation and two-dimensional FACS analysis. A = asynchronous, C = confluent for 6 days in 10% CS, CR = split into subconfluent conditions from C, D = differentiation mixture added to cells for 6 days (as described in Fig. 2A), DR = split into subconfluent conditions from D. Shown is a representative experiment because the rate at which the cells reenter the cycle varies slightly from experiment to experiment but the differences between the different cell lines remain the same. (B) TsA blocks differentiation of p107−/−/p130−/− cells if added before the differentiation mixture as indicated in the figure.
Protein Analysis.
Protein extracts from total cells were harvested in RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) at appropriate time points from cells during the differentiation process). Aliquots of protein samples were separated by SDS/10% PAGE, transferred to poly(vinylidene difluoride) membranes (Immobilon-P, from Millipore), and probed with relevant antisera. Polyclonal rabbit antiserum against C/EBPα was a kind gift from P. Rorth and S. L. McKnight (University of Texas Southwestern Medical School). The immunoreactive protein species were visualized by an ECL detection kit (Amersham Pharmacia). Immunolocalization was determined by staining cells with monoclonal antibodies directed against pRB (G3-245; PharMingen) or p107 (SD15). Fluorescein-labeled secondary antibodies were used to detect localization patterns. Cells were treated first for 10 min at room temperature with 4% paraformaldehyde, washed, and then treated with 0.5% Triton X-100 for 10 min at room temperature all in 1× PBS to prepare them for immunofluorescence. Primary and secondary antibodies were sequentially added to cells for 1 h at 37°C. Optical sections were obtained by using the CELLscan System (Scanalytics, Billerica, MA) equipped with a charge-coupled device camera, piezoelectric focus device, and computer-controlled excitation shutter. Deconvolution images shown are single sections through the cell nucleus for the images in Fig. 2C.
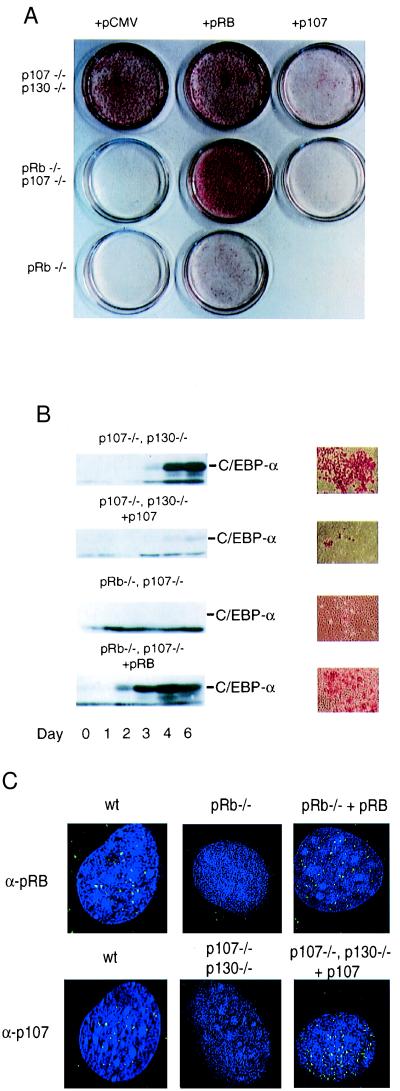
Figure 2.
Opposing roles of pRB and p107 in adipocyte differentiation. (A) Differentiation of p107−/−/p130−/−, pRB−/−/p107−/−, and pRB−/− cells stably transfected with pCMV, pCMV-p107, or pCMV-Rb, as indicated. Pools of cells were differentiated for 6 days and stained with oil red O followed by photography of the plates. (B) C/EBPα expression during the course of differentiation as marked in the figure, p107−/−/p130−/− cells transfected with pCMV or pCMV-p107 and pRB−/−/p107−/− cells transfected with pCMV or pCMV-Rb. At the end of each panel is the corresponding microscope view of the same cells. (C) Indirect immunofluorescence and deconvolution microscopy for pRB and p107. pRB (Upper) and p107 (Lower) were incubated with anti-pRB and anti-p107 antibodies followed by a fluorescein-labeled secondary antibody. The following cell lines were used: 3T3-L1 cell lines (wt); pRB−/− transfected with pCMV or pCMV-pRb, and p107−/−/p130−/− cells transfected with pCMV or pCMV-p107. Several cells were imaged with similar results.
