Abstract
The endoplasmic reticulum (ER) glycoprotein HMG-CoA reductase (HMGR) catalyzes the rate-limiting step in sterols biosynthesis. Mammalian HMGR is ubiquitinated and degraded by the proteasome when sterols accumulate in cells, representing the best example for metabolically controlled ER-associated degradation (ERAD). This regulated degradation involves the short-lived ER protein Insig-1. Here, we investigated the dislocation of these ERAD substrates to the cytosol en route to proteasomal degradation. We show that the tagged HMGR membrane region, HMG350-HA, the endogenous HMGR, and Insig-1-Myc, all polytopic membrane proteins, dislocate to the cytosol as intact full-length polypeptides. Dislocation of HMG350-HA and Insig-1-Myc requires metabolic energy and involves the AAA-ATPase p97/VCP. Sterols stimulate HMG350-HA and HMGR release to the cytosol concurrent with removal of their N-glycan by cytosolic peptide:N-glycanase. Sterols neither accelerate dislocation nor stimulate deglycosylation of ubiquitination-defective HMG350-HA(K89 + 248R) mutant. Dislocation of HMG350-HA depends on Insig-1-Myc, whose dislocation and degradation are sterol independent. Coimmunoprecipitation experiments demonstrate sterol-stimulated association between HMG350-HA and Insig-1-Myc. Sterols do not enhance binding to Insig-1-Myc of HMG350-HA mutated in its sterol-sensing domain or of HMG350-HA(K89 + 248R). Wild-type HMG350-HA and Insig-1-Myc coimmunoprecipitate from the soluble fraction only when both proteins were coexpressed in the same cell, indicating their encounter before or during dislocation, raising the possibility that they are dislocated as a tightly bound complex.
INTRODUCTION
A key mechanism for maintaining cholesterol homeostasis in mammalian cells involves modulating the stability of 3-hydroxy-3-methylglutaryl CoA reductase (HMGR). This enzyme catalyzes the rate-limiting step in the mevalonate (MVA) pathway in which essential sterols and nonsterol isoprenoids are synthesized (Goldstein and Brown, 1990). In cells deprived of sterols and/or MVA-derived isoprenoids, HMGR is a fairly stable protein that turns over with a half-life of 10–14 h. Conversely, in cells replenished with MVA pathway products, HMGR degradation is accelerated severalfold and its half-life drops to 1–4 h (Faust et al., 1982; Edwards et al., 1983; Sinensky and Logel, 1983), reflecting one of the highly regulated processes that prevent accumulation of these metabolites to toxic levels.
Similar to other proteins of the MVA pathway (Horton et al., 2002), HMGR is controlled also at the transcriptional level by the sterol regulatory element-binding protein (SREBP)-2, which binds to the promoter of the HMGR gene and activates transcription when demands for MVA-derived products increase (Osborne et al., 1985; Vallett et al., 1996). Like its closely related SREBP-1a and SREBP-1c factors, SREBP-2 is synthesized as a large endoplasmic reticulum (ER)-bound precursor whose transcription activation domain must be released from the membrane to function in the nucleus. On increased demand for MVA-derived metabolites, the SREBP precursor is transported by COP-II vesicles to the Golgi, where it is sequentially cleaved at two sites by resident site-1 and site-2 proteases. The liberated transcriptionally active domain enters the nucleus and stimulates the transcription of target genes, including HMGR (Sakai et al., 1996; Nohturfft et al., 1998a, 2000). When sterols are abundant and demand for MVA-derived products declines, the transport of the SREBP precursor out from the ER is prevented and transcription ceases (Nohturfft et al., 1999, 2000). This metabolically regulated traffic of the SREBP precursors critically depends on SREBP cleavage-activating protein (SCAP), a membrane protein with eight transmembranes spans (TMs), which tightly associates with the SREBP precursors and enables the packaging of both proteins in the COP-II vesicles and their transport from the ER to the Golgi (Hua et al., 1996; Sakai et al., 1997; Nohturfft et al., 1998b). In the presence of sterols, SCAP interacts with either one of two related resident ER membrane proteins, each with six TMs, designated Insig-1 and Insig-2 (Feramisco et al., 2004). This sterol-stimulated binding to Insig(s) masks the transport signal in SCAP and abrogates its packaging into the transport vesicles, thus indirectly immobilizes the SREBP precursor in the ER and prevents its movement to the Golgi (Yabe et al., 2002a; Yang et al., 2002). Mutations in SCAP that disrupt its binding to Insigs allow the transport of SREBP precursors to the Golgi even in sterol-replete cells (Yabe et al., 2002b).
Mammalian HMGR is an integral high mannose glycoprotein of the ER (Brown and Simoni, 1984; Liscum et al., 1985). Structurally, it is divided into two major domains: a C-terminal cytosol-facing domain that tetramerizes to form the active sites (Liscum et al., 1985; Istvan et al., 2000) and an N-terminal hydrophobic region that spans the ER membrane 8 times (Roitelman et al., 1992) and bears a single N-glycan (Liscum et al., 1983). This membrane region is dispensable for the enzymatic activity (Gil et al., 1985), but it is necessary for the metabolically controlled stability of the enzyme (Gil et al., 1985) and sufficient to confer sterol-accelerated degradation onto heterologous proteins (Skalnik et al., 1988; Cheng et al., 1999). Within this membrane region, TMs 2 through 6 bear significant sequence homology to the corresponding TMs in SCAP (Hua et al., 1996). This motif, designated sterol-sensing domain, also was recognized in other proteins that are involved in cholesterol-related processes (Kuwabara and Labouesse, 2002).
The regulated degradation of HMGR may be viewed as a special case of a more general process known as ER-associated protein degradation (ERAD). This process, a part of the cellular quality control machinery, ensures that proteins of the secretory pathway that fail to properly fold or assemble are retained in the ER and eliminated by the ubiquitin-proteasome system (for a recent review, see Nakatsukasa and Brodsky, 2008, and references therein). Indeed, the degradation of HMGR involves sterol-regulated conjugation of polyubiquitin chains to lysines 248 and 89 in the membrane region of HMGR and its eventual proteolysis by the 26S proteasome (Ravid et al., 2000; Doolman et al., 2004). DeBose-Boyd and colleagues have demonstrated that the Insig proteins play a crucial role also in HMGR degradation (Sever et al., 2003a,b). Their studies, which were carried out mostly with the short-lived Insig-1, depict a model in which the membrane region of HMGR binds to Insigs in a sterol-stimulated manner, akin to the association of SCAP with Insigs. Insig-1, but much less so Insig-2, interacts constitutively with gp78, a RING finger membrane E3 ubiquitin ligase (Fang et al., 2001; Kostova et al., 2007), which ubiquitinates Insig-1 and causes its rapid turnover (Song et al., 2005; Lee et al., 2006a). The sterols-stimulated association of Insig-1 with either SCAP or HMGR is mutually exclusive and leads to different consequences: although binding of SCAP displaces gp78 and thus stabilizes Insig-1 by preventing its ubiquitination, association of HMGR does not displace gp78 from Insig-1, but rather the bound HMGR becomes ubiquitinated and rapidly degraded (Lee et al., 2006a). Whether the Insig-1 that mediates this reaction is also ubiquitinated and degraded along with HMGR remains an open question.
To be fed into the digestive barrel of the cytosolic 26S proteasome, the polytopic HMGR must be extracted from the ER membrane. Our previous experiments with ER-derived microsomes have demonstrated that ubiquitinated HMGR becomes peripherally associated with the membrane, suggesting that it had been (at least partially) extracted and was no longer a membrane-embedded protein (Doolman et al., 2004). This extraction step, known as dislocation or retrotranslocation, is the hallmark of the ERAD pathway and it is applied to all ERAD substrates, luminal and membrane alike (Bar-Nun, 2005). The mechanism(s) that operate in dislocation and the nature of the putative channel through which ERAD substrates egress from the ER are only beginning to be elucidated, yet numerous studies have firmly established the critical role of the AAA-ATPase p97/VCP in this process (reviewed in Bar-Nun, 2005; Jentsch and Rumpf, 2007; Nakatsukasa and Brodsky, 2008). Together with its ERAD partners Ufd1p and Npl4p, and at the expense of ATP hydrolysis, this cytosolic complex is thought to drive the dislocation of most ERAD substrates and facilitate their delivery to the proteasome (Nakatsukasa and Brodsky, 2008). Indeed, p97 preferentially associates with the ubiquitinated HMGR (Doolman et al., 2004) and silencing p97 expression abrogates HMGR degradation (Song et al., 2005). However, the precise details of how HMGR is dislocated from the ER membrane remain unknown. Here, we demonstrate that, en route to proteasomal degradation, the polytopic HMGR and Insig-1 are dislocated to the cytosol as intact polypeptides. Moreover, our data indicate that these proteins tightly associate with each other before or during dislocation, suggesting that they are dislocated as a preformed complex.
