Abstract
Endoplasmic reticulum (ER) quality control mechanisms monitor the folding of nascent polypeptides of the secretory pathway. These are dynamic processes that retain folding proteins, promote the transport of conformationally mature proteins, and target misfolded proteins to ER-associated degradation (ERAD) pathways. Aided by the identification of numerous ERAD factors, late functions that include substrate extraction, ubiquitination, and degradation are fairly well described. By contrast, the mechanisms of substrate recognition remain mysterious. For some substrates, a specific N-linked glycan forms part of the recognition code but how it is read is incompletely understood. In this study, systematic analysis of model substrates revealed such glycans mark structural determinants that are sensitive to the overall folding state of the molecule. This strategy effectively generates intrinsic folding sensors that communicate with high fidelity to ERAD. Normally, these segments fold into the mature structure to pass the ERAD checkpoint. However, should a molecule fail to fold completely, they form a bipartite signal that comprises the unfolded local structure and adjacent enzymatically remodeled glycan. Only if both elements are present will the substrate be targeted to the ERAD pathway for degradation.
INTRODUCTION
Quality control pathways monitor protein folding and assembly throughout the cell. Although generally poorly understood, the best-described systems are those of the endoplasmic reticulum (ER). ER quality control (ERQC) describes an assortment of mechanisms that, collectively, retains unfolded or unassembled proteins and targets irreversibly misfolded proteins for destruction through the appropriate ER-associated degradation (ERAD) pathway.
Newly synthesized secretory and membrane proteins translocate across the ER membrane through the Sec61 translocon pore. The narrow pore size restricts passage to unfolded proteins so factors needed for their maturation reside in the ER (Rapoport, 2007). For most molecules, ER quality control serves to prevent their transport during the folding process. Once folding is complete, intrinsic export signals guide their transport via cargo sorting receptors concentrated at and in the membranes of ER exit sites marked by COPII coat proteins (Sato and Nakano, 2007).
Proteins that fail to fold are targeted to ERAD for turnover. Metazoans maintain multiple ERAD pathways with the total number unknown (Nakatsukasa and Brodsky, 2008). The best understood is linked to the calnexin/calreticulin folding cycle (for review, see Helenius and Aebi, 2004). Newly synthesized glycoproteins bind calnexin or calreticulin via mono-glucosylated N-glycans. The interaction assists in the folding process by preventing aggregation and by engaging other factors. Proteins failing to fold are directed to ERAD via the action of the ER degradation-enhancing α-mannosidase-like protein (EDEM; Molinari et al., 2003; Oda et al., 2003). Recently, EDEM1 and EDEM3 overexpression was shown to accelerate the demannosylation of substrates, and this effect is blocked by conserved catalytic mutations (Hirao et al., 2006; Olivari et al., 2006). In the next step, the substrate engages the Hrd1/SEL1L E3 ligase complex via the lectin-like proteins OS-9 and XTP3-B (Christianson et al., 2008; Hosokawa et al., 2008). Substrates then dislocate to the cytosol by Derlin-1 and the p97 complex (orthologues of the yeast Der1p and Cdc48p, respectively) where they are ubiquitinated and degraded by the 26S proteasome (Ye et al., 2001; Elkabetz et al., 2004; Lilley and Ploegh, 2004; Ye et al., 2004). Unassembled immunoglobulins lacking glycans can also be degraded via the Hrd1 complex (Okuda-Shimizu and Hendershot, 2007). It is unknown how these molecules are recognized but OS-9 and EDEM proteins presumably would not participate.
The framework of glycan-dependent luminal ERAD (termed ERAD-L) in budding yeast is conserved except for lack of a canonical calnexin/calreticulin cycle. Like its mammalian counterpart, the pathway is centered on the Hrd1p E3 ligase, which organizes a complex of membrane-associated factors that process substrates for degradation by the 26S proteasome (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006a). Most immediate is Hrd3p (orthologue of SEL1L), whose large luminal domain can directly engage substrates (Gardner et al., 2000; Denic et al., 2006; Gauss et al., 2006b). Next, a “proof-reading” function by its partner Yos9p (Yeast OS-9) is needed to advance the substrate for retro-translocation. Yos9p is a lectin that binds carbohydrates through its mannose 6-phosphate homology domain (Buschhorn et al., 2004; Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005; Denic et al., 2006). The membrane protein Der1p is linked to the complex via Usa1p and assists in substrate export (Knop et al., 1996a; Carvalho et al., 2006). The ubiquitin conjugating enzyme Ubc7p is the ubiquitination partner of Hrd1p (linked via Cue1p; Biederer et al., 1997), and the Cdc48 subcomplex (composed of Cdc48p, Ufd1p, Npl4p, and Ubx2p) mediates substrate extraction from ER membranes (Bays et al., 2001; Ye et al., 2001; Jarosch et al., 2002; Elkabetz et al., 2004).
For substrate recognition in the ER lumen, the structure of carbohydrate chains is essential for a subset of proteins. The enzymes glucosidase I, glucosidase II, and α-mannosidase I sequentially process the core GlcNAc2Man9Glc3 glycan to the GlcNAc2Man8 structure needed for efficient ERAD of the model substrate CPY* (Jakob et al., 1998; Hitt and Wolf, 2004). The relatively slow kinetics of α-mannosidase I activity prompted investigators to propose the “mannose timer” hypothesis. Simply put, proteins that do not fold within the enzymatically defined time window are processed for ERAD (Jakob et al., 2001). Despite the obvious appeal of the mechanism, it remained unclear how the GlcNAc2Man8 glycan signals ERAD when folded glycoproteins share the same structure. Recently, a subsequent and key processing step was proposed and confirmed experimentally (Hirao et al., 2006; Olivari et al., 2006; Clerc et al., 2009). The Htm1 protein (EDEM in mammals) cleaves a single mannose residue from the C-arm of the GlcNAc2Man8 glycan to expose a terminal α1,6-linked mannose. Remarkably, this moiety turns out to be the ligand for the substrate “proof-reading” factor Yos9p (Quan et al., 2008). Even with these important revelations, the mechanism was incomplete. Analyses of the model substrates CPY* and PrA* demonstrated that only single, specific glycans can signal ERAD (Kostova and Wolf, 2005; Spear and Ng, 2005). This suggested the requirement of additional determinants in the polypeptide chain. To understand how Htm1p-dependent substrates are recognized, it is necessary to fully decipher the nature of the determinant.
MATERIALS AND METHODS
Strains and Antibodies
Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table S1. Anti-hemagglutinin (HA) mAb (HA.11) was purchased from Covance Research Products (Princeton, NJ), anti-HA–conjugated agarose (sc-7392) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), anti-Kar2p and anti-Sec61p rabbit polyclonal antisera were gifts of Peter Walter (University of California, San Francisco, San Francisco, CA). Anti-CPY antibody was a generous gift of Reid Gilmore (University of Massachusetts, Worcester, MA). Anti-protein disulfide isomerase (PDI) antiserum was kindly provided by Karin Römisch (Saarland University, Germany).
Plasmids Used in This Study
Plasmids are listed in Supplementary Table S2 (Supplementary Materials). Plasmids were constructed using standard cloning protocols. All ERAD substrate constructs were confirmed by nucleotide sequencing over the entire length of their open reading frames. Unless otherwise noted in the text, ERAD substrates contained HA epitope tags at their C-terminus. Amino acid positions are designated using translation initiator methionines as position 1. Details of plasmid constructions are described in Supplementary Materials.
