Abstract
The transforming growth factor (TGF)-β superfamily regulates cell proliferation, apoptosis, differentiation, migration, and development. Canonical TGFβ signals are transduced to the nucleus via Smads in both major signaling branches, bone morphogenetic protein (BMP) or Activin/Nodal/TGFβ. Smurf ubiquitin (Ub) ligases attenuate these pathways by targeting Smads and other signaling components for degradation by the 26S proteasome. Here, we identify tumor necrosis factor (TNF)-receptor–associated factor-4 (TRAF4) as a new target of Smurf1, which polyubiquitylates TRAF4 to trigger its proteasomal destruction. Unlike other TRAF family members, which mediate signal transduction by TNF, interleukin, or Toll-like receptors, we find that TRAF4 potentiates BMP and Nodal signaling. In the frog Xenopus laevis, TRAF4 mRNA is stored maternally in the egg animal pole, and in the embryo it is expressed in the gastrula marginal zone, neural plate, and cranial and trunk neural crest. Knockdown of embryonic TRAF4 impairs signaling, neural crest development and neural folding, whereas TRAF4 overexpression boosts signaling and expands the neural crest. In human embryonic kidney 293 cells, small interfering RNA knockdown of Smurf1 elevates TRAF4 levels, indicating endogenous regulation of TRAF4 by Smurf1. Our results uncover new functions for TRAF4 as a Smurf1-regulated mediator of BMP and Nodal signaling that are essential for neural crest development and neural plate morphogenesis.
INTRODUCTION
Transforming growth factor (TGF)-β family of secreted cytokines, which include TGFβs, Activin, Nodal, and bone morphogenetic proteins (BMPs), regulate important processes during embryogenesis and adult tissue homeostasis. During early vertebrate embryogenesis, in particular, Nodal-related proteins induce the endoderm and mesoderm and help establish the primary body axes, whereas BMPs specify ventral cell fates in the ectoderm and mesoderm (De Robertis and Kuroda, 2004; Shen, 2007). In the vertebrate ectoderm, particularly in Xenopus, different doses of BMP signaling activity specify distinct cell types during gastrulation, in a dose-dependent manner: neural induction takes place in very low or absent BMP signaling, whereas epidermis is induced by high levels of BMP signaling. Early specification of the neural border cells such as the neural crest, sensory placodes and cement gland, however, requires intermediate levels of BMP signaling (LaBonne and Bronner-Fraser, 1998; Marchant et al., 1998; Nguyen et al., 1998; Barth et al., 1999).
The neural crest in particular is a unique group of multipotent cells that generate a broad variety of cell types, including pigment cells of the skin, neurons and glial cells of the peripheral nervous system, bone and cartilage of the face, and various components of the cardiovascular system (Crane and Trainor, 2006). Neural crest cells first emerge from the lateral border of the neural plate by the end of gastrulation, and after neural tube closure they begin to differentiate and migrate from their original location toward their final destinations in the embryo (Le Douarin and Dupin, 2003; Crane and Trainor, 2006; Harris and Erickson, 2007). Besides BMPs, other signaling molecules such as Notch/Delta, Wnts, and fibroblast growth factors (FGFs) are also involved in neural crest specification. Wnts and FGFs secreted by the ectoderm and paraxial mesoderm act on the ectoderm at the neural plate border, and these signals are essential for neural crest development in whole embryos, as well as in isolated ectodermal explants, such as Xenopus animal caps (Saint-Jeannet et al., 1997; Villanueva et al., 2002; Monsoro-Burq et al., 2003; Bastidas et al., 2004; Glavic et al., 2004a). Notch signaling seems to indirectly regulate neural crest formation through repression of BMP signaling (Glavic et al., 2004b).
The misregulation of TGFβ family signaling has detrimental consequences for embryos and adults, generating birth defects in the former and diseases such as cancer in the latter (Siegel and Massague, 2003; Waite and Eng, 2003). Therefore, understanding the mechanisms that ensure proper levels of TGFβ family signaling in receiving cells is of critical importance. Canonical TGFβ signaling is initiated by the binding of a dimeric ligand to two type I and two type II Ser/Thr kinase receptors. In this ligand–receptor complex, the type I receptor subunits become activated and phosphorylate receptor-associated Smads (R-Smads) that subsequently associate with Smad4 and become retained in the nucleus. Among the R-Smads, Smad1, -5, or -8 are specifically activated by the BMP receptors, whereas Smad2 and Smad3 are specifically activated by TGFβ-, Activin-, and Nodal-related receptors. In the nucleus, R-Smad/Smad4 complexes combine with sequence-specific transcriptional cofactors to bind DNA and induce or repress the expression of target genes (Attisano and Wrana, 2002; Shi and Massague, 2003). TGFβ family signaling can be regulated at the receptor, Smad, or transcription factor levels by two structurally similar E3 ubiquitin ligases, Smurf1 and Smurf2, which interact with TGFβ pathway components either directly (e.g., Smads) or indirectly (e.g., receptors via Smad6/7), and mediate their proteasomal degradation through polyubiquitylation (Zhu et al., 1999; Kavsak et al., 2000; Lin et al., 2000; Bonni et al., 2001; Ebisawa et al., 2001; Zhang et al., 2001; Suzuki et al., 2002; Murakami et al., 2003; Moren et al., 2005; Alexandrova and Thomsen, 2006; Shen et al., 2006).
We sought to isolate new proteins that bind to Smurf1, under the hypothesis that as targets or partners of Smurf1 they might participate in TGFβ family signaling and affect processes regulated by TGFβ pathways, particularly early developmental events in the Xenopus embryo. We performed a yeast two-hybrid screen to isolate Smurf1-interacting proteins and retrieved tumor necrosis factor receptor-associated factor (TRAF) 4. TRAF4 is a member of the TRAF family of receptor adaptor proteins, which contains seven members with known roles in immunity, inflammation, and apoptosis (Chung et al., 2002). In general, TRAFs function as scaffolds for receptors of the tumor necrosis factor receptor (TNFR) family, Toll-like receptors (TLRs), and interleukin (IL)-1 receptors, and they relay signals to downstream effectors that activate transcription factors such as nuclear factor-κB and activator protein-1 (Aggarwal, 2003; Dempsey et al., 2003).
When the primary protein structure is considered, TRAF4 shares the common features of other TRAF family members; however, TRAF4 does not fit the predominant signaling paradigm of other TRAFs, and its functions have remained rather elusive (Kedinger and Rio, 2007). Studies in cell culture provide some evidence that TRAF4 can associate with a few receptors of the TNFR and TLR families (Ye et al., 1999; Esparza and Arch, 2004; Takeshita et al., 2005) and positively regulate c-Jun NH2-terminal kinase (JNK) activity (Xu et al., 2002; Abell and Johnson, 2005); however, loss-of-function studies to support such roles are missing. Little information exists about the function of endogenous TRAF4. Wu et al. (2005) reported that knockdown of TRAF4 blocks endothelial cell migration. Knockout of the mouse TRAF4 gene resulted in partial embryonic lethality, and surviving TRAF4−/− pups exhibited neural tube, skeletal, and tracheal malformations (Regnier et al., 2002); however, a detailed cellular and molecular analysis of the developmental phenotypes was not performed, leaving the origin and exact nature of the defects unresolved. Unlike mice with other TRAF gene knockouts, TRAF4-deficient mice had normal immune responses, although dendritic cells from these mice exhibited reduced migration (Cherfils-Vicini et al., 2008).
Here, we describe several novel cellular and embryonic functions of TRAF4 by using Xenopus embryo and cultured mammalian cell assays. Our major findings show that endogenous TRAF4 potentiates signaling by the BMP and TGFβ/nodal/activin branches of the TGFβ superfamily, that TRAF4 levels can be regulated by the Smurf1 ubiquitin ligase, and that TRAF4 is an essential gene for neural crest development and neural folding. Our findings shed new light on this recalcitrant member of the TRAF family.
MATERIALS AND METHODS
Isolation of Xenopus TRAF4a and cDNA Constructs
A cDNA encoding a predicted full-length TRAF4 protein was retrieved by a yeast two-hybrid screen on a Xenopus oocyte cDNA library (Clontech, Mountain View, CA), by using a ubiquitin ligase-deficient mutant of Smurf1 (Smurf1-C699A) as bait. The retrieved cDNA encodes TRAF4a, and its sequence is identical to that of a X. laevis cDNA in the National Center for Biotechnology Information database, with accession number BC080018. Full-length TRAF4a was subcloned into pCDNA3.1, and we derived an expression construct (d5UTR-TRAF4/pCDNA3.1) lacking the 5′-untranslated region (UTR) and the target morpholino oligo (MO) binding site, but retaining the KOZAK sequence, for synthesis of mRNA to use in embryonic MO rescue experiments. For expression in cultured cells, TRAF4a was amplified by polymerase chain reaction (PCR) and subcloned into pCS2-hemagglutinin (HA) and pCMV-Tag3C, to create N-terminally tagged HA-TRAF4 and myc-TRAF4, respectively. All PCRs were performed using Platinum Pfx polymerase (Invitrogen, Carlsbad, CA) with low cycle number (<18 cycles), and all constructs were sequenced to confirm their identity.
Morpholino and mRNA Injections
Xenopus embryos were collected and microinjected as described previously (Alexandrova and Thomsen, 2006). Scrambled or antisense MOs were supplied by GeneTools (Philomath, OR), as follows: TRAF4a MO, 5′-ATCCTGCTCGGCGGGCTCCCCACTT-3′; and TRAF4b MO, 5′-TGCACCGACTCCCCGGCTCAAAAGA-3′. The scrambled morpholino from GeneTools was used as the negative control. The animal cap assay in Figure 5C used a slightly different morpholino to target TRAF4a, with a target site shifted downstream by three bases (5′-GGCATCCTGCTCGGCGGGCTCCCCA-3′). Synthetic mRNAs were synthesized with the mMessage kit (Ambion, Austin, TX). To test the efficiency and specificity of morpholinos, 2 ng of TRAF4a mRNA was coinjected with 50 ng of TRAF4a, TRAF4b, or control MO at the two-cell stage; and at stage 9, embryos were lysed and processed for Western blot analysis (described below). Overexpressed Xenopus TRAF4 protein was detected using a polyclonal antibody raised against human TRAF4 (Hypromatrix, Worcester, MA) and the Odyssey infrared imager (LI-COR, Lincoln, NE). To examine the effects on neural crest markers, eight–cell-stage embryos were injected with 10–12.5 ng of TRAF4a, TRAF4b, or control MOs, or with 1 ng of TRAF4a or green fluorescent protein (GFP) mRNA per cell.
