Abstract
Clathrin is involved in vesicle formation in the trans-Golgi network (TGN)/endosomal system and during endocytosis. Clathrin recruitment to membranes is mediated by the clathrin heavy chain (HC) N-terminal domain (TD), which forms a seven-bladed β-propeller. TD binds membrane-associated adaptors, which have short peptide motifs, either the clathrin-box (CBM) and/or the W-box; however, the importance of the TD binding sites for these motifs has not been tested in vivo. We investigated the importance of the TD in clathrin function by generating 1) mutations in the yeast HC gene (CHC1) to disrupt the binding sites for the CBM and W-box (chc1-box), and 2) four TD-specific temperature-sensitive alleles of CHC1. We found that TD is important for the retention of resident TGN enzymes and endocytosis of α-factor; however, the known adaptor binding sites are not necessary, because chc1-box caused little to no effect on trafficking pathways involving clathrin. The Chc1-box TD was able to interact with the endocytic adaptor Ent2 in a CBM-dependent manner, and HCs encoded by chc1-box formed clathrin-coated vesicles. These data suggest that additional or alternative binding sites exist on the TD propeller to help facilitate the recruitment of clathrin to sites of vesicle formation.
INTRODUCTION
The efficient delivery of proteins and lipids to specific cellular compartments of the secretory and endocytic pathways is achieved by transport in membrane-bound vesicles. Selective recruitment of cargo and formation of vesicles is mediated by coat proteins. The coat protein clathrin is present in all eukaryotic cells and is an important component in the membrane trafficking events of the trans-Golgi network (TGN)/endosomal system and in endocytosis at the plasma membrane (PM) (reviewed in Brodsky et al., 2001).
The assembly unit of the clathrin coat is the triskelion, which is composed of three 190-kDa clathrin heavy chains (HCs) and three 25- to 28-kDa clathrin light chains (LCs). Clathrin HCs trimerize at their C-termini, and each leg extends outward radially from a central vertex to generate distinct regions, including proximal and distal legs, a linker region, and finally a globular N-terminal domain (TD). A single LC noncovalently associates with each HC along the proximal leg region near the vertex, and proximal and distal regions from neighboring triskelions interdigitate to form the polygonal lattices characteristic of clathrin-coated membrane structures, with the TD facing inward toward the membrane surface (Fotin et al., 2004).
Clathrin assembly at sites of vesicle formation requires adaptor proteins to sort protein cargo into developing vesicles and additional assembly factors to facilitate lattice formation. Several adaptor proteins exist in eukaryotic cells, including the heterotetrameric adaptor protein complexes (APs), monomeric adaptor proteins (GGAs, epsins, and AP180s), and cargo-specific adaptor proteins. APs and monomeric adaptors have cargo-binding, phospholipid-binding, and clathrin-binding capabilities to help facilitate clathrin recruitment and coated vesicle formation. In addition, many accessory proteins also bind clathrin directly to facilitate lattice assembly (reviewed in Owen et al., 2004).
Adaptor proteins and assembly factors bind to the TD regions of clathrin triskelions via short, linear peptide sequences. The first consensus clathrin-binding sequence (SLLDLD) was identified in studies of the β3a subunit of the heterotetrameric adaptor complex AP-3 (Dell'Angelica et al., 1998), and subsequent alignment of sequences from the clathrin-binding regions of numerous adaptor proteins revealed the presence of a consensus clathrin-binding motif (LΦpΦp) called the clathrin-box motif (CBM), where Φ represents a hydrophobic residue and p represents a polar residue (reviewed by Dell'Angelica, 2001). More recently, a second clathrin-binding motif called the W-box motif (PWXXW), where X represents any amino acid, was identified in amphiphysin and sorting nexin 9 (SNX9), which are required for efficient endocytosis in mammalian cells (Ramjaun and McPherson, 1998; Drake and Traub, 2001; Lundmark and Carlsson, 2003; Miele et al., 2004).
X-ray crystallographic studies have revealed the location of the binding sites on the TD for the CBM and W-box motif peptides present in clathrin adaptors. The TD forms a seven-bladed propeller structure where the fold of each blade resembles a WD40 repeat, a protein–protein interaction module found in heterotrimeric G proteins that forms a four-strand β sheet (ter Haar et al., 1998) (Figure 1A). A binding pocket in the groove between blades 1 and 2 of the β-propeller structure accommodates the CBM peptides from the AP-3 β-subunit (LLDLD) and the G protein-coupled receptor-specific adaptor β-arrestin 2 (LIEFE) (ter Haar et al., 2000). A W-box motif peptide from amphiphysin (PWDLW), an accessory protein that facilitates membrane curvature during endocytic vesicle formation, fits into a deep pocket formed at the center of the top surface of the propeller structure (Miele et al., 2004). The contribution of these binding sites to clathrin-mediated protein trafficking events has not been tested in vivo.
Figure 1.
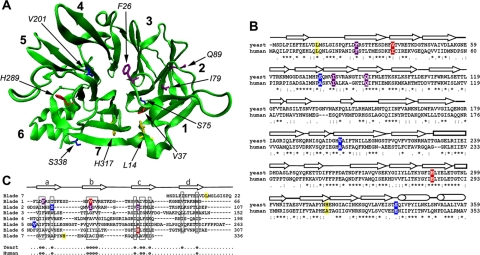
Structure of the clathrin heavy chain TD. (A) A homology model of the yeast TD (green) with blades 1–7 of the β-propeller numbered in bold. Side chains from amino acid residues that were mutated in the chc1-TD-ts alleles and the chc1-box allele are shown and color coded for each allele as follows: chc1-1 and chc1-3 (dark blue), chc1-2 (red), chc1-4 (yellow), and chc1-box (purple). (B) Sequence alignment of the yeast TD (residues 1-359) and human TD (residues 1-353) regions of clathrin heavy chain. Residues that were mutated in the chc1-TD-ts alleles and the chc1-box allele are highlighted according to the same color scheme used in A. An asterisk (*) denotes identical residues between the yeast and human sequence, a colon (:) denotes a conserved substitution between sequences, and a period (.) denotes a semiconserved substitution between sequences. (C) Alignment of the seven blades of the TD β-propeller structure. The approximate boundaries for every strand in each blade are denoted above the alignment. θ denotes conserved hydrophobic residues in both the yeast and human sequence. The S75P mutation of chc1-1 is also present in chc1-3.
Clathrin-mediated sorting is evolutionarily conserved, and many of the proteins that are critical for these pathways have homologues in lower eukaryotic cells. The yeast Saccharomyces cerevisiae has one clathrin heavy chain gene (CHC1), encoding a protein with nearly 50% identity to mammalian HC, and a light chain gene (CLC1). Clathrin-deficient yeast grow poorly, and in some strains a null allele is lethal (Lemmon and Jones, 1987; Munn et al., 1991). They redistribute Kex2 and dipeptidylaminopeptidase (Ste13), two enzymes involved in the processing and maturation of the mating pheromone α-factor, from the TGN to the plasma membrane (Payne and Schekman, 1989; Seeger and Payne, 1992). Clathrin is also important for endocytosis because the internalization of α-factor bound to its receptor Ste2, a G protein-coupled receptor, is reduced in cells lacking clathrin (Tan et al., 1993; Huang et al., 1997). There are many different TGN/endosomal adaptor proteins in yeast (AP-1, Gga1, and Gga2 and the epsin-related proteins Ent3 and Ent5), and disruption of these adaptor genes results in TGN/endosomal sorting phenotypes (Costaguta et al., 2001; Mullins and Bonifacino, 2001; Duncan et al., 2003; Friant et al., 2003; Eugster et al., 2004). However, in studies in which adaptor proteins containing CBM mutations were analyzed, no significant phenotypes were observed (Mullins and Bonifacino, 2001; Yeung and Payne, 2001). At least four endocytic adaptors have been studied in yeast—the epsin-related proteins Ent1 and Ent2 and the AP180 homologues Yap1801 and Yap1802—and all four possess C-terminal CBMs in which the carboxylic acid at the end of the polypeptide substitutes for the final polar residue in the motif. Yeast strains carrying a deletion in all four endocytic adaptor genes complemented with Ent1 lacking its CBM show no defects in uptake of the lipophilic dye FM4-64 or the a-factor receptor Ste3 (Baggett et al., 2003; Maldonado-Baez et al., 2008). The absence of major trafficking phenotypes in studies of CBM-deficient adaptor proteins could be due to the utilization of alternative binding site(s) on clathrin by these mutated adaptors, or other adaptors are able to compensate for the mutated adaptors and promote clathrin function.
Because no major TGN sorting or endocytic phenotypes have been observed upon removal of the CBMs from various yeast adaptor proteins, we decided to directly test the importance of their clathrin binding sites on the TD in vivo by introducing mutations into CHC1 that disrupt the conserved binding sites for both the CBM and the W-box motif (chc1-box). In addition, TD-specific, temperature-sensitive (ts) alleles of the CHC1 gene were generated and analyzed for their effects on clathrin function. In this report, we demonstrate that the TD is critical for overall clathrin function but that the previously identified adaptor binding sites on the TD are not necessary, suggesting that additional interactions on the propeller mediate adaptor association and facilitate clathrin assembly at sites of vesicle formation.
