Abstract
DNA damage activates the ataxia telangiectasia–mutated and Rad3-related (ATR) kinase signal cascade. How this system is restrained is not understood. We find that in estrogen receptor (ER)-positive breast cancer cells, UV or ionizing radiation and hydroxyurea rapidly activate ATR-dependent phosphorylation of endogenous p53 and Chk1. 17-β-estradiol (E2) substantially blocks ATR activity via plasma membrane-localized ERα. E2/ER reduces the enhanced association of ATR andTopBP1 proteins that follows DNA damage and strongly correlates to ATR activity. E2 inhibits ATR activation through rapid PI3K/AKT signaling: AKT phosphorylates TopBP1 at Serine 1159, thereby preventing the enhanced association of ATR with TopBP1 after DNA damage. E2 also inhibits Claspin:Chk1 protein association via AKT phosphorylation of Chk1, preventing Chk1 signaling to the G2/M checkpoint. ATR-phosphorylation of p53 induces p21 transcription, prevented by E2/ER. E2 delays the assembly and prolongs the resolution of γH2AX and Rad51 nuclear foci and delays DNA repair. E2/ER also increases the chromosomal damage seen from cell exposure to IR. Therefore, the restraint of ATR cascade activation may be a novel estrogen action relevant to breast cancer.
INTRODUCTION
In response to DNA damage, specific DNA repair mechanisms and cell cycle checkpoints are invoked, the latter to ostensibly provide adequate time to repair DNA (Sancar et al., 2004). Inherited or acquired mutations in epithelial cells are sometimes transforming and confer to cells growth (mutated Ras or ErbB2) and/or survival (inactivating p53 mutations) advantages. If a potentially transforming, acquired mutation is not repaired and is passed onto progeny, a tumor may result. Additional unrepaired mutations convey invasiveness and metastatic potential to established, less aggressive tumors. Thus, the functional loss of DNA repair mechanisms is important to cancer biology (Sancar et al., 2004; Bartkova et al., 2005).
Radiation damage in the cell evokes pathways that link DNA repair and cell cycle checkpoints. Ionizing radiation (IR) and UV radiation (UV) cause double-stranded DNA breaks and fork replication stalling, respectively (Kastan and Bartek, 2004). DNA damage is sensed, resulting in the activation of the serine/threonine kinases, ATM (ataxia-telangiectasia mutated) and ATR (ATM and Rad3-related; Bakkenist and Kastan, 2003; Kastan and Bartek, 2004). ATR activation results from replication stress, whereas ATM and ATR play roles in the cellular response to double-strand DNA breaks. Mutations of these and downstream substrate molecules (BRCA1 and 2, NBS, p53, and Chk2) can result in epithelial cell malignancies.
The activity of ATR requires its association with the ATRIP (Cortez et al., 2001) and TopBP1 proteins (Kumagai et al., 2006), and association with RPA-coated single-strand DNA activates the kinase. Inhibition of normal DNA repair signaling may simulate genetic loss of these molecules and predispose normal cells to acquire transforming mutations. However, endogenous inhibitory mechanisms that prevent the activation of ATR and downstream effectors are virtually unknown. For breast cancer, estradiol (E2), and estrogen receptors (ER) act as proliferation and survival factors that promote the development of this malignancy (Lippman and Dickson, 1989; Platet et al., 2004). Approximately 70% of breast cancers in women display the ER α isoform (ERα), and the major adjuvant therapies (to some form of surgery) usually involve anti-estrogen/receptor agents (Lippman and Dickson, 1989). Oncogenic actions of nuclear ER in breast tumors involve regulation of gene transcription (genomic actions; Keeton and Brown, 2005). Additionally, nongenomic actions of E2/ER mediated through membrane and mitochondrial ER pools and genomic actions resulting from kinase activation by membrane ER may also contribute to breast tumor biology (Levin, 2005; Levin and Pietras, 2007). Membrane ERα is the classical ERα that is found predominantly in the nucleus, but a smaller pool of this receptor traffics to the plasma membrane (Levin, 2005). The ligand-binding domain (E domain) of classical ERα is essential for the membrane pool of the sex steroid receptors to generate rapid signal transduction.
It is probable that E2/ER effect additional oncogenic processes (Naugler et al., 2007) and may alter DNA repair in target cells. Here, we describe the unexpected findings that E2/ERα inhibits ATR activation resulting from multiple DNA damage–inducing stimuli in breast cancer and normal mammary epithelial cells. As the first described endogenous inhibitors of ATR activation, the resulting E2/ER actions indicate potential novel functions that are germane to the impact of ATR inhibition for malignancy.
MATERIALS AND METHODS
Antibodies and Plasmids
TopBP1 (sc-22859) antibodies used for immunoprecipitation (IP) or immunoblot (IB) were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to Serine 216 of Cdc25C, Serine 10 of histone H3, Rad 51, and γH2AX (Serine139) were from Cell Signaling (Beverly, MA, or Santa Cruz Biotechnology). Wild-type (wt) ERα, membrane or nuclear-targeted E domain (ligand-binding domain) of ERα, pRK5-HERCD533 (dom-neg enhanced green fluorescent protein [EGFR] from Axel Ullrich (Institute of Molecular Biology, A*STAR, Singapore) (Redemann et al., 1992), and ERαS522A constructs were previously described (Razandi et al., 2003a; Pedram et al., 2006b). Wild-type and S280AChk1 plasmids and phospho-Serine 280 Chk1 antibodies were from Dr. Emma Shtivelman (Bionovo, Emeryville, CA) (King et al., 2004). Wtp53 and S15Ap53 were from Rainer Brockman via David Meek (University of Dundee, Dundee, United Kingdom) (Meek, 2002). PMT2-AHAKT (dominant-negative AKT) was provided by Dr. Julian Downward (London Research Institute, London, United Kingdom). Flag-wt, S1159A, and S1159D TopBP1 constructs were from Dr. Weei-Chin Lin (University of Alabama at Birmingham, Birmingham, AL) (Liu, K. et al., 2006b).
Cell Culture, Reagents, and Basic Experiments
All cell lines were from ATCC (Manassas, VA). MCF7 cells were grown using DMEM-F12, 10% FBS, and 1% antibiotic-antimycotic (Invitrogen, Carlsbad, CA; 15240-062) at 37°C to 75% confluency in 100-mm dishes. In experiments where the effects of estrogen were determined, cells were cultured for 24 h in phenol red–free, charcoal-stripped FBS to remove any estrogenic effects, before E2 addition.
For experiments using random cycling cells, culture was done in the absence of serum for 2 h, before DNA damage± E2 addition. For S-phase synchronization, MCF7 cells were grown to ∼60% confluence. Thymidine was added (final concentration 2 mM), and the cells were cultured at 37°C for 16 h. Medium was then removed, and cells were washed three times with PBS. Fresh medium without thymidine was added for 9 h at 37°C, followed by adding thymidine for 16 h. After washing, time-course studies of thymidine incorporation after release from the second thymidine incubation showed strong DNA synthesis at 6 h (S-phase). At this time ATR activity in response to DNA-damaging agents was determined.
Mouse mammary epithelial cells were isolated from 8-wk-old female C57/BL6 mice by enzymatic and mechanical methods and established as primary cultures for experiments. propyl-pyrazole-triol (PPT) and diarylpropionitrile (DPN) were from Tocris Chemicals (Ballwin, MO). Tyrphostin 1478 and LY294002 were from Calbiochem (La Jolla, CA). Small interfering RNAs (siRNAs) forTopBP1, and AKT were from Dharmacon (Boulder, CO). In general, E2 or other steroids or chemicals were added 20 min before DNA-damaging treatment. UV exposure (10 J/m2) was performed over 10 s using a Stratalinker 1800 (Stratagene, La Jolla, CA). Gamma radiation (IR) of cells was performed at our Radiation Therapy Center, after calculating the delivery of precise amounts of radiation (2 Gy) to cultured cells over 15 s. Hydroxyurea (HU), 1 mM, was added to the cells for 2 h, based on initial ATR kinase time-course studies.
The E domain of ERα was inserted into EGFP vectors (Clontech) targeting the E domain completely to the plasma membrane or nucleus as we described (Pedram et al., 2006b), and the constructs were transfected into ER null HCC-1569 cells. Cells were synchronized to S-phase and exposed to brief UV in the presence or absence of E2, and ATR activity was then determined.
Kinase Assays and 32Pi labeling
ATR kinase activity was determined in vitro. For kinase assays, both random cycling and S-phase–synchronized MCF7 cells were exposed to various treatments (e.g., brief UV±E2). MCF7 cells from each condition (∼1 million) were then washed with DMEM/F12 medium and lysed in buffer (50 mM Tris-HCL, pH 7.5,100 mM NaCl, 50 mM NaF, 5 mM EDTA, 0.5% Triton X-100, 40 mM β-glycerophosphate, 200 mM sodium orthovanadate, 40 mM PNPP, 100 mM PMSF, and protease inhibitor cocktail; Sigma, St. Louis, MO). The lysates were centrifuged at 14K rpm for 10 min, and the supernatants were exposed to 50 μl of suspended Protein A or G agarose beads in microcentrifuge tubes and rotated for 30 min at 4°C. This was done twice to ensure removal of endogenous IgG and nonspecific proteins from lysate. The cell lysate/protein G or A bead complex was centrifuged at 14,000 rpm for 30 s at 4°C. Without disturbing the pellet, the supernatant was transferred to a new tube. These precleared supernatants were stored on ice. For the ATR activity assay, 10 μl of ATR antibody (Santa Cruz Biotechnology, sc-1887or sc-21848) was conjugated to 50 μl of protein G beads (Sigma, P4691) for 2 h at room temperature, and the bead complex was washed. Then, 1 ml of precleared whole-cell extract from each experimental condition was added to the ATR antibody/protein G bead complex in lysis buffer (that also serves as the IP buffer), and rotated end-over-end overnight at 4°C. Beads were washed once with lysis buffer and twice with HEPES buffer (25 mM HEPES, 10 mM Mg-acetate). To the lysates/antibody/bead complexes, 40 μl of stock mixture, made up as 20 μl of 3 × kinase buffer (25 mM HEPES, pH 7.5, 10 mM MgAc, 2 mM DTT, 40 mM ATP), and 10 μl of H2O, was added to 2 mg glutathione S-transferase (GST)-Phas1 in 9 μl of water, and 1 μl [γ-32P]ATP. Each tube was vortexed and then incubated at 30°C for 30 min. SDS-PAGE loading buffer (40 μl) was then added, vortexed, boiled for 5 min, and centrifuged at 14K rpm for 1 min. Samples of 40 μl from each condition were loaded onto a 10% gel. After electrophoresis, the gel was fixed, dried, and subjected to autoradiography, reflecting 32P incorporated into the Phas-1 protein. ATR activity was also determined by specific phosphorylation of endogenous p53 (Serine 15) and Chk1 (Serine 345), as immunoblots with phospho-specific antibodies (Santa Cruz Biotechnology). Total protein immunoblots serves as loading controls for many experiments and reflect additional gel separation of the same cell protein lysates.