MATERIALS AND METHODS
Materials
Geneticin and Lipofectamine Plus reagent were obtained from Invitrogen (Carlsbad, CA), and hygromycin B was from Roche Applied Science (Indianapolis, IN). Mevalonolactone was from Fluka (Buchs, Switzerland), and 25-hydroxycholesterol was from Steraloids (Newport, RI). MG-132, N-acetyl-leucyl-leucyl-norleucinal (ALLN), and digitonin (high purity; catalog no. 300410) were purchased from Calbiochem (San Diego, CA), Z-VAD-fmk was from Enzo Life Sciences International (Plymouth Meeting, PA), and endoglycosidase H (EndoH) was from New England Biolabs (Ipswich, MA). 35S protein labeling mix was from PerkinElmer Life and Analytical Sciences (Boston, MA), and micro-bicinchoninic acid reagent from Thermo Fisher Scientific (Waltham, MA). All other reagents were bought from Sigma-Aldrich (St. Louis, MO). Compactin was a kind gift of R. Simoni (Stanford University, Stanford, CA). Fetal bovine lipoprotein-deficient serum (LPDS; d ≥ 1.25) was prepared by ultracentrifugation, as described previously (Goldstein et al., 1983).
Plasmids
Construction of HMG350-HA in pIRESneo2 vector was described previously (Doolman et al., 2004). Insig-1-Myc cloned in pcDNA3 (Yang et al., 2002) was kindly provided by J. Goldstein (University of Texas Southwestern Medical School, Dallas, TX) and was used directly or after subcloning into pcDNA3.1/hyg vector (Clontech, Mountain View, CA). FLAG-tagged wild-type p97 and K524A mutant (Kobayashi et al., 2002), subcloned in pcDNA3.1, were kindly provided by I. Shapira (Tel Aviv University, Tel Aviv, Israel), with permission from A. Kakizuka (Kyoto University, Kyoto, Japan). Mutageneses by polymerase chain reaction were performed by the overlap extension method (Ho et al., 1989), and the authenticity of all constructs and presence of desired mutations was verified by DNA sequencing.
Cells
Human embryonic kidney (HEK) 293 cells were maintained at 37°C in a 5% CO2 atmosphere in DMEM buffered with 10 mM Na-HEPES, pH 7.4, and supplemented with 10% (vol/vol) heat-inactivated fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (medium A). Insig-deficient SRD-15 cells (Lee et al., 2005), kindly provided by R. DeBose-Boyd (University of Texas Southwestern Medical School) were grown in minimal essential medium supplemented with 5% (vol/vol) LPDS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 μg/ml 25-hydroxycholesterol. Cells were transfected using Lipofectamine Plus reagent, and stable transfectants were initially selected in the presence of 1 mg/ml Geneticin and routinely maintained in 500 μg/ml drug. For doubly transfected cells, selection was also made in 400 μg/ml hygromycin, and maintenance of stable transfectants was in 200 μg/ml drug. Transiently transfected cells were analyzed 48–60 h after transfection. To up-regulate the levels of endogenous HMGR and stabilize transfected HMG350-HA, 12–16 h before the experiments the cells were depleted of sterols by refeeding them with medium supplemented with LPDS, 2 μM compactin, and 100 μM MVA (medium B), as described previously (Ravid et al., 2000). Metabolic labeling with [35S]methionine/cysteine and immunoprecipitation of labeled proteins were done as described previously (Roitelman and Simoni, 1992; Ravid et al., 2000; Doolman et al., 2004).
Cell Fractionation, Immunoprecipitation, and Immunoblotting
Cells were cooled on ice, washed twice with phosphate-buffered saline (PBS), permeabilized for 30–60 min on ice in solution C (PBS supplemented with 5 mM EDTA, 5 mM EGTA, 2 mM phenylmethanesulfonyl fluoride, 100 μM leupeptin, and 35 μM ALLN containing 0.025% [wt/vol] digitonin) and gently pipetted off the dish. The cells were centrifuged at 20,000 × g for 30 min at 4°C to obtain supernatant and pellet fractions. In Figure 1F, the 20,000 × g supernatant fraction was further centrifuged at 100,000 × g for 30 min in a 42.2 Ti rotor (Beckman Coulter, Fullerton, CA). Both 20,000 × g and 100,000 × g pellet fractions were solubilized in solution D (same as solution C but containing 1% [vol/vol] Igepal CA-630 [NP-40], and 1% [wt/vol] Na deoxycholate [DOC] replacing digitonin) and cleared by a 30-min centrifugation at 20,000 × g. After estimation of protein content, samples were incubated with the appropriate primary antibodies and precipitated with protein A-Sepharose beads. The bound immune complexes were washed three times in solution D, for 30 min each wash, while rotating at 4°C. Proteins were resolved by 5–15% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Optitran BAS-83 nitrocellulose membranes (Whatman Schleicher and Schuell, Keene, NH). The membranes were probed with the appropriate primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reaction.
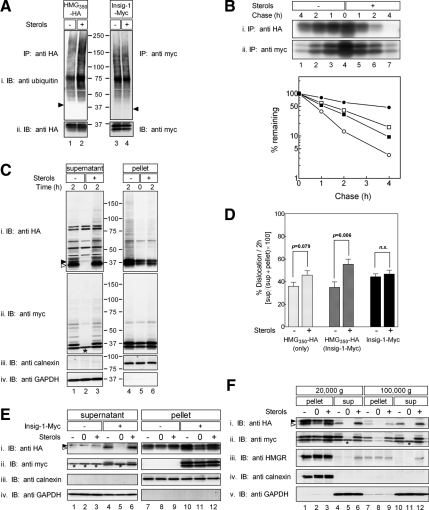
Figure 1.
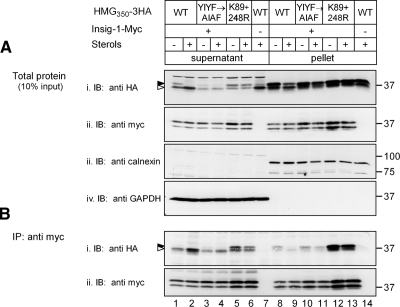
Ubiquitination, degradation, and dislocation of HMG350-HA, HMGR and Insig-1-Myc. (A) Sterol-depleted cells, stably expressing either HMG350-HA or Insig-1-Myc, were incubated for 2 h with ALLN (65 μM) with (+) or without (−) sterols (2 μg/ml 25-hydroxycholesterol plus 20 μg/ml cholesterol). Cells were lysed in solution D, and proteins from the postnuclear supernatant were immunoprecipitated (IP) with rabbit polyclonal anti-HA (lanes 1 and 2) or anti-myc (lanes 3 and 4) antibodies. The immune complexes were resolved by 5–15% SDS-PAGE, electroblotted onto nitrocellulose, and the membrane was first probed (IB) with a mouse anti-ubiquitin monoclonal antibody (top) and then reprobed with mouse anti-HA and anti-myc monoclonal antibodies (bottom). Arrowheads indicate the migration of the immunoprecipitated proteins. Mr markers (in kilodaltons) are shown. (B) Sterol-depleted cells, as in A, were pulse labeled for 30 min with 35S-protein label mix (100 μCi/dish; 300 μCi/ml) and chased for the indicated time in unlabeled medium in the absence or presence of sterols. Cells were lysed in solution D, and proteins were immunoprecipitated with the mouse monoclonal anti-HA (top) or anti-myc (bottom) antibodies. The immune complexes were resolved by SDS-PAGE followed by fluorography. Densitometric analysis of the gels is shown. •○, HMG350-HA; ■□, Insig-1-Myc; closed symbols, sterol-depleted cells; open symbols, sterol-treated cells. (C) Cells expressing both HMG350-HA and Insig-1-Myc, were incubated for the indicated time with ALLN with (+) or without (−) sterols, as described in A. Cells were permeabilized in solution C and fractionated by centrifugation, as described under Materials and Methods. Aliquots of the supernatant (60 μg of protein) and solubilized membranes (20 μg of protein) were IB and reprobed with the indicated antibodies. Closed and open arrowheads indicate glycosylated and un/deglycosylated forms of HMG350-HA, respectively. The asterisk (*) denotes a protein in the supernatant fraction that cross-reacted with the anti-myc antibody. Mr markers (in kilodaltons) are shown. (D) Densitometric analysis of experiments, as in C, of cells expressing only HMG350-HA (light gray bars; n = 12), or cells expressing both HMG350-HA and Insig-1-Myc or Insig-1-Myc alone probed with anti-HA (dark gray bars; n = 11) or with anti-Myc (black bars; n = 8) antibodies. The results (mean ± SEM) were evaluated by Student's t test, and p values are indicated; n.s., not significant. (E) On day 1, SRD-15 cells were transfected with 2 μg of pIRESneo2-HMG350-HA along with 0.5 μg of an empty vector (−) or pcDNA3.1-Insig-1-Myc (+), as indicated. On day 2, the cells were replated in triplicate dishes in medium lacking sterols but containing 50 μM lovastatin and 50 μM MVA. On day 3, the cells were harvested immediately (0) or incubated for 2 h with 65 μM ALLN in the presence (+) or absence (−) of sterols. Cells were permeabilized, and supernatant (45 μg of protein) and pellet (22 μg of protein) fractions were analyzed by immunoblotting with the indicated antibodies. Asterisks denote a protein in the supernatant fraction that cross-reacted with the anti-myc antibody. (F) Sterol-depleted cells expressing both HMG350-HA and Insig-1-Myc, were harvested immediately (0) or incubated for 2 h with 65 μM ALLN with (+) or without (−) sterols. Cells were permeabilized and fractionated by centrifugation, as described under Materials and Methods. The 20,000 × g supernatant was further centrifuged at 100,000 × g for 30 min. Samples of the pellet (100 μg of protein) and supernatant (30 μg of protein) fractions were analyzed by SDS-PAGE and IB with the indicated antibodies. Closed and open arrowheads indicate glycosylated and un/deglycosylated forms of HMG350-HA, respectively. The asterisk (*) denotes a protein in the supernatant fraction that cross-reacted with the anti-myc antibody.