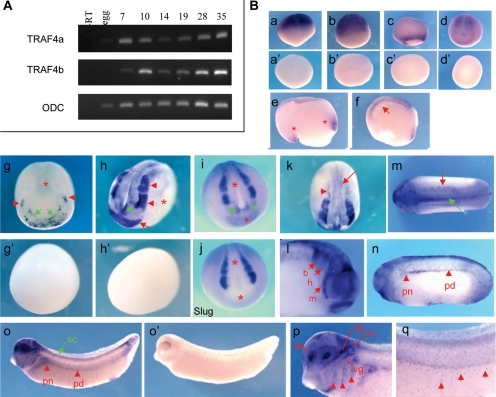
Figure 5.
Regional and tissue-specific expression of TRAF4 genes during Xenopus embryonic development. (A) Temporal expression of TRAF4a and TRAF4b analyzed by RT-PCR. Ornithine decarboxylase (ODC) was scored as a loading and RNA processing control. (B) In situ RNA hybridization showing spatial expression of TRAF4 (a–q). Side views of four-cell (a), stage 8 (b), and stage 10.5 (c) embryos (animal pole up). Dorsal-posterior view of a stage 13 embryo (d) reveals TRAF4 expression in the neural plate. Sense controls are shown in a′–d′. Sagittal sections of a stage 11 (e) and a stage 12.5 embryo (f) showing TRAF4 expression in the general mesoderm indicated with asterisk (e) and in the presumptive neural plate and the involuting mesoderm pointed with arrow (f). Dorsal-anterior view of a stage 14 embryo (g) and its sense control (g′). TRAF4 is expressed in the developing neural crest (red arrowheads), preplacodal and cement gland region (green arrows) and the posterior neural plate (asterisk). Anterior-lateral (h) and dorsal (k) views of a stage 17 embryo and sense control (h′). Strong expression of TRAF4 is detected in the cranial neural crest (red arrowheads), cement gland (red arrow) and the anterior neural fold hingepoints (green arrowhead) (h). Note the lack of expression in the nonneural ectoderm (asterisk) and weaker expression in the neural plate (h). TRAF4 is expressed in the lateral (arrowhead) and medial trunk neural crest (arrow) (k). Expression of TRAF4 (i) and Slug (j) in the anterior region of stage 17 embryos for comparison. Note that in the neural plate and the transverse neural fold TRAF4 is expressed, whereas Slug is not (asterisks). TRAF4 transcripts are enriched in the anterior neural hingepoints (green arrowheads) (i). Dorsal (anterior to the left) (m) and head (l) views of a stage 21 embryo. TRAF4 is expressed in the somites (red arrow) and the trunk neural crest (green arrow) (m) and in the mandibular (m), hyoid (h) and branchial (b) branches of the migrating cranial neural crest (arrowheads) (l). Lateral views of stage 25 (n) and stage 32 (o) embryos (anterior to the left). Red arrowheads point to TRAF4 expression in the pronephros (pn) and the pronephric duct (pd). TRAF4 also is expressed in the spinal cord (sc) (green arrow) (o). Sense control of a stage 32 embryo is shown in k′. In the head region, TRAF4 expression is detected in neural crest and sensory placode derivatives (p): pharyngeal arches (arrowheads), otic vesicle (ov), olfactory placodes (op), cranial ganglia (cg), vagal ganglia (vg). TRAF4 is expressed in the melanoblasts (arrowheads) (r), which are also neural crest derivatives.
Xenopus Animal Cap Assays and Quantitative Reverse Transcription (QRT)-PCR
Synthetic mRNAs or MOs were injected into the animal pole of two-cell stage embryos at the following doses: 1–1.5 ng of TRAF4 mRNA, 100 pg of BMP4 mRNA, 250 pg of C-terminally-truncated BMP receptor (tBMPR) mRNA, 50 ng of TRAF4a MO, and 50 ng of control MO. GFP mRNA was coinjected with TRAF4 mRNAs to normalize the total amount of injected mRNA. Animal caps were isolated at stage 8, cultured in 0.5× MMR until harvested at the appropriate stage. At least 12 animal caps per each treatment were pooled, and total RNA was extracted as described previously (Alexandrova and Thomsen, 2006), followed by cDNA synthesis with SuperScript II reverse transcriptase (Invitrogen) by using oligo d(T)16-20 primers (Invitrogen). The cDNA was purified by phenol:chloroform extraction, followed by ethanol/NH4OAc precipitation. Real-time quantitative PCR was performed with a LightCycler system (Roche Diagnostics, Indianapolis, IN). Primer sequences and conditions were as described previously (Kofron et al., 2001; Yokota et al., 2003). Standard curves were created in each run by using serial dilutions (1:1, 1:10, 1:100, and 1:1000) of stage-matched, whole embryo cDNA, and target gene expression levels in animal caps were normalized to the expression level of ornithine decarboxylase (ODC) in the same samples. That normalized value was plotted as percentage of the gene expression level in a stage-matched, uninjected whole embryo. For detection of TRAF4 paralogues by RT-PCR, primers were designed against short nonconserved sequences in the 3′UTRs of TRAF4a and TRAF4b cDNAs, as follows; U-TRAF4a: 5′-CTCTGTTCGAACTAGAAATTTGCTC-3′; D-TRAF4a: 5′-GCTGCTCAGATTTCTGTTTTAGG-3′; U-TRAF4b: 5′-CCGTTTGAACT TTGCTCTATG-3′; and D-TRAF4b: 5′-GACTTTGTATAATGCAAGAGGCTCC-3′. Quantitative RT-PCR reactions were performed on cDNA synthesized from total embryonic RNA (as described above) by using the Light Cycler system (Roche Diagnostics), with annealing at 55°C/5 s, elongation at 72°C/16 s, and acquisition at 78°C/3 s. To visualize amplification products directly, the QRT-PCR reactions were aborted during the linear phase of amplification and reaction aliquots were separated on a 2.5% agarose/TAE gel, stained with ethidium bromide, and imaged under UV illumination.
Lineage Tracing and In Situ RNA Hybridization
To trace injected cells, LacZ mRNA (50–100 pg/cell at 8-cell stage) was coinjected with MOs or mRNAs. Embryos were fixed in MEMPFA (1 × minimal essential medium [MEM] and 4% paraformaldehyde) for 40 min and stained with Magenta-Gal (LabScientific, Livingston, NJ) as described previously (Turner and Weintraub, 1994). Subsequently, embryos were refixed in MEMPFA for 1 h and processed for whole mount in situ RNA hybridization (WISH) as described previously (Harland, 1991). For double WISH, embryos were fixed in MEMPFA, and the second probe was labeled with fluorescein (Roche Diagnostics) and detected with an alkaline phosphatase-conjugated anti-fluorescein antibody (Roche Diagnostics) at 1: 10,000 dilution, followed by staining with 5-bromo-4-chloro-3-indolyl phosphate (BCIP) substrate (Roche Diagnostics). The following constructs used to synthesize probes were provided as gifts: Sox10 from Dr. J. P. Saint-Jeannet (University of Pennsylvania, Philadelphia, PA), Foxd3 from Dr. C. LaBonne (Northwestern University, Evanston, IL), and Slug and Sox2 from Dr. T. Bouwmeester (European Molecular Biology Laboratory, Heidelberg, Germany).
Western Blots, Cell Culture, Coimmunoprecipitation, and Ubiquitylation Assays
Human embryonic kidney (HEK) 293T and HEK293 cells were grown in 10% calf serum (HyClone Laboratories, Logan, UT), 45% F-12, and 45% DMEM (Invitrogen). HeLa cells were grown in 10% fetal calf serum (HyClone Laboratories) and DMEM (Invitrogen). Cells were transfected with DNA plasmids using poly(ethyleneimine) (a gift from Dr. J. C. Hsieh, Stony Brook University, Stony Brook, NY). Up to 1.5 μg DNA was transfected per well of a six-well plate, and empty vector (pCDNA3.1 or pCS2) was cotransfected to normalize the total amount of DNA applied per sample. Xenopus embryos or cultured cells were lysed in phosphate-buffered saline (PBS) containing 1% Triton X-100, 2 mM EDTA, 1 mM Na3VO4, and protease inhibitors. For coimmunoprecipitation of HA-TRAF4 and FLAG-Smurf1 (C699A), cells were lysed 24 h after transfection, HA-TRAF4 was pulled down with an affinity-purified polyclonal anti-HA antibody (Immunology Consultants Laboratory, Newberg, OR) bound to protein A-Sepharose beads (GE Healthcare, Chalfont St. Giles, United Kingdom) by preincubation for 2 h at 4C. These beads were incubated with cell lysates for 2 h at 4C, spun and washed with ice-cold lysis buffer several times, followed by suspension in Laemmli buffer, and resolved by SDS-polyacrylamide gel electrophoresis. Proteins were detected with either rat monoclonal anti-HA-conjugated horseradish peroxidase (HRP) (Roche Diagnostics) at (diluted 1:500) or mouse monoclonal anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO) (diluted 1:2000) followed by anti-mouse–conjugated HRP (Sigma-Aldrich) (1:5000). In ubiquitylation assays, the proteosome inhibitor MG132 (in dimethyl sulfoxide [DMSO]) was added at 20 mM, 16 h after DNA transfection, and cells were harvested 16–20 h thereafter. Addition of DMSO carrier served the negative control. Cells were lysed as above, but with addition of N-ethylmaleimide at 5 mM to inhibit deubiquitylating enzymes and MG132 at 100 mM to inhibit the proteasome. Myc-TRAF4 was immunoprecipitated as described above using affinity-purified rabbit polyclonal anti-c-Myc (Immunology Consultants Laboratory). HA-tagged ubiquitin was expressed from plasmid HA-Ub/pCGN, a gift from Dr. D. Bar-Sagi (New York University, New York, NY).
siRNA Treatment
Smurf1 Stealth siRNA (sf1-siRNA) was synthesized (Invitrogen) against the target sequence of ACUCAACCGACACUGUGAAAAACAC (based on that of Ozdamar et al., 2005). HEK293 cells were transfected with control- (Invitrogen) or sf1-siRNA by RiboMax (Invitrogen) according to the manufacturer's instructions. After siRNAs treatment (∼48 h), HEK293 cells were harvested and assessed for total Smurf1 and TRAF4 levels by Western blotting with mouse anti-Smurf1 (Wang et al., 2003), goat anti-TRAF4 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA) and mouse anti-β-tubulin (Sigma-Aldrich) antibodies.