Results and Discussion
Increased Differentiation Potential of p107/p130-Deficient Cells.
To investigate the effects of loss of various pRB family members on differentiation, we used adipogenesis as a model system. Adipocyte differentiation is a well-defined system that can be recapitulated in culture by the addition to fibroblasts of a mixture of factors (Fig. 1A), and is marked by cell cycle exit and the induction of a well characterized transcriptional cascade (21, 22, 24). During the differentiation process, members of the PPAR and C/EBP families occupy central roles both in cell fate determination and growth inhibition. Differentiation can be measured at the physiological level by oil-red-O staining of lipid accumulation in the cytoplasm, and at the molecular level by observing the induction of specific markers, such as C/EBPα. Although some sublines of 3T3 cells (for example 3T3-L1) differentiate well upon stimulation (25), most 3T3 cells do not differentiate unless coerced by exogenous expression of PPARγ and C/EBPα (26).
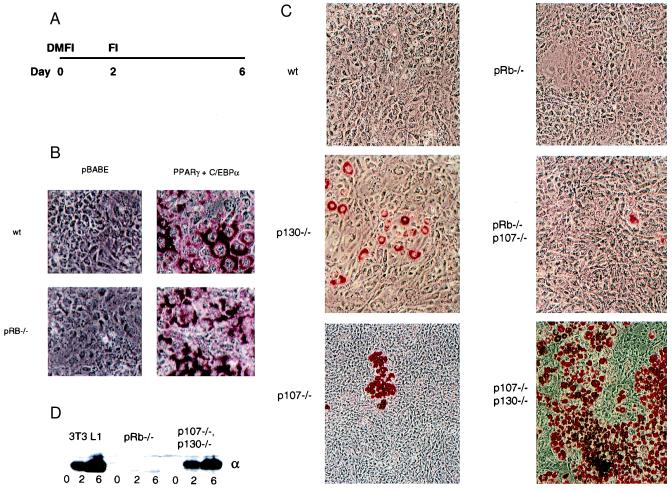
Figure 1.
Functional differences of pRB family members in adipocyte differentiation. (A) Differentiation protocol: cells are grown to confluence, treated with a differentiation mixture consisting of 10% FBS, insulin (10 μg/ml), dexamethasone (1 μM), and 3-isobutyl-1-methylxanthine (0.5 μM) for 2 days and then for an additional 4 days the cells are kept in DMEM containing 10% FBS and insulin (10 μg/ml). During this time, the cells exit the cell cycle and induce a regulatory cascade that leads to a differentiated phenotype that can be visualized by oil-red-O staining and the appearance of adipogenic markers. (B) The wt and pRB−/− 3T3 cells require exogenous expression of PPARγ and C/EBPα introduced by retroviral infection by using the viral vector as a control (pBabe) as indicated in the figure. The infected cells were selected with puromycin, pools of cells were grown to confluence, induced to differentiate for 6 days and stained with oil red O. (C) 3T3 lines of various genotypes were induced to differentiate for 6 days, stained with Oil-Red-O and representative fields of cells were photographed. Genotypes are as marked in the figure: wt, pRB−/−, p107−/−, p130−/−, p107−/−/p130−/−, pRB−/−/p107−/−. (D) C/EBPα protein expression in 3T3-L1, pRB−/− and p107−/−/p130−/− at confluence (day 0), day 2, and day 6 after treatment with the differentiation mixture.
As expected, 3T3 cells derived from wild-type (wt) embryos fail to differentiate when induced by the differentiation mixture, as measured by lipid accumulation (Fig. 1 B and C, Upper Left). Moreover, in agreement with previous studies of other 3T3 lines (26), these cells can be induced to differentiate by retrovirus-mediated expression of C/EBPα and PPARγ (27), followed by exposure to the differentiation mixture. Thus, the wt cells retain the intrinsic ability to differentiate in culture, but require exogenous gene expression (Fig. 1B, Upper).