Chk1activity was determined either using Serine 345 (Cell Signaling) phospho-Chk 1 antibodies for immunoblot, or as kinase activity using GST-Cdc25C as substrate. The in vitro kinase activity assay was similar to what was described for ATR. Immunoblots for Serine 280 Chk1 phosphorylation were also determined (antibody was a kind gift from Dr. Emma Shtivelman). Immunoprecipitated endogenous Cdc2 kinase from cells undergoing various conditions and p70 S6 kinase protein as exogenous substrate were used for in vitro Cdc2 kinase activity assays. Tyr15 phosphorylation of Cdc2 was determined by IB (antibody from Santa Cruz Biotechnology). Cdc2 kinase is also known as Cdk1 kinase.
32P-labeling of cellular protein was done in near confluent MCF7 cells transfected with Flag-wt or S1159ATopBP1 and recovered overnight. Transfected cells were placed for 18 h before labeling in fresh, phosphate-free DMEM (Invitrogen) supplemented with 5% charcoal-stripped FBS and incubated in 5% CO2 at 37°C. This medium was removed, and washed cells were pretreated with medium containing LY294002 for 20 min and then exposed to brief UV in the presence or absence of E2. Replacement with prewarmed medium containing 0.5 μCi/ml 32Pi (Perkin Elmer-Cetus) ± E2 ensued, and the cells were placed in a CO2 incubator inside a Plexiglas box for 2 h. Cells were washed to remove unincorporated 32Pi and lysed in the previously mentioned lysis buffer. Lysates were sonicated at 4°C for 10 min and then spun at 14K rpm for 5 min. Supernatants were placed in microcentrifuge tubes, and lysates were precleared with 50 μl of suspended agarose beads, for 30 min at 4°C while rotating. The lysate/bead complex in microcentrifuge tubes was centrifuged at 14K rpm for 30 s at 4°C. Without disturbing the pellet, the supernatant was transferred to a new tube and stored on ice. Flag antibody (20 μl, Santa Cruz Biotechnology) was conjugated to 50 μl of protein G beads (Sigma, P4691) for 2 h at room temperature. Then, 1 ml of precleared cell extract was added to the Flag antibody–protein G bead complex and rotated end-over-end overnight at 4°C. Beads were washed with PBS, and 50 μl of SDS-PAGE loading buffer was added. After boiling and centrifugation, 40 μl of each sample was loaded onto a 10% gel. Subsequently, the gel was fixed, dried, and subjected to autoradiography (using special intensifying screen for 32P).
Protein–Protein Interactions
IP of endogenous ATR, followed by immunoblot for TopBP1 was carried out in S-phase and in randomly cycling MCF7 cells under stated conditions. Whole-cell lysates were precleared by adding 1 ml of whole-cell lysate to 50 μl of protein A/G agarose beads at 4°C for 30 min. Beads were centrifuged at 3000 rpm for 30 s at 4°C. Supernatants (cell lysates) were transferred to a new microcentrifuge tube. To 1 ml of the above cell lysate, 10 μg of primary antibody (agarose conjugate) was added and incubated at 4°C overnight with end-to-end mixing. After centrifugation, pellets were washed three times with PBS. After final wash, the pellets were resuspended in 40 μl of 2× electrophoresis sample buffer, for separation by PAGE. Proteins from the gel were transferred to nitrocellulose and then exposed to TopBP1 antibodies for immunoblot, using the ExactaCruz kit (Santa Cruz Biotechnology) that features the elimination of light and heavy chain antibody detection. Similar techniques were used to assess Chk1:Claspin interactions. In some experiments, Flag antibodies were used. Cell localization of proteins and phosphorylation status was determined by immunofluorescent confocal microscopy after culturing the cells on glass coverslips before specific antibody staining, followed by FITC conjugated–second antibody.
DNA Repair and Viability Assays
S-phase and random cycling MCF7 cells were exposed to brief UV or IR±E2. The cells were allowed to proceed in DMEM-F12±E2 agarose, alkali-denatured, and subjected to electrophoresis. The cells were stained with DNA intercalating dye (SYBR green and visualized by epifluorescence (Trevigen, Gaithersburg, MD; UVDE Flare Assay Kit). Damaged, unrepaired DNA was seen as the “tail” of the Comet, quantified from 200 cells per condition by using Komet imaging software, version 5.5 (Andor Technology, South Windsor, CT). The experiment was repeated twice for mean ± SEM calculations of tail quantification. Cell viability was determined by MTT assay (Sigma) that measures mitochondrial dehydrogenase activity in viable cells. Random cycling MCF7 cells were exposed to DNA-damaging agents in the presence or absence of E2. At 24 h, the cells were incubated with medium containing MTT, 1 mg/ml, for 4 h at 37°C. MTT/formazan was extracted by overnight incubation at 37°C with 100 μl extraction buffer (20% SDS, 50% formamide adjusted to pH 4.7 with 0.02% acetic acid, and 0.025 N HCl). Optical densities at 570 nm were measured using extraction buffer as a blank.
P21 Transcription
Wild-type or S15Ap53 were expressed in HCC-1937 cells (p53 null, BRCA1 mutated, and ER negative), and the cells were recovered and then exposed to brief UV. DNA was obtained 6 h after treatment. In some cells, ERα was expressed, and the cells were incubated in the presence or absence of E2. RT-PCR for p21 was performed at 94°C for 3 min, followed by 26 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, using primers p21F (5′-CCAAGAGGAAGCCCTAATCC-3′) and p21R (5′-CCCTTCAAAGTGCCATCTGT-3′). As a control, the same reaction conditions were applied for PCR detection of the human GAPDH. The GAPDH primers were as follows: forward, 5′-AGCCACATCGCTCAGAACAC, and reverse, GAGGCATTGCTGATGATCTTG-3′, respectively.
CPD Quantification
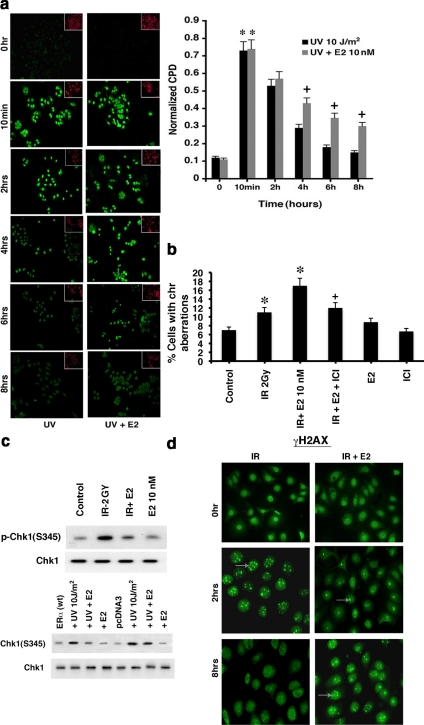
S-phase MCF7 cells were exposed to UV±E2 and then fixed at 10 min, 2, 4, 6, or 8 h and exposed to photoproduct specific CPD fluorescent antibody (Sigma). The nuclear fluorescent signal was quantified and normalized to nuclear area determined by propidium iodide (PI) staining. Insets show PI staining of the same fields.
γH2AX and Rad51 Foci Formation
MCF7 cells were cultured in glass plates with phenol red–free DMEM-F12 containing charcoal-treated FBS. The cells were synchronized with 1 μM nocodazole for 16 h and then released and irradiated with UV 10 J/m2 in the presence or absence of 10 nM E2. Cells were fixed with 4% freshly prepared paraformaldehyde at room temperature, at 10 min, 2, 4, 6, and 8 h after UV. Cells were then briefly treated with 0.5% Triton X-100, washed four times with PBS, and incubated with 2% BSA for 1 h at room temperature. Then the primary antibodies Rad 51 (Santa Cruz Biotechnology), or γH2AX (Upstate Biotechnology, Lake Placid, NY) were incubated with cells overnight at 4°C. Cells then were washed and incubated with secondary antibody conjugated with FITC (Vector Laboratories, Burlingame, CA) at 1:100 dilution for 1 h, and visualized under fluorescent microscopy.
Preparation of Metaphase Chromosome Spreads from Adherent Cells
MCF7 cells were grown in DMEM-F12 with estrogen-free serum 2 d before performing the experiments involving chromosome spreads. Nonsynchronized cells were exposed to 2 Gy IR in the presence or absence of 10 nM E2 ± ICI182780 (ICI), (Faslodex, AstraZeneca, Wilmington, DE) and then were allowed to recover at 37°C for 48 h. For the last 3 h of incubation, colcemid was added to a final concentration of 1 μg/ml and incubated at 37°C. The cells were washed with PBS (without Ca2+ and Mg2+) and then detached with trypsin-EDTA. The cell pellet was treated with 0.075 M KCl solution and pipetted up and down to break clumps. After centrifugation, the supernatant was aspirated, leaving 200 μl of the hypotonic solution. After centrifugation, the pellet was resuspended in1 ml of methanol:glacial acetic acid (3:1) solution, and the procedure was repeated. Resuspending the pellet then occurred in a small volume of fixative (<500 μl). A drop of mounting solution with 1% Giemsa stain was added to each slide. The slides were examined under oil immersion and viewed at 10 × 100 magnification. Chromosomal damage (chromatid breaks, dicentric chromosomes, or condensation [ring formation]) was scored from 200 cells in each condition, and the study was repeated.
Image Acquisition
The microscopic images were obtained using a Nikon Eclipse TE-200 scope with magnification from 200 × to 400 × at room temperature. A Diagnostic Instruments camera (model 3.2.0; Sterling Heights, MI) was used in conjunction with Spot Advance software, to capture and transfer images to the computer. Rhodamine- (red) and FITC (green)-conjugated secondary antibodies (Vector Laboratories) were used for fluorescent visualization.