MATERIALS AND METHODS
Strains, Media, and Growth Methods
Strains used in this study are listed in Table 1. YEPD and synthetic dropout media were prepared, and yeast mating, sporulation, and tetrad dissections were performed as described previously (Guthrie and Fink, 1991). Transformation of yeast was performed using the lithium acetate method of Gietz et al. (1995). For growth tests, log phase cultures were diluted to 107 cells/ml, and serial fourfold dilutions were spotted onto C-TRP plates and grown for 2–3 d at the indicated temperatures. To assess calcofluor sensitivity, cultures were diluted to 107 cells/ml, and serial fourfold dilutions were spotted onto YEPD plates with 50 μg/ml calcofluor white and grown for 3–5 d at 25 or 30°C where indicated. For the α-factor halo assay, a YEPD plate was first seeded with 5 × 105 BJ3556 cells in an agar overlay to generate a MATa sst1 lawn. Liquid cultures of MATα test cells and controls were diluted to 107 cells/ml and spotted onto the previously seeded plate and then incubated at 30°C for 48 h.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BJ3502 | MATaura3-52 his3 can1r pep4Δ::HIS3 prb1-Δ1.6R | |
| BJ3556 | MATasst1-2 ade2-1 his6 met1 cyh2 rme1 ura1 can1 | |
| PJ694α | MATα SL2793 | |
| SL12 | MATaleu2 ura3-52 trp1 chc1Δ::LEU2 scd1-v | |
| SL13 | MATα leu2 ura3-52 trp1 chc1Δ::LEU2 scd1-v pSL6 (URA3, CEN, CHC1) | |
| SL82 | MATα leu2 ura3-52 trp1 his1 ade6 chc1Δ::LEU2 scd1-i pAP4 (TRP1, CEN, CHC1) | |
| SL119 | MATα leu2 ura3-52 trp1 chc1Δ::LEU2 scd1-v | |
| SL1462 | MATaleu2 ura3-52 trp1 his3-Δ200 | |
| SL1463 | MATα leu2 ura3-52 trp1 his3-Δ200 | |
| SL2567 | MATα ura3-52 leu2,3-112 his3Δ200 trp-Δ901 lys2-801 suc2-Δ9 vac8-Δ::HIS3 | |
| SL2793 (PJ694a) | MATaleu2-3,112 ura3-52 his3-Δ200 trp1-901 gal4Δ ade2 gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | |
| SL3593 | MATaleu2 ura3-52 trp1 bar1-1 chc1Δ::LEU2 | |
| SL5089 | MATaleu2 ura3-52 trp1 his3-Δ200 ABP1-mRFP:KanMX | |
| SL5171 | MATα ura3 chc1Δ::LEU2 scd1-i pJRC3 (TRP1, CEN, chc1-2 [V37A,H289R]) | |
| SL5177 | MATaura3 his1 chc1Δ::LEU2 scd1-i pJRC2 (TRP1, CEN, chc1-1 [S75P]) | |
| SL5178 | MATaura3 chc1Δ::LEU2 scd1-i pJRC5 (TRP1, CEN, chc1-4 [L14P,H317R]) | |
| SL5181 | MATα ura3 chc1Δ::LEU2 scd1-i pJRC4 (TRP1, CEN, chc1-3 [S75P,V201A,S338P]) | |
| SL5538 | MATaura3-52 trp1 his3Δ200 leu2 chc1Δ::LEU2 ABP1-mRFP:KanMX GFP-CLC1 | |
| SL5634 | MATα leu2 ura3 trp1 chc1Δ::LEU2 scd1-i pJRC19 (TRP1, CEN, chc1-box [F26W,I79S,Q89M]) | |
| SL5719 | MATaleu2 ura3-52 trp1 his3-Δ200 ade2 chc1Δ::LEU2 chs6Δ::HIS3 | |
| YRV19 | MATachs6Δ::HIS3 ade2-101oc his3-Δ200 leu2-Δ1 trp1 ura3-52 | R. Schekman |
Generation of Mutant chc1 Alleles
To generate chc1-TD-ts alleles, the TD coding sequence (residues 1-363) of pAP4 (CEN, TRP1, CHC1, described in Lemmon et al., 1991) was replaced with the His3MX6 module flanked on either side by unique NotI restriction sites to generate pJRC1. Next, error-prone PCR mutagenesis (Bumbulis et al., 1998) was carried out with oligonucleotides (oligos) corresponding to 50 bp 5′ to the CHC1 ATG start codon and 50 bp 3′ to the end of the TD coding sequence (corresponding to amino acid 363), respectively, by using pAP4 as the template. NotI-digested pJRC1 and products of error-prone polymerase chain reaction (PCR) were cotransformed into SL13 (chc1Δ::LEU2 + pSL6 [CEN, URA3, CHC1]) (Lemmon and Jones, 1987) and repaired plasmids containing TD-specific point mutations were selected for on −URA-TRP–selective medium. To screen for ts alleles, colonies were replica plated onto two −TRP plates containing 5-fluoroorotic acid (5-FOA) to select for loss of the URA3, CHC1 plasmid and plates were incubated at 25 or 37°C. Plasmid DNAs from transformants that grew at 25°C but not at 37°C were isolated, retransformed into SL13, and retested for growth on −TRP + 5-FOA plates at 25 and 37°C to verify the ts nature of these alleles. DNA sequencing confirmed the generation of CEN, TRP1 plasmids pJRC2 (chc1-1), pJRC3 (chc1-2), pJRC4 (chc1-3), and pJRC5 (chc1-4).
Generation of the chc1-box allele was as follows. First, plasmid pRTH7 was generated using PCR amplification of the TD + ankle coding region (residues 1-500) from pAP4 with primers to allow for gap repair into the two hybrid GAL-binding domain plasmid pGBDU-C1 (James et al., 1996). Next, a BamHI/SalI fragment from pRTH7 was ligated into pGEX-4T1 (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) to generate pTMN7. Targeted mutagenesis of the TD to generate the chc1-box allele was performed using the QuikChange system (Stratagene, La Jolla, CA) with pTMN7 as the template. A TD fragment containing the three mutations (F26W, I79S, and Q89M) was amplified by PCR and cotransformed with Bsu36I and SexAI-digested pSL6 for gap repair in yeast to generate pSL6-box (CEN, URA3, chc1-box). NotI-digested pJRC1 and a TD fragment containing the chc1-box mutations amplified by PCR were cotransformed into yeast to generate pJRC19 (CEN, TRP1, chc1-box).
Plasmids
pTMN38 (2μ, URA3, GBD-ENT1) has been described in Newpher and Lemmon (2006). Plasmids pJRC6 (2μ, URA3, GBD-CHC1-TD[1-363]) and pJRC8 (2μ, LEU2, GAD-CHC1-TD[1-363]) were generated using PCR amplification of the TD coding region from pAP4 with primers to allow for gap repair into two-hybrid GAL-binding domain (pGBDU-C1) and GAL-activation domain (pGAD-C1) plasmids (James et al., 1996), respectively. Plasmids pJRC14 (2μ, LEU2, GAD-CHC1-TD[S75P]), pJRC15 (2μ, LEU2, GAD-CHC1-TD[L14P,H317R]), pJRC16 (2μ, LEU2, GAD-CHC1-TD[F26W,I79S,Q89M]), pJRC17 (2μ, LEU2, GAD-CHC1-TD[S75P,V201A,S338P]), and pJRC18 (2μ, LEU2, GAD-CHC1-TD[V37A,H289R]) were generated in identical manner to pJRC8 by using the appropriate template DNAs for each mutant allele. Plasmids pJRC20 (2μ, URA3, GBD-CHC1-TD[S75P]), pJRC21 (2μ, URA3, GBD-CHC1-TD[V37A,H289R]), pJRC22 (2μ, URA3, GBD-CHC1-TD[S75P,V201A,S338P]), pJRC23 (2μ, URA3, GBD-CHC1-TD[L14P,H317R]), and pJRC24 (2μ, URA3, GBD-CHC1-TD[F26W,I79S,Q89M]) were generated in identical manner to pJRC6 by using the appropriate template DNAs for each mutant allele. Cloning of yeast open reading frames into the GAL4 activation domain plasmid pOAD have been described in Hudson et al. (1997). To generate pOAD-Ent2(ΔCBM), a PCR product was generated using primers designed to introduce a stop codon following serine 609, immediately before the beginning of the C-terminal CBM, and to allow for gap repair into pOAD-Ent2 ORF. All PCR amplified regions were sequenced in resultant plasmids to verify the correct alleles and no other mutations were present.
Integration of GFP-CLC1
To generate cells expressing GFP-Clc1, a 3.5-kb EcoRI/NotI fragment from pTMN17 (Newpher et al., 2005) containing the coding sequence for GFP-Clc1 was ligated with EcoRI/NotI digested pRS306 to generate pJRC7. pJRC7 was digested with PacI to linearize the plasmid in the CLC1 region for targeted integration. This was transformed into a diploid strain resulting from the mating of SL13 with SL5089 in which the URA3, CHC1 plasmid had been dropped after mating. Cells were grown on synthetic complete medium + 5-FOA to select for excision of the URA3 gene and plasmid sequences, and candidate yeast colonies were screened for GFP-Clc1 expression by fluorescence microscopy. To verify removal of the URA3 gene, green fluorescent protein (GFP)-positive colonies were tested to confirm their inability to grow on −URA plates. Tetrad dissection resulted in the generation of SL5538.