Cell Staining
HeLa cells grown on coverslips coated with gelatin were transfected with plasmids encoding HA-TRAF4 and FLAG-Smurf1(CA). Cells were fixed with 4% paraformaldehyde/PBS and permeabilized with 0.1% Triton X/PBS. After blocking with 3% normal goat serum/PBS, cells were incubated with rabbit polyclonal anti-HA (Santa Cruz Biotechnology) and mouse monoclonal anti-FLAG (M2; Sigma-Aldrich) antibodies. Secondary antibodies were Alexa Fluor 594 goat anti-rabbit immunoglobulin G (IgG) and Alexa Fluor 488 goat anti-mouse IgG (Invitrogen). Images were taken by a Leica SP5 confocal microscope.
RESULTS
Identification and Sequence Analysis of Xenopus TRAF4 Homologues
To isolate potential new protein partners or substrates of Smurf1, we performed a yeast two-hybrid screen by using a ubiquitin ligase-deficient mutant of Xenopus Smurf1 (Smurf1C699A) harboring a Cys-to-Ala substitution at residue 699 in the catalytic HECT domain as bait (Alexandrova and Thomsen, 2006). One cDNA we retrieved encodes a predicted orthologue of mammalian TRAF4 (Figure 1), with a full-length open reading frame of 470 amino acids (corresponding to the X. laevis cDNA with accession no. BC080018 in the National Center for Biotechnology Information expressed sequence tag [EST] database). This clone was designated TRAF4a to distinguish it from another cDNA of high similarity present, which we designated as TRAF4b (National Center for Biotechnology Information accession no. BC076768; Figure 1). Within the open reading frames, the TRAF4a and TRAFb cDNAs share 90% identity at the nucleotide sequence level and 96% identity at the amino acid level. The cDNAs are significantly diverged in their 5′ and 3′UTR regions. Given that X. laevis is an allotetraploid species, in which two paralogues often exist for a given gene (Hayata et al., 1999; Oelgeschlager et al., 2003, 2004; Nelson and Nelson, 2004; Birsoy et al., 2006), it is very likely that these highly homologous cDNAs correspond to TRAF4 paralogs. Such a relationship is also supported by results of BLAST searches on the EST database and fully sequenced genome of X. tropicalis, a true diploid species of Xenopus, which seems to possess only one TRAF4 gene corresponding to the EST with accession no. BC076992. A sequence alignment of Xenopus and human TRAF4 proteins is shown in Figure 1. In this report, we focused on the functions of the TRAF4a paralogue in detail, and the TRAF4b paralogue to a lesser extent. However, results that follow indicate that the TRAF4b paralogue is expressed and functions similarly to TRAF4a. Hence, unless stated otherwise, the term “TRAF4” refers to TRAF4a throughout this report.
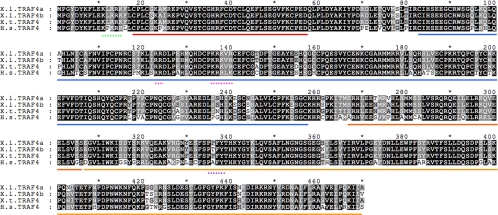
Figure 1.
Comparison of Xenopus and human TRAF4 proteins. Predicted protein sequences of X. laevis (X.l.), X. tropicalis (X.t.), and Homo sapiens (H.s.) TRAF4 proteins are aligned by using Genedoc software (National Resource for Biomedical Supercomputing, Pittsburgh, PA). Identical and similar amino acids conserved among all proteins are shown in black and dark gray boxes, respectively. Lighter shades of gray or no shading represent low levels of amino acid conservation and the lack of conservation, respectively. Domains of TRAF4 are underlined with solid lines: RING finger (red), zinc fingers (blue), coiled-coil or TRAF-N (dark orange), and TRAF-C (light orange). Specific sequence motifs are underlined with dashed lines: first putative nuclear localization signal (NLS; amino acids [a.a.] 11–15) (green), second putative bipartite NLS (a.a. 123–124 and a.a. 136–140) (pink), and PPXY or PY motif (a.a. 305–308) (purple). The predicted X. tropicalis TRAF4 protein shares 95% identity and 98% similarity with the proteins encoded by each X. laevis TRAF4 paralogue. Compared with human TRAF4, the predicted X. laevis TRAF4a and TRAF4b proteins are 77 and 76% identical, respectively, and both are 89% similar to human TRAF4 when conservative amino acid substitutions are considered.
TRAF4 Is a Substrate of Smurf1 Ubiquitin Ligase
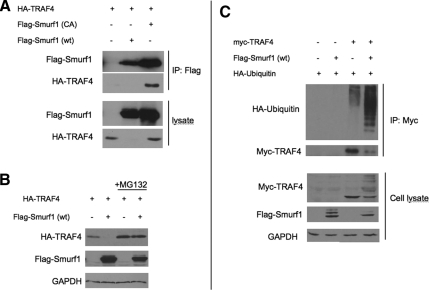
Because TRAF4 was retrieved as a Smurf1 binding protein, we first sought to verify whether Smurf1 and TRAF4 interact with each other in cells, by coimmunoprecipitation. HEK293T cells were transfected with cDNA expression plasmids for Xenopus TRAF4 (HA-TRAF4) alone or together with wild-type Xenopus Smurf1 [FLAG-Smurf1(wt)], or a ubiquitin ligase-deficient mutant of Xenopus Smurf1 [FLAG-Smurf1(CA) with a cysteine to alanine mutation introduced at residue 699; Zhu et al., 1999; Alexandrova and Thomsen, 2006]. FLAG-Smurf1 was immunoprecipitated from cell lysates and the presence of bound HA-TRAF4 was detected by Western blot with anti-HA antibodies (Figure 2A). We found that HA-TRAF4 readily coprecipitated with Smurf1(CA), indicating that two proteins physically interact. However, when HA-TRAF4 was coexpressed with wild-type Smurf1, we did not detect HA-TRAF4 in either the pull-down fraction or total cell lysate (Figure 2A). This suggests that HA-TRAF4 might be destroyed in the presence of wild-type, but not ubiquitin ligase-deficient, Smurf1 (see below).
Figure 2.
TRAF4 is ubiquitylated and degraded by Smurf1 in a 26S proteosome-dependent manner. (A) Western blot showing HA-TRAF4 interacts with FLAG-Smurf1-CA. HEK293T cells were transfected with plasmids containing HA-TRAF4, FLAG-Smurf1 wt, and FLAG-Smurf1-CA, as indicated in the figure panel. Lysates were subjected to immunoprecipitation using anti-FLAG antibody and bound HA-TRAF4 was detected by anti-HA antibody. (B) Western blot showing steady-state levels of HA-TRAF4 is regulated by FLAG-Smurf1 in HEK293T cells. HEK293T cells were transfected with plasmids containing HA-TRAF4 and FLAG-Smurf1 wt as indicated in the figure panel. HA-TRAF4 is undetectable in the presence of FLAG-Smurf1 but is restored when proteosome inhibitor MG132 is added. GAPDH is used as loading control (C) Western blot showing ubiquitylation of Myc-TRAF4 by FLAG-Smurf1 (wt) in HEK293T cells. HEK293T cells were transfected with plasmids containing Myc-TRAF4, FLAG-Smurf1 wt, and HA-ubiquitin as indicated in the figure panel. Cells were treated with MG132 before lysis. Lysates were subjected to immunoprecipitation using anti-Myc antibody and HA-ubiquitin was detected with anti-HA antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is used as loading control.
The interaction between Smurf1(CA) and TRAF4, and the apparent instability of TRAF4 caused by Smurf1(wt), prompted us to test more thoroughly whether Smurf1 can trigger degradation of TRAF4. Using transfected HEK293T cells, we examined whether Smurf1 can affect the steady-state levels of TRAF4 protein and found that, indeed, the presence of Smurf1(wt) eliminated TRAF4 protein (Figure 2B). Furthermore, this effect required 26S proteasome activity because TRAF4 degradation by Smurf1 was blocked by the proteasome inhibitor MG132 (Figure 2B). Because most proteasome-dependent protein degradation requires polyubiquitylation of the target, we tested whether Smurf1 catalyzes polyubiquitylation of TRAF4. HEK293T cells were cotransfected with myc-TRAF4 and HA-ubiquitin, together with or without FLAG-Smurf1(wt). After treatment with MG132, myc-TRAF4 was immunoprecipitated, and the pull-down fraction was analyzed by Western blot with an anti-HA antibody to detect ubiquitin. We observed the formation of an intense ubiquitin ladder on TRAF4 in the presence of FLAG-Smurf1 (wt), demonstrating that Smurf1 indeed polyubiquitylates TRAF4 (Figure 2C). Altogether, these results show that Smurf1 can target TRAF4 for proteasomal degradation via polyubiquitylation. Therefore, Smurf1 has the potential to regulate TRAF4 protein levels in vivo.
Smurf1 Regulates Physiological Levels of TRAF4
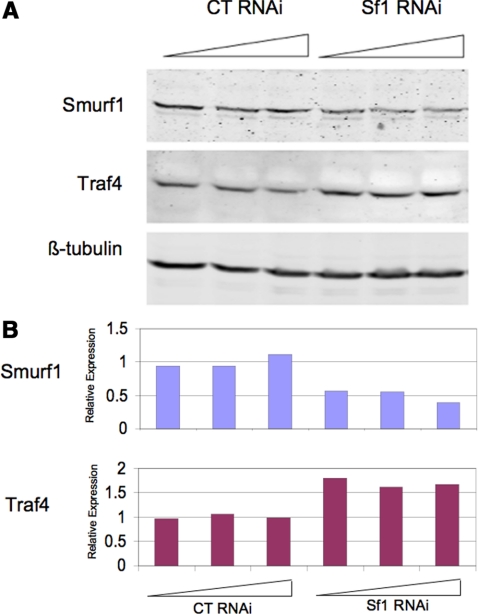
To determine whether Smurf1 regulates TRAF4 under endogenous conditions, we examined native TRAF4 protein levels in HEK293 cells transfected with control or Smurf1-specific siRNA and observed a significant increase in TRAF4 protein levels resulting from Smurf1 depletion compared with that in cells transfected with control siRNA (Figure 3A). Quantitation of protein on western blot shows that Smurf1 protein levels were reduced by half by the Smurf1-specific siRNA, resulting in more than a 50% increase in TRAF4 protein levels compared with control-treated cells (Figure 3B). These results demonstrate that endogenous TRAF4 protein is regulated by Smurf1.