Procedures for ERAD Substrate Analyses
Metabolic pulse-chase, cycloheximide chase, and preparation of cells for indirect immunofluorescence analyses were performed as described previously (Vashist et al., 2001). Imaging of cells in this study was performed using polyclonal rabbit anti-Kar2p and HA.11 mAb (Covance) as primary antibodies and Alexa Fluor 488 goat anti-mouse and Alexa Fluor 594 goat anti-rabbit (Invitrogen, Carlsbad, CA) as secondary antibodies. Cells were visualized using Zeiss LSM 510 META inverted microscope (Carl Zeiss MicroImaging, Thornwood, NY) with a Plan-Apochromat 100× Ph3 objective (1.4 NA; Carl Zeiss MicroImaging). Image acquisition was performed using standard PMT with LSM 510 (Carl Zeiss MicroImaging). Images were archived using LSM 5 Image Examiner (Carl Zeiss MicroImaging) and Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). No additional software adjustments were performed on images after acquisition. In pulse-chase experiments, quantification of labeled proteins in polyacrylamide gels was performed using a Typhoon phorphorimager (GE Healthcare Biosciences, Piscataway, NJ). Data points and error bars on graphs represent the SEM of three independent experiments.
Microsome Preparation and Native Coimmunoprecipitation Assay
Strains were grown to log phase in synthetic complete media lacking the appropriate component for plasmid selection. 50 OD600 equivalents of cell culture were harvested by centrifugation (3,000 × g, 5 min), washed twice in ice-cold water, and resuspended in 1 ml TN buffer (50 mM Tris, 50 mM NaCl, pH 7.4) containing 2 mM PMSF and 1.5% Protease Inhibitor Cocktail (P8215, Sigma, St. Louis, MO). Cell disruption was performed by bead beating (0.5-mm zirconium) on a vortex mixer 5 × 30 s, with 5 min on ice between each step. The supernatant was collected and pooled with 2× bead-wash with 1 ml TN buffer. The supernatant from a low speed spin (800 × g, 5 min) was subject to ultracentrifugation (30,000 × g, 30 min) to collect microsomes. The pellet was solubilized by adding 0.5 ml Tris-IP buffer (TN buffer containing 1% Triton X-100, 15% glycerol, and 2 mM PMSF) and incubated at 4°C for 1 h. The lysate was clarified by centrifugation (30,000 × g, 10 min) and a 20 μl portion was combined 500 μl of Tris-IP buffer, 5 μl anti-HA resin, and incubated at 4°C with rocking for 2 h. The beads were washed gently three times in ice-cold TN buffer containing 1% Triton X-100 and once in ice-cold TN buffer. Proteins were eluted from the beads by boiling in SDS-sample buffer without DTT for 5 min. Supernatants were transferred to a fresh tube, and DTT was added to 100 mM and boiled for another 5 min. Proteins were detected on immunoblots using anti-Kar2p or anti-HA antibody and visualized by enhanced chemiluminescence according to manufacturer's protocols (SuperSignal West Pico substrate, Pierce Biotechnology, Rockford, IL).
Preparation of Proteins for Mass Spectrometry
Strains were grown to late-log phase in synthetic complete media lacking the appropriate component for plasmid selection. Four thousand OD600 equivalents of cell culture were harvested by low-speed centrifugation (3000 × g, 10 min) and washed once in ice-cold water. Cells were resuspended in 100 ml spheroplasting buffer (10 mM KH2PO4, 40 mM K2HPO4, 1.4 M sorbitol, pH 7.5) containing 40 mg zymolyase 20T and incubated at room temperature for 90 min. Spheroplasts were collected by centrifugation (3000 × g, 5 min) and resuspended in 200 ml ice-cold TN buffer containing 1 mM PMSF. Spheroplasts were homogenized on ice in a Sartorius Potter S motorized homogenizer (1300 rpm, 15 strokes). The supernatant from a low-speed spin was subjected to ultracentrifugation (30,000 × g, 30 min) to pellet microsomes. Microsomes were solubilized by incubation in 20 ml of Tris-IP buffer at 4°C for 1 h, and insoluble material was removed by centrifugation (30,000 × g, 10 min). Fifty microliters of anti-HA–conjugated agarose beads were added to the supernatant and incubated rocked for 2 h at 4°C. The beads were washed gently three times in Tris-IP buffer and once in TN buffer. Proteins were eluted from the beads by boiling in nonreducing SDS loading buffer for 5 min. The supernatant was transferred to another tube, boiled for another 5 min in 100 mM DTT, resolved by SDS-PAGE, and visualized by Coomassie Blue staining. Proteins bands were excised from the gel and analyzed by MALDI-MS/MS Mass Spectrometry (National University of Singapore Proteins and Proteomics Centre). Tryptic fragments were analyzed by the 4800 MALDI TOF/TOF Analyzer (Applied Biosystems, Foster City, CA). Data were processed using Data Explorer v 4.9 (Applied Biosystems) and an S/N ratio of 10 was applied to the MS/MS mode for peak identification. Peptides were identified using MASCOT v 2.1 (Matrix Science, London, United Kingdom) as a search engine in NCBInr Protein Database.
Protease Sensitivity Assay
A microsome fraction was prepared as described above but in the absence of protease inhibitors (50 OD600 scale) and solubilized in 0.5 ml Tris-IP buffer lacking protease inhibitors. Ten microliters of the lysate was combined with 90 μl of ice-cold PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) containing freshly added trypsin at 5.0 μg/ml. After incubation at 4°C for times indicated in figures, 11.1 μl 100% TCA was added to the solution to stop the reaction. Proteins were precipitated by centrifugation (18,000 × g, 20 min), resuspended in 10 μl TCA resuspension solution (100 mM Tris, 3% SDS, pH 11.0), and boiled for 10 min. Samples were mixed with 10 μl 2× SDS-PAGE loading buffer with DTT and boiled for another 5 min, followed by immunoblotting.
RESULTS
A Bipartite Signal Targets Misfolded Glycoproteins to ERAD
CPY* and PrA* are well-studied model Htm1p-dependent ERAD substrates with known crystal structures for their folded counterparts carboxypeptidase Y (CPY) and proteinase A (PrA; Finger et al., 1993; Endrizzi et al., 1994; Aguilar et al., 1997). The prc1-1 (CPY*) allele contains a missense mutation that results in the G255R change at the core of the folded protein. PrA* differs from wild type by a short deletion near its N-terminus. These simple lesions cause complete folding failures that expose signals recognized by ERAD (Finger et al., 1993). Of what is currently known, only the C-terminal glycan of CPY* is required (Kostova and Wolf, 2005; Spear and Ng, 2005). The other three do not contribute. Similarly, PrA* relies on a single glycan, but near its N-terminus (Spear and Ng, 2005). The context specificity suggested that additional determinants embedded within the polypeptide chains, when unfolded, contribute to the recognition code. Short sequence-based signals or “degrons” have been identified for the ER-associated degradation of the folded proteins IgM heavy chain and cyclooxygenase (Mbonye et al., 2006; Shapira et al., 2007). By extension, if similar elements work with the glycan signals, disrupting them should destroy the signal.