In sharp contrast to the wt cells, cells deficient in both p107 and p130 display a very high differentiation potential (Fig. 1C, Lower Right). In fact, the degree of lipid accumulation, as well as the timing and level of C/EBPα induction in p107−/−/p130−/− fibroblasts, closely resemble that seen in differentiating 3T3-L1 cells (Fig. 1D, and data not shown). Cell lines lacking p107 or p130 individually exhibit an intermediate phenotype, each having a somewhat increased propensity to differentiate relative to wt cells (Fig. 1C, Left). As had been suggested by previous studies in fibroblasts generated from lung buds of pRB-deficient embryos (19), pRB−/− cells do not differentiate when exposed to the differentiation mixture (Fig. 1C, Top Right). Consistent with their reduced differentiation potential, C/EBPα is not induced in pRB−/− cells (Fig. 1D). Cells deficient in p107 and pRB differentiate to a low but detectable degree, which is greater than wt and pRB−/− cells but less than either p107−/− or p130-deficient cells. In addition, the low level of differentiation seen in pRB−/−/p107−/− cells also indicates that a pRB-negative status is not completely incompatible with differentiation. This was verified in experiments where exogenous expression of C/EBPα and PPARγ was found to induce differentiation in pRB−/− cells (Fig. 1B, Lower). Moreover, pRB−/−/p130−/− cells do not differentiate, even when coerced by C/EBP and PPAR (data not shown). These studies revealed that p107 may have a role directly opposed to that of pRB during adipocyte conversion of 3T3 cells. Significantly, p107 protein levels have been reported to decrease during differentiation of 3T3-L1 cells (28).
To confirm that the observed differences in differentiation potential result directly from the loss of pRB family members, and are not due to other mutations that have occurred during the establishment of these cell lines, we performed a rescue experiment in which pRB, p107, or a vector control were reintroduced into the various cell lines (Fig. 2). We determined that reintroduction of p107 substantially reduced the differentiation potential of the p107/p130-deficient cells (Fig. 2A, Upper). Similar results were observed when p130 was reintroduced into these cells (data not shown). Thus, the genetic loss of p107 and/or p130 is essential for the increased differentiation potential seen in p107−/−/p130−/− cells. In agreement with earlier studies (19), exogenous expression of pRB results in increased differentiation of pRB−/− cells (Fig. 2A, Middle), indicating a positive role for pRB in adipogenesis. Adipogenic induction by pRB is more dramatic in the pRB−/−/p107−/− genetic background (Fig. 2A, Lower), again consistent with an inhibitory effect of p107 on differentiation.
To verify the differentiation observed in the rescue experiments, we examined the levels of endogenous C/EBPα at various times after addition of differentiation mixture (Fig. 2B). Consistent with their degree of differentiation, as scored by oil-red-O staining, induction of C/EBPα expression is dramatically reduced in the p107−/−/p130−/− cells after reintroduction of p107, whereas C/EBPα induction is elevated in pRB−/−/p107−/− cells following expression of exogenous pRB. The expression and localization of the reintroduced pRB and p107 proteins was confirmed with indirect immunofluorescence (Fig. 2C), demonstrating that both the subnuclear localization and levels of the stably transfected proteins resemble those of the endogenous proteins. Taken together, these observations indicate that the genetic loss of pRB or p107/p130 results in a dramatically different phenotypic outcome in this differentiation model.
A Unique Role for pRB in Maintaining Exit from the Cell Cycle.
To further explore the basis for the differences in differentiation potential of cells lacking various members of the pRB family, we compared the requirement for pRB and p107/p130 in two aspects of adipogenesis that seemed particularly relevant; the cell cycle exit that is associated with differentiation and the transcriptional potential of C/EBP and PPAR. As shown previously (20), independent of genotype, all of the cell lines arrest at confluence, and remain arrested throughout the differentiation protocol, with a mainly G1 DNA content, and virtually no cells are found in S phase (Fig. 3A, and accompanying paper, ref. 20). Furthermore, the differentiated p107/p130-deficient cells do not reenter S phase when replated at lower density, whereas their contact-inhibited counterparts readily enter the cell cycle (Fig. 3A, Middle). Even though wt cells, when exposed to the differentiation mixture, do not display any indications of differentiation, such as lipid accumulation or C/EBPα expression, they exhibit a somewhat delayed reentry into S phase (Fig. 3A, Left). In sharp contrast, pRB−/− cells that were maintained in differentiation mixture for 6 days re-enter the cell cycle with kinetics similar to pRB−/− cells maintained at confluence under normal conditions (Fig. 3A, Right).