Antibodies
Anti-hemagglutinin (HA) (clone 12CA5), anti-myc (clone 9E10), and anti-HMGR (clone A9) ascites fluids were produced in mice. Antiserum against the membrane region of HMGR was described previously (Roitelman et al., 1992). Rabbit anti-HA, rabbit anti-myc, and anti-FLAG mouse monoclonal (clone M2) antibodies were purchased from Sigma-Aldrich. Mouse monoclonal antibodies against ubiquitin (clone P4D1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (0411) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-calnexin antiserum was a generous gift from Ron Kopito (Stanford University). Immobilized recombinant protein A was obtained from Repligen (Waltham, MA), and all horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
RESULTS
HMG350-HA and Insig-1-Myc Are Short-lived Proteins That Dislocate from the ER Membrane as Intact Polypeptides, but Sterols Accelerate the Degradation and Dislocation Only of HMG350-HA
To investigate events related to HMGR degradation, we used the model protein HMG350-HA, an HA-tagged membrane region of HMGR. This protein was demonstrated to faithfully reflect the fate of endogenous HMGR (Cheng et al., 1999; Doolman et al., 2004). To study the role of Insig-1 in the various steps of HMGR degradation, and for lack of suitable antibodies, a myc-tagged version of Insig-1 (Yang et al., 2002) was used.
Figure 1 shows that sterols stimulated the ubiquitination of HMG350-HA (Figure 1A, lanes 1 and 2) but had no effect on the ubiquitination of Insig-1-Myc (Figure 1A, lanes 3 and 4). Consistent with these results, radioactive pulse-chase experiments demonstrated that sterols accelerated the degradation of HMG350-HA (t½ decreased from ∼200 to ∼40 min), whereas the turnover of Insig-1-Myc remained relatively unchanged (t½ ∼70 min) (Figure 1B).
To specifically follow the dislocation step, cells stably expressing HMG350-HA and Insig-1-Myc were treated with ALLN to block proteasomal degradation and incubated for 2 h with or without sterols. The cells were permeabilized with digitonin, and supernatant and pellet fractions were separated by centrifugation (20,000 × g) and analyzed by immunoblotting. Probing with anti-calnexin and anti-GAPDH antibodies demonstrated the reliable separation between bulk cellular membranes, ER in particular, in the pellet fraction and soluble cytosolic proteins in the supernatant fraction (Figure 1C, iii and iv; E, iii and iv; and F, iv and v, respectively). In sterol-depleted cells under steady-state conditions (i.e., time 0), HMG350-HA was not found in the supernatant but was restricted to the pellet fraction (Figure 1Ci, lanes 2 and 5). After 2-h incubation in the presence of proteasome inhibitor in the absence of sterols, a marked increase in the level of full-length HMG350-HA was detected in the pellet, and a significant portion of this protein migrated as a high-molecular-weight smear indicative of polyubiquitinated species (Figure 1Ci, lane 4). Importantly, under these conditions, substantial amounts of HMG350-HA running as a doublet of bands (arrowheads; see below) were detected in the supernatant (Figure 1Ci, lane 1). Densitometric analysis of multiple such experiments showed that 35% of HMG350-HA were found in the supernatant fraction after 2-h incubation in the absence of sterols (Figure 1D). On incubation with sterols, the amount HMG350-HA in the pellet decreased compared with the sterol-depleted cells (Figure 1Ci, lanes 4 and 6). Notably, this decrease was compensated by a significantly greater proportion (55%) of HMG350-HA, trailed by a high-molecular-weight smear of its ubiquitinated species, that appeared in the supernatant (Figure 1Ci, lanes 1 and 3; and D). A more modest sterol-stimulated increase in the amounts of HMG350-HA that dislocated to the supernatant was noted in cells that did not overexpress Insig-1-Myc (Figure 1D). In these singly transfected stable cells, 36% of HMG350-HA was found in the supernatant in the absence of sterols and 46% in their presence (Figure 1D). The dependence of HMG350-HA dislocation on Insig-1-Myc was underscored in SRD-15 cells, which lack both Insig proteins (Lee et al., 2005). In these cells, the transiently transfected HMG350-HA did not dislocate to the supernatant unless Insig-1-Myc also was coexpressed (Figure 1Ei, compare lanes 1 and 3 with lanes 4 and 6, respectively), and HMG350-HA release was further stimulated by sterols (Figure 1Ei, compare lanes 4 and 6). Tracing the fate of the transfected Insig-1-Myc by reprobing the blots with an anti-myc antibody revealed that, similar to HMG350-HA, Insig-1-Myc resided exclusively in the pellet fraction of sterol-depleted cells (Figure 1Cii, lanes 2 and 5). After 2 h of ALLN treatment, full-length Insig-1-Myc and its derived ubiquitinated material accumulated in the supernatant (Figure 1Cii, lanes 1 and 3), and, to a lesser extent, in the pellet fraction (Figure 1Cii, lanes 4 and 6). Nonetheless, there was no difference in the distribution of Insig-1-Myc between the pellet and supernatant fractions whether or not the cells were incubated with sterols during this 2-h time period (46 and 44% in the supernatant fraction, respectively; Figure 1D). This distribution of HMG350-HA and Insig-1-Myc between the supernatant and pellet fractions is probably underestimated because during the 2-h incubation, the membrane pool of these proteins was continuously replenished with newly synthesized molecules. Yet, treating the cells with protein synthesis blockers during the experiment was specifically avoided because HMGR degradation is severely inhibited by such drugs (Chun et al., 1990; Roitelman and Simoni, 1992). Importantly, full-length endogenous HMGR also was dislocated to the supernatant fraction but only in sterol-treated cells (Figure 1Fiii, lane 6; ∼20%/2 h; n = 5). These results indicate that HMG350-HA shares a similar fate with the endogenous enzyme, lending further support to the use of HMG350-HA as a bona fide model to study the physiology and cell biology of HMGR. Finally, when the 20,000 × g supernatant was further spun at 100,000 × g, essentially identical results were observed (Figure 1F). The entire ER population was already pelleted at 20,000 × g, as indicated by calnexin, and the release of HMG350-HA to the 20,000 × g or the 100,000 × g supernatants was stimulated by sterols to a similar extent, although HMG350-HA and HMGR in the 100,000 × g supernatant appeared exclusively as the faster migrating band (Figure 1E, i and iii, lanes 6 and 12; also see Figure 3 and Discussion).
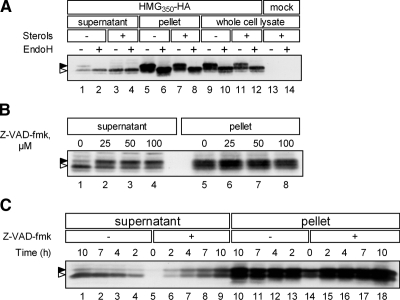
Figure 3.
Cytosolic PNGase removes N-glycan from dislocated HMG350-HA. (A) Mock-transfected cells or cells expressing HMG350-HA were incubated for 2 h with ALLN (65 μM) with (+) or without (−) sterols. Cells were permeabilized in solution C and fractionated (lanes 1–8), or solubilized in solution D (lanes 9–14). Aliquots of supernatant (75 μg of protein), solubilized pellet (20 μg of protein), or whole cell lysates (120 μg protein) were treated (+) or mock treated (−) with 1000 U of EndoH and analyzed. (B) Cells were preincubated for 1 h with the indicated concentrations of Z-VAD-fmk before addition of ALLN and sterols. The cells were incubated for additional 2 h and then permeabilized and fractionated. Aliquots of supernatant (60 μg of protein) and solubilized pellet (20 μg of protein) were analyzed. (C) Cells were incubated with ALLN and sterols, in the absence (−) or presence (+) of Z-VAD-fmk (50 μM). At the indicated time points, cells were permeabilized and fractionated and aliquots of supernatant (75 μg of protein) and solubilized pellet (20 μg of protein) were analyzed. All samples were resolved by 5–15% SDS-PAGE and immunoblotted with anti-HA monoclonal antibody. Closed and open arrowheads indicate glycosylated and un/deglycosylated forms of HMG350-HA, respectively.