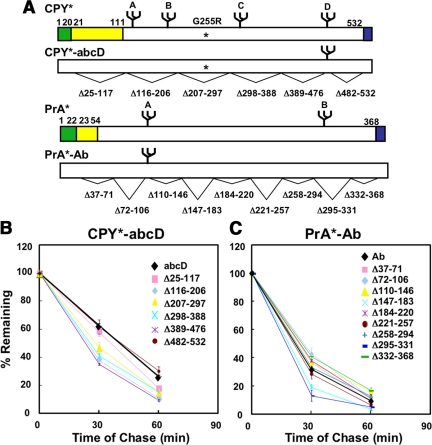
To test for degron-like elements, systematic deletion analysis was performed on CPY* and PrA*. The test molecules carry only their respective ERAD signal glycans (designated CPY*-abcD and PrA*-Ab, respectively) so that any effects can be attributed to them specifically. As shown in Figure 1, none of the variants are impaired for ERAD compared with full-length controls. Indeed, the turnover rates for most deletion constructs are slightly increased. This effect is likely attributable to their reduced sizes compared with control. These data suggest ERAD determinants do not integrate sequence-specific elements that would explain the stringent positional effects. The result is not entirely surprising since it has been proposed that a conformational sensor might needed in addition to the glycan timer to differentiate ER resident glycoproteins (Helenius and Aebi, 2004). We next queried whether the determinant might have a structural basis. For this, the three-dimensional structure of mature CPY was surveyed for possible clues.
Figure 1.
Deletion variants of CPY* and PrA* are degraded efficiently in wild-type cells. (A) Schematic representation of variants showing positions of individual deletions under each diagram. Amino acid numbers are ordered with the initiator methionine designated as “1.” CPY* and PrA* deletion variants in this series carry only their respective signal glycan as indicated by the extensions -abcD and -Ab, respectively (upper case signifies a functional glycosylation site and lower case a mutated site; see Figure 2 for the positions of each glycan). An asterisk indicates the position of the CPY* G255R mutation. PrA* lacks the sequences Leu55 through Tyr91 of PrA (Finger et al., 1993). Signal sequences are shaded green, prodomains shaded yellow, and HA-epitope tags shaded blue. Amino acid numbers in PrA* variants are based on their position on PrA*, which lacks the Leu55 to Tyr91 segment of PrA (see Figure 2C). Wild-type cells expressing CPY*-abcD variants (B) or PrA*-Ab variants (C) were pulse labeled for 10 min and chased for the times indicated. Proteins were immunoprecipitated using monoclonal HA antibody from detergent lysates and resolved by SDS-PAGE. Turnover rates were quantified by phosphorimager analysis. The data plotted reflect three independent experiments and the SEM.
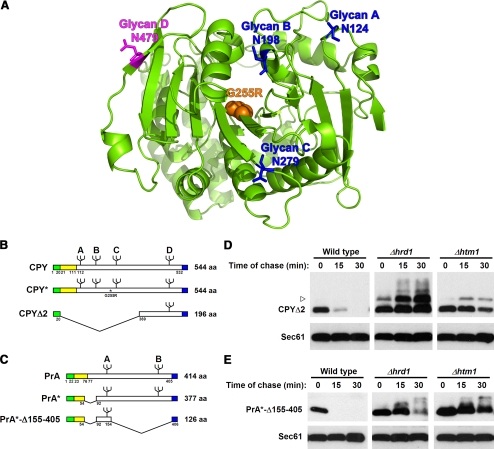
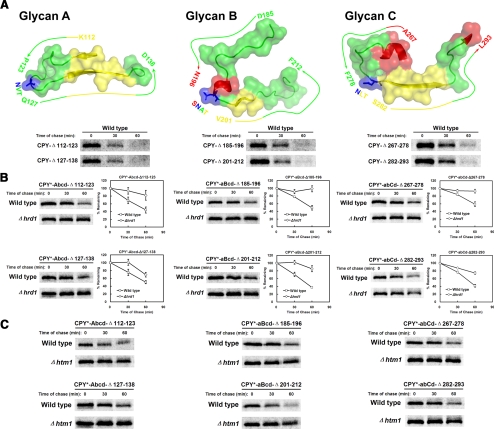
Depicted in Figure 2A is a ribbon representation of the CPY crystal structure with the positions of glycan asparagines indicated (Endrizzi et al., 1994). Notably, the D “signal” glycan is positioned on the end of an 11-stranded β-sheet, arranged mostly in parallel. Because of this configuration, stability of the β-sheet and vis-à-vis, the local environment of the D glycan, can be sensitive to the overall packing of the polypeptide. Thus, both short- and long-range folding failures could affect the correct ordering of the segment. The A, B, and C glycans, by contrast, are located on surface loops that form fewer contacts with other parts of the polypeptide. Because unfolded proteins can contain extensive secondary structure (Dyson and Wright, 2004), simple, ordered structures, if formed around these glycans in CPY*, may explain their failed recognition by ERAD.
Figure 2.
Signal glycans and adjacent peptide segments are sufficient to signal ERAD. (A) Ribbon diagram of mature CPY. The positions of glycans A, B, and C are shown in blue, and glycan D is shown in purple. Orange spheres mark Gly255, which is mutated to Arg in CPY*. Amino acid numbers are ordered with the initiator methionine at position 1. The diagram was generated using the PyMOL 0.99rc6 software (Delano Scientific, Palo Alto, CA) with coordinates from Endrizzi et al. (1994). (B) Schematic representation of preproCPY, CPY*, CPYΔ1, and CPYΔ2. Carbohydrates are represented by branch symbols. The asterisk indicates the CPY* G255R mutation. Signal sequences are shaded green, prodomains in yellow, and HA-epitope tags in blue. (C) Schematic representation of preproPrA, PrA*, and PrA*-Δ155-405. Carbohydrates are represented by branched symbols; A and B indicate Asn144 and Asn345, respectively. Shadings indicate the same elements as in B. (D) Wild-type, Δhrd1, or Δhtm1 cells expressing CPYΔ2 were grown to log phase, and cycloheximide was added to begin the chase for times shown. Whole-cell lysates were prepared, and proteins were resolved by SDS-PAGE. Substrates were detected on immunoblots using the anti-HA mAb (top panels). Blots were reprobed with anti-Sec61p antisera as a loading control (bottom panels). The arrowhead marks the position of a higher molecular weight form of CPYΔ2 that accumulates in the ERAD mutants. This form is most likely due to O-mannosylation, previously observed for other substrates (Harty et al., 2001; Vashist et al., 2001). (E) Turnover of PrA*-Δ155-405 in wild-type, Δhrd1, and Δhtm1 cells was analyzed as described in D.