Biochemical Procedures
For immunoblots of clathrin HC, extracts were prepared from 10 ml of log-phase cells (5 × 106 cells/ml) grown at the indicated temperatures by glass bead lysis in 250 μl of 0.1 M Tris, pH 7.5, containing 1 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Stepp et al., 1995). Lysates were centrifuged for 20 min at 10,000 × g, and the supernatants were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted using anti-Chc1 mouse monoclonal antibodies (Lemmon et al., 1988) and anti-α-tubulin rat monoclonal antibodies (ab6161) as a loading control (Abcam, Cambridge, MA). To monitor aminopeptidase I (Ape1) processing, extracts from cells grown at 30°C were prepared as described previously (Huang et al., 1999), and aminopeptidase was detected by immunoblotting using a rabbit-anti-Ape1 polyclonal antiserum at a 1:10,000 dilution (gift from Dan Klionsky, University of Michigan, Ann Arbor, MI). Clathrin-coated vesicles (CCVs) were isolated from SL5067 (CHC1) and SL5551 (chc1-box) yeast grown at 30°C by Sephacryl S-1000 column chromatography as described previously (Lemmon et al., 1988). Polyclonal rabbit antiserum to Apm1 has been described previously (Stepp et al., 1995). Detection of all proteins was performed with the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE) by using secondary antibodies labeled with infrared fluorophores.
α-Factor Endocytosis Assays
35S-α-factor uptake was performed as described previously (Dulic et al., 1991). Cells were incubated for 15 min at 37°C before the addition of radiolabeled α-factor. Plots show the best-fit trend lines generated in Excel (Microsoft, Redmond, WA). Data are the average of five independent experiments ± SD. Endocytic rates were determined by taking the average slope of the best-fit trend lines from the 3- to 9-min time points.
Microscopic Analysis of Cortical Clathrin Recruitment
Live cell imaging of GFP-Clc1p was performed using overnight cultures of cells grown to early to mid-log phase at 25°C in synthetic media. SL5538 (chc1Δ ABP1-RFP GFP-CLC1) transformed with expression plasmids for CHC1 or chc1-TD-mut alleles were treated with 250 μM latrunculin A (LAT-A) dissolved in dimethyl sulfoxide for 20 min. Actin disassembly was visualized in all cells using Abp1-red fluorescent protein (RFP) (data not shown). Clathrin accumulation was defined by the appearance of intense GFP-Clc1p cortical rim localization or by the appearance of discrete GFP-Clc1p cortical puncta. For each strain, 100–150 cells were counted, and data are from three experiments. Microscopy was performed using a fluorescence BX61 microscope (Olympus, Melville, NY) equipped with Nomarski differential interference contrast optics, a Uplan Apo 100× objective (numerical aperture [NA] 1.35), a CoolSNAP HQ camera (Photometrics, Tucson, AZ), Lambda 10-2 excitation and emission filters and Lambda LS 175-W xenon remote light source with liquid light guide (Sutter Instrument, Novato, CA).
To perform total internal reflection fluorescence microscopy (TIRFM), SL5538 (chc1Δ ABP1-RFP GFP-CLC1) transformed with CHC1 or chc1-TD-mut expression plasmids were grown to early to mid-log phase at 25°C in synthetic media, washed in phosphate-buffered saline (PBS), and 250 μl of cells was added to polylysine-coated dishes (MatTek, Ashland, MA) for 5 min. Excess cells were washed away, and 50 μl of fresh synthetic complete medium was added before coverslips were applied to prevent drying. TIRFM was performed using an IX-81 scope (Olympus), using a 100×/1.46NA PlanApo total internal reflection fluorescence (TIRF) objective (Olympus). Samples were visualized by illumination with 488 nm argon krypton and 543 nm helium neon lasers and captured by a three-color charge-coupled device (Hamamatsu, Bridgewater, NJ). Four-minute movies were taken at 2-s intervals by using 250-ms exposures for both RFP and GFP signals. For each strain, GFP-Clc1p patches were sorted into three categories: “mobile” patches that acquired the late endocytic actin marker protein Abp1-RFP before disappearing from the cell cortex; “immobile” patches that persisted at the cortex for >1 min and disappeared without acquiring actin; and “unproductive” patches, which appeared for <1 min but disappeared without receiving actin. Wide-field and TIRF images were acquired using SlideBook 4.01 (Intelligent Imaging Innovations, Santa Monica, CA).
Two-Hybrid Interaction Assays
For investigation of TD interaction with Ent1, overnight cultures of the two-hybrid reporter strain SL2793 cotransformed with bait and prey plasmids were diluted to 5 × 106 cells/ml and spotted onto complete synthetic medium lacking leucine and uracil (C-LEU-URA) to test for general growth and medium lacking leucine, uracil, and adenine (C-LEU-URA-ADE) to test for induction of the GAL2-ADE2 reporter. Cells were grown at 30°C for up to 5 d.
For investigation of TD interaction with Ent2 and Apl2, the PJ694α “bait” reporter strain transformed with each pGBD-CHC1-TD plasmid (or pGBD alone) was mated with the PJ694a “prey” reporter strain transformed with the corresponding pOAD-adaptor plasmid (or pOAD alone), and diploids were selected for on C-LEU-URA plates. Overnight cultures of each diploid strain were diluted to 5 × 106 cells/ml and spotted onto C-LEU-URA-ADE plates. Plates were incubated at 30°C for up to 5 d.
Homology Modeling of the Yeast TD
A homology model of the yeast TD (residues 1–363) was generated from the rat TD structure (PDB 1BPO) by using Swiss Model and visualized using the Visual Molecular Dynamic viewer (University of Illinois, Urbana-Champaign, IL). A sequence alignment of the yeast and human TD coding sequence was performed using ClustalW (European Molecular Biology Laboratory-European Bioinformatics Institute).
RESULTS
Generation of Heavy Chain N-terminal Domain-specific Alleles
Previous studies on clathrin function in S. cerevisiae have focused on observable phenotypes associated primarily with the null allele (chc1Δ), or the ts allele chc1-521 (chc1-ts) containing multiple point mutations in the C-terminal part of the protein that affect LC binding and HC trimerization (Pishvaee et al., 1997). To investigate the importance of the TD region on HC function, we sought to generate TD-specific chc1-ts alleles. Using random PCR mutagenesis and standard gap repair and plasmid shuffling techniques (see Materials and Methods for details), we isolated four chc1-TD-ts alleles [chc1-(1-4)]. A list of the mutation(s) present in each allele is provided in Table 2, and the locations of mutated residues within the TD structure are denoted in Figure 1A.
Table 2.
TD mutations in chc1 alleles
| chc1 allele | Mutation |
|---|---|
| chc1-1 | S75P |
| chc1-2 | V37A, H289R |
| chc1-3 | S75P, V201A, S338P |
| chc1-4 | L14P, H317R |
| chc1-box | F26W, I79S, Q89M |
In addition to the generation of the chc1-TD-ts alleles, we also generated a fifth HC allele (chc1-box) in which three point mutations (F26W, I79S, and Q89M) were introduced into the TD coding region. Glutathione transferase pulldown experiments have demonstrated previously that for the mammalian TD the F27W mutation, which corresponds to F26 in yeast TD, blocks the interaction of TD with a W-box motif-containing peptide from amphiphysin (Miele et al., 2004). The Q89M mutation has been shown previously to significantly disrupt binding of β-arrestin 2 to the TD (Goodman et al., 1997), and based on the x-ray crystal structures of the TD in complex with a clathrin-box peptide from either β-arrestin 2 or from the hinge region of the β-subunit of AP-3 (ter Haar et al., 2000), the I79S and Q89M mutations in combination should block clathrin-box motif-containing adaptor interaction with the TD. Although no W-box–containing adaptors in yeast have been identified, these mutations were all combined together to generate a TD that should have impaired binding to both motifs. Sequence alignment of the TD regions of yeast and human HC show that these residues are identical between species (Figure 1B).
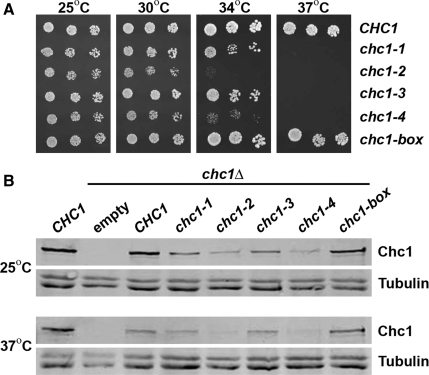
To compare the severity of the ts alleles and to test for any growth phenotype associated with the chc1-box allele, spot dilutions of cultures were grown on plates over a range of temperatures. Yeast whose only source of HC was from expression of any of the four chc1-TD-ts alleles were able to grow at the permissive temperature of 25°C (Figure 2A), and similar to chc1Δ cells, were inviable at 37°C. However, at the intermediate temperatures of 30 or 34°C, chc1-2 and chc1-4 yeast displayed more severe growth phenotypes than chc1-1 and chc1-3 yeast (Figure 2A). Surprisingly, yeast expressing HC from the chc1-box allele grew normally at all temperatures (Figure 2A).
Figure 2.
The chc1-box mutant is not temperature-sensitive for growth. (A) SL82 (chc1Δ + pAP4[CEN, CHC1]), SL5177 (chc1Δ + pJRC2[CEN, chc1-1]), SL5171 (chc1Δ + pJRC3[CEN, chc1-2]), SL5181 (chc1Δ + pJRC4[CEN, chc1-3]), SL5178 (chc1Δ + pJRC5[CEN, chc1-4]), and SL5634 (chc1Δ + pJRC19[CEN, chc1-box]) cells were diluted to 107 cells/ml, and fourfold serial dilutions were spotted onto plates and grown at the indicated temperatures for 3 d. (B) Immunoblots of extracts from chc1Δ yeast (SL12) transformed with pUN30 (CEN, empty), pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box) were detected with anti-Chc1 antibodies. Cells were grown continuously at 25°C, and then one-half of each sample was removed and incubated for 90 min at 37°C before harvesting. To verify equal protein loading, blots also were probed with antibodies to tubulin.