Dislocation of HMG350-HA and Insig-1-Myc Is Energy Dependent and Involves p97/VCP
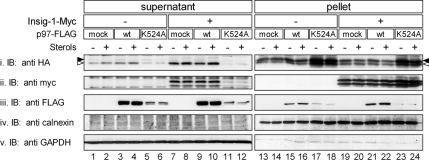
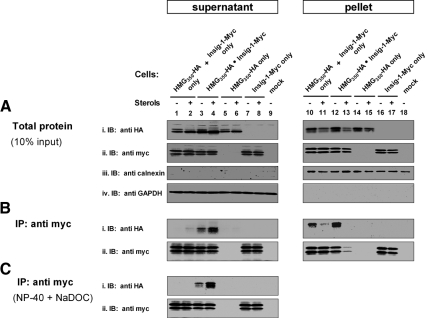
Dislocation of ERAD substrates and their extraction from the ER membrane is thought to be an active process involving multitude of ATPases, among which the role of the AAA-ATPase p97 is best established (Brodsky, 2007). Indeed, inhibition of cellular ATP production with cyanide and fluoride strongly inhibited the dislocation of both proteins to the supernatant (data not shown). To directly test whether p97 was involved in this process, cells were transiently transfected with plasmids encoding for HMG350-HA and Insig-1-Myc along with FLAG-tagged wild-type p97 or p97(K524A), a mutant in the essential D2 ATP-binding domain that acts in ERAD in a dominant-negative manner (Kobayashi et al., 2002). As clearly shown in Figure 2, expression of p97(K524A) abrogated the dislocation to the supernatant of HMG350-HA (Figure 2i, lanes 5 and 6), leading to HMG350-HA accumulation in the pellet fraction (Figure 2i, lanes 17 and 18). Importantly, although the release of HMG350-HA to the supernatant was hardly affected by the abundant coexpression of wild-type p97 (Figure 2i, compare lanes 1 and 2 with lanes 3 and 4 and lanes 7 and 8 with lanes 9 and 10), it was markedly enhanced by coexpression of Insig-1-Myc (Figure 2i, compare lanes 1 and 2 with lanes 7 and 8), further supporting the dependence of HMG350-HA dislocation on Insig-1 observed in SRD-15 cells (Figure 1E). Mutant p97(K524A) prevented the stimulated release of HMG350-HA even in these Insig-1-Myc–expressing cells (Figure 2i, lanes 11 and 12 and lanes 23 and 24). Blotting the membrane with the anti-myc antibody demonstrated that mutant p97(K524A) also blocked the appearance in the supernatant of Insig-1-Myc (Figure 2Bii, lanes 11 and 12) concomitantly with an impressive rise in its amounts in the pellet fraction (Figure 2Bii, lanes 23 and 24). These dominant-negative properties of the mutant p97(K524A) concerning dislocation of HMG350-HA and Insig-1-Myc were highly effective, even though the transfected protein was expressed at much lower levels than its wild-type version (Figure 2Biii, compare lanes 5, 6, 11, and 12 with lanes 3, 4, 9, and 10). It should be noted that, unlike differentiated PC12 cells (Kobayashi et al., 2002), within the time frame of the experiment, expression of this mutant p97 in HEK293 cells neither caused cytoplasmic vacuolization or cell death, nor did it affect the expression of cotransfected HMG350-HA or/and Insig-1-Myc (Figure 2, lanes 17, 18, 23, and 24). These results demonstrate that the release of HMG350-HA and Insig-1-Myc to the soluble fraction is an active, energy-requiring process that involves catalytically functional p97/VCP.
Figure 2.
Dislocation of HMG350-HA and Insig-1-Myc involves p97/VCP. On day 1, HEK293 cells were transfected with a plasmid mixture consisting of 3 μg of pIRESneo2-HMG350-HA, 0.5 μg of pcDNA3.1-Insig-1-Myc, and 0.5 μg of wild-type or K524A mutant of pcDNA3.1-p97-FLAG, as indicated. Total plasmid concentration was adjusted to 4 μg/dish with empty pcDNA3 vector. Forty-eight hours later, the cells were switched to medium B for additional 16 h after which they were treated for 2 h with ALLN (65 μM) with (+) or without (−) sterols. Cells were permeabilized, and supernatant (60 μg of protein) and pellet (20 μg of protein) fractions were analyzed by immunoblotting with the indicated antibodies. Closed and open arrowheads indicate glycosylated and un/deglycosylated forms of HMG350-HA, respectively.
Deglycosylation of Dislocated HMG350-HA Is Stimulated by Sterols
As noted in Figure 1, the dislocated HMG350-HA from sterol-depleted cells migrated in two distinct bands, whereas the protein in the supernatant fraction of sterol-treated cells ran largely as a single band whose migration coincided with that of the lower band from sterol-depleted cells (e.g., see Figure 1Fi, lanes 4 and 6). Similarly, a closer examination of the dislocated endogenous HMGR revealed that the migration of the protein in the supernatant was slightly faster than that of the protein in the pellet fraction (Figure 1Fiii, lane 6). These observations suggested that the faster migrating band represented species that was devoid of its single N-glycan. To directly test this possibility, we treated the supernatant and pellet fractions, as well as whole cell lysate, with EndoH, which cleaves high mannose N-glycans from glycoproteins. On EndoH digestion, the slower migrating HA immunoreactive band in the supernatant of sterol-depleted cells, which was also observed in the whole cell lysate, collapsed into the faster migrating HMG350-HA band (Figure 3A, lanes 1, 2, and 9–12), indicating its link to an N-glycan moiety. Such an EndoH-mediated electrophoretic mobility shift did not occur for the faster migrating band in the supernatant of sterol-replete cells (Figure 3A, lanes 3 and 4), indicating that this HMG350-HA was already devoid of its N-glycan. Sterols had no effect on the glycosylation status of HMG350-HA in the pellet as the same proportion of EndoH-sensitive and -resistant species was found in this fraction whether or not cells received sterols (Figure 3A, lanes 5–8).
These results indicated that sterols promoted the removal of the N-glycan from dislocated HMG350-HA and that this process was restricted to the supernatant fraction. A likely candidate for a cytosolic glycosidase is the peptide N:glycanase (PNGase) implicated in ERAD, which cleaves N-glycan chains from misfolded glycoproteins en route to proteasomal degradation (Suzuki et al., 2002). To test whether PNGase was involved in removing the N-glycan from cytosolic HMG350-HA, cells that were treated with ALLN and sterols to promote HMG350-HA deglycosylation were incubated, in addition, with increasing concentrations of Z-VAD-fmk. This cell-permeant peptide is a pan-caspase inhibitor that was shown to inhibit irreversibly also the activity of yeast and mammalian PNGase (Misaghi et al., 2004). Indeed, even at the lowest concentration tested (25 μM), Z-VAD-fmk strongly inhibited the removal of the N-glycan from HMG350-HA in the supernatant fraction (Figure 3B, lanes 1 and 2), without affecting the migration of HMG350-HA in the pellet. Together, these results indicate that cytosolic PNGase removes the N-glycan from HMG350-HA once it is dislocated to the cytosol. However, because Z-VAD-fmk did not prevent the appearance of the faster migrating HMG350-HA even at higher concentrations, we suspected that the glycan-less HMG350-HA in the supernatant was a mixture of glycosylated HMG350-HA that underwent PNGase-mediated deglycosylation and an unglycosylated species that also was dislocated. To address this issue, we followed the kinetics of HMG350-HA dislocation in sterol-treated cells in the absence or presence of Z-VAD-fmk. In the absence of the inhibitor (Figure 3C, lanes 4–1), most of the HMG350-HA accumulated in the supernatant in the glycan-less form (open arrowhead), and only trace amounts of the glycosylated protein (closed arrowhead) were detected. In the presence of Z-VAD-fmk, both forms of HMG350-HA accumulated with the same kinetics (Figure 3C, lanes 6–9). We conclude that both glycosylated and unglycosylated HMG350-HA dislocate from the ER equally well and that the rate of N-glycan removal from the dislocated species by cytosolic PNGase is enhanced in sterol-treated cells.