To begin testing our hypothesis, we wanted to determine whether peptide segments directly adjacent to signal glycans combine to form good substrates for ERAD. For this, mini-CPY and -PrA variants were constructed consisting only of the respective signal glycans and immediate peptide segments. The N-terminal 348 residues of proCPY were deleted to create CPYΔ2 (a 176-residue glycoprotein after signal sequence cleavage), and the C-terminal 251 residues of PrA* were deleted to create PrA*-Δ155-405 (a 104-residue glycoprotein after signal sequence cleavage; Figure 2, B and C). Cycloheximide chase experiments were performed to measure their turnover in wild-type and ERAD-L–deficient strains. Both variants degraded rapidly in wild-type cells and strongly stabilized in Δhrd1 and Δhtm1 strains (Figure 2, D and E). These mutants test for requirements of the Hrd1p complex and glycan remodeling functions of ERAD, respectively (Bordallo et al., 1998; Gardner et al., 2001; Carvalho et al., 2006; Clerc et al., 2009). These data demonstrate that luminal ERAD requires only the glycan and their local segments for recognition. Importantly, CPYΔ2, which lacks the G255R “star” mutation, shows that the loss of tertiary interactions through the large deletion is sufficient to activate the D glycan domain to function as an ERAD determinant. By contrast, the reciprocal variant that eliminates the D glycan region, called CPYΔ1, renders a misfolded glycoprotein retained by ERQC but not recognized by ERAD (Spear and Ng, 2005). Interestingly, the CPY C-terminal domain is sufficient to target nonsubstrate molecules to ERAD. When appended to dihydrofolate reductase, the Hrd1 pathway degrades the fusion protein in a glycan-dependent manner (Carvalho and Rapoport, personal communication).
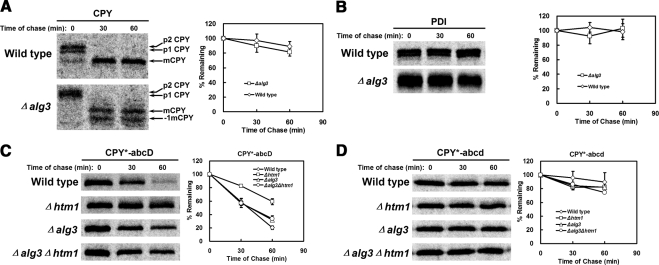
Aebi and coworkers demonstrate that the GlcNAc2Man7 structure generated by Htm1p is needed to trigger ERAD in budding yeast (see Figure 8; Clerc et al., 2009). The Yos9p receptor recognizes the terminal α1,6-mannose residue found in this structure (Quan et al., 2008). Other glycan structures containing the moiety (e.g., the N-linked GlcNAc2Man5 glycans in a Δalg3 strain) can also signal ERAD (Jakob et al., 1998; Clerc et al., 2009). The requirement for an unfolded local segment in our study raises the possibility that it combines with the glycan to form a bipartite signal read by ERAD. Alternatively, it could act upstream to recruit Htm1p to modify adjacent glycans, and it is this glycan structure alone that mark proteins for destruction. To distinguish between these possibilities, we compared the stability of folded endogenous proteins in wild-type and Δalg3 strains. Because all N-linked glycans in Δalg3 cells contain the terminal α1,6-mannose carbohydrate, all glycoproteins would be subject to ERAD should the glycan act alone as the signal. As shown in Figure 3, A and B, wild-type CPY and protein disulfide isomerase (PDI) are as stable in the Δalg3 strain as wild type. PDI is particularly notable because it is an ER glycoprotein continuously exposed to the ERAD machinery. As a control, the HTM1 requirement for CPY* degradation is bypassed in the Δalg3 background, which shows the efficacy of the artificial signal (Figure 3C). As importantly, this control demonstrates that ALG3 deletion did not disrupt other aspects of ERAD function. The effect is also restricted to misfolded glycoproteins since the turnover of a nonglycosylated form of CPY* is neither accelerated in a Δalg3 strain nor further stabilized in a Δhtm1 mutant (Figure 3D). These data show that signal glycans require the presence of adjacent unfolded peptide segments for recognition by ERAD.
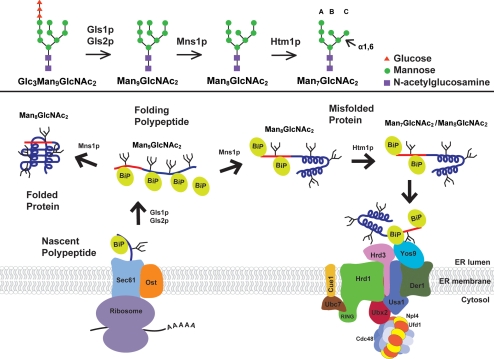
Figure 8.
Model of substrate recognition in luminal ERAD. Glycan processing by Gls1p, Gls2p, Mns1p, and Htm1p are shown in the top panel (Clerc et al., 2009). Bottom panel, polypeptide synthesis begins at membrane-bound ribosomes with translocation into the lumen via the Sec61 pore complex. The posttranslational translocation mechanism is not shown for simplicity. The substrate is N-glycosylated by the associated oligosaccharyl transferase (OST), and glycans are depicted as branched structures. Folding proceeds as chaperones (labeled “BiP”; other factors not shown for simplicity) engage the molecule. Molecules that correctly fold integrate ERAD determinants (red line) into the structure, silencing the signal and releasing BiP to allow export (left). Proteins that fail to fold are modified by Htm1p forming the glycan portion of the signal. BiP complexes bound to the ERAD determinant target to the Hrd1 complex at the ER membrane (right). Yos9p scans the substrate for the α1,6-mannose GlcNAc2Man7 structure. Molecules displaying the signal are retrotranslocated to the cytosol, ubiquitinated, and degraded by the 26S proteasome (not shown).
Figure 3.
Glycan structure alone is not sufficient for ERAD substrate recognition. (A and B) Wild-type and Δalg3 cells were pulse-labeled for 10 min and chased for times indicated. Proteins were immunoprecipitated from detergent lysates using polyclonal antisera and resolved by SDS-PAGE. Protein turnover was quantified by phosphorimager analysis. (A) Relative CPY turnover is plotted with representative phosphorimages shown. The Δalg3 mutant exhibits minor underglycosylation of proteins (Jakob et al., 1998). The positions of mature CPY bearing four glycans (mCPY) and three glycans (−1 mCPY) are indicated. (B) PDI stability was analyzed as described in A. (C and D) Turnover of CPY* variants were compared in wild-type, Δhtm1, Δalg3, and Δalg3Δhtm1 strains by metabolic pulse-chase analysis as in A and B. (C) Degradation of CPY*-abcD and (D) the nonglycosylated variant CPY*-abcd. The data plotted reflect three independent experiments and the SEM.
Local Conformational Perturbations Activate Nonsignal Glycans for ERAD
In CPY*, are nonsignal glycans inactive because they reside adjacent locally ordered structures? If so, deliberately disturbing the structures might create active ERAD signals. To test this idea, the segments directly adjacent to the A, B, and C glycans of CPY* were individually disrupted with 12 residue deletions (Figure 4A, top panels). These glycans do not participate significantly in ERAD signaling (Kostova and Wolf, 2005; Spear and Ng, 2005). To assess whether the lesions are indeed structurally disruptive, we applied a test that uses ERAD as the readout. For this, identical deletions were introduced in parallel in preproCPY and analyzed for their recognition by ERAD. ERAD of misfolded CPY is characterized by ER retention (lack of processing to the slower migrating p2 Golgi form, cf. Supplementary Figure S1) and rapid degradation (Stevens et al., 1982). As shown in Figure 4A, the absence of a p2 species followed by their rapid turnover demonstrates that the lesions are structurally disruptive, as defined by ERAD.
Figure 4.