Figure 3.
Smurf1 regulates endogenous TRAF4. (A) Western blot of Smurf1 and TRAF4 proteins from HeLa cells transfected with control or Smurf1-specific siRNA. Partial reduction of Smurf1 protein resulted in elevated TRAF4. (B) Quantitation of Smurf1 and TRAF4 proteins detected in the Western blot. Smurf1-specific siRNA reduced Smurf1 proteins levels by as much as 55% compared with control siRNA, resulting in a corresponding 55–75% increase in TRAF4. Expression levels are normalized to the average of control triplicates.
Smurf1(CA) and TRAF4 Colocalize in Cells
Because TRAF4 and Smurf1(CA) form stable, immunoprecipitable complexes, we tested whether and where these proteins interact in cells. HA-TRAF4 and FLAG-Smurf1(CA) were transfected into HEK293T cells either alone or together, and the transfected cells were stained using anti-HA (green) and anti-FLAG (red) antibodies. When HA-TRAF4 was expressed alone it localized primarily to the cytoplasm, although some cells exhibited speckle-like staining that might correspond to intracellular vesicles, as previously reported (Abell and Johnson, 2005; Li et al., 2005). Transfected FLAG-Smurf1-CA was found distributed within the cytoplasm as well as the plasma membrane, as observed previously and consistent with Smurf1 functioning in regulation of TGFβ receptors, epithelial cell polarity, and tight junction assembly (Ebisawa et al., 2001; Murakami et al., 2003; Wang et al., 2003; Ozdamar et al., 2005), but when both proteins were coexpressed, Smurf1 was found predominantly associated with HA-TRAF4 in the cytoplasmic island-like structures (Figure 4). We attempted to detect endogenous TRAF4 and Smurf1 in HEK293 and other cells, but were unable to do so. This might be due to many substrates of Smurf1 not being able to form stable, complexes in vivo, due to proteasomal targeting of substrates by Smurf1 (e.g., Smad1 and RhoA; Zhu et al., 1999; Wang et al., 2003). Our inability to detect endogenous complexes is consistent with a failure to detect complexes between overexpressed wild-type Smurf1 and TRAF4 by coimmunoprecipitation (Figure 2).
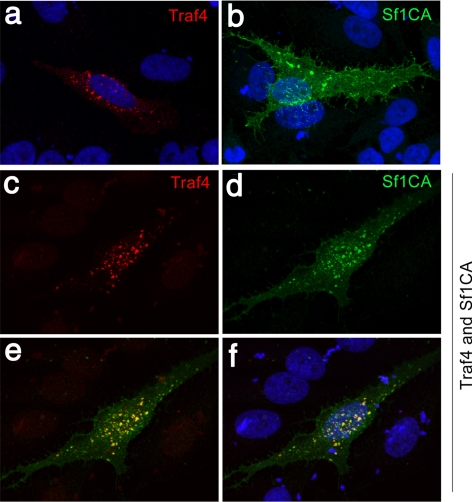
Figure 4.
Smurf1 colocalizes with TRAF4. HeLa cells were transfected with expression constructs for TRAF4, catalytically inactive Smurf1C699A (Sf1CA), or both genes, and then examined by immunofluorescent staining. TRAF4 alone localized to clusters of unknown identity (a), whereas Smurf1 alone localized to predominantly the cell surface, including filipodia (b). Coexpression of the two proteins caused Smurf1 and TRAF4 to colocalize, in what seem to be patches at the cell surface (c–f). 4,6-Diamidino-2-phenylindole (DAPI) staining in panels a, b and f.
Developmental Expression of X. laevis TRAF4
Because we are interested in the developmental roles of proteins implicated in TGFβ signaling, we examined the spatial and temporal dynamics of TRAF4 gene expression during Xenopus embryonic development. First, we determined the relative levels of TRAF4a and TRAF4b transcripts in staged embryos by RT-PCR, using primers that would specifically and exclusively detect each paralogue (see Materials and Methods for details). The results shown in Figure 5A demonstrate that transcripts of both TRAF4 paralogues are present at maternal (egg and stage 7 blastula) and zygotic (stage 10 onward) stages of embryonic development, through swimming tadpole (stage 35).
We next assessed whether there is spatial or tissue-specific TRAF4 expression during development, by WISH (Figure 5B). Because of the high degree of nucleotide conservation between the two TRAF4 paralogues, the probe we used is expected to hybridize with both TRAF4a and TRAF4b transcripts and therefore reveal the sum of TRAF4a/b expression. Our findings indicate a dynamically changing pattern of TRAF4 expression in both the ectoderm and mesoderm. At early cleavage (Figure 5Ba, 4-cell stage), and at midblastula (Figure 5Bb, stages 6–7) TRAF4 transcripts are present throughout the animal hemisphere of the embryo, which corresponds to the early ectoderm. In early gastrulation (stage 10.5) TRAF4 continues to be expressed the ectoderm, but it is also expressed in the involuting mesoderm encircling the blastopore, reflecting pan-mesodermal expression (Figure 5B, c and e). At the end of gastrulation (stage 12–13), TRAF4 become concentrated in the prospective neural plate and the underlying dorsal mesoderm (Figure 5B, d and f). In the early neurula (stage 14), TRAF4 expression is significantly reduced in all but the posterior portion of the neural plate, but TRAF4 transcripts become enriched in the anterior neural plate border, which gives rise to the neural crest, early placodes and the cement gland (Figure 5Bg). By midneurula (stage 17) TRAF4 is expressed strongly in the cranial and trunk neural crest, the cement gland and the anterior neural fold hingepoints (Figure 5Bh), which are rows of cells located where the neural plate bends to achieve closure.
The expression pattern we observed for TRAF4 in the cranial neural crest is essentially identical to the expression pattern found for the definitive neural crest marker Slug (Mayor et al., 1995; compare Figure 5B, i and j). Furthermore, we found TRAF4 expressed throughout the neural plate at a lower, but significant level than in the neural crest and cement gland (Figure 5B, h and k). After neural tube closure, TRAF4 expression continues to track with migrating cranial neural crest cells that will form the branchial, hyoid, and mandibular arches (stage 21 embryo; Figure 5Bl). TRAF4 expression at this stage is also apparent in the trunk neural crest, somites, optic vesicle, and the cement gland (Figure 5B, l and m). In the tailbud tadpole (stage 25) and hatching tadpole (stage 32), TRAF4 expression is notable in the head, spinal cord, pronephros, pronepric duct, and the epidermis in a salt and pepper pattern that likely corresponds to developing melanoblasts (pigment cells derived from the neural crest) (Figure 5B, n and o). In the hatching stage tadpole, TRAF4 is expressed in the head within discrete regions of the brain and neural crest derivatives (Figure 5Bp) that include the three pharyngeal arches (arrows) and the cranial and vagal ganglia. Consistent with its earlier expression in the pre-placodal zone, TRAF4 is also expressed at this stage in placodal derivatives that include the eye lens, otic vesicle, and olfactory placode.
TRAF4 Is a Positive Regulator of BMP Signaling
Smurf1 ubiquitylates proteins involved in all levels of TGFβ signal transduction, including receptors R-Smads and I-Smads and transcription factors such as Runx proteins (Zhu et al., 1999; Ebisawa et al., 2001; Murakami et al., 2003; Ying et al., 2003; Zhao et al., 2003; Alexandrova and Thomsen, 2006; Shen et al., 2006; Sapkota et al., 2007). Given our findings that TRAF4 is also a target of Smurf1 and that TRAF4 is highly expressed in Xenopus embryonic ectodermal and mesodermal cells engaged in TGFβ signaling, we hypothesized that TRAF4 might participate in the BMP or Nodal/Activin/TGFβ branches of the TGFβ superfamily. To test this hypothesis, we analyzed the effects of TRAF4 gain and loss of function on responses to TGFβ family ligands in Xenopus animal cap explants.
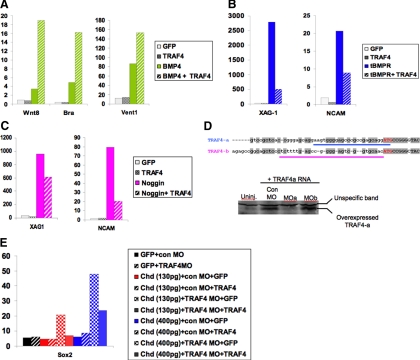
First, we tested whether TRAF4 affects BMP4 signaling by gain of function (Figure 5A) in Xenopus animal cap explants. We found that, compared with BMP4 overexpression alone, coexpression of BMP4 with TRAF4 significantly boosted induction of the general mesoderm marker Brachyury (Bra) and the ventral mesoderm marker Wnt8. TRAF4 also enhanced induction of the ventral ectoderm/mesoderm marker Vent1 (Figure 6A) in response to BMP4. Notably, TRAF4 overexpression on its own was not sufficient to activate the transcription of these BMP target genes. These results demonstrate that TRAF4 can exert a positive influence on BMP signaling, but its overexpression alone is not sufficient to activate the BMP pathway.
Figure 6.
TRAF4 is a positive regulator of BMP signaling. (A) TRAF4 enhances mesoderm induction by BMP4 in stage 10.5 animal caps. TRAF4 suppresses neural and cement gland induction (B) by a truncated, dominant-negative BMP receptor (tBR) and (C) by Noggin in stage 17 animal caps. (D) On the top is the alignment of 5′UTR regions of TRAF4a and TRAF4b immediately upstream of ATG start codon (red letters). Antisense morpholino oligo (MO) target sequences for TRAF4a (MOa) and TRAF4b (MOb) are indicated by a blue or pink underline, respectively. A western blot on the bottom shows translation of injected TRAF4a mRNA is blocked by MOa but not by MOb. Unspecific bands serve as loading control. (E) Treatment of animal caps with TRAF4 MOa promotes neural induction by a subthreshold dose of Chordin in stage 17 animal caps. In each graph, the y-axis shows the relative expression level of each gene in animal caps expressed as percentage of expression of the same gene in a stage-matched whole embryo. These expression levels are also normalized to an internal control “housekeeping” gene expressed in all cells, ornithine decarboxylase. Results are representative of two to six independent experiments.