HC proteins encoded by these chc1-TD-ts alleles might be unstable as the point mutations introduced into the TD region could result in aberrant folding of the β-propeller structure and destabilization of the HC. To determine whether full-length HC was produced, protein extracts from yeast encoding HC from each of the chc1-TD-ts alleles and the chc1-box allele were prepared, and HC protein levels were examined by immunoblot analysis using monoclonal antibodies to Chc1. When cells were grown at 25°C, all of the HC mutant proteins were expressed and detected as full-length proteins of 190 kDa (Figure 2B). The amount of HC expressed from each ts allele was significantly reduced relative to the wild-type allele (20–25% of wild-type for chc1-1 and chc1-3 alleles and 10–14% of wild-type for chc1-2 and chc1-4 alleles; n = 3), but HC expression from the chc1-box allele was comparable with wild type (∼90% of wild type; n = 3). Next, we tested whether these HC proteins would be stable after a shift to the nonpermissive temperature of 37°C for 90 min. The full-length protein was still present in cells expressing HC from all alleles (Figure 2B), although levels were significantly less in the more severe ts alleles (chc1-2 and chc1-4) suggesting protein destabilization might reduce their function.
TGN Sorting in chc1-TD Mutants
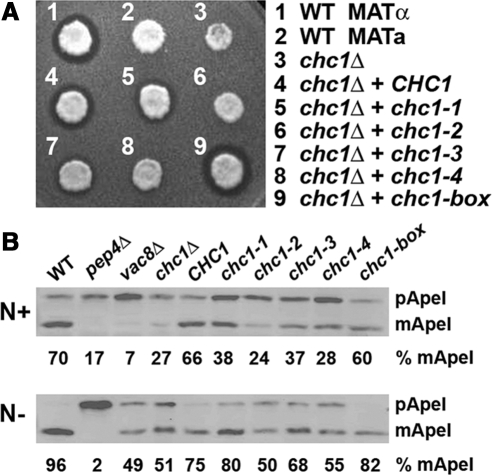
Clathrin-deficient MATα yeast are defective in the secretion of the mature form of the mating pheromone α-factor due to the mislocalization of α-factor processing enzymes, including Kex2, a subtilisin-like protease, and Ste13, a dipeptidyl-aminopeptidase, from the TGN to the plasma membrane (Payne and Schekman, 1989; Seeger and Payne, 1992). To determine whether HCs encoded by any of the chc1-TD mutant alleles could rescue the α-factor processing phenotype of chc1Δ cells, halo assays were performed in which MATα yeast expressing different HCs were tested for their ability to inhibit the growth of MATa cells supersensitive to mature α-factor at the semipermissive temperature of 30°C. Strong halos were observed in chc1Δ cells complemented with a plasmid carrying the CHC1 allele (Figure 3A, 4) indicative of the secretion of mature α-factor and restoration of proper TGN sorting. Yeast expressing HC from any of the four chc1-TD-ts alleles did not fully rescue the TGN sorting phenotype of chc1Δ (Figure 3A, 5–8) as the halos produced by each of these strains were smaller than the halo generated by cells complemented with wild-type HC. The most severe phenotypes were seen with chc1-2 and chc1-4 (Figure 3A, 6 and 8, respectively), consistent with the stronger growth phenotype observed at intermediate temperatures. chc1-box yeast produced a halo very comparable to the wild-type allele, indicating TGN retention/sorting seems normal (Figure 3A, 9).
Figure 3.
chc1-TD-ts alleles cause TGN sorting and Ape1 processing defects. (A) Halo assays for MATα chc1Δ yeast (SL119) transformed with pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), pSL6-box (CEN, chc1-box), and wild-type controls, SL1462 (MATa) and SL1463, (MATα) spotted onto a lawn of MATa (BJ3556) and grown at 30°C for 2 d. A zone of growth inhibition demonstrates secretion of mature α-factor. (B) Ape1 processing: immunoblots of wild-type (SL1463), pep4Δ (BJ3502), vac8Δ (SL2567), chc1Δ (SL12), and SL12 transformed with pUN30 (CEN, empty), pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pSL6-box (CEN, chc1-box), grown in normal medium (N+) or nitrogen starvation medium (N−) at 30°C and blotted with anti-Ape1 antibodies. The amounts of precursor (pApe1) and mature aminopeptidase (mApe1) were determined by densitometry, and the percentage of mApe1 is reported relative to total mApe1 + pApe1 for each.
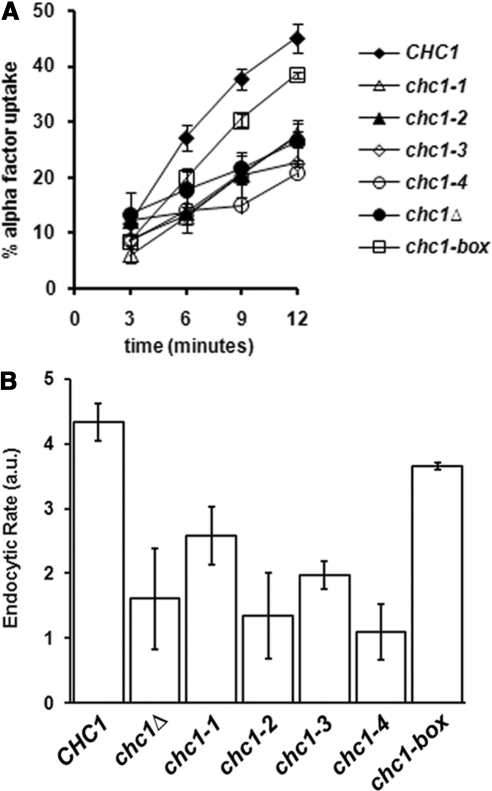
Figure 4.
The chc1-box allele, but not chc1-TD-ts alleles, rescues the endocytic phenotype of chc1Δ. MATa chc1Δ bar1-1 (SL3593) was transformed with pUN30 (CEN, empty), pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box). α-factor internalization was analyzed at 37°C as described in Materials and Methods.
Figure 5.
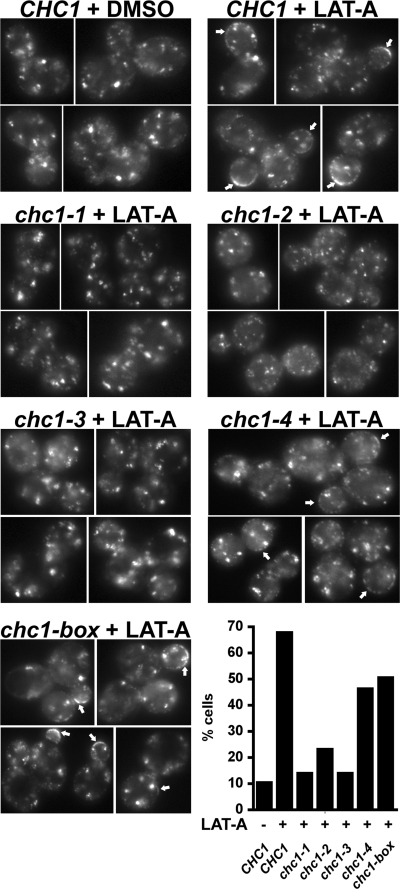
GFP-Clc1p accumulation at the cell cortex in chc1-TD alleles in the presence of LAT-A. GFP-CLC1 ABP1-mRFP chc1Δ (SL5538) was transformed with pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box). Cells grown to log phase at 25°C were treated with LAT-A (250 μM) for 20 min at 25°C before imaging. Abp1-mRFP was completely cytosolic after 20 min in LAT-A, indicating the disassembly of actin (data not shown). Arrows indicate the accumulation of clathrin at the cell cortex. Cells that showed strong cortical puncta or rim GFP staining were considered to have accumulated clathrin. The percentage of cells for each condition that contained cortical GFP-Clc1 accumulation is shown in the graph (n = 100–150).
Figure 6.
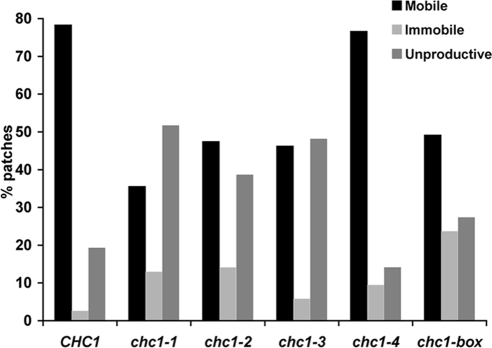
TIRFM of clathrin patch dynamics at the cell cortex Graph show percentages of GFP-Clc1 patch types observed in CHC1 and chc1-TD mutants at 25°C. Between 50 and 80 GFP-Clc1 patches for each strain were defined as mobile, immobile or unproductive as described in Materials and Methods. Strains are the same as in Figure 5.
Figure 7.