Glycan-proximal lesions can generate artificial ERAD determinants. (A) Top, surface rendering of segments adjacent to glycans A, B, and C of folded CPY using the PyMOL software described in Figure 2 (top panels). β-strands, loops, and α-helixes depicted in yellow, green, and red, respectively. Glycosylation sites are depicted in blue. Bottom, wild-type cells expressing CPY deletion variants were pulse-labeled for 10 min and chased for the times indicated. Proteins were immunoprecipitated using monoclonal HA antibody from detergent lysates, resolved by SDS-PAGE, and visualized on a phosphorimager. (B) The turnover of CPY* single-glycan variants in wild-type and Δhrd1 cells were analyzed by pulse-chase analysis as described in A. Decay was quantified by phosphorimager analysis and plotted with error bars indicating the SEM of three independent experiments. Representative phosphorimages of each experiment are shown. (C) Degradation of CPY* single-glycan variants were analyzed in wild-type and Δhtm1 cells.
Next, the deletions were introduced into the corresponding CPY* single-glycan variants. Unlike CPY*-abcD, variants containing only the A, B, or C glycan are not degraded by ERAD (Kostova and Wolf, 2005). As shown in Figure 4, B and C, all six variants are ERAD substrates dependent on HRD1 and HTM1. These experiments show that functional ERAD-L determinants can be created with nonsignalling glycans by introducing structurally disruptive lesions into adjacent segments.
These data show that ERAD-inactive glycan sites can be coaxed into mimicking ERAD signals by deliberately introducing structurally disruptive lesions into adjacent segments. In agreement with earlier experiments, the primary sequences surrounding signaling glycans appear to be unimportant but our analyses do not rule out amino acid preferences.
The Peptide Portion of the CPY ERAD Determinant Is Recognized by the BiP/Kar2p Chaperone
The studies thus far demonstrate that substrate recognition involves peptide segments to adjacent signaling glycans. To better understand how the determinant is recognized, we applied a biochemical approach. The whole length of the CPY* molecule is sensitive to trypsin digestion compared with the compactly folded CPY (Finger et al., 1993). We reasoned that application of the assay under conditions preserving protein–protein interactions might reveal regions recognized by ERAD factors. In turn, this information could be used to purify and identify the factors.
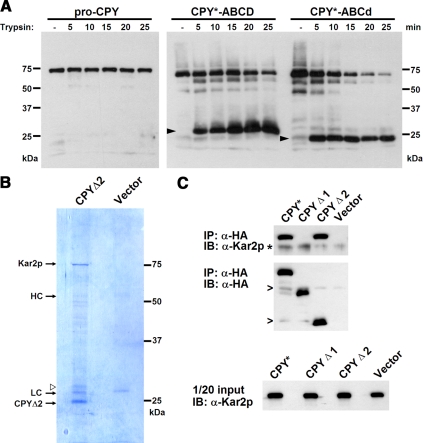
A microsomal fraction was prepared from cells expressing HA-tagged CPY* or CPY. Membranes were solubilized in detergent buffer under physiological conditions and subjected to limited trypsin digestion. Aliquots collected at various intervals were analyzed by immunoblotting for CPY. As shown in Figure 5A, proCPY is resistant to trypsin digestion during the time course as reported previously (Finger et al., 1993). By contrast, digestion of CPY* (the CPY* substrate includes the proregion of CPY) generates several fragments by 5 min, with the most prominent migrating near the 25-kDa marker. Unlike other fragments, it persists during the time course, suggesting protection by bound factors. The protected fragment represents the CPY* C-terminus because of the location of the epitope tag. Probing the blot using a polyclonal antiserum generated against proCPY, showed a similar pattern (except with higher background), so using the HA antibody did not limit the analysis (data not shown). Intriguingly, the protected fragment represents the area surrounding the D glycan—the region determined to be the CPY* ERAD determinant. We next tested whether the D glycan itself is required for protease protection. The assay was applied to the CPY*-ABCd variant, which lacks only the D glycan. As shown in Figure 5A (right), a nearly identical pattern was obtained except that the protected fragment migrated slightly faster at 23 kDa—the size expected without the glycan. These data show that the region that encompasses the CPY ERAD determinant can recruit factors with at least one that binds to the peptide portion independently of the D glycan.
Figure 5.
The peptide segments adjacent to the CPY* signal glycan are recognized by the chaperone BiP/Kar2p. (A) Microsomal fractions were prepared from Δcue1Δpep4Δprc1 cells expressing CPY-HA, CPY*, and CPY*-ABCd. Membranes containing the substrates solubilized in detergent under nondenaturing conditions and digested with 5 μg/ml trypsin for the times indicated. The proteins were separated by SDS-PAGE and detected on immunoblots with the anti-HA antibody. Arrowheads, the positions of protease-resistant fragments. (B) CPYΔ2 expressed in Δcue1Δpep4Δprc1 cells was immunopurified from microsomes under native conditions. Eluted proteins resolved by SDS-PAGE and visualized by Coomassie Brilliant Blue (left lane). The procedure was performed in parallel using cells lacking CPYΔ2 as a control (right lane). Open arrowhead, the position of the modified form of CPYΔ2 as observed in Figure 2D. (C) CPY*, CPYΔ1, and CPYΔ2 expressed in Δcue1Δpep4Δprc1 cells were immunoprecipitated from detergent lysates under were native conditions and resolved by SDS-PAGE, and immunoblots were probed for the substrate (middle panel) and BiP/Kar2p (top panel). Bottom panel, the load (1/20 of the total of each lysate probed for BiP/Kar2p on immunoblots). Asterisk and open arrowheads, cross-reactive background bands.
To identify factors associated with the CPY* ERAD determinant, the CPYΔ2 substrate was immunopurified from a microsomal fraction. CPYΔ2 was chosen because it encompasses the trypsin-protected segment and is a bona fide ERAD-L substrate (Figure 2D). Eliminating other parts of CPY* enables the purification of factors that are most relevant for ERAD. Purified complexes were resolved by SDS-PAGE (Figure 5B). Strongly staining bands were excised as gel slices, digested with trypsin, and analyzed by mass spectrometry. A prominent band migrating above the 75-kDa marker was identified as Kar2p (also known as BiP), a member of the Hsp70 family of chaperones (Supplementary Table S1). In yeast, KAR2 mutants are defective in ERAD and cause substrates to aggregate (Nishikawa et al., 2001; Kabani et al., 2003). Other bands include the IgG heavy and light chains partially released from the resin, CPYΔ2, and an in vivo modified form of CPYΔ2 that migrates slower (Figures 2D and 5B). Minor bands include BiP degradation products and other proteins that appear to be contaminants. BiP was absent from control lysates (Figure 5B, “vector”).
To determine the binding specificity to the ERAD determinant, CPY*, CPYΔ1, and CPYΔ2 were immunoprecipitated under nondenaturing conditions and probed for BiP on immunoblots. As shown in Figure 5C, the ERAD substrates CPY* and CPYΔ2 coimmunoprecipitated similar amounts of the chaperone. By contrast CPYΔ1, a deletion variant lacking the ERAD determinant, associated with BiP very poorly (though apparently absent in Figure 5C, longer exposures revealed a signal above the control). Notably, CPYΔ1 is a misfolded glycoprotein not degraded by ERAD (Spear and Ng, 2005). Taken together, these data show that BiP specifically binds the region of the CPY* ERAD determinant. It should be noted, however, that strong BiP binding alone is not sufficient to drive ERAD. The presence of the D glycan is critical. The nonglycosylated variant of CPY* that is not degraded by ERAD (Knop et al., 1996b) binds BiP as well as CPY* (Wei and Ng, unpublished results). Although the strategy revealed a key player in luminal ERAD and its substrate binding site, other ERAD factors that directly engage substrates including Htm1p (Clerc et al., 2009) and Hrd3p (Gauss et al., 2006a) were not recovered under the conditions used. It remains to be determined whether the bipartite signal is recognized by a single factor or whether it scaffolds multiple factors like BiP that activate ERAD when configured.