RESULTS
E2 and E2/ERα are Endogenous Inhibitors of ATR Kinase
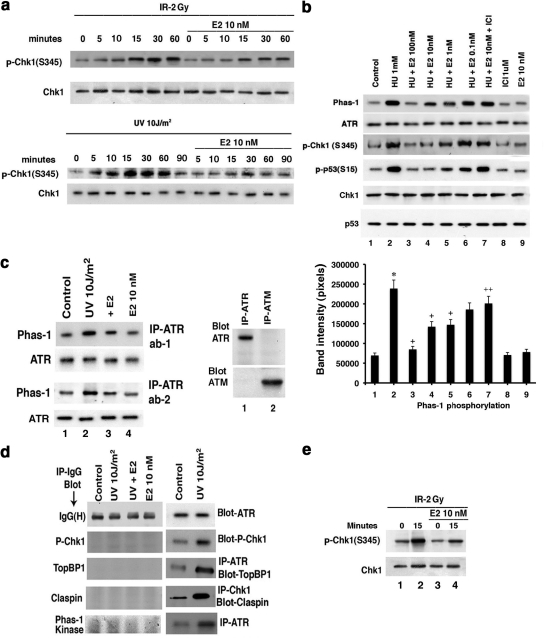
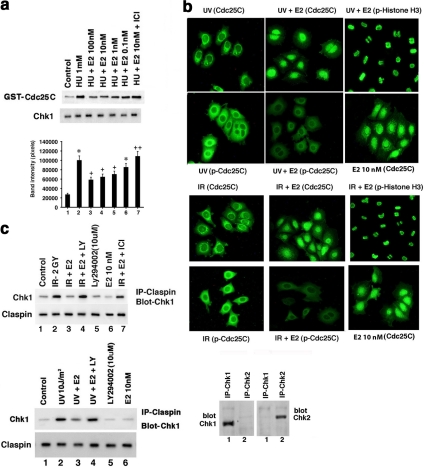
To determine if E2 restrains DNA damage–induced signaling, we exposed random cycling or S-phase–synchronized MCF7 cells to brief IR or UV or more prolonged HU. IR stimulated ATR kinase activity in 15–30 min, and UV activated ATR in 5 min (Figure 1a), whereas HU stimulated ATR activity by 1–2 h of cell exposure (Figure 1b). E2 substantially inhibited DNA damage–induced ATR activity, determined as endogenous Chk1 phosphorylation at Serine 345 (ATR site) or p53 Serine 15 phosphorylation (Figure 1, a and b). ICI, an ER antagonist and therapeutic agent used in treating breast cancer (Howell, 2006), partially but significantly prevented the effects of physiological levels of E2 (1–10 nM; Figure 1b, lanes 2, 4, and 7). We confirmed the specificity of the ATR activity assay using two different antibodies to N- or C-terminus ATR for IP, followed by in vitro kinase activity assay against Phas-1 as substrate (Figure 1c). ATR antibody did not identify ATM in our precipitated protein, and after IP with ATM antibody, only ATM was identified upon immunoblot with the ATM antibody. IP with IgG yielded no kinase activity in any study (Figure 1d). Activation of ATR in S-phase cells at 15 min after IR exposure was prevented by E2, 6 h after removal of the second thymidine pulse (Figure 1e).
Figure 1.
DNA damage–induced ATR is inhibited by estradiol (E2). (a) Time course of IR and UV activation of ATR, inhibited by 10 nM E2. Random cycling MCF7 cells were briefly exposed to DNA-damaging agents in the presence or absence of 10 nM E2. Endogenous Chk1 phosphorylation at Serine 345 was determined at various times after UV or IR exposure using a phospho-specific antibody for IB, after IP of total Chk1 protein and separation by SDS-PAGE. Total Chk1 serves as loading controls and was determined similarly by IB, reflecting an additional gel separation of the same cell lysates. IP with nonspecific, IgG antibody produced no kinase activity against Phas-1, the peptide used as the substrate for in vitro ATR activity assay described in Materials and Methods (Figure 1d). (b) Physiological E2 inhibits HU-induced ATR activity via ER in S-phase cells. Phas-1 peptide is the substrate for in vitro kinase activity, using ATR IP from MCF7 cells exposed for 2 h to 1 mM HU. Total ATR protein is the loading control. Endogenous p53 phosphorylation at Serine15 using phospho-specific antibody was also determined by IB, after total p53 IP. ICI182780 (ICI, 1 μM) is an ER antagonist. The bar graph represents three Phas-1 experiments combined, single determinations per condition in each experiment. * p < 0.05 by ANOVA plus Schefe's test for control vs HU, + p < 0.05 for HU vs HU+E2, ++ p < 0.05 for HU+10 nM E2 vs 10 nM E2+ICI. (c) Left, ATR in vitro assay in random cycling MCF7 cells using two different antibodies to the C- or N-terminus of ATR, respectively. Right, immunoblots of ATR or ATM, after IP of protein from MCF7 cells with ATR or ATM antibodies to show specificity of the ATR ab-1 used. (d) IgG antibody was used for IP produces and produced no kinase activation or protein–protein interactions under any conditions. This was shown by subsequent immunoblot or in vitro kinase activity assay. Left, IP with IgG, followed by IB for IgG (positive control), IB for phospho-Chk1, TopBP1, claspin, or after IP with the IgG antibody. Similarly, no ATR kinase activity (Phas-1 kinase) resulted from IP with IgG. Right, positive detection with appropriate antibodies for all studies and modulation by UV. (e) S-phase MCF7 cells were briefly exposed to IR±E2, and kinase activity was determined just before (time 0) and 15 min after radiation.
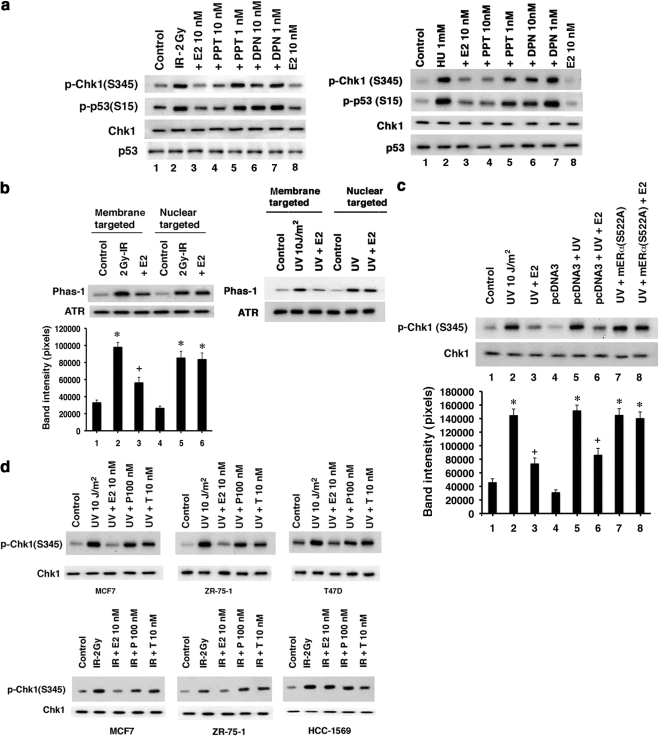
We then investigated which ER isoform (α or β; Greene et al., 1977; Kuiper et al., 1996) mediated these effects of E2 by comparing receptor specific agonists (Harrington et al., 2003). The ERα agonist PPT, but not the ERβ-specific agonist DPN inhibited ATR activation comparably to E2 in MCF7 cells (Figure 2a, lanes 2–4). All MCF7 cells show both membrane and nuclear ERα, and no dissociation of receptor pools is seen. Rapid modulation of kinases mainly results from actions of E2 at cell membrane-localized ER (Levin, 2005). We therefore targeted a functionally important region of ERα, the E domain/ligand-binding domain, to either the plasma membrane or nucleus of ER null breast cancer cells (Pedram et al., 2006b). Only the membrane-localized E domain of ERα supported E2 inhibition of radiation-activated ATR (Figure 2b). We also expressed a mutant ERα that specifically binds and sequesters endogenous ERα from localizing to the plasma membrane (Razandi et al., 2003a). S522AERα expression in S-phase MCF7 cells reversed E2-inhibition of UV-activated ATR (Figure 2c, lanes 2, 3, and 8). Because classical androgen and progesterone receptors are also expressed at the cell surface of MCF7 cells (Pedram et al., 2007), we asked whether these receptors also modulate ATR. In three steroid receptor positive breast cancer cell lines, only E2 and not progesterone or testosterone inhibited ATR activation, whereas E2 had no effect on ER-negative breast cancer cells (Figure 2d). The results indicate that E2 acts through membrane ERα to inhibit ATR activation by DNA damage but ligands for other sex steroid receptors in breast cancer do not inhibit ATR.
Figure 2.
E2 inhibits ATR activity through plasma membrane ERα. (a) E2, PPT (ERα-specific agonist), but not DPN (ERβ-specific agonist) inhibited endogenous p53 and Chk1 phosphorylation at ATR sites as stimulated by IR and HU. ER agonists in the absence of DNA damage had no effect (data not shown). (b) ER null HCC-1569 cells were transiently transfected to express the E domain of ERα targeted to the cell membrane or nucleus. After overnight cell recovery, ATR activity was determined in random cycling cells 15 or 5 min after IR or UV. Bar graph is from three experiments, * p < 0.05 for control vs IR or IR+E2 (nuclear), + p < 0.05 for IR vs IR+E2. (c) S522AERα expression blocks E2 inhibition of ATR. The mutant ER or pcDNA3 plasmids were transfected into MCF7, the cells recovered overnight, and S-phase cells were subjected to UV. Total Chk1 protein serves as loading normalization. * p < 0.05 for control vs various conditions, + p < 0.05 for UV vs UV+E2 or pcDNA3+UV vs pcDNA3+UV+E2. (d) Random cycling MCF7, ZR-75-1, and T47D breast cancer cells were exposed to E2, progesterone, or testosterone, and ATR activity after UV or IR was determined. E2 had no effect on HCC-1569 cells.
