Abstract
Switch (SWI)/sucrose nonfermentable (SNF) is an evolutionarily conserved complex with ATPase function, capable of regulating nucleosome position to alter transcriptional programs within the cell. It is known that the SWI/SNF complex is responsible for regulation of many genes involved in cell cycle control and proliferation, and it has recently been implicated in cancer development. The ATPase action of SWI/SNF is conferred through either the brahma-related gene 1 (Brg1) or brahma (Brm) subunit of the complex, and it is of central importance to the modification of nucleosome position. In this study, the role of the Brg1 and Brm subunits were examined as they relate to chromatin structure and organization. Deletion of the Brg1 ATPase results in dissolution of pericentromeric heterochromatin domains and a redistribution of histone modifications associated with these structures. This effect was highly specific to Brg1 and is not reproduced by the loss of Brm or SNF5/BAF47/INI1. Brg1 deficiency is associated with the appearance of micronuclei and aberrant mitoses that are a by-product of dissociated chromatin structure. Thus, Brg1 plays a critical role in maintaining chromatin structural integrity.
INTRODUCTION
The switch/sucrose nonfermentable (SWI/SNF) complex uses the energy of ATP hydrolysis to modulate nucleosome position and density across promoters and thus plays an important role in regulating gene expression. The SWI/SNF ATPase function is necessary for its chromatin remodeling activities (Laurent et al., 1993), and this function is manifested through one of two mutually exclusive subunits, brahma-related gene 1 (Brg1) or brahma (Brm). Although both Brg1 and Brm can function as the central ATPase in the SWI/SNF complex, each defines a discrete complex with unique biochemical activity. In addition, specific accessory factors interact with the ATPase subunits and play critical roles in chromatin remodeling activity. For example, SNF5/BAF47/INI1 is present in both Brg1- and Brm-associated complexes and is required for maximal chromatin remodeling activity, both in vitro and in yeast. Surprisingly, although Brg1 and Brm have >75% sequence homology, the impact of each individual subunit on chromatin biology and underlying cell biology is poorly understood (Phelan et al., 1999).
Due to its central function in chromatin remodeling, SWI/SNF function is required in many facets of gene regulation. Correspondingly, it is estimated that approximately 5% of the yeast genome is transcriptionally regulated by the SWI/SNF complex (Holstege et al., 1998; Sudarsanam et al., 2000). In mammalian cells, SWI/SNF plays a critical role in the activation of a diverse set of genes and has been shown to be recruited directly to transcriptional activation domains (Deroo and Archer, 2001; Hassan et al., 2001). In contrast, SWI/SNF is also required for transcriptional repression. For example, transcriptional repression of cell cycle genes mediated by the retinoblastoma tumor suppressor is dependent on SWI/SNF, implicating the involvement of SWI/SNF in cellular proliferation (Gunawardena et al., 2004, 2007).
Paradoxically, disruption of SWI/SNF function has alternatively been associated with tumorigenesis and lack of cellular/organismal viability. Specifically, loss or mutation of the SNF5/INI1 subunit is associated with rhabdoid tumorigenesis, and it has been convincingly demonstrated that the SNF5/INI1 gene acts as a tumor suppressor (Biegel et al., 1999; Sevenet et al., 1999; Roberts et al., 2002). Correspondingly, diminished expression or loss of Brg1, Brm, Baf57, Baf180, and several other chromatin-modifying genes has been observed in cancer cell lines and tumor specimens (Gregory and Shiekhattar, 2004). However, these implications in tumor development are seemingly at odds with important roles for SWI/SNF subunits in maintaining cellular viability and proliferative capacity (Bultman et al., 2000).
It is well known that chromosomal instability is a hallmark of cancer. Interestingly, loss of chromatin regulatory proteins can have deleterious effects on chromatin structure that can compromise genome stability and fuel tumorigenesis (David et al., 2003, 2006). Here, we used somatic cell culture models to define the impact of the SWI/SNF chromatin-modifying complex on chromatin biology and cell cycle progression.
MATERIALS AND METHODS
Cell Culture and Adenoviral Transduction
Primary and 3T3 cell lines were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin-streptomycin, and 2 mM l-glutamine at 37°C in 5% CO2. All cell lines were transduced with adenoviruses encoding green fluorescent protein (GFP), GFP-Cre recombinase (Cre), or Cre for 120 h.