These results suggest that one important feature of adipogenesis to which pRB, but not p107 and p130, might contribute is by affecting the establishment of a permanent exit from the cell cycle. However, pRB cannot be absolutely required to maintain cell cycle exit, because pRB−/− cells can be induced to differentiate by exogenous expression of C/EBPα and PPARγ (Fig. 1B). It is interesting to note that these two factors have themselves been suggested to have a role in maintaining cell cycle withdrawal during differentiation (29, 30) and might substitute for pRB in maintaining cell cycle exit. Indeed, we have found that expression of C/EBPα and PPARγ in pRB−/− cells bypasses the requirement for pRB in cell cycle exit and differentiation (Fig. 1B, and data not shown).
To determine whether the initiation of adipogenic differentiation requires the repression of E2F target genes by pRB, we used the histone deacetylase inhibitor trichostatin A (TsA), which has previously been shown to result in derepression of pRB target genes (31–33). Confluent p107−/−/p130−/− cells, where pRB-mediated repression is still intact, were treated with TsA, and then subjected to the differentiation protocol. Differentiation is blocked if TsA is added before the addition of differentiation inducing agents but not if the differentiation protocol has already been initiated (Fig. 3B). This experiment further supports the notion that pRB repression of E2F transcription is required early in the differentiation program.
pRB and p107 Have Opposing Effects on Differentiation-Specific Transcription Factors.
Adipocyte-specific gene expression is controlled by a transcriptional regulatory cascade in which C/EBPα and PPARγ play key roles. pRB has been previously reported to promote the activity of differentiation-associated transcription factors, such as MyoD and C/EBP (16, 17, 19, 34). In some cases, pRB-mediated activation of these factors appears to occur through direct mechanisms. For example, previous studies have suggested that pRB can bind to C/EBP family members (19). Moreover, we found that pRB and p107 partially colocalize with C/EBP and PPAR in differentiating 3T3 L1 cells (data not shown). However, the mechanism by which pRB stimulates transcription remains unclear. Therefore, we examined the ability of pRB and p107 to regulate the transcriptional activities of C/EBP and PPAR. First, we analyzed the effect of expressing pRB and p107 on C/EBPα-mediated transcription in cells lacking both proteins. As previously reported members (19), we observed that C/EBPα-induced transcription is stimulated by pRB (Fig. 4A, Left). The pRB-dependent increase in C/EBP-mediated transcription requires an intact C/EBP binding site (data not shown). In contrast, p107 has no effect on C/EBPα-mediated transcription even at concentrations where repression of E2F transcription is dramatic (Fig. 3A, Right). These experiments suggest that a special role for pRB in stimulating differentiation is because of its ability to activate C/EBPα-mediated transcription, a property that is not shared by p107.
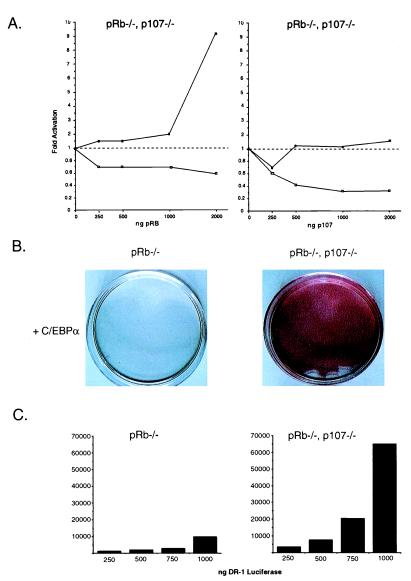
Figure 4.