The identity of BiP at the D glycan site has other important implications. Structural and biochemical analysis established that the Hsp70 chaperone family, and BiP specifically, bind peptide substrates in extended conformations (Fourie et al., 1994; Zhu et al., 1996). These data reinforce the evidence that the ERAD determinant is a glycan attached to a disordered structure.
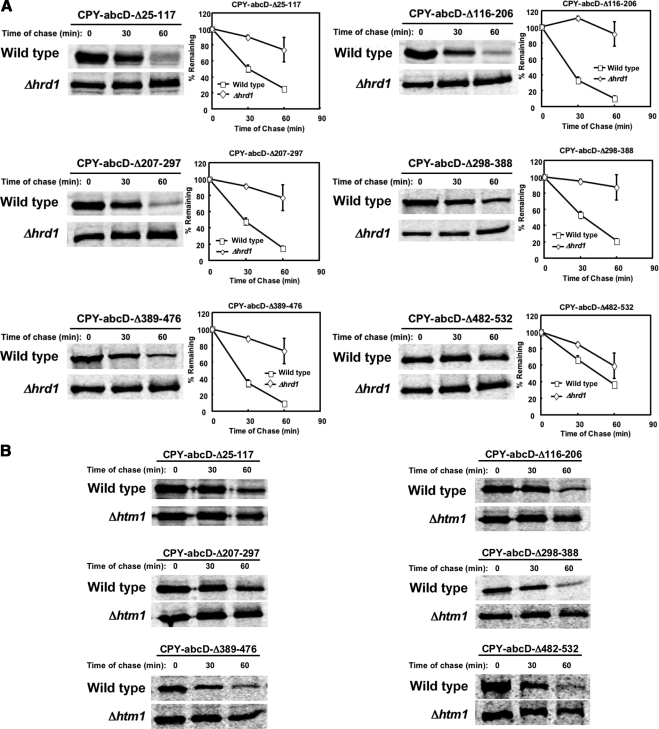
Substrate Signaling Domains Act as Reporters of Protein Misfolding
The D glycan determinant is required for the ERAD of CPY*. However, it was not known whether its use is specifically associated to the G255R “star” mutation or reflects a universal signal for CPY misfolding. To answer this question, we introduced lesions at locations throughout CPY systematically. To attribute signaling to the D glycan domain, all mutants are variants of CPY-abcD (CPY lacking the A, B, and C glycans), which itself folds efficiently (see Figure 7C). By pulse-chase analysis, all of the variants were degraded by ERAD-L (Figure 6, A and B) confirming that the D glycan domain can detect structural perturbations throughout the polypeptide. The slightly less efficient degradation of CPY-abcD-Δ482-532 (a C-terminal deletion) is likely due to the deletion being less structurally disruptive than the others. A similar variant, CPY*-abcD-Δ482-532, differing only by its G255R “star” mutation, was degraded as efficiently as CPY* (Figure 1B).
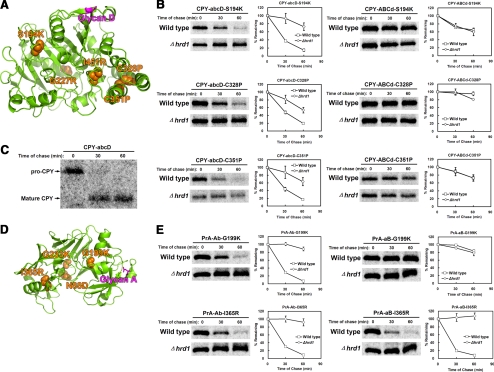
Figure 7.
The CPY and PrA signal glycans mark domains broadly sensitive to structural defects. (A) Ribbon diagram of the CPY structure showing position of the D glycan to introduced lesions. The D signal glycan site is in purple, and the positions of mutated residues are in orange. (B) Misfolded CPY variants bearing only the D glycan (left panels) or bearing the A, B, and C glycans (right panels) were analyzed for degradation efficiency in wild-type and Δhrd1 strains as described in Figure 1. (C) CPY-abcD was pulse labeled for 10 min and chased for the times indicated. Proteins were immunoprecipitated using polyclonal CPY antisera (the strain lacks endogenous CPY) from detergent extracts and resolved by SDS-PAGE. (D) Ribbon diagram of the PrA structure showing the position of the A glycan (purple) to mutated residues (orange spheres; Parr et al., 2007). (E) PrA variants bearing only the A glycan (left panels) or only the B glycan were analyzed by pulse-chase analysis as in B.
Figure 6.
The CPY ERAD determinant can detect lesions throughout the polypeptide. A systematic deletion series was constructed using CPY-abcD as was done for CPY*-abcD (see Figure 1). (A) Turnover profiles were determined for each variant in wild-type and Δhrd1 cells as described for Figure 1. The data represent three independent experiments; error bars, SEM. Representative images from phosphorimager scans of each experiment are shown. (B) Degradation of variants in panel A in wild-type and Δhtm1 cells.
We next tested the sensitivity of the D glycan determinant in reporting less severe perturbations. CPY surface positions structurally distant from the D glycan site were selected for disruption (Figure 7A, mutations marked on the ribbon diagram). Nonconservative amino acid substitutions were first introduced into fully glycosylated CPY to assess their effects to folding and transport. Two alleles, G227R and I451R, did not affect maturation (Supplementary Figure S1A). Three other mutations, S194K, C328P, and C351P, prevented maturation indicating defects in folding (Supplementary Figure S1B). The mixed result was not unexpected because protein surfaces are more structurally tolerant to amino acid substitution than core regions (Cunningham and Wells, 1989; Lawrence et al., 2007).
To assess the effectiveness of the glycan D determinant to report misfolding, the S194K, C328P, and C351P mutations were introduced singly into CPY-abcD. As shown in Figure 7B (left panels), all three variants were efficiently degraded by ERAD. Because these are novel CPY misfolding mutants, we tested whether other glycans could report on the lesions. For this, the mutations were introduced into CPY-ABCd, which lacks only the D glycan. In no case did the three glycans support ERAD (Figure 7B, right panels). The result was surprising because one mutation, S194K, is positioned only four residues away from the B glycan. Taken together, these data show that the D glycan determinant serves as a sensitive reporter of CPY misfolding. Other glycan sites do not perform this function unless their attached segments are directly and severely altered.