Immunoblotting
Cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon-P membrane (Millipore, Billerica, MA). Membranes were incubated with the following primary antibodies: Brg1 (sc-17796; Santa Cruz Biotechnology, Santa Cruz, CA), Brm (sc-17828; Santa Cruz Biotechnology), vimentin (gift from Dr. Wallace Ip, University of Cincinnati, Cincinnati, OH), Cre (69050; Novagen, Madison, WI), p53 (CM5p; Novocastra, New Castle, United Kingdom), and lamin B (sc-6217; Santa Cruz Biotechnology).
Immunofluorescence and 5-Bromo-2′-deoxyuridine (BrdU) Incorporation
Cells were subjected to the indicated treatment, fixed with methanol:acetone (1:1) or 3.7% formaldehyde, and permeabilized with 0.3% Triton X-100. Cells were then stained for the indicated proteins. For BrdU, cells were subjected to the indicated treatment and incubated with BrdU reagent (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) for 1 h before fixation in 3.7% formaldehyde. Cells were then permeabilized with 0.3% Triton X-100 in phosphate-buffered saline, and incubated with rat anti-BrdU. Cells were then stained with rhodamine anti-rat immunoglobulin G, and mounted on slides. In assays wherein abnormal heterochromatin was quantified, “abnormal” was determined by disorganization or loss of clearly identifiable, normally discrete and punctate (round) heterochromatin domains.
Flow Cytometry
BrdU reagent was added to cells for 1 h before harvest. Cells were then harvested by trypsinization, fixed with ethanol, and incubated with propidium iodide and fluorescent anti-BrdU secondary antibody. Histograms represent 15,000 gated events. Histograms were analyzed using FlowJo software, version 8.7 (Tree Star, Ashland, OR).
Polymerase Chain Reaction (PCR)
Total genomic DNA was extracted using DNeasy blood and tissue kit (69504; QIAGEN, Valencia, CA). Indicated portions of DNA were amplified via polymerase chain reaction; amplified DNA was separated using a 1.5% agarose gel. For mouse genotyping (Brm), mouse tail DNA was extracted and analyzed by PCR using a combination of three different primers: sense 5′-CCTGAGTCATTTGCTATAGCCTGTG-3′, antisense 5′-GGACTGCCAGCTGCA-GAG-3′, and 5′-CATCGCCTTCTAT-CGCCTTC-3′. For mouse genotyping (Brg1), mouse tail DNA was extracted and analyzed by using the following primers: sense 5′-GATCAGCTCATGCCCTAAGG-3′ and antisense 5′-CCTACAGTTCCATG-CAGCTGG-3′.
Retroviral Transduction, Plasmid Transfection, and Selection
Retroviral Transduction.
Cells were transduced for 7 h with a retrovirus containing a p53 dominant-negative allele (p53DD). The transduced cells were selected for 14 d in 0.2 mg/ml G418 sulfate (Calbiochem, San Diego, CA).
Plasmid Transfection.
Cells were transfected for 18 h using Lipofectin transfection reagent (Invitrogen, Carlsbad, CA) following manufacturer-provided protocol. After 48 h, cells were passaged into 2 μg/ml puromycin for 7 d. These cells were passaged again into 2 μg/ml puromycin for another 7 d before beginning experimental procedures.