E2/ER Inhibits ATR through AKT
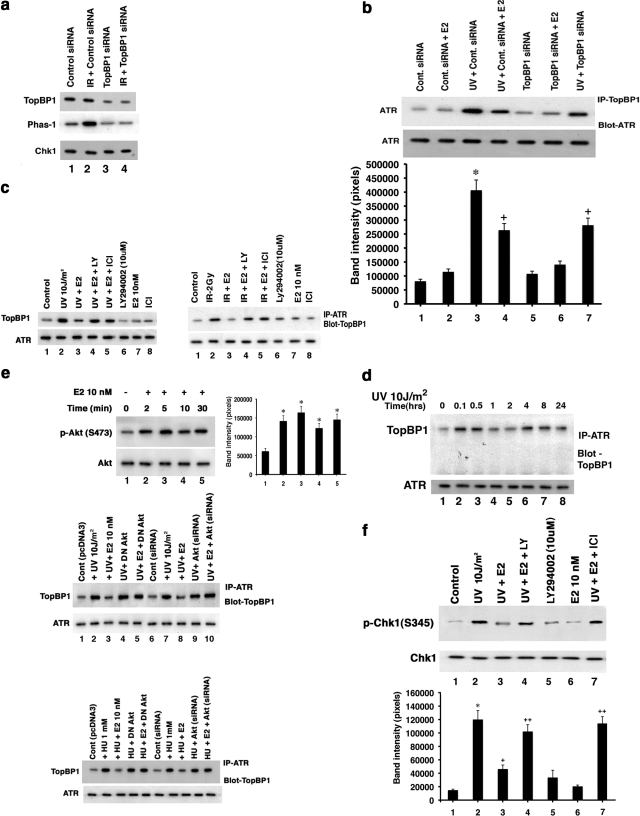
ATR activity depends on the physical association of ATR with ATRIP (Cortez et al., 2001) and TopBP1 (Kumagai et al., 2006) proteins. Using siRNA knockdown, we confirmed that TopBP1 is required for IR-induced ATR activation (Figure 3 a, lanes 2 and 4). Protein knockdown also diminished basal ATR activity (Figure 3a, lanes 1 and 3), but some activity remained because protein elimination was not complete. This may reflect diminished TopBP1 that associates with ATR, resulting from TopBP1 protein knockdown (Figure 3b, lanes 3 and 7).
Figure 3.
E2 modulates TopBP1 protein association with ATR. (a) siRNA knockdown of the TopBP1 proteins in MCF7 cells (top lanes). Specific immunoblots of TopBP1 protein was determined after IP, 48 h after MCF7 cells were transfected with siRNA. IP with IgG antibody yielded no proteins on subsequent specific immunoblot (Figure 1d). TopBP1 knockdown resulted in the loss of basal or IR-induced ATR activity, determined in S-phase–synchronized cells (middle lanes). Kinase assay was carried out as described in Materials and Methods. Chk1 protein serves as loading/protein normalization control (bottom lane), and the study was repeated. (b) ATR:TopBP1 associations are inhibited by TopBP1 siRNA after UV exposure. Top: WB reveals less protein–protein interaction (lane 3 vs lane 7) upon TopBP1 knockdown or E2 addition (lanes 4) in the setting of UV exposure. ATR immunoblots serve as loading/normalization controls. Bottom, experiments were done in random cycling MCF7 cells, as well as S-phase cells. Bar graph is from three experiments. * p < 0.05 for either control vs UV+control siRNA, + p < 0.05 for UV+control siRNA vs UV+control siRNA+E2 or UV+siRNATopBP1. (c) E2 inhibits ATR:TopBP1 associations. MCF7 cells were exposed to brief UV or IR±E2, and the respective cell lysates were obtained at 5 and 15 min after radiation treatment. Lysates were immunoprecipitated with antibody for ATR, and precipitates were subjected to gel electrophoresis then IB with antibodies for TopBP1. In some conditions, the PI3K inhibitor, LY294002, was added. Total ATR IB was the loading control, and IgG antibody used for IP showed no associations of ATR with TopBP1 (Figure 1d). (d) Time course of TopBP1:ATR associations after DNA damage. S-phase MCF7cells were exposed to brief UV, and IP of ATR was followed by IB for TopBP1 at various time points after UV. IP with IgG antibody resulted in no bands (Figure 1d). The study was repeated. (e) Top, E2 rapidly stimulates activation of AKT in MCF7 cells exposed to UV, determined by IB with specific antibody to phospho-Serine 473. Total AKT IB serves as normalization control. Bottom, UV- or HU-induced ATR:TopBP1 associations are inhibited by E2, the steroid effect being dependent on AKT. Cells were transfected with control or specific siRNA to AKT or a dominant-negative AKT plasmid and then recovered and synchronized in S-phase. The cells were then exposed to radiation, without or with E2, and specific IP:IB ensued from cell lysates obtained 5 min after UV. Bar graph is from three experiments. * p < 0.05 for control vs E2. (f) E2 inhibits ATR activation through PI3K. S-phase cells were exposed to UV±E2, sometimes in the presence of LY294002 (PI3K inhibitor). ATR activity as Chk1 phosphorylation was determined in this representative study of two experiments. Bar graph is from three experiments. * p < 0.05 for control vs UV or UV+E2+LY, + p < 0.05 for UV vs UV+E2.
Constitutive and low association of ATR with TopBP1 was noted in cells not exposed to DNA damage (Figure 3, b and c, controls). This level of TopBP1 association correlated to a low level of ATR activity before DNA insult (Figures 1, a–c, controls or time 0). However, when cells were subjected to multiple causes of DNA damage, we detected rapidly enhanced association of TopBP1 with ATR (Figure 3, b and c). This correlated to enhanced ATR activity after DNA damage. The association of ATRIP and ATR is reported as constitutive after DNA damage (Cortez et al., 2001), so ATR:ATRIP interactions were not determined.
We speculated that in these settings, E2 prevents complexing of enhanced endogenous proteins, thereby inhibiting ATR activity. Supporting this idea, UV at 5 min and IR at 15 min each induced a stronger association of ATR with TopBP1, inhibited by E2 (Figure 3, b and c). To further understand the kinetics of ATR andTopBP1 protein complexing, we carried out association studies over 24 h after a single episode of exposure to UV. Association of TopBP1 with ATR rapidly increased (0.1 h), persisting for 30 min to 1 h (Figure 3d). A second association was seen at 4–8 h, followed by diminished association at 24 h. These results indicate a dynamic process of protein–protein interactions after DNA damage alone.
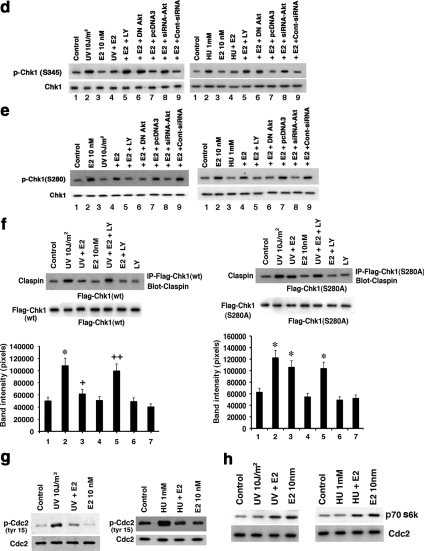
What modulates the dynamic interactions of ATR:TopBP1 proteins especially as regulated by E2? TopBP1 is a phosphoprotein but the significance of phosphorylation is not well understood. Recently, the serine/threonine kinase, AKT, was reported to phosphorylate TopBP1 at Serine1159, promoting a functional interaction with E2F1 (Liu, K. et al., 2006). We found that E2 activates phosphatidylinositol 3′-kinase (PI3K)-dependent AKT by 2 min (Figure 3e, top). Importantly, the ability of E2 to prevent ATR:TopBP1 association at 5 min was reversed by the PI3 kinase inhibitor LY294002 (Figure 3c). Reversal was also seen by expression of dominant negative AKT or siRNA to AKT (21) (Figure 3e, middle and bottom, lanes 2 and 3 vs 4, 5, and 10); this also occurred in the setting of HU.
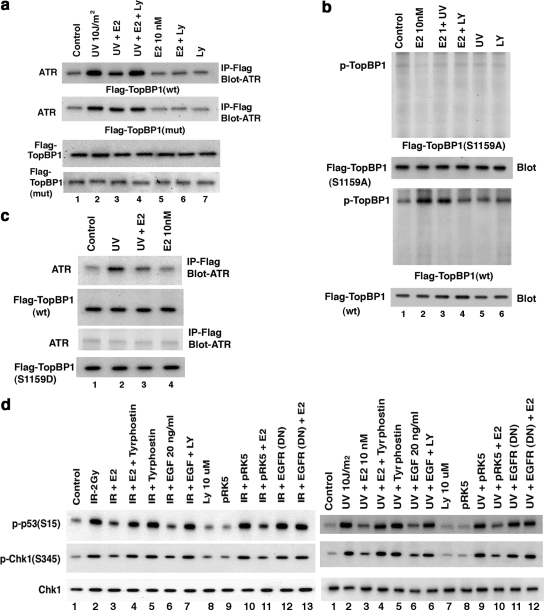
To further support AKT-inhibition of ATR:TopBP1 association, we expressed wt or S1159A mutated, Flag-Top BP1 in MCF7 cells. Both forms of expressed TopBP1 increasingly associated with endogenous ATR upon DNA damage (Figure 4a). E2 inhibited Flag-wtTopBP1 association with ATR, in a PI3K-dependent manner, but could not prevent the association of Flag-S1159ATopBP1 with ATR (Figure 4a, lanes 1–4). This suggests that Serine 1159 is the functional site by which E2/ER-induced AKT prevents enhanced ATR:TopBP1 association upon DNA damage. To reinforce this idea, we expressed Flag-wt or S1159ATopBP1 and labeled the MCF7 cells with 32P. E2 stimulated the strong phosphorylation of expressed wtTopBP1 via PI3K signaling, but this was completely lost in the S1159A mutant (Figure 4b). Thus, the AKT site at Serine1159 of TopBP1 is the key amino acid that is a targeted for E2-stimulated phosphorylation via PI3K, and UV did not affect this phosphorylation. We also expressed a Serine1159 phosphomimetic mutant (Flag-S1159DTopBP1) and determined its association with ATR (Figure 4c). As expected, Flag-wtTopBP1 was recruited to complex with ATR upon UV exposure, significantly prevented by E2 (lanes 1–3). In contrast, no UV-induced association of Flag-S1159DTopBP1 and ATR was detected. These data strongly support the idea that E2 induces the phosphorylation of TopBP1 at the AKT site. S1159 phosphorylation prevents the increased association of TopBP1 and ATR that occurs upon DNA damage. The ability of E2 to block ATR activation after DNA damage occurs through a PI3K/AKT-mediated mechanism (Figure 3f). It is likely to occur at least in part through AKT, preventing the increased association of TopBP1 with ATR, shown here.
Figure 4.