To further test the hypothesis that TRAF4 affects BMP signaling, we checked whether TRAF4 could rescue BMP responses in animal caps cells challenged with BMP inhibitors. In isolated animal cap explants, BMP signaling naturally occurs between cells of this tissue to specify the epidermis and inhibit neural differentiation, but if these endogenous BMP signals are blocked, the cap cells acquire neural fates (Vonica and Brivanlou, 2006). To lower the level of endogenous BMP signaling, we injected animal caps with mRNA encoding either a tBMPR, which acts as a dominant-negative inhibitor (Graff et al., 1994), or the secreted BMP inhibitor Noggin. Both of these agents triggered expression of neural (neural cell adhesion molecule [NCAM]) and cement gland (XAG-1) markers, which are well known responses to BMP inhibition in animal caps (Figure 6, B and C). These effects were reversed, however, by providing exogenous TRAF4 together with either inhibitor. This demonstrates that BMP signaling can be reestablished by raising the levels of TRAF4 in cap cells. The ability of TRAF4 to rescue the effects of BMP ligand or receptor inhibitors provides additional evidence that TRAF4 is a positive effector of BMP signal transduction, acting downstream of BMP receptors.
During normal development, TRAF4 is expressed in the animal pole ectoderm at blastula and gastrula stages (Figure 5B). Therefore, we tested whether TRAF4 might function to affect BMP responses by these cells. To test this possibility, we asked whether blocking endogenous TRAF4 in animal caps would alter cap responses to BMP signals. To block each TRAF4 paralogue, we designed antisense MOs to interfere with translation of either the TRAF4a or TRAF4b mRNA, by targeting sequences in the 5′UTR, just upstream of the ATG start codon of each transcript (Figure 6D). These target sequences are sufficiently divergent (sharing <50% identity) to allow specific knockdown of each paralogue with the corresponding MOs. This is demonstrated by the ability of the TRAF4a-specific MO, but not the TRAF4b-specific MO, to inhibit translation of TRAF4a mRNA when coinjected into embryos (Figure 6D, right).
To determine whether endogenous TRAF4 functions in BMP signaling in animal cap tissue, we attempted to knock down TRAF4 by injecting MOa (or a control, random sequence MO) into animal caps, and subsequently assaying whether general neural (NCAM and Sox2) or cement gland (XAG-1) marker genes were activated. We found that injection of either control MO or TRAF4 MOa did not trigger expression of these neural markers (Figure 6E; data not shown). However, when the TRAF4 MOa was combined with a limiting dose of Chordin (a secreted BMP inhibitor) that alone was insufficient to neuralize the caps, significant neuralization was observed, as indicated by activation of the Sox2 (Figure 6E). To check whether this effect was specific for the TRAF4 target, we coinjected the TRAF4 MOa and a synthetic TRAF4 mRNA lacking the 5′UTR and MOa target site sequences (dUTR-TRAF4 or dUTR). This MOa-resistant TRAF4 mRNA significantly reversed the effects of the MO, confirming MOa targeting specificity.
The results mentioned above demonstrate that TRAF4 potentiates BMP signaling in animal pole ectoderm and that when endogenous TRAF4 is inhibited, the ectoderm becomes sensitized to the neuralizing effects of BMP inhibition. Because the latter effect in particular requires Smad1/5/8 signaling, TRAF4 most likely acts in the canonical (Smad1/5/8) BMP signaling pathway.
TRAF4 Is a Positive Regulator of Nodal Signaling
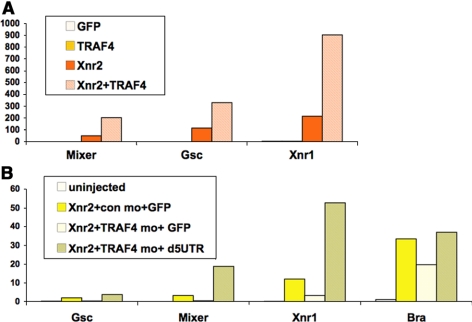
After finding that TRAF4 affects BMP responses, and is expressed in the marginal zone of the early gastrula, we next asked whether TRAF4 might affect the Nodal-related signaling branch of the TGFβ pathway, particularly because Nodals are the principal mesoderm-inducing signals acting on the marginal zone of the Xenopus embryo. As with the BMP4 gain-of-function experiments described above, we injected Xenopus Nodal-related 2 (Xnr2) mRNA and TRAF4, alone or in combination, into animal caps and scored for mesoderm induction. Figure 7A demonstrates that at a very low dose of Xnr2 (10 pg), sufficient to elicit a significant yet limited mesoderm induction response, coinjection of TRAF4 significantly enhanced induction of genes that mark mesoderm (goosecoid, Xenopus Nodal-related 1 or Xnr1) and mesendoderm (mixer). Overexpression of TRAF4 alone, however, did not induce these marker genes at any dose tested up to 4 ng. We also performed loss-of-function tests in animal caps to determine whether endogenous TRAF4 is required for mesoderm induction by Xenopus Nodal-related ligands. Animal caps were injected with control or TRAF4 MOs and treated with Xnr2. Figure 7B shows that whereas a 5-pg dose of Xnr2 induced mesoderm (goosecoid, Xnr1, brachyury) and mesendoderm (mixer) genes, addition of the TRAF4 Moa, but not a control MO, eliminated or significantly reduced induction of these markers. Therefore, we conclude that TRAF4 can enhance signaling activity in both major branches of canonical TGFβ signaling: BMP and nodal/activin/TGFβ, and furthermore that endogenous TRAF4 is necessary for animal caps to respond to these pathways.
Figure 7.
TRAF4 is a positive regulator of Nodal signaling. (A) TRAF4 enhances mesoderm induction by Xenopus nodal 2 (Xnr2) in Xenopus animal caps. Xnr2 mRNA (10 pg) was injected at the two-cell stage into the animal pole of Xenopus embryos. Animal caps were cut at stage 8, and mesoderm marker expression was scored in animal caps harvested at early gastrula, stage 10.5. (B) Treatment of animal caps with TRAF4 MOa, but not a control MO (con mo), reduces mesoderm induction by Xnr2 (5 pg). Coinjection of TRAF4 mRNA lacking the MO target sequence (d5UTR), rescues the response of animal caps to Xnr2. The y-axis shows relative expression levels and normalization was done as explained in Figure 5 legend.
TRAF4 Has Negligible Effects on Wnt and FGF Pathways
Having found that TRAF4 enhances gene expression responses to both major branches of TGFβ signaling, we wondered whether TRAF4 might affect other signaling pathways, particularly the Wnt and FGF pathways because, in addition to TGFβ signals, these govern germ layer induction and early patterning in the Xenopus embryo. We used animal cap assays to test whether TRAF4 affected responses to Wnt8 and enhanced embryonic FGF (eFGF) and found that expression of Wnt target genes Xnr3 and Siamois were not altered by TRAF4 overexpression, and eFGF responses (induction of brachyury, a direct FGF target gene) were at most slightly enhanced (1.5-fold) by TRAF4, compared with eFGF alone (Supplemental Figure 2). In contrast, TRAF4 significantly boosted BMP4 and Xnr2 target gene expression some 4- to 20-fold. The results demonstrate that TRAF4 is predominantly specific to TGFβ pathways in the context of early Xenopus embryonic tissue.
MO Knockdown of TRAF4 Causes Neural Tube Closure, Anterior and Axial Defects
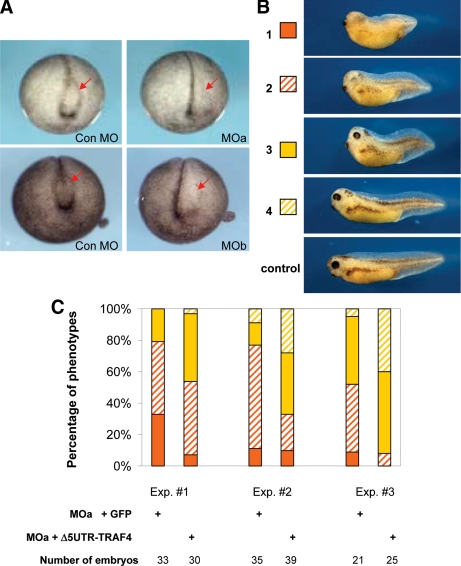
We investigated the requirement for TRAF4 in Xenopus development by targeting both TRAF4 paralogues in developing embryos, with paralogue-specific MOs. We found that early cleavage stage embryos injected into the dorsal marginal zone with MOs for either TRAF4a (MOa) or TRAF4b (MOb) developed normally through gastrulation. However, anterior defects were apparent at the beginning of neurulation in embryos that were injected at the four-cell stage into the two dorsal/anterior-fated blastomeres. Similar effects are shown in Figure 8A, in which either MOa or MOb was injected into the left side of the embryo, in one dorsal marginal zone blastomere at the four-cell stage, or one dorsal-animal pole blastomere at the eight-cell stage. Either type of targeting resulted in failed neural fold formation on the injected side in >90% of accurately targeted embryos (n ≥ 200; targeting verified by LacZ lineage tracing). Embryos injected with the control MO (con MO) were normal (Figure 8A).
Figure 8.
General effects of TRAF4 loss of function and rescue by TRAF4 mRNA. (A) Unilateral (left-sided) injection of TRAF4a MO (MOa) or TRAFb MO (MOb), but not the control MO (Con MO), causes loss of the neural fold, particularly the neural fold hingepoints on the injected side, which is indicated by arrows (B) Classification of phenotypes caused by injection of TRAF4a MO into dorsal marginal zone, from most severe to minor (1–4) at stage 34: 1, almost complete loss of eyes and pigment, very short axis; 2, rudimentary eyes, severe loss of pigments, short axis; 3, moderate loss of eyes and pigments, short axis; and 4, minor defects in eyes, axis, and pigmentation. Refer to the graph in C for quantification of phenotype frequencies. Note reduced pigments (derivatives of the neural crest) in phenotypes 1 and 2. (C) Developmental defects caused by TRAF4a MO (MOa) were partially rescued by MO-resistant TRAF4a (ΔUTR-TRAF4) mRNA. The graphs show percentage of embryos with phenotypes from 1 to 4, represented by colors next to embryo pictures in B. Results from three independent rescue experiments are shown. Note the shift in percentage of severe phenotypes toward minor phenotypes, when dUTR-TRAF4 mRNA is coinjected with MOa. Injection of GFP mRNA served as a negative control.
In addition to neural plate folding defects, we also observed general head and dorsal axial defects when either MOa or MOb was injected bilaterally into the dorsal marginal zone at the four-cell stage, which would target both the mesoderm and neural plate (Figure 8B). At tadpole stages, these embryos exhibited eye loss, reduced head size, a shortened anteroposterior axis, and loss of pigment cells (a neural crest derivative). In any gene knockdown experiment, it is important to demonstrate specificity by a rescue experiment. We achieved this by successfully reversing the head, neural plate, and neural crest phenotypes caused by the TRAF4 MOa (Figure 8, B and C) by coinjection of the same synthetic TRAF4a mRNA lacking the MO binding site (dUTR-TRAF4) that was used in the animal cap rescue experiments in Figures 6 and 7.