We next extended the analysis to PrA. In the context of the PrA* mutation (a 37-amino acid deletion in its prodomain), the N-terminal A glycan signals ERAD, whereas the C-terminal B glycan does not. Unlike CPY, folded PrA has a bilobed structure connected by single peptide segment (Figure 7D). Thus, it seemed possible that additional ERAD determinants might be used if the domains could fold independently. Four nonconservative point mutations were introduced singly into wild-type PrA. The G199K and I365R alleles caused misfolding, whereas the N88D and G233K changes were benign (Supplementary Figure S1, C and D). To assess how individual glycans contribute to ERAD signaling, we altered the singly glycosylated PrA-Ab and PrA-aB molecules with the folding disruptive mutations. In pulse-chase experiments, PrA-Ab-G199K was degraded efficiently by ERAD, but PrA-aB-G199K was stable in both wild-type and Δhrd1 cells (Figure 7E, top panels). This could be expected because the mutation, like that of PrA*, is located in the N-terminal lobe. The I365R mutation, located in the other lobe, did not generate the reciprocal pattern. The PrA-aB-I365R protein was efficiently degraded by ERAD showing that a B glycan determinant can be activated if folding is disrupted nearby (Figure 7E, bottom right panel). Surprisingly, the A glycan determinant was just as effective even though it is located on the other lobe (Figure 7E, bottom left panel). These data show that the A glycan determinant, like the D glycan signal of CPY, can function as a general reporter of PrA folding.
DISCUSSION
Folded substrates of the ubiquitin-proteasome system (UPS) are recognized through signals displayed on protein surfaces (Hershko et al., 2000). Usually, the E3 ligases catalyzing ubiquitin attachment play the most direct role in recognition. Unfortunately, this basic concept is not easily applied to ERAD. Foremost, the stringency of ERAD must be calibrated. The ideal “sieve” should catch severely misfolded molecules but permit the passage of minor genetic variants as not to impede the evolutionary process. Proteins also have the potential to misfold in numerous ways so some features prominent in one form can be absent in another. Compounding the problem is the likelihood that many features found on actively folding proteins are also present on “misfolded” proteins. Lastly, the sheer number of potential substrates rules out a motif-based mechanism without putting structural constraints on proteins. With this view, it was difficult to envision how the ERAD mechanism can follow the UPS paradigm. However, indications of substrate signals abound.
In budding yeast, different ERAD pathways screen misfolded substrates based on the topological location of lesions (Hill and Cooper, 2000; Taxis et al., 2003; Huyer et al., 2004; Vashist and Ng, 2004). These observations implied that specific physical hallmarks are used for recognition. By extension, different ERAD pathways suggest a diversity of ERAD signals. The strongest clues to the existence of actual degradation signals or “degrons” emerged from mutational studies of the CPY* model. Eliminating its glycans completely abolished degradation (Knop et al., 1996b). The subsequent discovery of lectin-like ERAD factors firmly planted the idea that the glycans themselves can act as signals. One of the factors, Htm1/Mnl1p (EDEM in mammals), is essential for the degradation of misfolded glycoproteins (Hosokawa et al., 2001; Jakob et al., 2001; Nakatsukasa et al., 2001). It was originally proposed to act as a lectin-like receptor for ERAD (Hosokawa et al., 2003; Molinari et al., 2003). However, recent work has shown that Htm1/Mnl1p is a mannosidase that converts the GlcNAc2Man8 (GlcNAc, N-acetylglucosamine; Man, mannose) structure to GlcNAc2Man7 by removing the terminal α1,2-linked mannose residue from the C-arm of the glycan (Clerc et al., 2009). This reaction leaves a new terminal mannose residue that is α1,6-linked, the ligand of the ERAD lectin-like receptor Yos9p (Quan et al., 2008). The combined activities of Mns1p (converts GlcNAc2Man9 to GlcNAc2Man8) and Htm1/Mnl1p generate the glycan portion of the bipartite ERAD signal (Figure 8, top panel). By activating the signal for ERAD, these enzymes act sequentially as a molecular timer to differentiate “misfolded” proteins (GlcNAc2Man7) from folding intermediates (GlcNAc2Man9 and GlcNAc2Man8).
The finding that only specific glycans on model substrates could act as signals suggested additional determinants embedded in the polypeptide chain. Thus, an understanding of the complete determinant is required before the mechanism of substrate recognition can be solved. The determinant itself—a single glycan attached to an unfolded segment—turned out to be deceptively simple given the strict context specificity of the glycan. The benefit of the design became clear as we sought to understand why other glycan sites in CPY* fail to signal. By considering the established principle that unfolded proteins can sustain extensive secondary structures (Dyson and Wright, 2004), we postulated that inactive glycans might be positioned in segments that maintain localized ordered structures. This notion was supported experimentally. Only when structurally disruptive lesions were introduced in direct proximity to nonsignal glycans were they recognized by ERAD (Figure 4). These results emphasize the idea that ERAD determinants play a specialized role in ER quality control. In the case of the D glycan, it is entirely dispensable for CPY's normal function. It is not required for the maturation or transport of proCPY and eliminating the glycan actually increases its enzymatic activity (Winther et al., 1991).
By incorporating protein disorder into the ERAD signal, the region harboring it effectively functions as a sensor that reports directly on local folding. If the determinant remains unfolded by the time it can be processed by Mns1p and Htm1p, the activated signal is displayed for the ERAD machinery. Our data show that glycoproteins use this mechanism, but in a more sophisticated way. The CPY D glycan appears to be positioned so formation of the local structure is dependent on the overall packing of the polypeptide (Figure 2A). This would extend the reach of the sensor to include all long-range perturbations affecting tertiary structure. This mechanism was confirmed experimentally and its importance for quality control is underscored by the observation that other glycan sites fail to substitute in most instances (Figures 6 and 7). These data support the earlier proposal that the glycan timer mechanism might require an associated conformational sensor to spare ER resident proteins (Helenius and Aebi, 2004). Although the concept of an intrinsic sensor domain was not previously demonstrated, the structural basis of ordered/disordered segments in unfolded proteins is well documented. After tertiary structure disruption, the barnase protein continues to maintain secondary structures, whereas chymotrypsin inhibitor 2 is largely devoid of ordered structures (Fersht and Daggett, 2002). Similarly, “intrinsically disordered proteins” make up a large family whose members have some or all of their peptide segments disordered in solution (Dyson and Wright, 2005). For some, assembly with partner proteins converts disordered segments into folded structures, usually α-helixes. In principle, any domain containing a segment whose structure depends on the correct packing of the overall structure can be used as an ERAD folding sensor simply by attaching an N-linked glycan.
The proposed mechanism, though remarkably simple, satisfies the broad requirements of ERAD. It supports a reliable disposal system for misfolded proteins and is tolerant for the emergence of minor genetic variants. This dynamic was observed even within the confines of this study. Misfolded forms were efficiently degraded (Figures 1, 6, and 7), whereas variants bearing nonconservative changes, but not grossly misfolded, were transported and processed normally (Supplementary Figure S1). By employing a single determinant independent of sequence, the mechanism places fewer structural demands on proteins. However, this might be at the cost of reducing the range of aberrant forms recognized. Contrary to this notion, the CPY and PrA ERAD determinants were observed to function flawlessly regardless of the perturbation. By contrast, other glycan sites, which would certainly have other functions, are generally incapable of signaling ERAD.