RESULTS
Targeted Deletion of SWI/SNF ATPases in Cell Culture
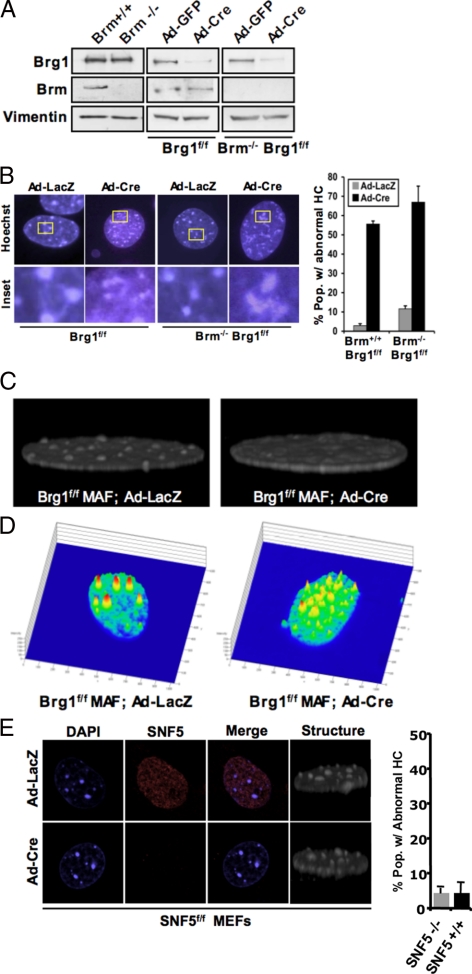
The SWI/SNF complex is implicated in a wide range of highly cancer-relevant processes, such as transcriptional regulation, cell cycle control, and DNA repair. Here, models of Brg1 and Brm knockout were used to directly define the impact of deficiency of these subunits on cellular function. To perform these studies, we utilized mouse adult fibroblasts that were derived from Brg1f/f, Brm−/−, and compound Brm−/−Brg1f/f mice. By employing Cre-recombinase in vitro, any combination of core ATPase deficiency could be created (Supplemental Figure S1A). Cell cultures were transduced with adenoviruses encoding either GFP or GFP-Cre, and analyses of GFP-fluorescence revealed >95% of cells were infected. Correspondingly, the Brg1lox allele was efficiently recombined in those cultures subjected to infection with GFP-Cre encoding adenoviruses (Supplemental Figure S1B). Consistent with the genetic recombination, the expression of Brg1 protein was significantly reduced at 120 h after infection (Figure 1A). As expected, the Brm knockout cell cultures exhibited the appropriate genomic organization of the targeted allele and failed to express Brm protein (Figure 1A). Thus, each combination of ATPase deficiency could be modeled in these cell cultures.
Figure 1.
Loss of core ATPases causes appearance of abnormal heterochromatin in primary fibroblasts. (A) Primary MAFs were transduced with adenovirus encoding either GFP or Cre-recombinase. At 120 h after infection, cells were harvested, and protein lysates were resolved by SDS-PAGE. The indicated proteins were detected by immunoblotting. (B) Cells were cultured on glass coverslips and transduced with adenovirus encoding either LacZ or Cre. At 120 h after infection, coverslips were fixed and stained with Hoechst dye and visualized by fluorescence microscopy. Percentage of cells with abnormal heterochromatin (HC) was counted. (C) Cells were prepared as described in B, stained with 4,6-diamidino-2-phenylindole (DAPI), and visualized by confocal microscopy. Serial sections were imaged and stacked to create a three-dimensional image of chromatin structure. (D) Cells were visualized by confocal microscopy, and relative chromatin staining intensity was quantitatively graphed. Staining intensity represents chromatin condensation/organization, with red indicating the highest level of organization and blue indicating the least. (E) Cells were stained for SNF5, and with DAPI, they were visualized by confocal microscopy. Three-dimensional images of chromatin density were taken. The percentage of cells with abnormal heterochromatin was counted.
Loss of Brg1 Leads to Nuclear Malformations
Because SWI/SNF subunits have been shown to play important roles in chromatin dynamics, nuclear structure and organization were examined in the context of ATPase deficiency. At 120 h after infection, cells were stained with Brg1 antibodies to unequivocally define Brg1-deficient cells, and chromatin was labeled with Hoechst 33258. Primary murine fibroblasts harbor highly discrete pericentromeric heterochromatin domains, which are readily identified through DNA staining. In cultures with intact ATPase function, these pronounced domains were clearly identifiable in >90% of cells (Figure 1B). The deletion of Brg1 yielded a dramatic disruption of these chromatin domains, as evidenced by lack of defined borders and an overall dispersion of heterochromatin (Figure 1B). Importantly, this effect occurred in the majority of cells lacking Brg1. In contrast, Brm deficiency lead to only a modest impact on the fidelity of such heterochromatin domains. To further explore the effect of Brg1 deletion on chromatin organization, nuclei were visualized by confocal microscopy. Optical sectioning revealed that heterochromatin domains have a discrete three-dimensional structure (Figure 1C). However, the deletion of Brg1 results in the loss of this structure and a corresponding disorganization of heterochromatin in the nucleoplasm (Figure 1C). Consistent with this assessment, the discrete heterochromain domains are abnormally dispersed in quantitative monitoring of chromatin intensity (Figure 1D). To illustrate the specificity of this phenomenon to Brg1 loss, the acute deletion of SNF5 was achieved by Cre-recombinase expression in SNF5f/f mouse embryonic fibroblasts. Strikingly, deletion of this SWI/SNF core-complex component yields no change in the structure or presence of heterochromatin domains (Figure 1E). Furthermore, these observed affects on heterochromatin are not an artifact of Cre-recombinase expression, because intact chromatin domains are observed in other models that have been exposed to Cre-recombinase (Figure 1E). Thus, Brg1 specifically plays a critical role in the maintenance of heterochromatin domains.