Sterol-stimulated Formation and Dislocation of HMGR · Insig-1-Myc Complexes
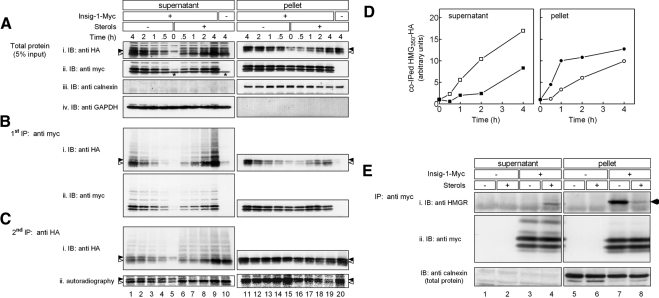
Because association with Insig is a necessary step in HMGR degradation (Sever et al., 2003b), we next studied the relationships between Insig-1-Myc binding and HMG350-HA dislocation in cells that stably coexpressed both tagged proteins. The cells were pulse labeled with [35S]methionine/cysteine mix, chased with ALLN in the absence or presence of sterols, permeabilized with digitonin, and fractionated. Potential complexes in the supernatant and solubilized pellet fractions were first immunoprecipitated with an anti-myc antibody and blotted successively with an anti-HA antibody, to detect coprecipitating HMG350-HA, and with an anti-myc antibody to detect the precipitated Insig-1-Myc. The remaining unbound HMG350-HA was directly precipitated with an anti-HA antibody, immunoblotted with an anti-HA antibody, and exposed to a PhosphorImager screen (Figure 4). As shown, immunoprecipitation of Insig-1-Myc pulled down HMG350-HA in a specific manner both from the supernatant and pellet fractions, because the anti-myc antibody did not precipitate any HA-tagged material from cells that did not express Insig-1-Myc (Figure 4Bi, lanes 10 and 20). Insig-1-Myc · HMG350-HA complexes also could be specifically immunoprecipitated with antibodies against the membrane region of HMGR (data not shown). Compared with sterol-depleted cells, addition of sterols resulted in significantly higher amounts of HMG350-HA that were associated and coprecipitated with Insig-1-Myc from the supernatant at any given time point (Figure 4Bi, lanes 6–9; and D). The coprecipitated material, especially from the sterol-treated cells, was heavily polyubiquitinated, as evidenced by the high-molecular-weight species trailing the major bands of HMG350-HA (Figure 4Bi, lanes 6–9). Moreover, both glycosylated and un/deglycosylated forms of HMG350-HA associated with Insig-1-Myc, but, as in Figure 3, in the supernatant of sterol-treated cells mostly the un/deglycosylated species associated with Insig-1-Myc (Figure 4Bi, lanes 1–9). These findings were corroborated by analyzing the remaining HMG350-HA that did not coprecipitate with Insig-1-Myc (Figure 4C). Again, compared with untreated cells, more polyubiquitinated and un/deglycosylated HMG350-HA was found in the supernatant fraction of sterol-treated cells (Figure 4Ci, lanes 1–9). A direct autoradiography of this unbound material demonstrated that sterols promoted nearly instantaneously the deglycosylation of the newly synthesized HMG350-HA that dislocated to the supernatant (Figure 4Cii, lanes 6–9). Concomitant with the sterol-stimulated buildup of HMG350-HA · Insig-1-Myc in the supernatant, the accumulation of this complex in the pellet fraction was markedly attenuated in sterol-treated cells relative to sterol-depleted cells (Figure 4Bi, lanes 11–19; and D). This indicated again that more HMG350-HA was extracted from the pellet of sterol-treated cells. Notably, Insig-1-Myc in the pellet fraction associated equally well with the glycosylated and the un/deglycosylated forms of HMG350-HA. Reprobing the nitrocellulose with an anti-myc antibody demonstrated that Insig-1-Myc also dislocated to and gradually accumulated in the supernatant but at a constant rate that was not affected by sterols (Figure 4Bii, lanes 1–9). Throughout the time course of the experiment, Insig-1-Myc was not depleted from the pellet fraction and its amount remained relatively constant (Figure 4, Aii and Bii, lanes 11–19). This suggests that the accumulation of the short-lived Insig-1-Myc in the pellet upon proteasome inhibition by ALLN was offset by its dislocation to the supernatant. Similar to HMG350-HA, Insig-1-Myc associated with full-length endogenous HMGR both in the supernatant and pellet fractions (Figure 4E). However, immunoprecipitation of Insig-1-Myc pulled down HMGR only from the supernatant of sterol-treated cells (Figure 4Ei). This is consistent with Figure 1F, in which endogenous HMGR dislocated to the supernatant only when cells were incubated with sterols. Together, these results demonstrate that, at any given moment, more HMGR molecules were associated with Insig-1-Myc in the supernatant of sterol-treated cells.
Figure 4.
Sterol-stimulated formation and dislocation of HMGR · Insig-1-Myc complex. Sterol-depleted cells that express HMG350-HA together (+) or without (−) Insig-1-Myc were pulse-labeled for 30min with of 35S–cell-labeling reagent (100 μCi). Cells were chased in the presence of ALLN with (+) or without (−) sterols, permeabilized, and fractionated. (A) Aliquots of supernatant and solubilized pellet (5% of samples) were analyzed by SDS-PAGE and immunoblotting (IB) with the indicated antibodies. (B) The rest 95% of the samples were first subjected to immunoprecipitation (first IP) of Insig-1-Myc with a mouse monoclonal anti-myc antibody, and immune complexes were analyzed by SDS-PAGE and IB with rabbit antibodies for the presence of Insig-1-Myc (ii) and coprecipitated HMG350-HA (i). (C) The supernatants from the first IP were cleared with protein A-Sepharose beads and reprecipitated (second IP) with a mouse monoclonal anti-HA antibody and immune complexes were analyzed by SDS-PAGE for the presence of HMG350-HA by IB with a rabbit anti-HA antibody (i) and by direct autoradiography (ii). Closed and open arrowheads indicate glycosylated and un/deglycosylated forms of HMG350-HA, respectively. Asterisks (Aii, lanes 5 and 10) denote a protein in the supernatant fraction that cross-reacted with the anti-myc antibody. (D) Densitometric analysis of the blots. The HA immunoreactive signal in Bi (including trailing smear) was divided by the Myc-immunoreactive signal in Bii (including trailing smear), setting the ratio at time zero as one. Results are the mean of two independent experiments. Closed symbols, sterol-depleted cells; open symbols, sterol-treated cells. (E) Sterol-depleted naïve or Insig-1-Myc–expressing cells were incubated for 2 h with ALLN and sterols, as indicated. Cells were permeabilized in solution C and fractionated. Proteins from the supernatant and pellet fractions were IP with rabbit polyclonal anti-myc antibodies. The immune complexes were resolved and IB with a mouse anti-HMGR monoclonal antibody (i; arrow) and then reprobed with mouse anti-myc monoclonal antibodies (ii). Aliquots of the fractions, representing 5% of protein input, were immunoblotted with rabbit anti-calnexin antibody.
Interaction of Insig-1-Myc with HMG350-HA: Effects of Mutations on Dislocation
It has been suggested that sterol-enhanced interaction of the membrane region of HMGR with Insig is a prerequisite for reductase polyubiquitination and eventual proteasomal degradation. Sterol-regulated binding of HMGR to Insig critically depends on the sequence YIYF in the second TMs of HMGR, which constitutes the N-terminal span of the sterol-sensing domain. An identical tetrapeptide also is found in the sterol-sensing domain of SCAP (Hua et al., 1996), and a mutation in this sequence abrogates complex formation between the mutant SCAP and Insig-1 (Yang et al., 2002). Sterol-regulated conjugation of polyubiquitin chains to HMGR takes place at residues 89 and 248, the only lysines residues in the membrane region that face the cytosol (Roitelman et al., 1992). We examined whether and how these critical elements in HMGR affected its dislocation to the soluble fraction.
Similar to results shown above, treatment with sterols of cells expressing the wild-type protein resulted in higher amounts of un/deglycosylated HMG350-HA that were released to the supernatant and coimmunoprecipitated with Insig-1-Myc (Figure 5, Ai and Bi, lanes 1 and 2) concomitant with a marked decrease in amounts of complex that remained in the pellet fraction (Figure 5Bi, lanes 8 and 9). By contrast, sterols had a much smaller effect on complex formation between the YIYF→AIAF mutant and Insig-1-Myc either in the supernatant or the pellet fractions (Figure 5Bi, lanes 3, 4, 10, and 11). This was also indicated by the poor release of the HMG350-HA(YIYF→AIAF) to the supernatant and the inability of sterols to stimulate this release (Figure 5Ai, lanes 3 and 4). This indicates that the interaction of HMG350-HA with Insig-1-Myc plays a crucial role in the dislocation of HMG350-HA to the soluble fraction. Despite its poor release and deglycosylation (Figure 5Ai, lanes 3 and 4), the dislocated HMG350-HA(YIYF→AIAF) that was associated with Insig-1-Myc was predominantly un/deglycosylated, similar to the wild-type protein (Figure 5Bi, lanes 1–4). Radioactive pulse-chase experiments confirmed that the turnover of HMG350-HA(YIYF→AIAF) mutant was not regulated by sterols (data not shown).
Figure 5.
Interactions with Insig-1-Myc and dislocation of HMG350-HA mutants. Cells expressing the indicated constructs of HMG350-HA, together (+) or without (−) Insig-1-Myc, were incubated for 2h with ALLN with (+) or without (−) sterols, permeabilized in solution C and fractionated. (A) Aliquots of supernatant and solubilized pellet (10% of samples) were analyzed by SDS-PAGE and IB with the indicated antibodies. (B) The rest 90% of the samples were subjected to IP with an anti-myc monoclonal antibody, and immune complexes were analyzed by SDS-PAGE and immunoblotting (IB) with rabbit anti-myc and anti-HA antibodies for the presence of Insig-1-Myc (ii) and coprecipitated HMG350-HA (i), respectively. Closed and open arrowheads indicate glycosylated and un/deglycosylated forms of HMG350-HA, respectively. Mr markers (in kilodaltons) are shown.
A mutant HMG350-HA in which lysines 89 and 248 were replaced by arginines does not undergo sterol-regulated polyubiquitination and degradation (Sever et al., 2003a; Doolman et al., 2004). In response to sterols, the amounts of HMG350-HA(K89+248R) that were released to the supernatant and pulled down along with Insig-1-Myc did not increase but were even slightly reduced (Figure 5, Ai and Bi, lanes 5 and 6). Moreover, the HMG350-HA(K89+248R) mutant that dislocated to the supernatant and interacted with Insig-1-Myc was not deglycosylated in response to sterols (Figure 5, Ai and Bi, lanes 5 and 6). Contrary to a previous report (Sever et al., 2003a), we were unable to observe any enhanced association between the mutated HMG350-HA(K89+248R) and Insig-1-Myc in the pellet fraction of sterol-replete cells in comparison to sterol-depleted cells. Relative to cells expressing wild-type protein, the apparently greater proportion of HMG350-HA(K89+248R) that associated with Insig-1-Myc in the pellet fraction was not increased by sterols (Figure 5Bi, lanes 12 and 13) and resulted from higher amounts of immunoprecipitated Insig-1-Myc (Figure 5Bii, lanes 12 and 13).