Mechanisms of E2 inhibition of ATR activity. (a) Mutant TopBP1 does not dissociate from ATR. Flag-tagged wtTopBP1 or S1159ATopBP1 was expressed in MCF7 cells. After recover, random cycling cells were used for ATR:TopBP1 association determined by IP:IB under UV±E2 and LY conditions. Western blot of total Flag wt and S1159A (mutant) TopBP1 proteins are shown. Experiments were also repeated in S-phase cells. (b) E2 stimulates the phosphorylation of TopBP1 at Serine 1159 through PI3K/AKT signaling. MCF7 cells were transfected to express Flag-wt or S1159ATopBP1, and the cells recovered, labeled with 32P, and subjected to E2, UV, LY, E2+UV, or E2+LY. IP using Flag antibody followed by PAGE, and autoradiography ensued. In parallel studies, total Flag-wt or S1159ATopBP1 protein was IB as expression/loading controls. Results are representative of two experiments. (c) S1159DTopBP1 does not associate with ATR after DNA damage. Flag-wt TopBP1 or AKT site, phospho-mimetic mutant S1159DTopBP1 were expressed in MCF7 cells, and the cells were recovered and then exposed to UV±E2. IP with Flag antibody was followed by IB with ATR antibody. No association of wtTopBP1 and ATR was seen when nonspecific IgG was substituted (data not shown). (d) ER requires EGFR to inhibit ATR activation. MCF7 were first transfected with dominant-negative EGFR(DN) (pRK5-HERCD533; lane 13) or empty vector (pRK5), and the cells were recovered and then synchronized in S-phase. The cells were then exposed to IR or UV (±E2) or UV+EGF, and in some cells, UV+tyrphostin 1478 (EGFR tyrosine kinase inhibitor; lane 4) or UV+EGF+LY. p53 and Chk1 phosphorylation at the ATR sites were determined from cell lysates obtained at 15 and 5 min after IR or UV, respectively, by specific IB. Total Chk1 protein serves as loading control.
Previously, others and we showed that membrane ER signaling to PI3K and other kinases requires transactivation of the epidermal growth factor receptor (EGFR) in breast cancer (Filardo et al., 2000; Razandi et al., 2003b). We therefore determined whether EGFR was necessary for the ATR-modulating actions of E2/ER. In the setting of IR or UV, ATR activation was inhibited by E2, but this was prevented by a soluble inhibitor of the EGFR tyrosine kinase or by expressing a kinase deficient mutant EGFR (Redemann et al., 1992; Figure 4d, lanes 2–4 and 13). Interestingly, EGF independently inhibited DNA damage–induced ATR activation, prevented by LY294002 (lanes 2, 6, and 7). Thus, cross-talk between membrane ER and EGFR activates PI3K/AKT to restrain ATR signaling in breast cancer. Furthermore, we identify EGF as an additional novel inhibitor of DNA-damage signaling through ATR. Both ER and the EGFR family of receptors play important roles in promoting breast cancer biology (Lippman and Dickson, 1989; Linggi and Carpenter, 2006).
E2/ER Inhibits Chk1 Activation via AKT
We then examined Chk1 kinase activity directed against the Cdc25C phosphatase substrate protein (Stanford and Ruderman, 2005). HU-induced Chk1 activity was inhibited by E2 at 1 nM, reversed by ICI (Figure 5a). Chk1 phosphorylates Cdc25C at Serine 216 and thereby promotes binding to 14-3-3 proteins, resulting in sequestration and degradation of the phosphatase in the cytoplasm (Sanchez et al., 1997; Boddy et al., 1998). This prevents Cdc25C from removing the inhibitory phosphorylation on Tyr15 of the Cdc2 kinase (Cdk1), resulting in the G2/M checkpoint after DNA damage. Nocodazole-synchronized MCF7 were released for 6 h and then exposed to brief IR or UV±E2. After IR or UV alone, abundant Cdc25C protein was phosphorylated at Serine 216 and found with total Cdc25C protein in cytoplasm (Figure 5b, left panels). The lack of nuclear localization suggests that most Cdc25C was phosphorylated and retained in the cytosol. In contrast, cells coexposed to IR or UV and E2 showed a nuclear distribution of Cdc25C protein, and a marked reduction of phosphorylation (middle panels). Because Cdc25C is probably active against Cdk1 in the nucleus, we expected that E2-exposed cells would progress through mitosis. Supporting this idea, all E2-exposed cells showed intense nuclear histone H3 Serine10 phosphorylation, an important marker of mitosis (Xu and Kastan, 2004; Figure 5b, right panels).
Figure 5.
Chk1 activity is inhibited by E2. (a) S-phase MCF7 cells were exposed for 2 h to HU±E2 and HU+E2+ICI at which time the cells were lysed. Endogenous Chk1 protein was immunoprecipitated from cell lysates for in vitro kinase activity assays using exogenous GST-Cdc25C as substrate. Total Chk1 protein serves as the loading control for each condition. Bar graph is from three experiments. * p < 0.05 for control vs HU, + p < 0.05 for HU vs HU+E2, ++ p < 0.05 for HU+E2 10 nM vs HU+E2 10 nM+ICI. (b) Cdc25C is retained in cytoplasm after phosphorylation induced by UV and inhibited by E2. MCF7 were cultured on glass-bottom dishes, synchronized with 16-h exposure to nocodazole, and then released from nocodazole for 6 h. The cells were then exposed to brief UV or IR±E2, and immunostaining with primary antibodies to total (nonphosphorylated) Cdc25C protein, phospho-Serine 216 Cdc25C, and phospho-Serine 10 of Histone H3 ensued. The second antibody was FITC-conjugated, goat-anti mouse IgG. Fluorescent microscopy was then performed. E2 alone–treated cells show Cdc25C localized mainly in the nucleus. (c) Chk1:Claspin association is inhibited by E2. Cell lysates underwent IP with a Claspin antibody, followed by IB of the precipitates using a Chk1 antibody. Lysates were from MCF7 cells exposed to IR or UV±E2 for 5 or 15 min, respectively. Some cells were also exposed to LY294002. The study was repeated, and reverse order for IP:IB was also done (data not shown). Total Claspin protein immunoblots serve as loading controls, whereas IP with nonspecific IgG antibody showed no association of Chk1 and Claspin (Figure 1d). IP with Chk-1 antibody followed by IB with Chk-1 antibody shows the Chk-1 protein, whereas IP with a Chk-2 antibody did not show IB with Chk-1 ab. Specificity of Chk-2 antibody is also shown for comparison. (d) Chk1 Serine345 phosphorylation is inhibited by E2 in PI3K/AKT-dependent manner. Some MCF7 cells were transfected to express DN-AKT or siRNA to AKT, recovered, and investigated during random cycling. Most cells were exposed to UV or HU±E2 in the absence or presence of LY294002. The cell lysates were used for phospho-Chk1 (S345) immunoblot, and the study was repeated. (e) Chk1 Serine280 phosphorylation is stimulated by E2 in PI3K/AKT-dependent manner. Experiments were carried out as described in panel d, and IB was performed with phospho-Serine 280 Chk1 antibody. (f) S280AChk1 mutation prevents E2-induced Claspin:Chk1 protein dissociation upon DNA damage. MCF7 cells were transiently transfected to express Flag-wt or S280AChk1 and then subjected to UV in the absence or presence of E2 and LY. Chk1:Claspin association was determined by IB of claspin after IP with Flag antibody. Western blot of total Flag-tagged wt and S280AChk1 proteins are shown. Bar graph is from three experiments; * p < 0.05 for control vs UV (left) or UV+E2, UV+E2+LY (right), + p < 0.05 for UV vs UV+E2 (left), ++ p < 0.05 for UV+E2 vs UV+E2+LY. (g) Cdc2 (Cdk1) inhibitory phosphorylation is blocked by E2. MCF7 cells were synchronized with nocodazole for 16 h, released for 6 h, and then exposed to UV or HU±E2. IB of cell lysates with specific antibody determined Tyr15 phosphorylation of Cdc2. Total Cdc2 protein was immunoblotted as control. (h) E2 stimulates Cdc2 kinase activity in the setting of DNA damage. Cdc2 was immunoprecipitated after brief UV±E2 exposure from nocodazole-synchronized and released MCF7 cells. Cells were also exposed to E2 in the absence of UV. In vitro kinase activity was determined using p70S6 kinase protein as exogenous substrate.
How is Chk1 activity inversely modulated by DNA damage–induced signaling and E2/ER? One potential mechanism we demonstrated is that E2 blocks ATR-induced phosphorylation of Chk1 at Serine 345 (Figure 1). In addition, Chk1 activity is up-regulated by the physical interaction between Claspin and Chk1 proteins (Yoo et al., 2006). Linking these observations, phosphorylation of Claspin by ATR may help present Chk1 to ATR for Serine 345 phosphorylation (Kumagai et al., 2004; Liu S. et al., 2006; Yoo et al., 2006). We found that IR and UV induced a strong association between endogenous Claspin and Chk1, inhibited by E2 in PI3 kinase–related manner (Figure 5c). AKT inhibits the phosphorylation of Chk1 at Serine 345, perhaps by phosphorylating Serine 280 on Chk1 (King et al., 2004; Puc et al., 2005), leading to cytoplasmic sequestration. We found that UV- or HU-induced Chk1 phosphorylation at Serine 345 was inhibited by E2 in AKT-dependent manner (Figure 5d, lanes 2, 4, 5, and 8). Importantly, E2 stimulated Chk1 Serine 280 phosphorylation through PI3K/AKT, unaffected by UV or HU (Figure 5e, lanes 4, 5, and 8) or IR (data not shown).
This suggested a second AKT-related mechanism by which E2/ER blocks Chk1 Ser345 phosphorylation. To support this, we expressed a Flag-tagged wt or Serine280Ala mutant Chk1 in MCF7 cells. On DNA damage, Claspin comparably and increasingly associated with either expressed Chk1 (Figure 5f). Thus, this serine does not affect Chk1: Claspin association after DNA damage. However, upon mutant expression, E2 could not significantly prevent S280AChk1:Claspin association. In contrast, Flag-wt Chk1 interactions with Claspin were perturbed by E2 signaling through PI3K/AKT. Thus, E2 inhibition of 1) ATR activity and 2) Claspin:Chk1 association (via AKT phosphorylation of Chk1) reduces Serine 345 Chk1 phosphorylation that correlates to Chk1 activity.