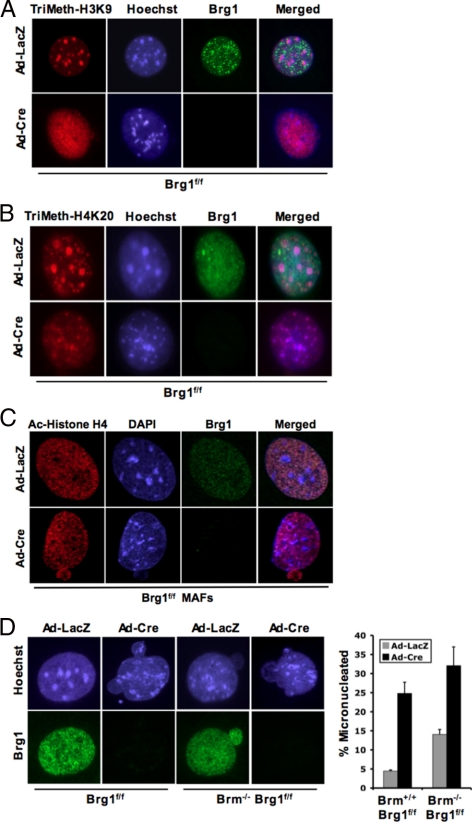
Pericentromeric heterochromatin is characterized by specific histone modifications. To delineate how Brg1 deficiency influenced the distribution of these histone modifications, cells were costained for Brg1 (to define negative cells) and histone modifications. As shown in Figure 2A, histone H3-trimethyl lysine 9 modification is largely confined to pericentromeric heterochromatin domains in cells harboring SWI/SNF ATPase activity. In contrast, there was a significant dispersion and redistribution of histone H3-trimethyl lysine 9 species with the deletion of Brg1. Similar results were observed with the histone H4-trimethyl lysine 20 modification, which was altered in the absence of Brg1 (Figure 2B). Interestingly, this result was specific to Brg1 loss, because no changes in histone modifications were observed in models of Brm and SNF5 loss (Supplemental Figure S2). Furthermore, this phenomenon is relatively specific to modifications associated with heterochromatin, because acetylation of histone H4 and histone H3 were not significantly altered with the deletion of Brg1 (Figure 2C; data not shown). Thus, loss of Brg1 impinges on both the structural fidelity of pericentromeric heterochromatin and modifications that define these distinct chromatin structures.
Figure 2.
Brg1 is required to maintain specific histone methylation. Cells were cultured on glass coverslips and transduced with adenovirus encoding either LacZ or Cre. (A–C) At 120 h after infection, coverslips were fixed and stained for the indicated modifications or proteins and visualized via immunofluorescence microscopy. (D) At 120 h after infection, coverslips were fixed and stained as indicated. The percentage of cells containing micronuclei was counted.
Role of Brg1/Brm in Maintenance of Genomic Structure
Multiple studies have suggested that aberrations in chromatin organization can have significant effects on proliferation and genome stability. In the investigation of chromatin structure, it was apparent that deficiency of SWI/SNF subunits was associated with a corresponding increase in cells harboring micronuclei (Figure 2D). Cells lacking Brm exhibited a modest yet significant increase in micronuclei formation (Figure 2D), which is consistent with a previous report (Coisy-Quivy et al., 2006). However, the acute loss of Brg1 resulted in a striking increase in micronuclei. This level of micronuclei was subtly increased in the context of combined Brg1 and Brm deficiency (Figure 2D). Thus, the percentage of cells harboring micronuclei largely correlated with the aberrations in heterochromatin organization, wherein Brg1 deficiency was responsible for a more significant impact.