Interaction between HMG350-HA and Insig-1-Myc in the Cytosol Depends on Their Prior Association in the ER Membrane
The results described thus far demonstrated that HMG350-HA and Insig-1-Myc associated with each other in the soluble supernatant fraction. Such an interaction between these two ER membrane polytopic proteins could have been formed after each protein has dislocated from the ER membrane independently of the other, or, possibly, by dislocation from the ER membrane as a preformed complex. To distinguish between these two possibilities, we performed the experiment shown in Figure 6. One set of dishes was plated with cells that stably expressed both HMG350-HA and Insig-1-Myc (“HMG350-HA · Insig-1-Myc”). A second set of dishes was plated with a mixture of cells, each of which stably expressed either HMG350-HA or Insig-1-Myc (“HMG350-HA only + Insig-1-Myc only”). The HMG350-HA only and Insig-1-Myc only cells were also plated in separate dishes as controls. After an overnight incubation under sterol-depleting conditions, the cells were incubated for 2 h with ALLN, with or without sterols. The cells were washed, permeabilized with digitonin, fractionated by centrifugation, and the proteins were immunoprecipitated, as detailed below.
Figure 6.
Interaction between HMG350-HA and Insig-1-Myc requires their expression in the same cell. Sterol-depleted cells expressing either HMG350-HA, Insig-1-Myc, or both, and a mixture of cells expressing either proteins, were incubated for 2h with ALLN with (+) or without (−) sterols, and cells were permeabilized and fractionated. (A) Aliquots of supernatant (5%) and solubilized pellet (10%) (see Note) were analyzed by SDS-PAGE and immunoblotting (IB) with the indicated antibodies. The rest 95% of the supernatant fraction was divided into two equal portions. (B) One portion was directly immunoprecipitated (IP) with an anti-myc monoclonal antibody and immune complexes were analyzed by SDS-PAGE and immunoblotting with rabbit anti-myc and anti-HA antibodies, for the presence of Insig-1-Myc (ii) and coprecipitated HMG350-HA (i), respectively. (C) To the other supernatant portion, NP-40 and NaDOC were added to 1% each, and the samples were immunoprecipitated and immunoblotted as described in B. Note: Because the supernatant was divided into two portions, the 5% sample of the initial fraction actually represents 10% of total protein input in each portion used for the analyses in B and C.
As can be clearly seen, HMG350-HA coimmunoprecipitated with Insig-1-Myc from the supernatant fraction only when both proteins were coexpressed in the same cell (Figure 6Bi, lane 3) and, again, the amounts of HMG350-HA that were pulled down markedly increased when the cells were treated with sterols (Figure 6Bi, lane 4). There were traces of the complex in the supernatant fraction from the sterol-treated mixed cells expressing each protein individually (Figure 6Bi, lane 2), but rather significant amounts in the pellet fraction of the mixed cells (Figure 6Bi, lanes 10 and 11). Because the pellet fractions were solubilized in NP-40 and NaDOC, to control for their possible effects, these detergents were added also to the supernatant fraction before immunoprecipitation. Under these conditions, no HMG350-HA was coprecipitated with Insig-1-Myc from the supernatants of the mixed cells (Figure 6Ci, lane 2). Importantly, whether or not present during immunoprecipitation, these detergents did not interfere with the interaction in the HMG350-HA · Insig-1-Myc complex that persisted in the supernatant of cells coexpressing both proteins (Figure 6, Bi and Ci, lanes 3 and 4). These data indicate that the association between HMG350-HA and Insig-1-Myc that is enhanced by sterols and detected in the supernatant occurs in vivo only when these two polytopic proteins reside in the same ER membrane. Hence, HMGR and Insig-1 are dislocated as a tightly bound preformed complex.
DISCUSSION
Dislocation of proteins from the ER to the cytosol is the hallmark of ERAD. This step allows unrestricted access of doomed ER proteins to the cytosolic 26S proteasome (Bar-Nun, 2005). Our current study on the eight TMs HMGR and six TMs Insig-1 ER resident proteins provides new insight into the dislocation of polytopic ER membrane proteins that undergo ERAD. By fractionating permeabilized proteasome-inhibited cells, we demonstrate that endogenous HMGR and a tagged version of its membrane region, as well as Insig-1-Myc, are released to the cytosol as full-length intact polypeptides. These results complement reports on the dislocation of luminal ERAD substrates (Rodighiero et al., 2002; Elkabetz et al., 2004; Afshar et al., 2005) and of membrane proteins with a single TMs (Huppa and Ploegh, 1997; Tortorella et al., 1998; Ye et al., 2003). Together with recent in vitro evidence for dislocation of ubiquitinated polytopic Ste6*p (Nakatsukasa et al., 2008) and Hmg2p (Garza et al., 2009) from yeast microsomes, it seems that, among the possible models suggested for dislocation of polytopic membrane ERAD substrates (Nakatsukasa and Brodsky, 2008), complete extraction to the cytosol of intact polypeptide chains is a general feature of the delivery to the proteasome of this class of proteins.
In accordance with sterols' activity in enhancing ubiquitination and rapid turnover of HMGR and HMG350-HA (Ravid et al., 2000; Doolman et al., 2004), here the effect of sterols also is manifested in stimulating the release of these proteins to the cytosol. The specificity of this process is underscored by the finding that dislocation of Insig-1-Myc is not affected by sterols. We have previously shown in microsomes from lovastatin-resistant cells that ubiquitinated HMGR becomes a peripherally associated protein inasmuch as it can be stripped from the membranes by high pH carbonate wash (Doolman et al., 2004). Here, we demonstrate in naive cells that HMGR, and more so HMG350-HA, are fully dislocated to a 100,000 × g supernatant cytosolic fraction. This dislocation requires metabolic energy and is facilitated by the AAA-ATPase p97/VCP, probably with its ERAD partners Ufd1p and Npl4p. These results are consistent with previous findings that p97 preferentially associates with ubiquitinated HMG350-HA (Doolman et al., 2004) and HMGR (data not shown) and that HMGR degradation is abrogated upon siRNA-mediated p97 depletion or abolishment of the ubiquitin binding capacity of Ufd1p (Song et al., 2005; Cao et al., 2007).
In agreement with previous reports (Lee and Ye, 2004; Gong et al., 2006; Lee et al., 2006a), we also find that Insig-1-Myc is a short-lived protein that becomes ubiquitinated and degraded by the proteasome. Moreover, here we demonstrate that intact Insig-1-Myc dislocates to the cytosol in an energy-requiring process that involves p97/VCP. It was demonstrated by Lee et al. (2006a) that, unlike HMGR, sterols stabilized Insig-1 in a manner dependent on SCAP. It has been proposed that sterols, which promote the mutually exclusive binding of HMGR and SCAP to Insigs, cause Insig-1 population to partition between HMGR-bound and SCAP-bound species. The SCAP-bound Insig-1 is stabilized because SCAP displaces gp78; hence, it prevents Insig-1 ubiquitination (Lee et al., 2006a). This was observed when Insig-1 and SCAP were both transiently expressed from the weak thymidine kinase promoter, conditions that presumably preserve a correct stoichiometry between these two proteins (Gong et al., 2006). However, in our hands, neither ubiquitination nor dislocation or degradation of Insig-1-Myc was affected by sterols. Possibly, the levels of Insig-1-Myc, which was overexpressed from a strong promoter, overwhelmed endogenous SCAP, thus obscured any stabilizing effect of the latter.
The single N-glycan of HMGR, which plays no role in reductase degradation (Jingami et al., 1987; Sekler and Simoni, 1995), provided a valuable tool to examine events during and after dislocation. In sterol-depleted cells, both glycosylated and unglycosylated species of HMG350-HA were released to the 20,000 × g supernatant. Sterols not only increased the total amount of dislocated protein but also promoted removal of its N-glycan by cytosolic PNGase. It is not known whether PNGase is directly activated by sterols. A likely explanation to these results is increased substrate availability for the enzyme. Namely, that the luminal side of HMGR membrane region, in particular the N-glycan-bearing loop between TMs 7 and 8 (Roitelman et al., 1992), gains better access to PNGase in the cytosol of sterol-treated cells. High-speed centrifugation demonstrated that the cleared cytosol (100,000 × g supernatant) contained deglycosylated HMG350-HA exclusively (Figure 1F), whereas both glycosylated and un/deglycosylated species were found in the 20,000 × g supernatant and pellet. The latter fraction represents bulk cellular membranes as it exclusively stains positive for calnexin. The nature of the 100,000 × g pellet remains to be determined, but it was devoid of calnexin, indicating that it was not generated by artifactual ER fragmentation. Moreover, with the exception of the glycosylation state, the release to the 20,000 × g or 100,000 × g supernatants was almost identical and was stimulated by sterols to a similar extent. Thus, the 20,000 × g fractions faithfully represent the starting point and the finish line of the dislocation process.