Loss of TRAF4 Disrupts Neural Crest Formation
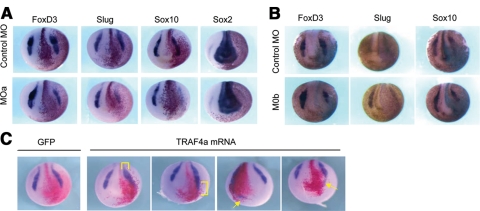
The distinct expression of TRAF4 in both cranial and trunk neural crest, combined with the head and pigmentation defects observed in TRAF4 MO-injected embryos, implicate TRAF4 in neural crest formation. To test whether TRAF4 is required for neural crest development, we targeted prospective neural crest cells with the TRAF4 MOs, by injecting adjacent dorsal and ventral animal pole blastomeres on the left side, at the eight-cell stage (10–12.5 ng of MO/cell). LacZ mRNA was included as a lineage tracer, and the resulting embryos were scored for neural crest gene expression at midneurula stages. Our results show that MO targeting of either TRAF4a or TRAF4b significantly reduced the expression of neural crest markers Slug, Foxd3, and Sox10 in descendents of the injected cells (Figure 9, A and B). In contrast, the expression level of a pan-neural marker, Sox2, was not affected, although the territory of Sox2 sometimes seemed slightly expanded due to the neural plate remaining wide and flat as a result of disrupted neural folding. These results were observed in 60–80% of the embryos in multiple independent experiments, in each of which 20 embryos or more were scored. Therefore, we conclude that both of the TRAF4 paralogues are essential for normal neural crest cell differentiation and neural plate folding.
Figure 9.
TRAF4 depletion and overexpression affects neural crest markers. (A) Unilateral (left-sided) injection of TRAF4a MO or (B) TRAF4b MO decreases expression of neural crest markers (Foxd3, Slug, and Sox10) at stage 17 on the injected side, which is designated by LacZ staining, red (A) and blue (B) stains. (C) Unilateral injection of TRAF4a mRNA produces ectopic (arrows) and expanded neural crest (brackets), demonstrated by in situ hybridization for the neural crest marker Slug. GFP mRNA was injected as control, red stain corresponds to lineage tracer LacZ which indicates the injected cells.
TRAF4 Overexpression in the Neural Plate Expands the Neural Crest
Next, we tested whether TRAF4 might cause a gain of function phenotype when overexpressed in the ectoderm. Because the two TRAF4 paralogues encode proteins that are 96% identical and thus anticipated to function identically, we only tested TRAF4a in these experiments. Adjacent dorsal and ventral animal pole blastomeres on one side of eight–cell-stage embryos were injected with either TRAF4 mRNA or control GFP mRNA, together with LacZ mRNA as a lineage tracer. Neural crest formation was monitored by in situ expression of the neural crest-specific gene Slug. When TRAF4 injections were targeted to the region of the neural plate and adjacent ectoderm, we observed expanded Slug expression in lateral and anterior regions adjacent to the normal Slug expression domain (Figure 9C). In some cases, we also observed Slug expression at ectopic locations at the rim of the anterior neural plate. We did not observe ectopic neural crest development, however, in every ectodermal cell that received TRAF4 mRNA, and we never observed ectopic neural crest marker expression within the neural plate. Ectopic crest formation was triggered by TRAF4 only in what seems to be “crest competent” ectodermal cells that are spatially restricted to the border of the anterior neural plate (Figure 9C). Consistent with these findings, overexpression of TRAF4 on its own, in isolated animal cap explants, did not induce any key neural crest marker genes (Msx1, Foxd3, and Twist; data not shown). Therefore, we conclude that although TRAF4 is not capable of functioning independently to initiate neural crest differentiation in the ectoderm, its ectopic expression in regions adjacent to the neural crest may affect signaling pathways that do so.
TRAF4 and Smurf1 Both Are Expressed in the Xenopus Neural Plate and Neural Crest
The expression pattern of TRAF4, its requirement for normal neural crest development, and the biochemical interactions between Smurf1 and TRAF4, prompted the question of whether a regulatory relationship exists between these two proteins during embryonic development, particularly in the neural crest and neural plate. To ascertain whether such a relationship might be possible, we directly compared the embryonic expression patterns of TRAF4 and Smurf1 by single and double-probe WISH (Figure 10). Previous studies have shown that Smurf1 is expressed in the nascent mesoderm before gastrulation, the axial and paraxial mesoderm at gastrula and neurula stages, and the neural plate where it is required for anterior neural patterning and neural fold formation (Zhu et al., 1999; Alexandrova and Thomsen, 2006). Whether Smurf1 is expressed in the neural crest, however, was not specifically investigated.
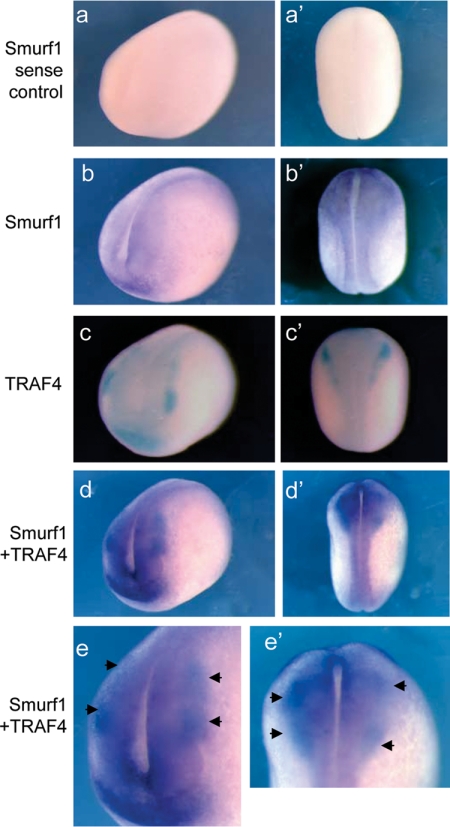
Figure 10.
Expression patterns of TRAF4 and Smurf1 overlap during midneurula stage. Antero-lateral (a–d) and dorsal (a′–d′) views of stage 17 embryos processed by WISH. Single-probe WISH shows expression of Smurf1 in the neural plate and the surrounding region including the neural crest and the cement gland (b and b′). Sense controls for Smurf1 are shown in a and a′. Single-probe WISH using BCIP as substrate shows TRAF4 expression (c and c′). Double-probe WISH shows expression of Smurf1 (purple) and TRAF4 (blue) overlap in the neural plate, neural crest and cement gland (d and d′). Enlarged anterior and dorsal views of the overlapping expression regions are shown in e and e′, respectively. Arrows point to expression of TRAF4 in the neural crest (blue), which lies within the regions of Smurf1 expression (purple) (e and e′).
We re-examined the pattern of Smurf1 expression in midneurula stage embryos and found that it displays a graded expression pattern, with strongest expression in the neural plate and immediately adjacent ectoderm where the neural crest arises. Outside of those areas, in the territory that will form epidermis, Smurf1 expression is significantly lower (Figure 10, b, b′, d, and d′). This pattern of Smurf1 expression contrasts that of TRAF4, which is expressed predominantly in the neural crest and at lower levels in the neural plate (Figure 10, c and c′; also see Figure 5). Like Smurf1, TRAF4 is not expressed significantly in the late neurula ectoderm. Single and double WISH analysis revealed that TRAF4 expression in neural plate and neural crest cells overlap with the expression domain of Smurf1 (Figure 10, d, e, d′, and e′). In addition, TRAF4 and Smurf1 are coexpressed in the region of anterior ectoderm that will form the cement gland (compare Figure 10, a–d). These findings are consistent with the possibility that TRAF4 protein levels in the neural plate and neural crest could be regulated by the ubiquitin ligase activity of Smurf1, a possibility that requires further testing.
DISCUSSION
In this study, we report new signaling and developmental activities for TRAF4, in the context of the Xenopus embryo. Although it is a member of the TRAF family of receptor adaptors that characteristically function in cytokine signaling, the role of TRAF4 in endogenous situations has eluded definition. At the biochemical level, we have identified TRAF4 as a new substrate of the E3 ubiquitin ligase, Smurf1, which we have shown binds to and polyubiquitylates TRAF4 to trigger its destruction by the 26S proteasome. In the context of signal transduction, we have found that TRAF4 is a positive effector of BMP and Nodal-related signaling branches of the TGFβ superfamily, and in the Xenopus embryo we have shown that TRAF4 is expressed within the neural plate and neural crest and that TRAF4 is essential for normal neural plate folding and neural crest development.
We have found that TRAF4 acts as a positive effector of BMP and Nodal/Activin/TGFβ signaling in the context of the Xenopus embryo, and particularly the animal cap, which consists of multipotent cells capable of differentiating into ecto-, meso- or endodermal tissues in response to particular types and doses of TGFβ ligands (including Nodals, Vg1, Activin, and BMPs). We found that TRAF4 can significantly enhance mesoderm induction by BMP4 and reverse the neuralizing effects of BMP antagonists. Conversely, when endogenous TRAF4 is reduced by morpholino knockdown, cap neuralization by BMP inhibitors is significantly enhanced. We also observed that TRAF4 enhanced mesoderm induction by overexpressed Smad1 (data not shown). These lines of evidence argue that TRAF4 most likely functions in canonical Smad1/5/8 signaling, rather than a side pathway. We also found that TRAF4 functions in Nodal signaling, as adding TRAF4 to animal caps boosts mesoderm induction by Nodal-related 2 ligand (Xnr2), and knockdown of endogenous TRAF4 reduces responses to Xnr2. The effects of TRAF4 in Xenopus embryonic assays are rather specific to TGFβ pathways, as we observed only minor effects of overexpressed TRAF4 on animal cap responses to eFGF, and no effects of TRAF4 on cap responses to Wnt8. The results of our signaling tests demonstrate that TRAF4 is necessary for full responses of Xenopus ectodermal cells to BMP and nodal signals, revealing a novel function for this member of the TRAF family.