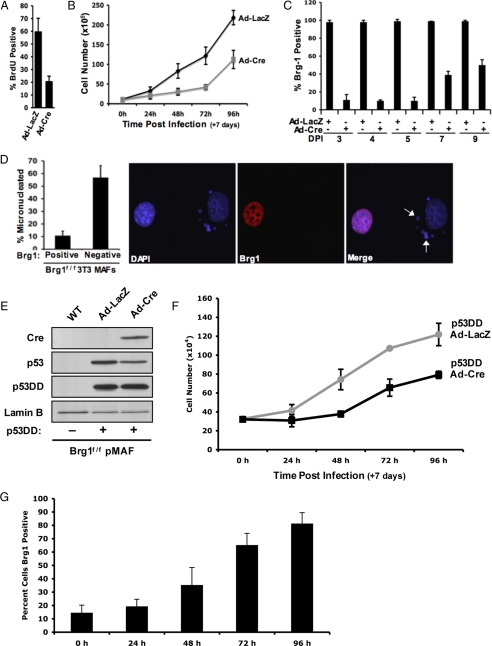
Although many attempts were made to establish long-term primary cultures lacking Brg1, these cells were ultimately selected against in culture. Because primary cells have a finite proliferative capacity, the Brg1f/f cells were subjected to spontaneous immortalization by using a 3T3 protocol. Due to the unlimited proliferative capacity of this model, we could readily assess the effect of Brg1 loss on cell proliferation. Initially, BrdU incorporation was analyzed after the deletion of Brg1 (Figure 3A). These analyses showed a significant reduction in BrdU incorporation with Brg1 loss. These effects were further confirmed by analyses of cellular proliferation, wherein the deletion of Brg1 initially suppressed proliferation (Figure 3B). However, direct analyses of Brg1 by immunostaining of these cultures showed the emergence of Brg1-positive cells that, over time, began to represent the majority of the population (Figure 3C). This inhibition of proliferation was also associated with the formation of micronuclei in the immortalized Brg1-compromised cultures. Nearly 60% of all 3T3 cells in these Brg1-compromised cultures harbored micronuclei (Figure 3D). These analyses imply that loss of Brg1 has a deleterious impact on proliferation.
Figure 3.
Brg1 deficiency causes micronuclei formation and reduces cellular proliferative capacity in vitro. Brg1f/f 3T3 MAFs were cultured and transduced with adenovirus encoding either LacZ or Cre for 120 h. (A) One hour before harvest, cells were pulse labeled with BrdU. Coverslips were fixed and stained for BrdU incorporation, visualized, and quantified. (B) Cells were seeded at equal density 7 d after infection. At indicated the times, plates were fixed and stained with 1% crystal violet. Five random fields per plate were counted, per time point—results are indicative of three separate experiments. (C) Coverslips were harvested at the indicated times, fixed, and stained for Brg1 expression. Cells were visualized by immunofluorescence, and results are quantified at different days postinfection (DPI). (D) Coverslips were harvested 120 h after infection, fixed, and costained for DAPI and Brg1. The percentage of Brg1-positve or -negative cells containing micronuclei was quantified. (E–G) Brg1f/f primary MAFs were cultured and transduced with retrovirus encoding p53DD, resulting in immortalization of the cultures. (E) Cells were transduced with adenovirus encoding either LacZ or Cre. At 120 h after infection, cells were harvested and protein lysates were resolved by SDS-PAGE. Immunoblotting was performed for indicated proteins. (F) Cells were seeded at equal density 7 d after infection. At the indicated times, plates were fixed and stained with 1% crystal violet. Five random fields per plate were counted, per time point—results are indicative of three separate experiments. (G) Coverslips were harvested at the indicated times, fixed, and stained for Brg1 expression. Cells were visualized by immunofluorescence, and results are quantified at different DPI.
Because aberrant nuclear structure could function to induce a DNA damage checkpoint, we evaluated the impact of disabling p53 function on proliferation in the absence of Brg1. Primary Brg1f/f mouse adult fibroblasts (MAFs) were transduced with a retrovirus encoding p53DD. This results in aberrant stabilization of the endogenous p53 (Figure 3E) and immortalization of the cultures (data not shown), consistent with a previous report (Bosco et al., 2007). As with the 3T3 cultures, Brg1 deletion in this context resulted in a suppression of proliferation (Figure 3F). Furthermore, single cell analyses demonstrated that Brg1 deletion was selected against over extended culture (Figure 3G). These data suggest that the defects in chromatin organization experienced with loss of Brg1 result in the inhibition of proliferation, even with p53 function compromised.