By coimmunoprecipitation, we detected in the cytosol complexes between Insig-1-Myc and HMG350-HA as well as with endogenous HMGR. These complexes were stable enough to withstand extensive washes in NP-40 and NaDOC. Kinetic analysis demonstrated that sterols stimulated the formation of these complexes and accelerated their dislocation to the soluble fraction, where both HMG350-HA and Insig-1-Myc accumulated as polyubiquitinated species. Consequently, much less HMG350-HA · Insig-1-Myc and HMGR · Insig-1-Myc complexes remained in the membrane fraction of sterol-treated cells. Importantly, HMG350-HA and HMGR in the dislocated complexes were predominantly in their nonglycosylated form, indicating that the N-glycan was neither required for association with Insig-1-Myc, nor did this association impede subsequent removal of the N-glycan by PNGase. It should be noted that a considerable portion of HMG350-HA that did not coprecipitate with Insig-1-Myc remained in the cytosol. Probably, this population was complexed with endogenous Insig-1, represented HMG350-HA that dissociated from the complex after dislocation, or both. In cells not exposed to sterols, this apparently uncomplexed dislocated HMG350-HA was relatively enriched for the glycosylated species, whereas in sterol-treated cells, deglycosylation was stimulated and both complexed and uncomplexed HMG350-HA was devoid of N-glycan. Together with the observation that Insig-1-bound HMG350-HA in the soluble fraction is preferentially nonglycosylated, these results suggest that Insig(s) binding also may promote deglycosylation by PNGase. Indeed, it was shown that gp78, along with p97/VCP, form a ternary complex with PNGase (Li et al., 2006). Thus, sterol-enhanced interaction of HMGR with Insig and its associated E3 ligase gp78 not only facilitates HMGR ubiquitination but also promotes recruitment of PNGase. Interestingly, inhibiting PNGase with Z-VAD-fmk affected neither dislocation of HMG350-HA (Figure 3) nor degradation of HMGR (data not shown). This is in line with the reported ability of the proteasome to degrade glycoproteins without prior removal of their N-glycans (Kario et al., 2007).
Studies have demonstrated that mutations in the sterol-sensing domain impede HMGR regulated degradation (Sever et al., 2003a; Lee et al., 2006b). In particular, mutation of the conserved tetrapeptide YIYF abrogated sterol-regulated complex formation with Insig-1, as assessed by blue-native PAGE (Sever et al., 2003a). Consistent with these results, we confirmed that mutation of this sequence markedly inhibited sterol-accelerated degradation of HMG350-HA (data not shown) as well as sterol-stimulated association with Insig-1-Myc (Figure 5). The dependence of HMG350-HA dislocation on Insig-1-Myc in transiently transfected SRD-15 cells (Figure 1E) together with the stimulatory effect of transfected Insig-1 (Figure 2), the poor release of the HMG350-HA(YIYF→AIAF) mutant and the inability of sterols to stimulate its deglycosylation attest to the key role of Insig-1 in HMGR dislocation and recruitment of PNGase.
Unexpectedly, we found that the (K89+248R) substitution, which markedly blunted (but not completely eliminated) HMG350-HA ubiquitination and blocked its sterol-accelerated degradation (Doolman et al., 2004), also abolished sterol-stimulated association with Insig-1-Myc. This result is at odds with the data of Sever et al. (2003a) who showed sterol-enhanced binding to Insig-1 of a similar ubiquitination-defective K248R mutant. These differences may stem from their analysis by blue-native PAGE of proteins that were transiently expressed in a SCAP-deficient Chinese hamster ovary-derived cell line (Sever et al., 2003a). Nevertheless, in our hands, the inability of sterols to promote Insig-1 association with the (K89+248R) mutant was echoed in the nonregulated release to the soluble fraction and inefficient N-glycan removal by cytosolic PNGase. Interestingly, Insig-1 binds glycosylated and unglycosylated (K89+248R) mutant as efficiently, indicating that the N-glycan does not interfere with this association. Combined, the results with the HMG350-HA mutants support the role of Insig in recruiting PNGase, albeit indirectly presumably through associated gp78, to the dislocating HMGR. It is also possible that PNGase may be recruited via its interacting ubiquitin-binding protein HR23 (Park et al., 2001; Kim et al., 2006). Such a mechanism may be hindered by the hypo-ubiquitination of the (K89+248R) mutant.
Our results demonstrate that the interaction in the supernatant between HMG350-HA and Insig-1-Myc mandates that the two proteins are coexpressed in the same cell. Because these proteins associate with each other also in the pellet fraction, this strongly suggests that they are extracted form the ER membrane together, forming a stable complex before or during dislocation. Inasmuch as Insig-1-Myc dislocates at a constant rate, the increased levels of coprecipitated HMG350-HA in the soluble fraction indicate that sterols stimulate the formation of this HMG350-HA · Insig-1-Myc complex, in agreement with Sever et al. (2003a). It is, therefore, tempting to hypothesize that Insig-1 facilitates HMGR degradation consequently to its own destruction, by acting as a “suicide chaperone.” Once HMGR and Insig-1 reach the cytosol as a soluble complex of ubiquitinated polypeptides, they probably dissociate from one another, as each protein is degraded by the proteasome at its own intrinsic rate. This mechanism is reminiscent of the targeted elimination of major histocompatibility complex class I heavy chain by the human cytomegalovirus US11 protein (Lilley and Ploegh, 2005). This virally encoded type I ER membrane glycoprotein associates with nascent class I chains and promotes their dislocation to the cytosol, where they are deglycosylated by PNGase and rapidly degraded by the proteasome in a process that depends on active ubiquitination system (Kikkert et al., 2001). US11 itself follows the fate of its victim: it is extracted from the ER to the cytosol, deglycosylated by PNGase, and degraded in the ubiquitin–proteasome pathway (Baker and Tortorella, 2007). It is not yet clear, however, whether class I and US11 egress to the cytosol separately or together as a complex.
When a soluble ERAD substrate dislocates, it moves from one aqueous milieu, within the ER lumen, to another at the cytosolic side of the membrane. However, dislocation of a membrane protein mandates its complete extraction from the lipid bilayer, a situation that is exacerbated with polytopic ERAD substrates with multiple TMs. Not only is this process energetically expensive but also it challenges the cell to prevent these proteins from forming insoluble aggregates when the hydrophobic TMs are exposed to the hydrophilic environment of the cytosol. Indeed, specific AAA-ATPases, such as p97, and other molecular chaperones provide the driving force for dislocation and prevention of protein aggregation (Nishikawa et al., 2001; Ye et al., 2001; Elkabetz et al., 2004; Huyer et al., 2004; Bar-Nun, 2005; Buck et al., 2007; Lipson et al., 2008). The current study describes a system that should allow dissecting the steps in dislocation of polytopic ERAD substrates en route to proteasomal degradation and identification of cellular components that take part in this process.
ACKNOWLEDGMENTS
We thank Joseph Goldstein for kindly providing Insig-1-Myc plasmid, Russell DeBose-Boyd for the SRD-15 cells, Ilana Shapira for the FLAG-tagged wild-type and p97(K524A) plasmids, Ron Kopito for anti-calnexin antiserum and constructive advice, and Shoshana Bar-Nun for many helpful discussions and critical reading of the manuscript. This work was performed in partial fulfillment of the requirements for the Ph.D. degree of G.S.L. at the Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
Abbreviations used:
- ALLN
N-acetyl-leucyl-leucyl-norleucinal
- DOC
deoxycholate
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated protein degradation
- EndoH
endoglycosidase H
- GAPDH
glyceraldehydes-3-phosphate dehydrogenase
- HMGR
3-hydroxy-3-methylglutaryl coenzyme A reductase
- LPDS
lipoprotein-deficient serum
- MVA
mevalonate
- PAGE
polyacrylamide gel electrophoresis
- PNGase
peptide N:glycanase
- PBS
phosphate-buffered saline
- SREBP
sterol regulatory element-binding protein
- SCAP
sterol regulatory element-binding protein cleavage-activating protein
- TMs
transmembrane span(s).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0953) on May 20, 2009.