In developing Xenopus embryos, endogenous Nodal-related ligands function to induce mesoderm within the marginal zone, a region where we have shown TRAF4 is normally expressed. The shorter body-axis and reduced head structures we observed in some TRAF4 morphants are consistent with a possible reduction in mesoderm development; however, an examination of morphant embryos injected in the marginal zone with the TRAF4a morpholino did not reveal notable alterations in general or organizer-specific mesodermal gene expression (Supplemental Figure 1). One possibility is that the TRAF4 morpholino has subtle, yet aggregate, long-term effects on the mesoderm during gastrula and neurula stages, resulting in foreshortened embryos. It is also possible that the axial defects are secondary to morphogenetic defects in the neural plate, discussed next.
The neural plate folding abnormalities we observed in TRAF4 morphant embryos support a role for TRAF4 in morphogenesis of the neural plate. In the early neurula, TRAF4 is expressed at a low but detectable level in the neural plate and is enriched at neural plate hingepoints. With respect to the latter, TRAF4 knockdown inhibited formation of hingepoints and also interfered with neural plate folding and dorsal neural tube closure. These neural tube closure defects resemble the phenotypic effects seen in TRAF4 knockout mice, in which midfetal to full-term TRAF4 mutants showed defective neural tube closure (Regnier et al., 2002). These mouse embryonic defects occurred in the absence of overt changes in neural plate gene expression, similar to our observation of normal Sox2 expression in the neural plate of TRAF4 morphant Xenopus embryos. Thus, TRAF4 likely regulates neural tube morphogenesis rather than specification of neural fate. The phenotypes of the TRAF4 knockout mice were scored after day E13.5, when organogenesis was well underway, so the early embryonic basis of the reported defects were not revealed, and no links to molecular pathways were investigated in that study.
Whether reduced of BMP/Smad1 or nodal/Smad2 signaling in TRAF4 knockdown Xenopus or knockout mouse embryos is directly responsible for abnormal neural plate morphogenesis remains to be determined, but other alternatives are also possible. TRAF4 might regulate neural fold formation and neural plate closure through interactions with various potential partners such as mitogen-activated protein kinase kinase kinase (MEKK) 4, a mitogen-activated protein kinase (MAPK) kinase that can be activated by TRAF4 (Abell and Johnson, 2005) and is required for mouse neural plate closure (Abell et al., 2005). TRAF4 also has been shown essential for epithelial tight junction formation (Kedinger et al., 2008), which also may explain why TRAF4 knockdown interferes with neural plate hingepoint formation and folding. In frog and mouse embryos, cell shape changes and cytoskeletal rearrangements associated with neural tube closure are controlled by actin-binding or actin-regulating proteins, such as Shroom, RhoGAP190, Arg/Abl, Vinculin, and Mena (Koleske et al., 1998; Xu et al., 1998; Lanier et al., 1999; Brouns et al., 2000; Haigo et al., 2003). Physical interactions have been reported between TRAF4 and proteins that regulate cytoskeletal organization, including ArgBP2 (an Arg/Abl binding protein), Hic5 (a focal adhesion protein), Pyk2 (a focal adhesion kinase), myosin heavy chain 9, and actinin (Xu et al., 2002; Wu et al., 2005; Rozan and El-Deiry, 2006). Also, we have shown previously that Smurf1 is essential for Xenopus neural plate folding (Alexandrova and Thomsen, 2006), and its link to TRAF4 we report here could be relevant to that function.
Our expression and knockdown analyses uncovered an unexpected role for TRAF4 the neural crest developmental program. Knockdown of either paralogue of TRAF4 in the cranial and trunk neural crest domains significantly inhibited anterior neural crest marker gene expression and differentiation of mature trunk melanocytes, revealing that TRAF4 is essential for neural crest development. A key question is, which process of neural crest differentiation does TRAF4 affect? Several signaling pathways are known to operate in neural crest specification and differentiation, namely, BMP, Wnt, FGF, and Notch, with BMP and Wnt featuring predominantly in early specification (Labonne and Bronner-Fraser, 1998; Bastidas et al., 2004). Because we found that TRAF4 enhances BMP signaling in Xenopus ectoderm, the most parsimonious hypothesis is that TRAF4 affects neural crest induction, differentiation, or both by modulating BMP signals in the nonneural ectoderm bordering the neural plate, which is a territory that is competent to form neural crest (Bastidas et al., 2004). Interestingly, overexpression of TRAF4 in this territory caused expansion of the neural crest. A possible explanation for this effect is that elevation of TRAF4 in the neural plate border leads to establishment of a larger domain that can attain the dose of BMP signaling that is permissive for neural crest formation. Although some earlier studies showed that overactivation of BMP signaling caused a reduction of the neural crest, it should be noted that in those studies the mRNA injections were done at the two-cell stage, which most likely disrupted the formation of the dorsal mesoderm, a tissue that provides essential signals (such as BMP inhibitors) for induction of the neural crest. Conversely, injection of BMP inhibitors at the one- or two-cell stage was shown to expand the neural crest, presumably as a secondary effect of expanded dorsal mesoderm (LaBonne and Bronner-Fraser, 1998; Wawersik et al., 2005). In contrast to those studies, in our work we targeted TRAF4 mRNA specifically into the neural crest region by injecting animal blastomeres at the 8- or 16-cell stage, which minimizes, if not completely eliminates, exposure of the underlying mesoderm to exogenous TRAF4 mRNA.
Alternatively, TRAF4 could affect neural crest formation by modifying other signaling pathways or by acting independently as a neural crest inducer. Elevating Wnt signals in the neurectoderm will expand neural crest. Also, FGF signals are known to affect neural crest induction in Xenopus by inducing and cooperating with Wnt signals (Monsoro-Burq et al., 2005; Hong et al., 2008). It is formally possible that TRAF4 affects those pathways in crest development. However, we did not observe an effect of TRAF4 on animal cap responses to Wnt8 ligand, but we did find that overexpressed TRAF4 has a slight, positive effect on animal cap responses to eFGF. If not BMP signaling, it is possible that TRAF4 affects crest development by boosting FGF signaling, which indirectly stimulates Wnt signaling. Furthermore, TRAF4 might affect neural crest development through MAPK kinase 4 or other partner proteins and pathways, but because TRAF4 overexpression alone is insufficient to trigger neural crest differentiation in naïve nonneural ectoderm, TRAF4 probably functions as a obligate partner in neural crest signaling and differentiation pathways, as opposed to being a “master regulator” or “inducer” of neural crest.
This study commenced with our isolation of TRAF4 as an interacting partner of Smurf1 in a yeast two-hybrid screen, and we showed that this interaction can result in polyubiquitylation of TRAF4 and its destruction by the 26 proteasome. We also showed that this interaction is physiologically relevant in a human kidney cell line (HEK293), because knockdown of Smurf1 with siRNA results in elevated TRAF4 protein levels. Thus, Smurf1 can regulate TRAF4 when the two proteins are coexpressed in cells. In the Xenopus embryo, Smurf1 and TRAF4 are coexpressed in several tissues, including the nascent mesodermal germ layer and derived notochord, somite and muscle tissues, as well as the neural plate and neural crest (Alexandrova and Thomsen, 2006; this study). Although we demonstrate a physiological relationship between Smurf1 and TRAF4 in a mammalian cell line, the molecular mechanism of BMP and nodal/Activin/TGFβ signal enhancement by TRAF4 remains to be determined and is presently being investigated. As an interacting target of Smurf1, one possibility is that TRAF4 competes for Smad binding to Smurf1, reducing Smad ubiquitylation and degradation, thereby raising Smad levels and signaling output. Another possibility is that TRAF4 interacts with receptors, affecting their activity. Recently, TRAF6 was shown to bind TGFβ receptors to affect TAK1-p38/JNK signaling, but not Smad signaling (Sorrentino et al., 2008; Yamashita et al., 2008).
Posttranslational modifications such as SUMOylation are known to regulate key neural crest regulatory proteins (Taylor and LaBonne, 2007), and we suggest that the overlapping patterns of Smurf1 and TRAF4 result in Smurf1 regulating TRAF4 in the neural crest and neural plate.
More specifically, we postulate that Smurf1 functions to govern TRAF4 protein levels through ubiquitin-dependent proteasomal degradation, thereby limiting the positive influence of TRAF4 on BMP, and possibly FGF signals. This has yet to be demonstrated directly and remains a hypothesis to be tested. The capacity of Smurf1 to target TRAF4 degradation also might influence other functions of TRAF4, such as in vivo interactions with various protein partners (discussed above) or in MEKK4 signaling in mammalian cells (Abell and Johnson, 2005). The functional relationship between Smurf1 and TRAF4 in diverse cell signaling, differentiation and embryonic scenarios warrants further investigation. Furthermore, the ability of Smurf1 to target TRAF4 for degradation raises the possibility that Smurf1 might target other members of the TRAF family to regulate their actions in other cellular contexts, such as TNF and Toll-like/IL-1 signaling. Recently, Smurf2 was shown to interact with TRAF2 to affect TNFR ubiquitylation and destruction (Carpentier et al., 2008).
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Bar-Sagi, C. LaBonne, T. Bouwmeester, J. Saint-Jeannet, and J. C. Hsieh for gifts of plasmids or other reagents. We thank members of the Thomsen laboratory, B. Holdener, and H. Sirotkin for helpful comments and discussion. This work was supported by National Institutes of Health grant R01 GM-076599 (to G.H.T.) and a NYSTEM Institutional Development Award to Stony Brook University.
Abbreviations used:
- BMP
bone morphogenetic protein
- FGF
fibroblast growth factor
- MO
morpholino oligonucleotide
- TGF
transforming growth factor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0325) on May 20, 2009.