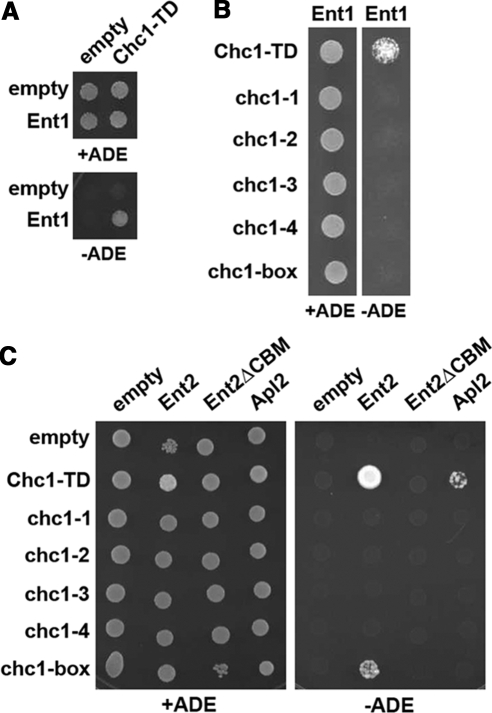
HC-TD mutations disrupt interactions with clathrin adaptors Ent1, Ent2, and Apl2. (A) A two-hybrid reporter strain (SL2793) was transformed with either the empty GAL4 binding domain (GBD) plasmid pGBDU-C1 or pGBD-ENT1 in combination with either the empty GAL4 activation domain (GAD) plasmid pGAD-C1 or pGAD-CHC1-TD(1-363). Cells from each set of transformants were diluted to 5 × 106 cells/ml, spotted onto C-LEU-URA (total growth) and C-LEU-URA-ADE (activation of the GAL2:ADE2 reporter) plates, and grown at 30°C for 3 d. (B) SL2793 was cotransformed with pGBD-ENT1 and each of the following GAD-CHC1-TD plasmids: pJRC8 (GAD-CHC1-TD), pJRC14 (GAD-CHC1-TD[chc1-1]), pJRC15 (GAD-CHC1-TD[chc1-2]), pJRC16 (pGAD-CHC1-TD[chc1-3]), pJRC17 (GAD-CHC1-TD[chc1-4]), or pJRC18 (GAD-CHC1-TD[chc1-box]) and plated and grown as described in A. (C) PJ694α was transformed with pGBDU-C1 or the following plasmids: pJRC6 (GBD-CHC1-TD), pJRC20 (GBD-CHC1-TD[chc1-1]), pJRC21 (GBD-CHC1-TD[chc1-2]), pJRC22 (GBD-CHC1-TD[chc1-3]), pJRC23 (GBD-CHC1-TD[chc1-4]), pJRC24 (GBD-CHC1-TD[chc1-box]) and mated with PJ694a transformed with either the empty GAD plasmid pOAD, pOAD-Ent2, pOAD-Ent2(ΔCBM), or pOAD-Apl2. Diploids were selected on C-LEU-URA plates, and then activation of the ADE2 reporter was assayed as described above.
Figure 8.
Clathrin HC encoded by the chc1-box allele forms coated vesicles. Clathrin-coated vesicles were prepared from chc1Δ cells (SL12) transformed with pAP4 (CEN, CHC1) or pJRC19 (CEN, chc1-box), grown at 30°C, and fractionated on a Sephacryl S-1000 column. Column fractions 22–48 for each preparation were immunoblotted with anti-Chc1 and anti-Apm1 antibodies.
Figure 9.
The chc1-box allele restores calcofluor resistance to chs6Δ chc1Δ (ΔΔ) cells. (A) SL119 (chc1Δ), YRV19 (chs6Δ), SL5719 (chs6Δ chc1Δ[ΔΔ]), and ΔΔ + pAP4 (CEN, CHC1) cells were diluted to 1 × 107 cells/ml, and fourfold serial dilutions were plated onto both YEPD plates and YEPD plates containing 50 μg/ml calcofluor white and incubated at 30°C for 3–5 d. (B) ΔΔ cells transformed with pAP4 (CEN, CHC1), pJRC2 (CEN, chc1-1), pJRC3 (CEN, chc1-2), pJRC4 (CEN, chc1-3), pJRC5 (CEN, chc1-4), or pJRC19 (CEN, chc1-box) were diluted to 1 × 107 cells/ml, and fourfold serial dilutions were plated as described in A and incubated at 30°C (or 25°C where indicated).
Ape1 Processing in chc1-TD Mutant Cells
Whereas most resident vacuolar hydrolases are delivered to the vacuole via the secretory pathway, at least one hydrolase, Ape1, is delivered through the cytoplasm-to-vacuole (Cvt) targeting pathway (Harding et al., 1995). In this pathway, Ape1 is initially synthesized as a precursor protein in the cytoplasm, and upon delivery to the vacuole is processed in a Pep4-dependent manner to generate the active protease. Many of the genes involved in the Cvt pathway also are involved in autophagy (Khalfan and Klionsky, 2002), and our lab showed previously that clathrin deficiency causes Ape1-processing defects (Huang et al., 1999). To determine whether the TD mutations affect Ape1 maturation, chc1-TD-ts and chc1-box cells were grown at the semipermissive temperature of 30°C in nitrogen-rich medium (Cvt pathway growth conditions) or nitrogen starvation medium (which induces autophagy), and Ape1 processing was monitored by the blotting of extracts with aminopeptidase I antibodies.
In wild-type cells, the majority of Ape1 protein (∼70%) is present as the 50-kDa mature protease (mApe1) under Cvt conditions (N+) (Figure 3B), but in chc1Δ cells only 27% was mature, similar to previous observations (Huang et al., 1999). In strains expressing HC from any of the chc1-TD-ts alleles, more Ape1 was present in the precursor form compared with that from CHC1 cells. Ape1 processing in the null (chc1Δ) and all chc1-TD-ts mutants was more complete upon stimulation of the autophagy pathway; however, chc1-2 and chc1-4 cells more closely resembled the chc1Δ phenotype, whereas chc1-1 and chc1-3 cells more closely resembled CHC1. chc1-box cells yielded mature protease comparable to CHC1 under both Cvt (N+) and autophagy (N−) conditions (Figure 3B), indicating that Ape1 processing and Cvt/autophagy are normal in the box mutant.
Endocytic Analysis in chc1-TD Mutants
Studies of endocytosis in yeast have focused on the internalization of the mating pheromone α-factor bound to the G protein-coupled receptor Ste2. Clathrin-deficient yeast have partial defects in endocytosis, as α-factor internalization is reduced by ∼ 50% (Tan et al., 1993; Huang et al., 1997). To determine whether the TD is critical for HC function in endocytosis, we performed α-factor internalization assays with cells expressing HC-TD mutations after shifting to 37°C. In chc1Δ yeast transformed with an empty CEN plasmid, the amount of internalized α-factor after 12 min of uptake was roughly 50% of that seen in chc1Δ yeast complemented with wild-type CHC1 (Figure 4A), similar to previous studies of clathrin-deficient yeast (Tan et al., 1993; Huang et al., 1997). Yeast cells expressing HC from any of the four chc1-TD-ts alleles were not able to suppress the α-factor internalization defect of chc1Δ cells (Figure 4A), but the chc1-box allele provides near complete rescue, with an endocytic rate that was ∼ 85% of wild type (Figure 4B).
Visualization of Clathrin and Endocytic Dynamics in TD Mutants
GFP-LC is a suitable marker for localizing HC within yeast cells as GFP-Clc1 has been shown to completely colocalize with the fluorescent signal of Chc1-RFP (Newpher et al., 2005). GFP-LC localizes to the TGN, endosomes, and small cortical patches. Cortical actin patches are the sites of endocytosis in yeast, and clathrin recruitment to these sites is dependent upon the endocytic adaptors Ent1/2 and Yap1801/2 (Newpher et al., 2005), which have C-terminal clathrin-box motifs. However, the clathrin in these cortical patches is often masked by the intense fluorescent clathrin signal on TGN/endosomal structures.
To visualize clathrin and to enhance detection of cortical clathrin, we treated cells expressing GFP-Clc1, Abp1-RFP, and the TD mutant HCs with LAT-A. LAT-A is an actin monomer-sequestering drug that inhibits actin assembly, causing immobilization and accumulation of cortical patches containing early endocytic proteins including clathrin (Kaksonen et al., 2003; Newpher et al., 2005). In all TD mutants, GFP-LC was observed on internal structures (with and without LAT-A, Figure 5; data not shown), indicating that the HC from all alleles is able to bind endosomal/TGN compartments at 25°C. In cells expressing wild-type HC, ∼70% of cells showed either discrete GFP-Clc1 puncta or rim GFP-Clc1 staining at the cortex after LAT-A treatment (Figure 5, CHC1 + LAT-A, arrows). However, cortical clathrin accumulation in LAT-A–treated cells was significantly reduced (∼10–23% of cells) in cells expressing HC from the chc1-1, chc1-2, and chc1-3 alleles (Figure 5), consistent with the inability of these alleles to suppress the chc1Δ endocytic phenotype. Surprisingly, GFP-Clc1 was visualized at the cortex in a large percentage of cells (∼50% of cells) expressing HC from the chc1-4 allele (Figure 5), but consistent with its endocytic defect at 37°C, its recruitment to cortical sites was ablated at elevated temperature (Supplemental Figure 1). The cortical accumulation of clathrin in chc1-2 was also decreased to only a few percent of cells at 37°C as well (Supplemental Figure 1). Note that at elevated temperature there were still visible internal patches of clathrin, even though cortical accumulation was impaired; however, we cannot rule out that these are assemblies that form spontaneously and are not associated with membrane. In contrast, in the chc1-box mutant ∼50% of cells had cortical clathrin, but this recruitment was not temperature dependent (Figure 5 and Supplemental Figure 1). This is consistent with the α-factor internalization assays where the box mutant internalized clathrin nearly as well as wild type at 37°C.