E2/ER Inhibition of ATR and Chk1 Signaling Impacts Substrate Functions
On DNA damage, the Cdc25C phosphatase is phosphorylated and inactivated by Chk1. Loss of Cdc25C phosphatase activity enhances the inhibitory tyr15 phosphorylation of Cdc2 (Cdk1) that promotes the G2/M checkpoint (Hutchins and Clarke, 2004). We found that UV and HU enhanced the inhibitory phosphorylation of Cdc2 at tyr15 that reflects Cdc25C inactivation by DNA damage and was prevented by E2 (Figure 5g). This indicates a potential mechanism by which E2 overcomes the G2/M checkpoint. In the absence of UV or HU, E2 stimulated Cdc2 activity, as well as overcoming the restraint of kinase activity imposed by DNA damage (Figure 5h).
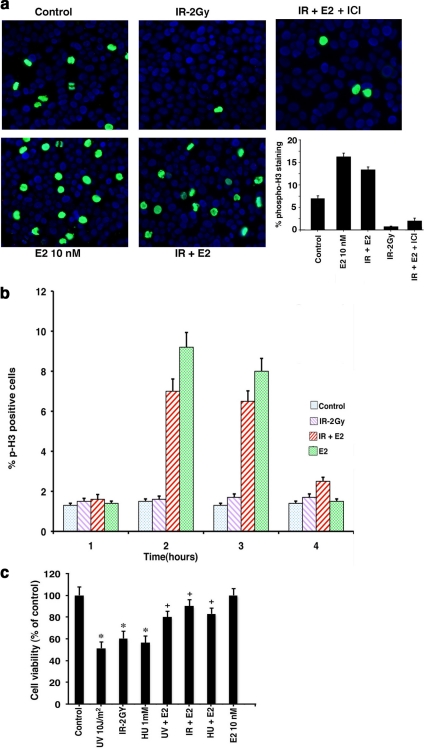
At least partially as a result of these actions, E2 promoted bypass of the G2/M checkpoint. IR-induction of the G2/M checkpoint in randomly cycling MCF7 cells was prevented by E2, seen as phospho-H3 (Serine 10) staining (Figure 6a). The E2 effect was reversed by ICI. The sex steroid has previously been shown to stimulate H3 phosphorylation, in vitro and in vivo (Brenner et al., 2003). We also determined that E2 promoted a bypass of the G2/M checkpoint induced by IR, as seen in Figure 6b. Here, nocodazole-synchronized cells were released from nocodazole, exposed to IR±E2, and the cells were assayed for phospho-H3 over 6 h. Additionally, E2 significantly reversed the loss of cell viability caused by DNA damage (Figure 6c).
Figure 6.
E2 overcomes the G2/M checkpoint and promotes cell viability after DNA damage. (a) G2/M checkpoint was determined by cells showing nuclear phospho-histone H3 (Serine 10) staining. Randomly cycling MCF7 cells were exposed to brief IR±E2 or E2 alone, and at 4 h after IR the cells were fixed and permeabilized for antibody staining. A representative study of two is shown, and nuclei are identified by DAPI stain. Bar graph is from 200 cells counted per condition in each of two experiments for phospho-H3 staining. (b) Estrogen promotes bypass of the G2/M checkpoint. MCF7 cells were synchronized with nocodazole for 16 h to induce a G2/M block. Nocodazole-containing media was then removed and replaced with fresh DMEM-F12 media alone (control or IR alone conditions) or with E2 10 nM in DMEM-F12, and then cells were exposed to IR. The cells were stained with antibody to detect phospho-H3 over 6 h, and the bar graph represents results from 200 cells per condition from each of two experiments. (c) Cell viability of MCF7 exposed briefly to DNA-damaging agents in the absence or presence of E2 was determined by the MTT assay, as described in Materials and Methods. Bar graph is from the mean ± SEM of three experiments. * p < 0.05 by ANOVA+Schefe's test for control vs DNA-damaging agents; + p < 0.05 for UV-damaging agents vs UV-damaging agents+E2.
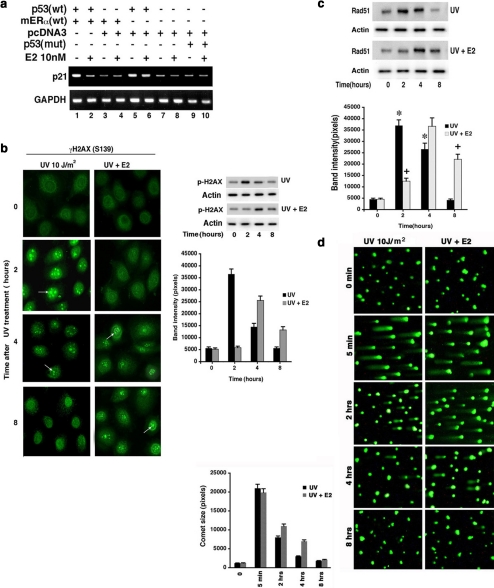
What additional and important outcomes of DNA damage–induced ATR signaling are impacted by E2/ER? p53 activation stimulates transcription of the p21 cyclin-dependent kinase inhibitor, an important contributor to the G1/S checkpoint after DNA damage (Meek, 2002). Phosphorylation of p53 at Serine 15, a known ATR (and Chk1 and ATM) site, contributes to activating this p53 transcriptional function (Meek, 2002). We found that DNA damage stimulates ATR-dependent phosphorylation of Serine15 in endogenous p53, inhibited by E2 (Figures 1b and 2a). We therefore examined whether E2 blocks p21 transcription through this mechanism. HCC-1937 breast cancer cells (BRCA1 mutated, p53 mutated, and ERα null) were transfected to express either wt or S15Ap53 and ERα, then exposed to UV±E2. Expression of wtp53 resulted in strong endogenous p21 expression, inhibited by the coexpression of ERα and requiring exposure to E2 (Figure 7a, lanes 1 and 2 vs control lane 3). In contrast, expression of S15Ap53 did not stimulate p21 (lane 9). Our results suggest that the ability of E2/ERα to block ATR-induced Serine 15 phosphorylation of p53 importantly contributes to the abrogation of p21 induction that would likely impact the G1/S checkpoint. By blocking DNA damage–induced signaling, E2/ER inhibits a key function of intact p53 in breast cancer cells.
Figure 7.
Outcomes of DNA damage signaling. (a) Serine 15 phosphorylation of p53 is required for up-regulated p21 gene expression, inhibited by E2. HCC1937 cells were transfected to express wt or S15Ap53, and cells were recovered and synchronized in S-phase and then exposed to brief UV±E2. p21 was determined by RT-PCR 6 h later, and GAPDH expression served as control. (b) γH2AX complex formation in response to UV and E2. Left, MCF7 cells were assessed at various times after UV±E2 as shown by immunofluorescent microscopy. Right, immunoblot for γH2AX. Assay is described in Materials and Methods. The bar graph is the mean ± SEM of two immunoblot experiments combined. (c) Rad 51 complex formation is delayed by E2. Immunoblots of Rad51 protein in the nucleus from MCF7 cells exposed to UV or UV±E2 in a representative experiment is shown. The bar graph is the mean ± SEM of immunoblot density from three experiments. * p < 0.05 for control vs UV at 2 and 4 h; + p < 0.05 for UV vs UV+E2 at 2 and 8 h. (d) Single-cell DNA repair. Experiments were carried out as described in Materials and Methods. The trailing tail of the DNA reflects unrepaired DNA and was quantified over 8 h in two experiments using Komet imaging software, version 5.5 (Andor Tech). Data are mean ± SEM from 200 cells assessed per condition, in each of two experiments.
DNA Repair Is Delayed by E2/ER
Interfering with ATR signaling should alter the kinetics of DNA repair. Repair occurs after key regulatory proteins complex at the site(s) of histone damage, and we determined the DNA repair response of MCF7 cells. Assemblage of nuclear repair foci containing Serine 139-phosphorylated histone 2 variant (γH2AX) was fully evident at 2 h and was barely seen by 8 h after UV exposure (Figure 7b). E2 delayed this response in that the peak occurred at 4 h, and complex formation was very evident at 8 h. E2 also diminished the magnitude of UV-induced phosphorylation of γH2AX at the ATR site. Similarly, nuclear Rad 51 focus formation was delayed by 2 h, compared with UV alone (Figure 7c). Also, resolution of the foci was prolonged where at 8 h, fivefold more foci remained in the E2-treated cells (Figure 7c).
We also assessed DNA repair using the Comet assay that mainly reflects nucleotide excision repair in this setting. UV-induced DNA damage was substantially repaired by 4 h and completed by 8 h (Figure 7d). E2 delayed this, with unrepaired DNA persisting most noticeably at 4 h. Additionally, specific resolution of cyclobutane pyrimidine dimer photo adducts was delayed at 4–8 h of E2 exposure (Figure 8a). Although the Comet DNA repair assay showed a kinetic “catch-up” at 8 h in the setting of E2 (Figure 7d), the other assays of DNA damage foci or photo adduct repair consistently showed continuing delay induced by sex steroid at the latest times assessed (Figures 7, b and c, and 8a). Additionally, when the assays were carried out with IR as the DNA-damaging agent, the Comet assay showed about two times the amount of persisting DNA damage at 8 h from cells also exposed to E2 (compared with IR alone; data not shown).
Figure 8.
E2 modulates DNA damage complexes in breast cancer and normal breast epithelial cells. (a) Cyclobutane pyrimidine dimers form after exposure to UV. Spectrophotometry quantified CPD immunofluorescence of S-phase MCF7cells exposed to brief radiation ± E2, and cells were fixed with 4% paraformaldehyde at the times indicated and incubated with CPD-specific antibody. PI fluorescence from the nuclei of the cells was used to normalize all readings. Data are mean ± SEM from three experiments; * p < 0.05 for control vs UV or UV+E2 at 2 h; + p < 0.05 for UV vs UV+E2 at 4–8 h. (b) E2 stimulates increased chromosomal damage in the setting of IR. MCF7 were exposed to 2 Gy IR and then incubated with or without E2 and ICI for 48 h. Metaphase spreads were conducted as described in Materials and Methods. Chromosome analysis was conducted from 200 metaphase spreads per condition in each of two experiments. * p < 0.05 for control (sham irradiation) vs IR or IR+E2; + p < 0.05 for IR+E2 vs IR+E2+ICI. (c) E2 modulates ATR activation after IR exposure of S-phase mouse mammary epithelial cells (top), and ATR activation in ERα-transfected human breast epithelial cells (MCF10A; bottom). Primary cultures of mouse epithelial cells were from 8-wk-old female mice. ERα or pcDNA3 (control) was transfected into the ER-negative MCF10A cells for experiments, the cells recovered and synchronized in S-phase. (d) E2 delays γH2AX foci formation and resolution after IR in S-phase normal breast epithelial cells. Studies were carried out as described in Materials and Methods.