REFERENCES
- Abell A. N., Johnson G. L. MEKK4 is an effector of the embryonic TRAF4 for JNK activation. J. Biol. Chem. 2005;280:35793–35796. doi: 10.1074/jbc.C500260200. [DOI] [PubMed] [Google Scholar]
- Abell A. N., et al. Ablation of MEKK4 kinase activity causes neurulation and skeletal patterning defects in the mouse embryo. Mol. Cell Biol. 2005;25:8948–8959. doi: 10.1128/MCB.25.20.8948-8959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal B. B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- Alexandrova E. M., Thomsen G. H. Smurf1 regulates neural patterning and folding in Xenopus embryos by antagonizing the BMP/Smad1 pathway. Dev. Biol. 2006;299:398–410. doi: 10.1016/j.ydbio.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L., Wrana J. L. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- Barth K. A., Kishimoto Y., Rohr K. B., Seydler C., Schulte-Merker S., Wilson S. W. Bmp activity establishes a gradient of positional information throughout the entire neural plate. Development. 1999;126:4977–4987. doi: 10.1242/dev.126.22.4977. [DOI] [PubMed] [Google Scholar]
- Bastidas F., De Calisto J., Mayor R. Identification of neural crest competence territory: role of Wnt signaling. Dev. Dyn. 2004;229:109–117. doi: 10.1002/dvdy.10486. [DOI] [PubMed] [Google Scholar]
- Birsoy B., Kofron M., Schaible K., Wylie C., Heasman J. Vg 1 is an essential signaling molecule in Xenopus development. Development. 2006;133:15–20. doi: 10.1242/dev.02144. [DOI] [PubMed] [Google Scholar]
- Bonni S., Wang H. R., Causing C. G., Kavsak P., Stroschein S. L., Luo K., Wrana J. L. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- Brouns M. R., Matheson S. F., Hu K. Q., Delalle I., Caviness V. S., Silver J., Bronson R. T., Settleman J. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–4903. doi: 10.1242/dev.127.22.4891. [DOI] [PubMed] [Google Scholar]
- Carpentier I., Coornaert B., Beyaert R. Smurf2 is a TRAF2 binding protein that triggers TNF-R2 ubiquitination and TNF-R2-induced JNK activation. Biochem. Biophys. Res. Commun. 2008;374:752–757. doi: 10.1016/j.bbrc.2008.07.103. [DOI] [PubMed] [Google Scholar]
- Cherfils-Vicini J., Vingert B., Varin A., Tartour E., Fridman W. H., Sautes-Fridman C., Regnier C. H., Cremer I. Characterization of immune functions in TRAF4-deficient mice. Immunology. 2008;124:562–574. doi: 10.1111/j.1365-2567.2008.02810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J. Y., Park Y. C., Ye H., Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- Crane J. F., Trainor P. A. Neural crest stem and progenitor cells. Annu. Rev. Cell Dev. Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- De Robertis E. M., Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey P. W., Doyle S. E., He J. Q., Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/s1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Esparza E. M., Arch R. H. TRAF4 functions as an intermediate of GITR-induced NF-kappaB activation. Cell Mol. Life Sci. 2004;61:3087–3092. doi: 10.1007/s00018-004-4417-0. [DOI] [PubMed] [Google Scholar]
- Glavic A., Maris Honore S., Gloria Feijoo C., Bastidas F., Allende M. L., Mayor R. Role of BMP signaling and the homeoprotein Iroquois in the specification of the cranial placodal field. Dev. Biol. 2004a;272:89–103. doi: 10.1016/j.ydbio.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Glavic A., Silva F., Aybar M. J., Bastidas F., Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004b;131:347–359. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Graff J. M., Thies R. S., Song J. J., Celeste A. J., Melton D. A. Studies with a Xenopus BMP receptor suggest that ventral mesoderm-inducing signals override dorsal signals in vivo. Cell. 1994;79:169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- Haigo S. L., Hildebrand J. D., Harland R. M., Wallingford J. B. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 2003;13:2125–2137. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- Harland R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harris M. L., Erickson C. A. Lineage specification in neural crest cell pathfinding. Dev. Dyn. 2007;236:1–19. doi: 10.1002/dvdy.20919. [DOI] [PubMed] [Google Scholar]
- Hayata T., Eisaki A., Kuroda H., Asashima M. Expression of Brachyury-like T-box transcription factor, Xbra3 in Xenopus embryo. Dev. Genes Evol. 1999;209:560–563. doi: 10.1007/s004270050289. [DOI] [PubMed] [Google Scholar]
- Hong C. S., Park B. Y., Saint-Jeannet J. P. Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development. 2008;135:3903–3910. doi: 10.1242/dev.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Kedinger V., Alpy F., Baguet A., Polette M., Stoll I., Chenard M. P., Tomasetto C., Rio M. C. Tumor necrosis factor receptor-associated factor 4 is a dynamic tight junction-related shuttle protein involved in epithelium homeostasis. PLoS ONE. 2008;3:e3518. doi: 10.1371/journal.pone.0003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedinger V., Rio M. C. TRAF4, the unique family member. Adv. Exp. Med. Biol. 2007;597:60–71. doi: 10.1007/978-0-387-70630-6_5. [DOI] [PubMed] [Google Scholar]
- Kofron M., Klein P., Zhang F., Houston D. W., Schaible K., Wylie C., Heasman J. The role of maternal axin in patterning the Xenopus embryo. Dev. Biol. 2001;237:183–201. doi: 10.1006/dbio.2001.0371. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- LaBonne C., Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Lanier L. M., Gates M. A., Witke W., Menzies A. S., Wehman A. M., Macklis J. D., Kwiatkowski D., Soriano P., Gertler F. B. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Le Douarin N. M., Dupin E. Multipotentiality of the neural crest. Curr. Opin. Genet. Dev. 2003;13:529–536. doi: 10.1016/j.gde.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Li J. M., Fan L. M., Christie M. R., Shah A. M. Acute tumor necrosis factor alpha signaling via NADPH oxidase in microvascular endothelial cells: role of p47phox phosphorylation and binding to TRAF4. Mol. Cell Biol. 2005;25:2320–2330. doi: 10.1128/MCB.25.6.2320-2330.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Liang M., Feng X. H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Marchant L., Linker C., Ruiz P., Guerrero N., Mayor R. The inductive properties of mesoderm suggest that the neural crest cells are specified by a BMP gradient. Dev. Biol. 1998;198:319–329. [PubMed] [Google Scholar]
- Mayor R., Morgan R., Sargent M. G. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A. H., Fletcher R. B., Harland R. M. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq A. H., Wang E., Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Moren A., Imamura T., Miyazono K., Heldin C. H., Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 2005;280:22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- Murakami G., Watabe T., Takaoka K., Miyazono K., Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. K., Nelson R. W. Cdc42 Effector Protein 2 (XCEP2) is required for normal gastrulation and contributes to cellular adhesion in Xenopus laevis. BMC Dev. Biol. 2004;4:13. doi: 10.1186/1471-213X-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. H., Schmid B., Trout J., Connors S. A., Ekker M., Mullins M. C. Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Dev. Biol. 1998;199:93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager M., Kuroda H., Reversade B., De Robertis E. M. Chordin is required for the Spemann organizer transplantation phenomenon in Xenopus embryos. Dev. Cell. 2003;4:219–230. doi: 10.1016/s1534-5807(02)00404-5. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager M., Tran U., Grubisic K., De Robertis E. M. Identification of a second Xenopus twisted gastrulation gene. Int. J. Dev. Biol. 2004;48:57–61. doi: 10.1387/ijdb.15005575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B., Bose R., Barrios-Rodiles M., Wang H. R., Zhang Y., Wrana J. L. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- Regnier C. H., Masson R., Kedinger V., Textoris J., Stoll I., Chenard M. P., Dierich A., Tomasetto C., Rio M. C. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc. Natl. Acad. Sci. USA. 2002;99:5585–5590. doi: 10.1073/pnas.052124799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozan L. M., El-Deiry W. S. Identification and characterization of proteins interacting with Traf4, an enigmatic p53 target. Cancer Biol. Ther. 2006;5:1228–1235. doi: 10.4161/cbt.5.9.3295. [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet J. P., He X., Varmus H. E., Dawid I. B. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc. Natl. Acad. Sci. USA. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G., Alarcon C., Spagnoli F. M., Brivanlou A. H., Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Shen M. M. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Shen R., Chen M., Wang Y. J., Kaneki H., Xing L., O'Keefe R. J., Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J. Biol. Chem. 2006;281:3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Siegel P. M., Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N., Zhang S., Heldin C. H., Landström M. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- Suzuki C., Murakami G., Fukuchi M., Shimanuki T., Shikauchi Y., Imamura T., Miyazono K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 2002;277:39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- Takeshita F., et al. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur. J. Immunol. 2005;35:2477–2485. doi: 10.1002/eji.200526151. [DOI] [PubMed] [Google Scholar]
- Taylor K. M., LaBonne C. Modulating the activity of neural crest regulatory factors. Curr. Opin. Genet. Dev. 2007;17:326–331. doi: 10.1016/j.gde.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Turner D. L., Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Villanueva S., Glavic A., Ruiz P., Mayor R. Posteriorization by FGF, Wnt, and retinoic acid is required for neural crest induction. Dev. Biol. 2002;241:289–301. doi: 10.1006/dbio.2001.0485. [DOI] [PubMed] [Google Scholar]
- Vonica A., Brivanlou A. H. An obligatory caravanserai stop on the silk road to neural induction: inhibition of BMP/GDF signaling. Semin. Cell Dev. Biol. 2006;17:117–132. doi: 10.1016/j.semcdb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Waite K. A., Eng C. From developmental disorder to heritable cancer: it's all in the BMP/TGF-beta family. Nat. Rev. Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., Wrana J. L. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wawersik S., Evola C., Whitman M. Conditional BMP inhibition in Xenopus reveals stage-specific roles for BMPs in neural and neural crest induction. Dev. Biol. 2005;277:425–442. doi: 10.1016/j.ydbio.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Wu R. F., Xu Y. C., Ma Z., Nwariaku F. E., Sarosi G. A., Jr, Terada L. S. Subcellular targeting of oxidants during endothelial cell migration. J. Cell Biol. 2005;171:893–904. doi: 10.1083/jcb.200507004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Baribault H., Adamson E. D. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Xu Y. C., Wu R. F., Gu Y., Yang Y. S., Yang M. C., Nwariaku F. E., Terada L. S. Involvement of TRAF4 in oxidative activation of c-Jun N-terminal kinase. J. Biol. Chem. 2002;277:28051–28057. doi: 10.1074/jbc.M202665200. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Fatyol K., Jin C., Wang X., Liu Z., Zhang Y. E. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., et al. TRAF family proteins interact with the common neurotrophin receptor and modulate apoptosis induction. J. Biol. Chem. 1999;274:30202–30208. doi: 10.1074/jbc.274.42.30202. [DOI] [PubMed] [Google Scholar]
- Ying S. X., Hussain Z. J., Zhang Y. E. Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2-induced osteoblast conversion by targeting Smad5 for degradation. J. Biol. Chem. 2003;278:39029–39036. doi: 10.1074/jbc.M301193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota C., Kofron M., Zuck M., Houston D. W., Isaacs H., Asashima M., Wylie C. C., Heasman J. A novel role for a nodal-related protein; Xnr3 regulates convergent extension movements via the FGF receptor. Development. 2003;130:2199–2212. doi: 10.1242/dev.00434. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Qiao M., Oyajobi B. O., Mundy G. R., Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J. Biol. Chem. 2003;278:27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.