We next performed TIRF microscopy experiments to monitor the behavior of cortical/endocytic clathrin in these cells. In time-lapse movies of CHC1 cells, three different types of GFP-Clc1 patch dynamics were observed. First, a majority of the patches at the cell surface recruited Abp1-RFP before moving away from the evanescent field and into the cell. These mobile patches represented 78% of the total number of events analyzed in CHC1 cells (n = 63; Figure 6), in good agreement with previous observations (Newpher et al., 2005). Second, occasional immobile GFP-Clc1 patches (∼3%) would persist at the surface for an extended period (≥1 min), display very limited lateral movement, and never acquire Abp1-RFP before their disappearance (Figure 6). Third, ∼19% of events in wild-type cells consisted of shorter lived patches (<1 min) where clathrin puncta formed but also did not acquire Abp1/actin before disappearing. We refer to these as unproductive events. When clathrin patch dynamics were observed in chc1-TD-ts and chc1-box cells at the permissive temperature, three different patterns emerged (Figure 6). Strikingly, chc1-4 resembled the wild type, with ∼77% of events scored as mobile, i.e., receiving Abp1 before disappearing. This pattern is consistent with the LAT-A analysis in Figure 5, showing substantial cortical recruitment of clathrin in chc1-4 cells at 25°C. The three other ts alleles (chc1-1, chc1-2, and chc1-3) formed a second class, with ∼40–50% unproductive patches, whereas the number of longer lived immobile patches was a smaller percentage (5–14%) of the total. The large percentage of unproductive patches may be reflected in the decreased recruitment in the presence of LAT-A (Figure 5). Significantly, the chc1-box allele forms a third class. It has fewer unproductive patches (∼25%) compared with the second class (chc1-1, chc1-2, and chc1-3), but it also has an increased percentage of immobile patches that persist for >60 s but disappear without receiving Abp1. Perhaps the immobile clathrin patches contribute to the cortical signal seen in LAT-A, thus explaining why chc1-box retains fairly strong cortical recruitment of clathrin.
Using wide field imaging, we also analyzed the dynamics of Sla2, an endocytic coat protein that is recruited after clathrin in the immobile phase of endocytosis. In each of the mutants, nearly all Sla2 cortical patches (>95%) had normal lifetimes and acquired Abp1 and internalized (data not shown). Also, there were very few of the Abp1-marked actin comet tails often seen in cells without clathrin. We infer from this that immobile and unproductive clathrin patches may abort before acquiring other later coat module factors, like Sla2, perhaps because of impaired ability to bind adaptors, engage other components of the endocytic machinery and/or cluster receptors. Abortive events during endocytic clathrin coat assembly have been clearly documented in mammalian cells (Ehrlich et al., 2004; Loerke et al., 2009).
Adaptor binding to HC-TD mutants
To investigate whether the TD regions encoded by these alleles could interact with endocytic adaptors, two-hybrid interaction assays between the endocytic adaptors Ent1 and Ent2, each containing a C-terminal CBM, and the HC-TD encoded by the chc1-TD-ts and chc1-box alleles were performed. In addition, two-hybrid interactions were examined between HC-TD proteins and Apl2, the large (β) subunit of the Golgi adaptor AP-1 that contains an internal CBM.
Although a wild-type TD prey interacted with an Ent1 bait (Figure 7A), none of the mutant TD prey, including one containing the chc1-box allele mutations, were able to bind (Figure 7B). Similar results were seen between HC-TD and Apl2 as only the wild-type TD bait interacted with the Apl2 prey, although the interaction with wild-type TD was fairly weak to begin with (Figure 7C). In this assay, the most robust interaction was observed between the wild-type TD and Ent2 (Figure 7C), but similar to Ent1, TDs containing any of the point mutations from the chc1-TD-ts alleles still disrupted this interaction. Surprisingly, a TD containing the chc1-box allele mutations was still able to interact with Ent2, although the interaction was much weaker compared with the wild-type TD (Figure 7C). The binding results were not caused by differences in stability of GBD-TD mutant proteins, as all were expressed to similar steady-state levels (see Supplemental Figure 2).
To assess whether Ent2 association with TD containing the chc1-box allele mutations was mediated by the Ent2 C-terminal CBM, a truncated version of Ent2 lacking the final four residues (amino acids 610–613) that constitute the C-terminal CBM was generated (Ent2ΔCBM) and tested in the two-hybrid assay. Ent2ΔCBM was unable to interact with either wild-type TD or a TD containing chc1-box mutations (Figure 7C), indicating that the CBM is required for efficient Ent2 association with the TD.
Heavy Chain Encoded by chc1-Box Forms Clathrin-coated Vesicles
Recruitment of soluble clathrin triskelions to the membrane surface is mediated in part by interactions between the TD and clathrin-box motif containing adaptor proteins. The I79S and Q89M mutations of the chc1-box allele should disrupt TD-clathrin box adaptor protein interactions, but the lack of major growth, TGN sorting, α factor uptake, or Ape1 processing phenotypes for the chc1-box allele suggests that clathrin function is not significantly impaired in these cells. The microscopy data also indicates that Chc1-box protein is recruited to membranes even in the absence of the CBM-binding region. To further test for clathrin function, we subjected chc1-box cells to our standard protocol for isolation of CCVs. Cells were spheroplasted, lysed, and after differential centrifugation, a 100,000 × g microsomal membrane pellet was resuspended and fractionated on a Sephacryl S-1000 column (Lemmon et al., 1988). Column fractions were analyzed by SDS-PAGE followed by immunoblotting with monoclonal antibodies to Chc1. In CHC1 cells, HC peaked in column fractions 34–36 (Figure 8). The heterotetrameric Golgi adaptor AP-1 also cofractionated with HC as shown by blotting for the μ chain Apm1 (Figure 8). When microsomal membranes from chc1-box cells were similarly fractionated, the HC protein peak was located in similar column fractions as was observed for CHC1 cells (Figure 8), indicating that the HC protein encoded by chc1-box could form clathrin-coated vesicles. However, Apm1 did not cofractionate with clathrin isolated from chc1-box cells, but instead eluted in later fractions presumably as free AP-1 complex (Figure 8). This suggests that TD binding to AP-1 is critical for stable incorporation of this adaptor into clathrin-coated structures.
Chitin Synthase Trafficking in chc1-TD Mutant Strains
Because AP-1 was not stably associated with CCVs in the chc1-box mutant, we tested whether an AP-1–dependent pathway was affected in the TD mutants. The AP-1/clathrin pathway is involved in the transport of proteins, including chitin synthase III (Chs3) from early endosomes back to the TGN (Valdivia et al., 2002). Chs3 can be found both intracellularly in Kex2-containing compartments and at the PM where it localizes to the mother-bud neck to deliver a ring of chitin around developing buds (Chuang and Schekman, 1996). The delivery of Chs3 from the TGN to the cell surface is dependent upon the Chs5/Chs6 coat complex, as chs5Δ or chs6Δ cells show reduced chitin deposition at the bud site and intracellular retention of Chs3 (Santos and Snyder, 1997; Ziman et al., 1998). chs6Δ yeast are therefore resistant to the toxic effects of the chitin-binding compound calcofluor white (Figure 9A) (Ziman et al., 1998). Deletion of clathrin or AP-1 subunit genes eliminates intracellular Chs3 retention and restores calcofluor sensitivity in the chs6Δ background (Figure 9A) (Valdivia et al., 2002).
Therefore, calcofluor sensitivity growth assays were performed with chc1Δ chs6Δ (ΔΔ) cells transformed with plasmids encoding wild-type or each of the chc1-TD mutants. ΔΔ cells complemented with wild-type CHC1 restored intracellular Chs3 retention and calcofluor resistance (Figure 9, A and B). Expression of HC from each chc1-TD-ts allele was not able to fully restore calcofluor resistance to ΔΔ cells (Figure 9B), and consistent with the results obtained in the TGN sorting, mature α-factor secretion halo assay, the chc1-2 and chc1-4 mutants were more sensitive to calcofluor, although chc1-4 retained some residual function at 25°C. The chc1-box allele restored calcofluor resistance in a manner similar to wild-type HCs (Figure 9B), indicating that Chs3 retrieval from early endosomes to the TGN is efficient in these cells even though AP-1 association with CCVs is impaired.