To determine the impact for chromosome damage, MCF7 cells were exposed to IR±E2 or E2+ICI. At 48 h after IR, chromosomal damage was scored from metaphase spreads (Figure 8b). Radiation of the breast cancer cells increased chromosome damage but this was strongly further augmented by E2. ICI almost completely reversed the effects of E2, indicating the E2 effect was ER mediated. These results suggest an important new mechanism by which E2/ER may promote the tumor biology.
Effects of E2/ER in Nontransformed Breast Epithelial Cells
Inhibition of DNA damage signaling to cell cycle checkpoints and DNA repair could promote both the transformation of normal cells to malignancy and alter subsequent biology of established tumor. We therefore asked whether E2/ER inhibits DNA damage–induced signaling in normal breast epithelial cells. To determine this, we isolated adult mouse breast epithelial cells and established acute primary cultures. Approximately 50% of the cells expressed ERα by immunofluorescent staining (data not shown). These cells responded to IR with rapid ATR activation (Figure 8c, top) and γH2AX foci formation (Figure 8d). E2 substantially blocked ATR activation and delayed the assembly and resolution of the foci. We also found that expression of ERα in nonmalignant, ER null, human MCF10A breast epithelial cells supported E2 inhibition of ATR activation (Figure 8c, lower figure). Thus, E2 acts similarly in nontransformed and transformed breast epithelial cells.
DISCUSSION
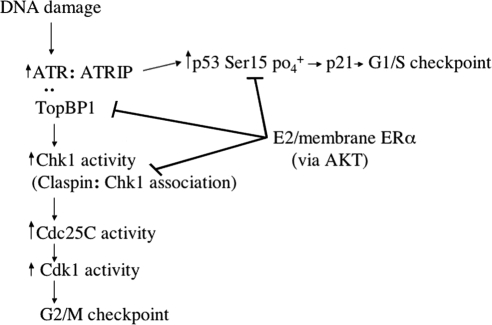
DNA-damage induced signaling stimulates prompt DNA repair and cell cycle checkpoints and has been proposed to suppress tumor formation (Kastan and Bartek, 2004; Bartkova et al., 2005). Supporting this idea, loss of such signaling consequent to mutations of ATM, ATR, and Chk1 and 2, or downstream substrates promotes the development of tumors (Deng and Brodie, 2001; Bartkova et al., 2005). It is likely that functional inhibition of intact kinase and substrate proteins in the setting of DNA damage has similar consequences. However, endogenous proteins that restrain damage-induced signaling and the mechanisms involved are virtually unknown. We identify the carcinogenic steroid E2 and membrane-associated ERα as endogenous inhibitors of ATR cascade signaling to the G2/M cell cycle checkpoint and prompt DNA repair in breast cancer cells (Figure 9). Interestingly, the therapeutic ER antagonist, ICI inhibits this action of E2/ER potentially preserving DNA-damage signaling as a novel function to prevent reoccurrence or progression of breast cancer. Whether Fulvestrant fails to prevent these actions in breast cancers that are resistant to endocrine therapy is an important question that is under evaluation.
Figure 9.
Schematic of estrogen and membrane ERα-inhibiting ATR pathway signaling after DNA damage. E2 acting at membrane-localized ERα rapidly activates PI3K/AKT to phosphorylate and dissociate TopBP1 from ATR in the setting of DNA damage. This leads to inhibition of ATR activity and downstream signaling to the G2/M checkpoint and p53-induced p21 transcription. Signaling through PI3K and AKT also inhibits Chk1 activity by phosphorylating claspin.
Although UV, one of the DNA damage–inducing stimuli used here, is not important to breast cancer pathogenesis, it serves as a useful model for understanding the rapid activation of ATR (Cortez et al., 2001). Comparable results were found in several ER-positive breast cancer cell lines as a response to HU, UV, or IR. In the setting of rapid ATR kinase stimulation, we demonstrated that physiological levels of E2 act at plasma membrane-localized ERα (and not nuclear ERα) to activate PI3K/AKT and inhibit ATR signaling and Chk1 phosphorylation. These novel findings add to the growing body of data implicating rapid signaling by membrane ER in the development of breast cancer (Levin and Pietras, 2007). It has recently been shown that in human breast cancer cells, membrane and nuclear ERα are the same protein (Pedram et al., 2006a).
We report that AKT activation by E2/ERα reduces DNA damage–enhanced ATR:TopBP1 association and ATR activity. This is important because the interaction of TopBP1 and ATR is required for ATR activity (Kumagai et al., 2006). Mechanistically, we found that an AKT-site phosphomimetic mutant TopBP1 protein was unable to associate with ATR after DNA damage, in contrast to expressed wild-type TopBP1 or endogenous TopBP1 in MCF7 cells. Further, E2/ER stimulated the phosphorylation of TopBP1 only at the AKT site (Serine 1159), and mutation of this serine to alanine prevented sex steroid block of the enhanced complexing of ATR:TopBP1 proteins after DNA damage. E2/ERα and AKT also prevented the enhanced association of Claspin and Chk1 proteins, leading to inhibition of Chk1 phosphorylation at the ATR substrate site. These findings suggest potential new mechanisms by which AKT contributes to breast carcinogenesis (Carpten et al., 2007), ones that may also underlie EGF/EGFR-inhibition of ATR activation that we demonstrated.
Inhibition of ATR-Chk1 signaling would be expected to increase sensitivity to DNA-damaging agents and decrease cell survival. In the absence of DNA damage, E2/ER activation of PI3K and AKT contributes to the survival and growth effects of the steroid in breast cancer (Ahmad et al., 1999; Marquez and Pietras, 2001, Fernando and Wimalasena, 2004; Rodrik et al., 2005). When E2/ER functions during DNA damage, cell survival effects appear to dominate, as we showed. This may form the basis for the clinical recommendation that women with breast cancer should not take estrogen during radiation or endocrine therapies, therapies that severely damage DNA to induce breast cancer apoptosis.
Consistent with E2/ER preventing ATR and Chk1 activation, the G2/M cell cycle checkpoint is overcome by the sex steroid. E2/ER inhibition of Chk1 phosphorylation of Cdc25C prevented DNA-damage restraint of Cdc2 kinase activity, promoting the passage of these cells prematurely into mitosis. Additionally, we found that E2/ER stimulates Cdc2 (Cdk1) activity in the absence of DNA damage. Acting as a growth factor in breast cancer (Cordera and Jordan, 2006), E2 stimulates cyclin gene transcription, Cdk2 activity, and down-regulates cyclin-dependent kinase inhibitors (e.g., p27), resulting in part from nuclear ERα action (Foster et al., 2001; Doisneau-Sixou et al., 2003; Eeckhoute et al., 2006). We propose that in the setting of DNA damage, E2/ER has multiple effects to promote cell cycle progression. We identify novel mechanisms where plasma membrane ER inhibits crucial DNA damage–induced signal transduction that invokes the G2/M checkpoint. Additionally, p53-induced p21 transcription is important to the G1/S checkpoint after DNA damage (Meek, 2002; Sancar et al., 2004) and is lost in our cell model as a result of E2/ER blocking ATR-dependent Serine 15 phosphorylation of p53. In the majority of breast cancers, p53 is not mutated and appears to function normally. We propose that E2/ER may inhibit several important tumor suppressor functions of intact p53 by preventing posttranslational modifications (e.g., phosphorylation) that augment the stability and transcriptional functions of this protein. E2 has been reported to stimulate p53 transcription but the importance of this is unknown (Horvath et al., 1996; Qin et al., 2002).
E2/ER inhibition of DNA damage–induced cell cycle checkpoints and prompt DNA repair might allow passage of unrepaired mutations to cell progeny. As a result, E2/ER could promote early development of breast cancer and additional functions (e.g., tissue invasion) through mutation acquisition at later stages. Regarding this, we showed E2/ERα blocks ATR activation and downstream functions in both normal breast epithelial and breast cancer cells. Additionally, three assays (γH2AX and Rad51 foci and cyclobutane pyrimidine dimer photo adduct resolutions) all suggest persisting DNA damage from UV, although the Comet results did not show this (but did show kinetic delay of repair). However, approximately twice the DNA damage persisted by Comet assay in cells exposed to IR+E2, compared with IR alone. Most importantly, we found enhanced chromosomal damage in the presence of IR+E2, compared with radiation alone. IR is a more relevant stimulus to breast neoplasia compared with UV, probably because IR exposure frequently causes double-strand breaks.
Further supporting the idea that E2 promotes persisting DNA damage and hence mutation acquisition, female August/Copenhagen/Irish (ACI) rats treated with physiological E2 develop mammary carcinomas that exhibit aneuploidy, chromosomal amplifications, and instability, all prevented by tamoxifen (Li et al., 2004). Aneuploidy has increasingly been implicated as causing human cancer (Pellman, 2007). Amplification of c-myc and cyclin E genes and chromosome instability in ACI tumors is also characteristic of most human mammary ductal carcinomas in situ and primary invasive ductal breast cancers that usually express ER (Li et al., 2002). E2 metabolites (quinone forms of hydroxy-catecholestrogens) have also been reported to directly damage DNA in breast epithelium, promoting tumorigenesis (Cavalieri et al., 1997). These metabolites act independently of the estrogen receptor and can react with DNA to form depurinating adducts. Such adducts when released from DNA generate apurinic sites, and error-prone base excision repair of this damage may lead to mutations (Cavalieri et al., 2006). Our results indicate that it is likely E2 itself acts through ERα to further damage chromosomes in the setting of radiation, but this does not rule out an added contribution from estrogen metabolites.
The DNA damage response (DDR) involves both ATR and ATM to prevent cancer formation (Bartkova et al., 2005). IR primarily activates ATM and then ATR, and we recently found that E2/ER also inhibits rapid ATM activation by γ-radiation (data not shown). Oncogenes such as c-myc stimulate the DDR in human cells and suppress tumorigenesis mainly through p53 activation (Gorgoulis et al., 2005). Because ER functionally inhibits ATR and ATM, and an important target of the p53 arm of the DDR, we propose the propensity toward mutation acquisition is increased. This may occur in both premalignant and transformed breast epithelial cells.
ACKNOWLEDGMENTS
We thank Wen Hwa Lee for helpful discussions. The work was supported by grants from the Research Service of the Department of Veteran's Affairs and National Institutes of Health Grant RO1 CA10036 to E.R.L.