REFERENCES
- Afshar N., Black B. E., Paschal B. M. Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell Biol. 2005;25:8844–8853. doi: 10.1128/MCB.25.20.8844-8853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. M., Tortorella D. Dislocation of an ER membrane glycoprotein involves the formation of partially dislocated ubiquitinated polypeptides. J. Biol. Chem. 2007;282:26845–26856. doi: 10.1074/jbc.M704315200. [DOI] [PubMed] [Google Scholar]
- Bar-Nun S. The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast. Curr. Top. Microbiol. Immunol. 2005;300:95–125. doi: 10.1007/3-540-28007-3_5. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation) Biochem. J. 2007;404:353–363. doi: 10.1042/BJ20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., Simoni R. D. Biogenesis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase, an integral glycoprotein of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1984;81:1674–1678. doi: 10.1073/pnas.81.6.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck T. M., Wright C. M., Brodsky J. L. The activities and function of molecular chaperones in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2007;18:751–761. doi: 10.1016/j.semcdb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wang J., Qi W., Miao H.-H., Wang J., Ge L., DeBose-Boyd R. A., Tang J.-J., Li B.-L., Song B.-L. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab. 2007;6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Cheng H. H., Xu L., Kumagai H., Simoni R. D. Oligomerization state influences the degradation rate of 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 1999;274:17171–17178. doi: 10.1074/jbc.274.24.17171. [DOI] [PubMed] [Google Scholar]
- Chun K. T., Bar-Nun S., Simoni R. D. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J. Biol. Chem. 1990;265:22004–22010. [PubMed] [Google Scholar]
- Doolman R., Leichner G. S., Avner R., Roitelman J. Ubiquitin is conjugated by membrane ubiquitin ligase to three sites, including the N-terminus, in transmembrane region of mammalian 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for sterol-regulated enzyme degradation. J. Biol. Chem. 2004;279:38184–38193. doi: 10.1074/jbc.M405935200. [DOI] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Tanaka R. D., Fogelman A. M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in rat hepatocytes. J. Biol. Chem. 1983;258:7272–7275. [PubMed] [Google Scholar]
- Elkabetz Y., Shapira I., Rabinovich E., Bar-Nun S. Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplasmic reticulum-bound p97/Cdc48p and proteasome. J. Biol. Chem. 2004;279:3980–3989. doi: 10.1074/jbc.M309938200. [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J. R., Luskey K. L., Chin D. J., Goldstein J. L., Brown M. S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc. Natl. Acad. Sci. USA. 1982;79:5205–5209. doi: 10.1073/pnas.79.17.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco J. D., Goldstein J. L., Brown M. S. Membrane topology of human Insig-1, a protein regulator of lipid synthesis. J. Biol. Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- Garza R. M., Sato B. K., Hampton R. Y. In vitro analysis of Hrd1p-mediated retrotranslocation of its multi-spanning membrane substrate HMG-CoA reductase. J. Biol. Chem. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985;41:249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Basu S. K., Brown M. S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Gong Y., Lee J. N., Lee P.W.C., Goldstein J. L., Brown M. S., Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Ho S. N., Hunta H. D., Horton R. M., Pullena J. K., Peasea L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Horton J. D., Goldstein J. L., Brown M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Nohturfft A., Goldstein J. L., Brown M. S. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Huppa J. B., Ploegh H. L. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble lumenal protein. J. Biol. Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Rumpf S. Cdc48 (p97): a ‘molecular gearbox’ in the ubiquitin pathway? Trends Biochem. Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Jingami H., Brown M. S., Goldstein J. L., Anderson R.G.W., Luskey K. L. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J. Cell Biol. 1987;104:1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kario E., Tirosh B., Ploegh H. L., Navon A. N-linked glycosylation does not impair proteasomal degradation, but affects class I MHC presentation. J. Biol. Chem. 2007;283:244–254. doi: 10.1074/jbc.M706237200. [DOI] [PubMed] [Google Scholar]
- Kikkert M., Hassink G., Barel M., Hirsch C., van der Wal F. J., Wiertz E. Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol. Biochem. J. 2001;358:369–377. doi: 10.1042/0264-6021:3580369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. The Png1-Rad23 complex regulates glycoprotein turnover. J. Cell Biol. 2006;172:211–219. doi: 10.1083/jcb.200507149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Tanaka K., Inoue K., Kakizuka A. Functional ATPase activity of p97/Valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 2002;277:47358–47365. doi: 10.1074/jbc.M207783200. [DOI] [PubMed] [Google Scholar]
- Kostova Z., Tsai Y. C., Weissman A. M. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell Dev. Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara P. E., Labouesse M. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- Lee J. N., Song B., Debose-Boyd R. A., Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase, gp78. J. Biol. Chem. 2006a;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- Lee J. N., Ye J. Proteolytic activation of SREBP induced by cellular stress through depletion of Insig-1. J. Biol. Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- Lee P.C.W., Nguyen A. D., DeBose-Boyd R. A. Mutations within membrane domain of HMG CoA reductase confer resistance to sterol-accelerated degradation. J. Lipid Res. 2006b;48:318–327. doi: 10.1194/jlr.M600476-JLR200. [DOI] [PubMed] [Google Scholar]
- Lee P.C.W., Sever N., DeBose-Boyd R. A. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J. Biol. Chem. 2005;280:25242–25249. doi: 10.1074/jbc.M502989200. [DOI] [PubMed] [Google Scholar]
- Li G., Zhao G., Zhou X., Schindelin H., Lennarz W. J. The AAA ATPase p97 links peptide N-glycanase to the endoplasmic reticulum-associated E3 ligase autocrine motility factor receptor. Proc. Natl. Acad. Sci. USA. 2006;103:8348–8353. doi: 10.1073/pnas.0602747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol. Rev. 2005;207:126–144. doi: 10.1111/j.0105-2896.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Lipson C., Alalouf G., Bajorek M., Rabinovich E., Atir-Lande A., Glickman M., Bar-Nun S. A proteasomal ATPase contributes to dislocation of ERAD substrates. J. Biol. Chem. 2008;283:7166–7175. doi: 10.1074/jbc.M705893200. [DOI] [PubMed] [Google Scholar]
- Liscum L., Cummings R. D., Anderson R.G.W., DeMartino G. N., Goldstein J. L., Brown M. S. 3-Hydroxy-3-methylglutaryl-CoA reductase: a transmembrane glycoprotein of the endoplasmic reticulum with N-linked “high-mannose” oligosaccharides. Proc. Natl. Acad. Sci. USA. 1983;80:7165–7169. doi: 10.1073/pnas.80.23.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 1985;260:522–530. [PubMed] [Google Scholar]
- Misaghi S., Pacold M. E., Blom D., Ploegh H. L., Korbel G. A. Using a small molecule inhibitor of peptide:N-glycanase to probe its role in glycoprotein turnover. Chem. Biol. 2004;11:1677–1687. doi: 10.1016/j.chembiol.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J. L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S.-I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Brown M. S., Goldstein J. L. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc. Natl. Acad. Sci. USA. 1998a;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Brown M. S., Goldstein J. L. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J. Biol. Chem. 1998b;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- Nohturfft A., DeBose-Boyd R. A., Scheek S., Goldstein J. L., Brown M. S. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl. Acad. Sci. USA. 1999;96:11235–11240. doi: 10.1073/pnas.96.20.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Osborne T. F., Goldstein J. L., Brown M. S. 5′ end of HMG CoA reductase gene contains sequences responsible for cholesterol-mediated inhibition of transcription. Cell. 1985;42:203–212. doi: 10.1016/s0092-8674(85)80116-1. [DOI] [PubMed] [Google Scholar]
- Park H., Suzuki T., Lennarz W. J. Identification of proteins that interact with mammalian peptide:N-glycanase and implicate this hydrolase in the proteasome-dependent pathway for protein degradation. Proc. Natl. Acad. Sci. USA. 2001;98:11163–11168. doi: 10.1073/pnas.201393498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T., Doolman R., Avner R., Harats D., Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-Hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- Rodighiero C., Tsai B., Rapoport T. A., Lencer W. I. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 2002;3:1222–1227. doi: 10.1093/embo-reports/kvf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A., Jr, Simoni R. D. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 1992;117:959–973. doi: 10.1083/jcb.117.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitelman J., Simoni R. D. Distinct sterol and nonsterol signals for the regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 1992;267:25264–25273. [PubMed] [Google Scholar]
- Sakai J., Duncan E. A., Rawson R. B., Hua X., Brown M. S., Goldstein J. L. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- Sekler M. S., Simoni R. D. Mutation in the lumenal part of the membrane domain of HMG-CoA reductase alters its regulated degradation. Biochem. Biophys. Res. Commun. 1995;206:186–193. doi: 10.1006/bbrc.1995.1026. [DOI] [PubMed] [Google Scholar]
- Sever N., Song B.-L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 2003a;278:52479–52490. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. Accelerated degradation of HMG CoA reductase mediated by binding of Insig-1 to its sterol-sensing domain. Mol. Cell. 2003b;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- Sinensky M., Logel J. Inhibition of degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase by mevinolin. J. Biol. Chem. 1983;258:8547–8549. [PubMed] [Google Scholar]
- Skalnik D. G., Narita H., Kent C., Simoni R. D. The membrane domain of 3-hydroxy-3-methylglutaryl-coenzyme A reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto β-galactosidase. J. Biol. Chem. 1988;263:6836–6841. [PubMed] [Google Scholar]
- Song B.-L., Sever N., DeBose-Boyd R. A. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Park H., Lennarz W. J. Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions. FASEB J. 2002;16:635–641. doi: 10.1096/fj.01-0889rev. [DOI] [PubMed] [Google Scholar]
- Tortorella D., Story C. M., Huppa J. B., Wiertz E.J.H.J., Jones T. R., Bacik I., Bennink J. R., Yewdell J. W., Ploegh H. L. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 1998;142:365–376. doi: 10.1083/jcb.142.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallett S. M., Sanchez H. B., Rosenfeld J. M., Osborne T. F. A direct role for sterol regulatory element binding protein in activation of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene. J. Biol. Chem. 1996;271:12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- Yabe D., Brown M. S., Goldstein J. L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA. 2002a;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D., Xia Z.-P., Adams C. M., Rawson R. B. Three mutations in sterol-sensing domain of SCAP block interaction with Insig and render SREBP cleavage insensitive to sterols. Proc. Natl. Acad. Sci. USA. 2002b;99:16672–16677. doi: 10.1073/pnas.262669399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H., Rapoport T. A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H., Rapoport T. A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]