DISCUSSION
Most previous studies examining the role of clathrin function in yeast have used either the null allele (chc1Δ) or a temperature-sensitive allele (chc1-521, also referred to as chc1-ts) that disrupts LC binding and/or trimerization of HC (Seeger and Payne, 1992; Pishvaee et al., 1997). Additional clathrin HC mutations have been described previously (Lemmon et al., 1991; Chen and Graham, 1998), but they also were localized in the C-terminus close to the trimerization domain. To directly assess the role of the TD in clathrin function, we generated a HC allele, chc1-box, containing substitutions in the binding sites for CBMs and W-box motifs present in known clathrin adaptor proteins. In addition, we isolated four chc1-TD-ts alleles in which random mutation(s) were introduced throughout the TD coding region. The TD seems to be necessary for overall clathrin function as the chc1-TD-ts alleles all demonstrated impairment of several sorting pathways that require clathrin. Surprisingly, HC expression from the chc1-box allele resulted in no major growth or trafficking phenotypes, suggesting that additional or alternative binding sites might exist on the TD for clathrin adaptor proteins.
chc1-TD-ts Alleles and the Location of Point Mutations
Although clathrin-dependent functions were perturbed in cells expressing HC from each chc1-TD-ts allele, the chc1-1 and chc1-3 mutants displayed less severe growth, TGN enzyme retention, endosome-to-TGN retrieval of Chs3, and Ape1 processing phenotypes compared with the chc1-2 and chc1-4 mutants. The severity of the phenotypes caused by each of these alleles may be due in part to differences in HC protein expression. In fact using siRNA-mediated knockdown of HC, Moskowitz et al. (2005) observed dramatic changes in endocytic rates of transferrin receptor with a narrow range of clathrin concentrations, and reduction of HC levels by as little as 30–40% significantly impaired internalization (Moskowitz et al., 2005). The amount of HC expressed in chc1-1 and chc1-3 cells was reduced four- to fivefold compared with HC expression from the CHC1 allele, and expression of HC from the chc1-2 and chc1-4 alleles was reduced 7- to -10-fold relative to CHC1. However, at the permissive temperature of 25°C, each mutant HC protein facilitated robust growth and clathrin was associated with internal membranes as visualized by fluorescence microscopy. In addition, though the chc1-2 and chc1-4 caused similar, more severe phenotypes in the several tests for clathrin function, strains with these two alleles behaved differently in a number of assays. For example, the chc1-4 allele, but not the chc1-2 allele, was able to partially restore clathrin function in TGN sorting halo assays and calcofluor resistance at 25°C (Figure 9B; data not shown). Also, chc1-4, but not chc1-2, showed significant cortical clathrin recruitment in LAT-A and near normal clathrin patch dynamics by TIRFM at 25°C. Moreover, we found that complete removal of the TD, causes a null-like phenotype, but this HC is expressed at two- to threefold higher levels than the HC encoded by chc1-4 (unpublished observations). Therefore, protein expression levels cannot fully account for the differences observed for each chc1-TD-ts allele.
Phenotypic differences caused by the chc1-TD-ts alleles may arise from specific alterations in the TD β-propeller structure and/or binding ability caused by each set of mutations. Both the chc1-1 and chc1-3 alleles possess the S75P mutation. Serine 75 is located at the beginning of blade 2 near the central axis of the propeller structure (Figure 1, A and B), and the substitution of proline may result in aberrant folding. The chc1-3 allele encodes two additional substitutions (V201A and S338P); however, because HC protein levels were similar in cells expressing these alleles and the chc1-3 mutation did not confer more severe phenotypes relative to chc1-1, these two residues probably do not contribute to major structural alterations.
The chc1-2 allele contains two mutations (V37A and H289R) that could contribute to destabilization of the propeller. Valine 37 is found in propeller blade 1, but the side chain does not participate in the formation of either hydrophobic pocket that accommodates the hydrophobic side chains of CBM residues. Replacing one hydrophobic residue with another hydrophobic residue is not very likely to alter protein folding and result in TD destabilization. Histidine 289, identical between yeast and humans (Figure 1B), is located within a region of propeller blade 6 with a high degree of sequence identity between yeast and mammalian TDs. The histidine side chain lines the groove formed between blades 5 and 6, and mutating this residue to arginine might affect the folding of blade 6 or alter the packing of blades 5 and 6 with respect to one another. Alternatively, it is possible that the groove formed between blades 5 and 6 functions as a binding pocket for CBM-containing adaptors. Evidence to suggest this possibility comes from studies on tachylectin-2, whose five-blade β-propeller has binding sites for N-acetylglucosamine between each blade of its propeller (Beisel et al., 1999), but further characterization of this allele is necessary to distinguish between these two possibilities.
Two missense mutations (L14P and H317R) were encoded by the chc1-4 allele. Leucine 14 is identical between yeast and human HC (Figure 1B) and resides in a short α-helical segment of the loop connecting the final β-strand of blade 7 to the first β-strand of blade 1. Mutating this leucine residue to proline might affect the orientation of blade 1 with respect to the other six propeller blades. Histidine 317 is located in blade 7 with its side chain facing the groove between blades 7 and 1 of the TD propeller. An arginine residue at this position might affect the folding of blade 7 or alter the packing of blades 7 and 1. The GBD-chc1-4 TD fusion exhibited a slower mobility on gels, suggesting there is some structural alteration in this TD.
The chc1-4 allele is unusual among the ts alleles in that clathrin recruitment to the cortex in LAT-A was relatively strong at 25°C, and the percentages of clathrin patch types seen by TIRFM were most similar to wild-type. Thus, for endocytosis at least, the chc1-4 allele has properties of a true ts allele, and its unique properties will make it interesting for further study.
chc1-box Allele Mutations and Clathrin Function
HC expression from the chc1-box allele was expected to produce significant phenotypes in clathrin-dependent processes as the mutations introduced in this allele have been shown to disrupt interaction with adaptor proteins in in vitro binding assays (Goodman et al., 1997; Miele et al., 2004). However, chc1-box cells showed robust growth, TGN/endosomal sorting (α factor processing and calcofluor sensitivity), Ape1 processing (an indicator of Cvt/autophagy function), and α factor internalization. HCs encoded by chc1-box were also able to form clathrin-coated vesicles. In more refined analysis, we did notice partial impairment of clathrin recruitment to cortical sites in LAT-A, but compared with chc1-1, chc1-2, and chc1-3, as well as chc1-4 at 37°C, this recruitment was quite strong and was not temperature-dependent. Also the TIRFM showed these cells have a higher percentage of immobile clathrin patches and unproductive patches that disappear without acquiring Abp1 and completing internalization than CHC1 or chc1-4 cells. Nevertheless, although more events with these altered dynamics were observed, the box mutant still internalized α factor at near wild-type rates. Together, these data indicate that the critical residues of the TD that constitute the known binding site for CBM and W-box motif containing adaptors are dispensable for clathrin function in yeast.
One possible explanation of these data is that other clathrin binding motifs in adaptor proteins might be able to compensate for the inability of the CBM to associate with the TD. Multiple copies of a short peptide motif ([D/S]LL) were identified in AP180, a synapse-specific adaptor protein that is critical for synaptic vesicle endocytosis, and in the large subunits of heterotetrameric adaptors AP-1, AP-2, AP-3, and AP-4 (Morgan et al., 2000). The dileucine sequence of the (D/S)LL motif may participate in hydrophobic interactions with other grooves of the TD β-propeller, in which the hydrophobic nature of each groove is conserved (ter Haar et al., 1998); however, the exact interaction site(s) for this motif have not been defined. Many yeast adaptor proteins, including the large β and γ subunits of AP-1 (Apl2 and Apl4, respectively), possess one or more copies of the (D/S)LL sequence, and each motif might have different groove specificities because there is greater degeneracy in the surrounding sequence compared with the CBM.
Alternatively, a recently identified binding site within the ankle region of HC for appendage domains from three subunits of clathrin adaptor protein complexes (tetrameric adaptor subunits AP-1γ, AP-2α, and AP-2β) and the appendage domain from the monomeric adaptor protein Gga1 (Knuehl et al., 2006) could compensate for the disrupted TD-binding site and facilitate clathrin function. Modeling and mutagenesis studies indicated that the HC ankle–appendage interaction, unlike clathrin box and W-box association with the TD, occurred over an extended interface, and two surface residues within the HC ankle (C682 and G710) were essential in the appendage–HC ankle interaction (Knuehl et al., 2006). Sequence alignment of the ankle domain regions from yeast and mammalian HC shows that G710 is present in the yeast HC ankle (residue G716), whereas C682 is not conserved in the yeast sequence. This interaction could be important for some adaptors, suggested by the differences in interaction of Ent1 and Apl2 compared with Ent2 with our TD fragment that lacked the ankle region.
We favor the idea that these chc1-box allele data could be explained by an additional binding site(s) within the yeast TD that facilitates CBM-mediated adaptor binding. A TD fragment containing the chc1-box mutations was able to interact with Ent2, but this interaction was not nearly as strong as the interaction observed between the wild-type TD and Ent2. Interaction of both the wild-type TD and TD(box) domains with Ent2 was dependent upon the C-terminal CBM of Ent2, suggesting that one or more other sites on the TD, possibly other grooves between the propeller blades, are being used by the Ent2 CBM for its association: 1) a higher affinity interaction with the blade 1/2 groove and 2) a much lower affinity interaction with a second TD binding site. TD fragments with chc1-box mutations did not interact with either Ent1 or Apl2, but because the strength of the interactions between the wild-type TD and these two adaptors was much weaker when compared with the TD-Ent2 interaction, it is possible that utilization of a second TD binding site in the TD(chc1-box) fragment was not strong enough to detect using this assay. Alternatively, we cannot exclude that the blade 1/2 interaction was not completely ablated by the chc1-box mutations.