Abbreviations used:
- ATR
ataxia telangiectasia-mutated and Rad3-related
- ATM
ataxia telangiectasia-mutated
- Chk1
checkpoint kinase1
- Cdk1
cyclin-dependent kinae 1
- DPN
diarylpropionitrile
- UV
ultraviolet
- EGF
epidermal growth factor
- ER
estrogen receptor
- IR
ionizing radiation
- HU
hydroxyurea
- PPT
propyl-pyrazole-triol.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0085) on May 28, 2009.
REFERENCES
- Ahmad S., Singh N., Glazer R. I. Role of AKT1 in 17-beta-estradiol- and insulin-like growth factor I (IGF-1)-dependent proliferation and prevention of apoptosis in MCF-7 breast carcinoma cells. Biochem. Pharmacol. 1999;58:425–430. doi: 10.1016/s0006-2952(99)00125-2. [DOI] [PubMed] [Google Scholar]
- Bakkenist C. J., Kastan M. B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bartkova J., et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Furnari B., Mondesert O., Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Brenner R. M., Slayden O. D., Rodgers W. H., Critchley H. O., Carroll R., Nie X. J., Mah K. Immunocytochemical assessment of mitotic activity with an antibody to phosphorylated histone H3 in the macaque and human endometrium. Hum. Reprod. 2003;18:1185–1193. doi: 10.1093/humrep/deg255. [DOI] [PubMed] [Google Scholar]
- Carpten J. D., et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Cavalieri E. L., et al. Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc. Natl. Acad. Sci. USA. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri E., Chakravarti D., Guttenplan J., Hart E., Ingle J., Jankowiak R., Muti P., Rogan E., Russo J., Santen R., Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim. Biophys. Acta. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Cordera F., Jordan V. C. Steroid receptors and their role in the biology and control of breast cancer growth. Semin. Oncol. 2006;33:631–641. doi: 10.1053/j.seminoncol.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S. J. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Deng C. X., Brodie S. G. Knockout mouse models and mammary tumorigenesis. Semin. Cancer Biol. 2001;11:387–394. doi: 10.1006/scbi.2001.0394. [DOI] [PubMed] [Google Scholar]
- Doisneau-Sixou S. F., Sergio C. M., Carroll J. S., Hui R., Musgrove E. A., Sutherland R. L. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr. Relat. Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J., Carroll J. S., Geistlinger T. R., Torres-Arzayus M. I., Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando R. I., Wimalasena J. Estradiol abrogates apoptosis in breast cancer cells through inactivation of BAD: Ras-dependent nongenomic pathways requiring signaling through ERK and Akt. Mol. Biol. Cell. 2004;15:3266–3284. doi: 10.1091/mbc.E03-11-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R. Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, gpr30, and occurs via transactivation of the epidermal growth factor receptor through release of HB-EGF. Mol. Endocrinol. 2000;14:1649–1660. doi: 10.1210/mend.14.10.0532. [DOI] [PubMed] [Google Scholar]
- Foster J. S., Henley D. C., Bukovsky A., Seth P., Wimalasena J. Multifaceted regulation of cell cycle progression by estrogen: regulation of Cdk inhibitors and Cdc25A independent of cyclin D1-Cdk4 function. Mol. Cell. Biol. 2001;21:794–810. doi: 10.1128/MCB.21.3.794-810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V. G., et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Greene G., Closs L. E., Fleming H., DeSoimbre E. R., Jensen E. Antibodies to estrogen receptor: immunochemical similarity of estrophillin from various mammalian species. Proc. Natl. Acad. Sci. USA. 1977;74:3681–3685. doi: 10.1073/pnas.74.9.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington W. R., Sheng S., Barnett D. H., Petz L. N., Katzenellenbogen J. A., Katzenellenbogen B. S. Activities of estrogen receptor alpha- and beta-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol. Cell Endocrinol. 2003;206:13–22. doi: 10.1016/s0303-7207(03)00255-7. [DOI] [PubMed] [Google Scholar]
- Howell A. Pure oestrogen antagonists for the treatment of advanced breast cancer. Endocr. Relat. Cancer. 2006;13:689–706. doi: 10.1677/erc.1.00846. [DOI] [PubMed] [Google Scholar]
- Horvath G., Leser G., Karlsson L., Delle U. Estradiol regulates tumor growth by influencing p53 and bcl-2 expression in human endometrial adenocarcinomas grown in nude mice. In Vivo. 1996;10:411–416. [PubMed] [Google Scholar]
- Hutchins J. R., Clarke P. R. Many fingers on the mitotic trigger: post-translational regulation of the Cdc25C phosphatase. Cell Cycle. 2004;3:41–45. [PubMed] [Google Scholar]
- Kastan M. B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Keeton E. R., Brown M. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol. Endocrinol. 2005;19:1543–1554. doi: 10.1210/me.2004-0395. [DOI] [PubMed] [Google Scholar]
- King F. W., Skeen J., Hay N., Shtivelman E. Inhibition of Chk1 by activated PKB/Akt. Cell Cycle. 2004;3:634–637. [PubMed] [Google Scholar]
- Kuiper G.G.J.M., Enmark E., Pelto-Huiko M., Nilsson S., Gustafsson J.-A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Kim S. M., Dunphy W. G. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J. Biol. Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H. Y., Dunphy W. G. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Levin E. R. Integration of the extra-nuclear and nuclear actions of estrogen. Mol. Endocrinol. 2005;19:1951–1959. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E. R., Pietras R. J. Estrogen receptors outside the nucleus in breast cancer. Breast Cancer Res. Treat. 2007;108:351–361. doi: 10.1007/s10549-007-9618-4. [DOI] [PubMed] [Google Scholar]
- Li J. J., Papa D., Davis M. F., Weroha S. J., Aldaz C. M., El-Bayoumy K., Ballenger J., Tawfik O., Li S. A. Ploidy differences between hormone- and chemical carcinogen-induced rat mammary neoplasms: comparison to invasive human ductal breast cancer. Mol. Carcinog. 2002;33:56–65. doi: 10.1002/mc.10022. [DOI] [PubMed] [Google Scholar]
- Li J. J., Weroha S. J., Lingle W. L., Papa D., Salisbury J. L., Li S. A. Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc. Natl. Acad. Sci. USA. 2004;101:18123–18128. doi: 10.1073/pnas.0408273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linggi B., Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Lippman M. E., Dickson R. B. Mechanisms of growth control in normal and malignant breast epithelium. Recent Prog. Horm. Res. 1989;45:383–435. doi: 10.1016/b978-0-12-571145-6.50012-1. [DOI] [PubMed] [Google Scholar]
- Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell. Biol. 2006a;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Paik J. C., Wang B., Lin F. T., Lin W. C. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006b;25:4795–4807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez D. C., Pietras R. J. Membrane-associated binding sites for estrogen contribute to growth regulation of human breast cancer cells. Oncogene. 2001;20:5420–5430. doi: 10.1038/sj.onc.1204729. [DOI] [PubMed] [Google Scholar]
- Meek D. W. p53 Induction: phosphorylation sites cooperate in regulating interaction. Cancer Biol. Ther. 2002;1:284–290. doi: 10.4161/cbt.82. [DOI] [PubMed] [Google Scholar]
- Naugler W. E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A. M., Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- Pedram A., Razandi M., Levin E. R. Nature of functional estrogen receptors at the plasma membrane. Mol. Endocrinol. 2006a;20:1996–2009. doi: 10.1210/me.2005-0525. [DOI] [PubMed] [Google Scholar]
- Pedram A., Razandi M., Wallace D. C., Levin E. R. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol. Biol. Cell. 2006b;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A., Razandi M., Sainson R.C.A., Kim J. K., Hughes C. C., Levin E. R. A conserved mechanism for steroid receptor translocation to the plasma membrane. J. Biol. Chem. 2007;282:2278–2288. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- Pellman D. Aneuploidy and cancer. Nature. 2007;446:38–39. doi: 10.1038/446038a. [DOI] [PubMed] [Google Scholar]
- Platet N., Cathiard A. M., Gleizes M., Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit. Rev. Oncol. Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Puc J., et al. Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Qin C., Nguyen T., Stewart J., Samudio I., Burghardt R., Safe S. Estrogen up-regulation of p53 gene expression in MCF-7 breast cancer cells is mediated by calmodulin kinase IV-dependent activation of a nuclear factor kappaB/CCAAT-binding transcription factor-1 complex. Mol Endocrinol. 2002;16:1793–1809. doi: 10.1210/me.2002-0006. [DOI] [PubMed] [Google Scholar]
- Razandi M., Alton G., Pedram A., Ghonshani S., Webb D., Levin E. R. Identification of a structural determinant for the membrane localization of ERα. Mol. Cell. Biol. 2003a;23:1633–1646. doi: 10.1128/MCB.23.5.1633-1646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razandi M., Pedram A., Parks S., Levin E. R. Proximal events in ER signaling from the plasma membrane. J. Biol. Chem. 2003b;278:2701–2712. doi: 10.1074/jbc.M205692200. [DOI] [PubMed] [Google Scholar]
- Redemann N., Holzmann B., von Ruden T., Wagner E. F., Schlessinger J., Ullrich A. Anti-oncogenic activity of signalling-defective epidermal growth factor receptor mutants. Mol. Cell. Biol. 1992;12:491–498. doi: 10.1128/mcb.12.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik V., Zheng Y., Harrow F., Chen Y., Foster D. A. Survival signals generated by estrogen and phospholipase D in MCF-7 breast cancer cells are dependent on Myc. Mol. Cell. Biol. 2005;25:7917–7925. doi: 10.1128/MCB.25.17.7917-7925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;273:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Wong C., Thoma R. S., Richman R., Wu Z., Piwnica-Worms H., Elledge S. J. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Stanford J. S., Ruderman J. V. Changes in regulatory phosphorylation of Cdc25C Ser287 and Wee1 Ser549 during normal cell cycle progression and checkpoint arrests. Mol. Biol. Cell. 2005;16:5749–5760. doi: 10.1091/mbc.E05-06-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Kastan M. B. Analyzing cell cycle checkpoints after ionizing radiation. Methods Mol. Biol. 2004;281:283–292. doi: 10.1385/1-59259-811-0:283. [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Jeong S. Y., Dunphy W. G. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 2006;20:772–783. doi: 10.1101/gad.1398806. [DOI] [PMC free article] [PubMed] [Google Scholar]