pRB and p107 differentially affect C/EBP and PPAR activities. (A) Stimulation of C/EBPα activity (■) by pRB, but not p107, at concentrations where they both repress E2F-mediated transcription (□). Cells deficient in pRB and p107 (pRB−/−/p107−/−) were transfected with 100 ng of a C/EBPα reporter driving luciferase (aP2-luciferase) and 100 ng of an E2F reporter driving the expression of chloramphenicol acetyltransferase [(E2F)4-CAT]. Transcriptional activity of these two reporters was measured using 200 ng of pBabe-C/EBPα and increasing concentrations of pCMV-Rb or pCMV-p107 as shown in the figure. All values were corrected to a β-actin-β-galactosidase internal control and the result of multiple experiments is presented as average fold induction as compared with vector alone. (B) C/EBPα is sufficient to induce differentiation of pRB−/−/p107−/− cells but not pRB−/− cells as indicated in the figure. The experiment was carried out as previously described in Fig. 2B. (C) The transcriptional activity from a PPAR reporter driving luciferase (DR-1 luciferase) is higher in pRB−/− cells that lack p107 as shown in the bar graphs bellow the differentiated phenotypes. Increasing amounts of reporter were transfected into pRB−/− and pRB−/−/p107−/− cells and luciferase values were corrected to a β-actin–β-galactosidase internal control. A representative example of many experiments is shown.
A potential explanation for the inhibitory effect of p107 on differentiation was revealed when we examined the effects of exogenous expression of C/EBPα and PPARγ on differentiation potential (Figs. 1B and 4B, and data not shown). In these experiments, we consistently observed a lower requirement for PPARγ in cells deficient for p107. For example, exogenous expression of C/EBPα alone is sufficient to induce differentiation of pRB−/−/p107−/− cells (Fig. 4B), whereas pRB−/− cells require exogenous expression of both C/EBPα and PPARγ to achieve a highly differentiated phenotype (Figs. 1B and 4B). One possible explanation for this finding is that p107 inhibits PPARγ activity. Therefore, we examined PPARγ activity in pRB−/− and pRB−/−/p107−/− cells that have similar endogenous levels of PPARγ protein. PPAR transcriptional activity is significantly increased in the pRB/p107-deficient cells as compared with pRB−/− cells (Fig. 4C). These results suggest that the distinct roles for pRB family members in differentiation are due, at least in part, to differences in their abilities to regulate the activities of differentiation-associated transcription factors.
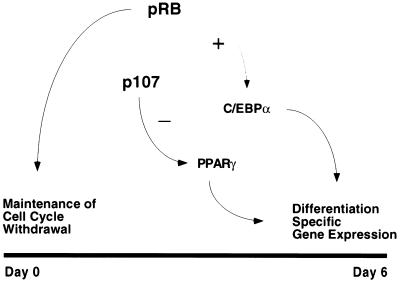
To explain our overall observations, we suggest a dual role for pRB in cell cycle withdrawal and differentiation, and an inhibitory effect of p107 on transcriptional regulators involved in the differentiation process (Fig. 5). Although pRB and p107 exhibit opposing functions in adipocyte differentiation, the role of p130 is less clear. We observed that loss of p130 in a pRB-deficient cell results in a decreased differentiation potential, whereas loss of p130 in p107-deficient cells significantly increases their differentiation potential (Fig. 1C, and data not shown). Such context-specific activity of p130 further supports the notion that combinatorial roles for the pRB family members can significantly influence cellular properties, and that it may not be possible to establish their complete spectrum of functions when studied in isolation.
Figure 5.
Model.
Given the results described in this report, it is necessary to broaden our definition of the functions of pRB family members. Although their ability to control the timing of G1 remains undisputed, it is becoming increasingly clear that they play additional cellular roles. Although cell culture experiments have potential drawbacks, our results demonstrate that the combined use of mutant 3T3 cell lines and phenotypic rescue by gene re-introduction provides an important setting in which to compare and contrast the functions of these proteins and their upstream and downstream targets.