Although this study focused on two well-studied model substrates, it is also clear that other signals exist for luminal ERAD substrates. For example, the mammalian cyclooxygenase isoform COX-2 contains a 19-amino acid segment and an N-linked glycan near its C-terminus that greatly reduces its half-life compared with COX-1 (Mbonye et al., 2006). Both are ER-resident proteins and regulating the stability of folded COX-2 appears to be the primary function of the degron. In a second example, the secreted form of the IgM heavy chain, μs, contains a C-terminal 20-amino acid segment containing an N-linked glycan responsible for ER retention and degradation in pre-B-cells (Shapira et al., 2007). Transplanting the sequence to yellow fluorescent protein and thyroid peroxidase is sufficient to cause their retention and degradation. Interestingly, elimination of the glycan does not prevent degradation but instead accelerates it. Thus, the degron can function in the absence of glycosylation. Similarly, unassembled kappa light chains, which are nonglycosylated, are also degraded by ERAD (Okuda-Shimizu and Hendershot, 2007). How these substrates signal ERAD remains yet to be determined.
By combining our findings with the recent studies of Aebi, Weissman, and coworkers (Quan et al., 2008; Clerc et al., 2009), a detailed model of substrate recognition in the ERAD-L pathway was constructed (Figure 8). After translocation from the Sec61 translocon and core glycosylation by oligosaccharyl transferase, the nascent polypeptide begins the process of folding assisted by chaperones and folding catalysts. If the protein folds correctly, the glycopeptide determinant folds into its ordered state, releases BiP, and transported if directed by export signals (Figure 8, left). If the protein fails to fold after “mannose timer” enzymes process the signal glycan, the resulting Man7GlcNAc2 structure combined with adjacent disordered segment combines to form a degradation signal recognized by ERAD. The identity of ERAD factors and how they decode this signal remain to be determined. However, BiP might play an role in targeting since it can bind directly to the Hrd1 complex, probably through Yos9p (Denic et al., 2006). The glycan is likely not required for the targeting step because nonglycosylated CPY* associates with the Hrd1 complex but not degraded (Denic et al., 2006; Gauss et al., 2006a). Yos9p scans for the terminal α1,6-mannose component of the Man7GlcNAc2 glycan. If present, the substrate is translocated across the membrane though an unidentified pore. On the cytosolic face, the substrate is ubiquitinated through the concerted effort of Hrd1p and Ubc7p. The substrate is fully extracted from the membrane by the Cdc48p/Npl4p/Ufd1p complex and finally, degraded by the 26S proteasome (not shown; Bays et al., 2001; Ye et al., 2001; Jarosch et al., 2002; Elkabetz et al., 2004).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Ng Lab for discussion and comments. We thank Chia Sing Tan, Yu Jun Tan, Jeremy Brodhead, and the TLL and NUS core facilities for providing excellent technical support. We thank Adam Yuan for assistance using the PyMOL software. We are grateful to the laboratories of Markus Aebi and Jonathan Weissman for sharing their data before publication. This work was supported by funds from the Temasek Trust and by grants from the Japan Society for the Promotion of Science to K.K. (Postdoctoral Fellowship).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0231) on May 20, 2009.
REFERENCES
- Aguilar C. F., Cronin N. B., Badasso M., Dreyer T., Newman M. P., Cooper J. B., Hoover D. J., Wood S. P., Johnson M. S., Blundell T. L. The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae. J. Mol. Biol. 1997;267:899–915. doi: 10.1006/jmbi.1996.0880. [DOI] [PubMed] [Google Scholar]
- Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati A., Denic V., Quan E. M., Weissman J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Biederer T., Volkwein C., Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Bordallo J., Plemper R. K., Finger A., Wolf D. H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn B. A., Kostova Z., Medicherla B., Wolf D. H. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 2004;577:422–426. doi: 10.1016/j.febslet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. C., Wells J. A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y., Shapira I., Rabinovich E., Bar-Nun S. Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplasmic reticulum-bound p97/Cdc48p and proteasome. J. Biol. Chem. 2004;279:3980–3989. doi: 10.1074/jbc.M309938200. [DOI] [PubMed] [Google Scholar]
- Endrizzi J. A., Breddam K., Remington S. J. 2.8-Angstrom structure of yeast serine carboxypeptidase. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Daggett V. Protein folding and unfolding at atomic resolution. Cell. 2002;108:573–582. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- Finger A., Knop M., Wolf D. H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Fourie A. M., Sambrook J. F., Gething M. J. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- Gardner R. G., Shearer A. G., Hampton R. Y. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell. Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Jarosch E., Sommer T., Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 2006a;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Gauss R., Sommer T., Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006b;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty C., Strahl S., Romisch K. O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 2001;12:1093–1101. doi: 10.1091/mbc.12.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hill K., Cooper A. A. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 2000;19:550–561. doi: 10.1093/emboj/19.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao K., et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- Hitt R., Wolf D. H. DER7, encoding alpha-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res. 2004;4:815–820. doi: 10.1016/j.femsyr.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Tremblay L. O., You Z., Herscovics A., Wada I., Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Jakob C. A., Bodmer D., Spirig U., Battig P., Marcil A., Dignard D., Bergeron J. J., Thomas D. Y., Aebi M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001;2:423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C. A., Burda P., Roth J., Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Spear E. D., Ng D. T. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Knop M., Finger A., Braun T., Hellmuth K., Wolf D. H. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996a;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop M., Hauser N., Wolf D. H. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast. 1996b;12:1229–1238. doi: 10.1002/(sici)1097-0061(19960930)12:12<1229::aid-yea15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kostova Z., Wolf D. H. Importance of carbohydrate positioning in the recognition of mutated CPY for ER-associated degradation. J. Cell Sci. 2005;118:1485–1492. doi: 10.1242/jcs.01740. [DOI] [PubMed] [Google Scholar]
- Lawrence M. S., Phillips K. J., Liu D. R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Mbonye U. R., Wada M., Rieke C. J., Tang H. Y., Dewitt D. L., Smith W. L. The 19-amino acid cassette of cyclooxygenase-2 mediates entry of the protein into the endoplasmic reticulum-associated degradation system. J. Biol. Chem. 2006;281:35770–35778. doi: 10.1074/jbc.M608281200. [DOI] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J. L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Nishikawa S., Hosokawa N., Nagata K., Endo T. Mnl1p, an alpha -mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem. 2001;276:8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Hosokawa N., Wada I., Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Okuda-Shimizu Y., Hendershot L. M. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari S., Cali T., Salo K. E., Paganetti P., Ruddock L. W., Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- Parr C. L., Keates R. A., Bryksa B. C., Ogawa M., Yada R. Y. The structure and function of Saccharomyces cerevisiae proteinase A. Yeast. 2007;24:467–480. doi: 10.1002/yea.1485. [DOI] [PubMed] [Google Scholar]
- Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Sato K., Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- Shapira I., Charuvi D., Elkabetz Y., Hirschberg K., Bar-Nun S. Distinguishing between retention signals and degrons acting in ERAD. J. Cell Sci. 2007;120:4377–4387. doi: 10.1242/jcs.011247. [DOI] [PubMed] [Google Scholar]
- Spear E. D., Ng D. T. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J. Cell Biol. 2005;169:73–82. doi: 10.1083/jcb.200411136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Szathmary R., Bielmann R., Nita-Lazar M., Burda P., Jakob C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther J. R., Stevens T. H., Kielland-Brandt M. C. Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivo stability, or activity. Eur. J. Biochem. 1991;197:681–689. doi: 10.1111/j.1432-1033.1991.tb15959.x. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H., Rapoport T. A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.