HCs encoded by the chc1-box allele were able to form CCVs, but these were not enriched for the AP-1 adaptor. Unlike mammalian AP-1 β, yeast Apl2 lacks the β-appendage domain and could not interact with the HC ankle binding site even if present in the yeast HC. Yeast Apl4, the AP-1 γ chain, does contain a relatively conserved γ-appendage domain that could mediate binding to the HC ankle (Hirst et al., 2000; Knuehl et al., 2006). However, this potential interaction must not be sufficient for stable incorporation of AP-1 into CCV in the box mutant. Likewise, if the TDs containing chc1-box mutations only possess a lower affinity binding site for adaptor-associated CBMs, this low-affinity interaction between AP-1 and clathrin was easily disrupted during the cell fractionation procedure. In previous studies, AP-1 incorporation into CCVs was almost abolished in cells containing mutations to the two CBM sequences in the β-subunit (Apl2) and a C-terminal deletion of the clathrin-binding region in the γ-subunit (Apl4) (Yeung and Payne, 2001). Thus, the reduced AP-1 association with CCVs isolated from chc1-box cells is consistent with these observations and suggests that stable incorporation of AP-1 into CCVs is mediated by the interaction of CBM-containing AP-1 subunits with the TD. Nevertheless, AP-1 interaction with clathrin seems to operate in vivo to abolish calcofluor sensitivity in the box mutant, indicating that the AP-1/clathrin-dependent, early endosome-to-TGN pathway used for the intracellular retention of Chs3 was functional in these cells. Likely, the AP-1 interaction occurs in the context of other coat components that might stabilize or augment its function during cargo selection steps and membrane invagination. In general, the clathrin box mutant analysis presented here provides additional support for the prevailing view that clathrin-mediated function is facilitated by the cooperative interactions of multiple accessory factors. Further work will be directed toward identification of additional HC adaptor CBM binding sites that are predicted from our studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Randy Schekman for providing the chs6Δ strain, Dan Klionsky for providing aminopeptidase 1 antiserum, and Onaidy T. Torres for technical assistance. J.R.C. was supported by National Institutes of Health training grant T32-HL07188 and American Heart Association postdoctoral fellowship 6-6253Y. R.J.C. was supported by National Institutes of Health training grant T32-HL07188. D.R.B. was supported by National Institutes of Health National Research Service Award fellowship F32-GM084677. This work was funded by a Ministerio de Educación y Ciencia grant BUF2005-04089 (to M. G.) and by National Institutes of Health grants DK-53249 (to L. T.) and GM-084677 (to S.K.L.).
Abbreviations used:
- 5-FOA
5-fluoroorotic acid
- AP
heterotetrameric adaptor protein complex
- Ape1
aminopeptidase I
- CBM
clathrin-box motif
- CCV
clathrin-coated vesicle
- Cvt
cytoplasm-to-vacuole transport
- HC
heavy chain
- LAT-A
latrunculin-A
- LC
light chain
- PM
plasma membrane
- TD
N-terminal domain
- TGN
trans-Golgi network
- ts
temperature sensitive
- TIRFM
total internal reflection fluorescence microscopy.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-10-1082) on May 20, 2009.
REFERENCES
- Baggett J. J., D'Aquino K. E., Wendland B. The Sla2p talin domain plays a role in endocytosis in Saccharomyces cerevisiae. Genetics. 2003;165:1661–1674. doi: 10.1093/genetics/165.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel H. G., Kawabata S., Iwanaga S., Huber R., Bode W. Tachylectin-2, crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C., Wakeham D. E. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Bumbulis M. J., Wroblewski G., McKean D., Setzer D. R. Genetic analysis of Xenopus transcription factor IIIA. J. Mol. Biol. 1998;284:1307–1322. doi: 10.1006/jmbi.1998.2285. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Graham T. R. An arf1Delta synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics. 1998;150:577–589. doi: 10.1093/genetics/150.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta G., Stefan C. J., Bensen E. S., Emr S. D., Payne G. S. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica E. C. Clathrin-binding proteins: got a motif? Join the network! Trends Cell Biol. 2001;11:315–318. doi: 10.1016/s0962-8924(01)02043-8. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Klumperman J., Stoorvogel W., Bonifacino J. S. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Drake M. T., Traub L. M. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J. Biol. Chem. 2001;276:28700–28709. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- Dulic V., Egerton M., Elguindi I., Raths S., Singer B., Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Duncan M. C., Costaguta G., Payne G. S. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M. L., Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Eugster A., Pecheur E. I., Michel F., Winsor B., Letourneur F., Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Friant S., Pecheur E. I., Eugster A., Michel F., Lefkir Y., Nourrisson D., Letourneur F. Ent3p Is a PtdIns(3,5)P2 effector required for protein sorting to the multivesicular body. Dev. Cell. 2003;5:499–511. doi: 10.1016/s1534-5807(03)00238-7. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Goodman O. B., Jr, Krupnick J. G., Gurevich V. V., Benovic J. L., Keen J. H. Arrestin/clathrin interaction. Localization of the arrestin binding locus to the clathrin terminal domain. J. Biol. Chem. 1997;272:15017–15022. doi: 10.1074/jbc.272.23.15017. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Harding T. M., Morano K. A., Scott S. V., Klionsky D. J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. M., D'Hondt K., Riezman H., Lemmon S. K. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 1999;18:3897–3908. doi: 10.1093/emboj/18.14.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. M., Gullberg L., Nelson K. K., Stefan C. J., Blumer K., Lemmon S. K. Novel functions of clathrin light chains: clathrin heavy chain trimerization is defective in light chain-deficient yeast. J. Cell Sci. 1997;110:899–910. doi: 10.1242/jcs.110.7.899. [DOI] [PubMed] [Google Scholar]
- Hudson J. R., Jr, Dawson E. P., Rushing K. L., Jackson C. H., Lockshon D., Conover D., Lanciault C., Harris J. R., Simmons S. J., Rothstein R., Fields S. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 1997;7:1169–1173. doi: 10.1101/gr.7.12.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D. G. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Khalfan W. A., Klionsky D. J. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr. Opin. Cell Biol. 2002;14:468–475. doi: 10.1016/s0955-0674(02)00343-5. [DOI] [PubMed] [Google Scholar]
- Knuehl C., Chen C., Manalo V., Hwang P. K., Ota N., Brodsky F. M. Novel binding sites on clathrin and adaptors regulate distinct aspects of coat assembly. Traffic. 2006;7:1688–1700. doi: 10.1111/j.1600-0854.2006.00499.x. [DOI] [PubMed] [Google Scholar]
- Lemmon S., Lemmon V. P., Jones E. W. Characterization of yeast clathrin and anticlathrin heavy-chain monoclonal antibodies. J. Cell Biochem. 1988;36:329–340. doi: 10.1002/jcb.240360403. [DOI] [PubMed] [Google Scholar]
- Lemmon S. K., Jones E. W. Clathrin requirement for normal growth of yeast. Science. 1987;238:504–509. doi: 10.1126/science.3116672. [DOI] [PubMed] [Google Scholar]
- Lemmon S. K., Pellicena-Palle A., Conley K., Freund C. L. Sequence of the clathrin heavy chain from Saccharomyces cerevisiae and requirement of the COOH terminus for clathrin function. J. Cell Biol. 1991;112:65–80. doi: 10.1083/jcb.112.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke D., Mettlen M., Yarar D., Jaqaman K., Jaqaman H., Danuser G., Schmid S. L. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R., Carlsson S. R. Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J. Biol. Chem. 2003;278:46772–46781. doi: 10.1074/jbc.M307334200. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L., Dores M. R., Perkins E. M., Drivas T. G., Hicke L., Wendland B. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol. Biol. Cell. 2008;19:2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A. E., Watson P. J., Evans P. R., Traub L. M., Owen D. J. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat. Struct. Mol. Biol. 2004;11:242–248. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- Morgan J. R., Prasad K., Hao W., Augustine G. J., Lafer E. M. A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J. Neurosci. 2000;20:8667–8676. doi: 10.1523/JNEUROSCI.20-23-08667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz H. S., Yokoyama C. T., Ryan T. A. Highly cooperative control of endocytosis by clathrin. Mol. Biol. Cell. 2005;16:1769–1776. doi: 10.1091/mbc.E04-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C., Bonifacino J. S. Structural requirements for function of yeast GGAs in vacuolar protein sorting, alpha-factor maturation, and interactions with clathrin. Mol. Cell Biol. 2001;21:7981–7994. doi: 10.1128/MCB.21.23.7981-7994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A. L., Silveira L, Elgort M., Payne G. S. Viability of clathrin heavy-chain-deficient Saccharomyces cerevisiae is compromised by mutations at numerous loci–implications for the suppression hypothesis. Mol. Cell Biol. 1991;11:3868–3878. doi: 10.1128/mcb.11.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Lemmon S. K. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic. 2006;7:574–588. doi: 10.1111/j.1600-0854.2006.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T. M., Smith R. P., Lemmon V., Lemmon S. K. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- Pishvaee B., Munn A., Payne G. S. A novel structural model for regulation of clathrin function. EMBO J. 1997;16:2227–2239. doi: 10.1093/emboj/16.9.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjaun A. R., McPherson P. S. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J. Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- Santos B., Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M., Payne G. S. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J. Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp J. D., Pellicena-Palle A., Hamilton S., Kirchhausen T., Lemmon S. K. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol. Biol. Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P. K., Davis N. G., Sprague G. F., Payne G. S. Clathrin facilitates the internalization of seven transmembrane segment receptors for mating pheromones in yeast. J. Cell Biol. 1993;123:1707–1716. doi: 10.1083/jcb.123.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E., Harrison S. C., Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc. Natl. Acad. Sci. USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E., Musacchio A., Harrison S. C., Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. H., Baggott D., Chuang J. S., Schekman R. W. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Yeung B. G., Payne G. S. Clathrin interactions with C-terminal regions of the yeast AP-1 beta and gamma subunits are important for AP-1 association with clathrin coats. Traffic. 2001;2:565–576. doi: 10.1034/j.1600-0854.2001.20806.x. [DOI] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Tsung M., Hamamoto S., Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.