Acknowledgments
We thank W. S. Pear for the BOSC retroviral packaging cell line, B. M. Spiegelman for PPAR and C/EBP retroviral constructs, the PPAR-reporter construct, and PPAR polyclonal antibodies, P. Rorth and S. L. McKnight for C/EBP antibodies, and D. M. Lane for ap-2 reporter construct. We also express our sincere gratitude to A. McClatchey, J. Settleman, S. R. Salama, B. S. Schulman, J. Zhao, M. Walhout, M. Vidal, D. Barbie, and all of the members of the Harlow and Dyson Laboratories for helpful discussions and reading of this manuscript in its various forms. M.C. has been sponsored by a MBRC Tosteson Award and a fellowship from the Medical Foundation, and B.K.K. by a fellowship from Leukemia Society of America. This work was supported by grants from the National Institutes of Health (to E.H.).
Abbreviations
- PPAR
peroxisome proliferator-activated receptor
- C/EBP
CCAAT enhancer binding protein
- wt
wild type
- TsA
trichostatin A
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190343597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190343597
References
- 1.Sparkes R S, Murphree A L, Lingua R W, Sparkes M C, Field L L, Funderburk S J, Benedict W F. Science. 1983;219:971–973. doi: 10.1126/science.6823558. [DOI] [PubMed] [Google Scholar]
- 2.Lee W H, Bookstein R, Hong F, Young L J, Shew J Y, Lee E Y. Science. 1987;235:1394–1399. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 3.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C, Lai C-C, Herrup K, Lee W-H, Bradley A. Nature (London) 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 4.Jacks T, Fazeli A, Schmitt E, Bronson R, Goodell M, Weinberg R. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 5.Jacks T. Annu Rev Genet. 1996;30:603–636. doi: 10.1146/annurev.genet.30.1.603. [DOI] [PubMed] [Google Scholar]
- 6.Mulligan G, Jacks J. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 7.Cobrinik D, Lee M-H, Hannon G, Mulligan G, Bronson R T, Dyson N, Harlow E, Beach D, Weinberg R A, Jacks T. Genes Dev. 1996;10:1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 8.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 9.Lukas J, Parry D, Aagarrd L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Nature (London) 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 10.Qin X Q, Chittenden T, Livingston D M, Kaelin W G., Jr Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 11.Nevins J R. Curr Opin Genet Dev. 1994;4:130–134. doi: 10.1016/0959-437x(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 12.Dyson N, Buchkovich K, Whyte P, Harlow E. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 13.Ewen M E, Ludlow J W, Marsilio E, DeCaprio J A, Millikan R C, Cheng S H, Paucha E, Livingston D M. Cell. 1989;58:257–267. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 14.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Nature (London) 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 15.Whyte P, Williamson N M, Harlow E. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, Schneider J W, Condorelli G, Kaushai S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 17.Novitch B G, Mulligan G J, Jacks T, Lassar A B. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacksenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 19.Chen P-L, Riley D J, Chen Y, Lee W-H. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 20.Classon M, Salama S, Gorka C, Mulloy R, Braun P, Harlow E. Proc Natl Acad Sci USA. 2000;97:10820–10825. doi: 10.1073/pnas.190343497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh W-C, McKnight S L. Curr Opin Cell Biol. 1995;7:885–890. doi: 10.1016/0955-0674(95)80074-3. [DOI] [PubMed] [Google Scholar]
- 22.Tontonoz P, Hu E, Spiegelman B M. Curr Opin Genet Dev. 1995;5:571–576. doi: 10.1016/0959-437x(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 23.Yeh W C, Cao Z, Classon M, McKnight S L. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 24.Mandrup S, Lane M D. J Biol Chem. 1997;272:5367–5370. doi: 10.1074/jbc.272.9.5367. [DOI] [PubMed] [Google Scholar]
- 25.Green H, Kehinde O. Cell. 1974;1:113–116. [Google Scholar]
- 26.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 27.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richon V M, Lyle R E, McGehee R E., Jr J Biol Chem. 1997;272:10117–10124. doi: 10.1074/jbc.272.15.10117. [DOI] [PubMed] [Google Scholar]
- 29.Umek R, Friedman A, McKnight S L. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 30.Altiok S, Xu M, Spiegelman B M. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T E. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 32.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 33.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 34.Sellers R S, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]