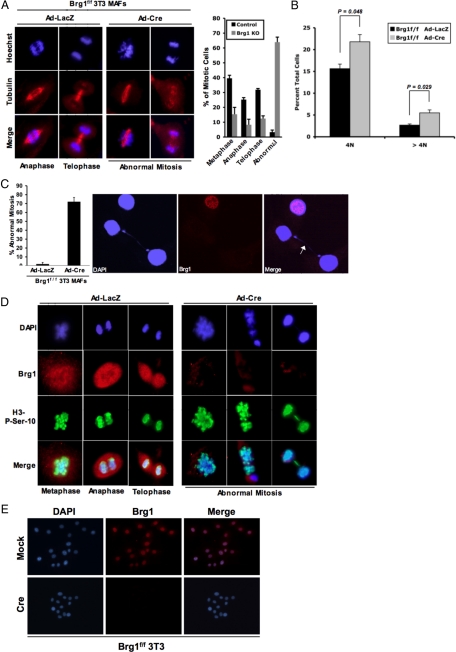
To determine the basis for the inhibition of proliferation, we initially analyzed the cell cycle distribution by flow cytometry. With acute deletion of Brg1 there was an increase in cells harboring 4N and >4N DNA content (Figure 4B). To explore whether Brg1 deletion induced alterations in mitotic entry or progression, the presence and stage of mitotic nuclei were evaluated. In immortalized cultures, mitotic cells were readily apparent with the absence of Brg1; however, these cells expressed hallmarks of aberrant mitosis that could be easily identified. This finding was even apparent in mixed cultures in which not all cells had lost Brg1 (Figure 4A). Importantly, >70% of cells harboring Brg1 deficiency exhibited some form of abnormal mitotic cell division (Figure 4C). Subsequently, we determined whether loss of Brg1 influenced the phosphorylation of Ser10 on histone H3, which is associated with mitotic entry. These analyses showed that loss of Brg1 did not preclude this histone modification (Figure 4D). However, there was a significant selection against cells in anaphase or telophase. Those few Brg1-deficient cells in anaphase or telophase (Figure 4A) manifested mitotic bridges, lagging chromosomes, and evidence of mitotic catastrophes (Figure 4D). Together, these studies indicate that although Brg1 is not required for mitotic entry, appropriate chromatin structure is requisite for proper and effective execution of mitosis. To rigorously identify whether cells lacking Brg1 could proliferate, a vector expressing Cre with a selectable marker was used. After selection, the majority of cells still expressed Brg1; however, rare Brg1-deficient cells were detected. Passaging these cells at low density enabled us to specifically interrogate whether single cells could proliferate into microcolonies. From >150 colonies analyzed, only two demonstrated expansion in the absence of Brg1. Therefore, although there is a strong selection against Brg1 loss, few cells can ultimately proliferate under this subversive condition (Figure 4E and Supplemental Figure S3).
Figure 4.
Brg1 deficiency causes mitotic abnormality and aneuploidy. 3T3 cells were cultured and transduced with adenovirus encoding either LacZ or Cre. (A) At 120 h after infection, coverslips were fixed and stained as indicated. The percentage of cells in each phase of mitosis was quantified based on chromatin/tubulin organization. (B) At 120 h after infection, cells were harvested and stained with propidium iodide and counted via flow cytometry. The percentage of total gated cells with 4N and >4N DNA content was quantified. (C) At 120 h after infection, coverslips were fixed and stained as indicated; the percentage of cells exhibiting abnormal mitosis was counted based on DAPI staining. (D) At 120 h after infection, coverslips were fixed and stained as indicated. (E) Brg1f/f 3T3 MAFs were transfected with either a pBABE-puromycin selectable Cre-encoding plasmid, or a pBABE-puromycin selectable control plasmid. Cells were then passaged into media containing puromycin for 14 d. Cells (1 × 103) were plated onto glass coverslips in a 10-cm dish and allowed to grow for 48 h, and then the cells were harvested and stained for Brg1 and DAPI.
DISCUSSION
It is known that the SWI/SNF complex employs one of two different core ATPases, Brg1 or Brm, to remodel chromatin structure by repressing and facilitating transcription. In this study, the individual function of Brg1 and Brm on chromatin organization, nuclear structure, and mitotic division was determined.
The SWI/SNF complex is thought to use the Brg1 and Brm subunits interchangeably to mediate the ATPase function critical for chromatin remodeling (Roberts and Orkin, 2004). In terms of biochemical activity, there is significant functional redundancy between these core ATPases (Phelan et al., 1999). Furthermore, several transcriptional processes can be mediated via the activity of either ATPase. However, there are clear distinctions between Brg1 and Brm related to tissue-specific dependence and overall organismal survivability (Muchardt and Yaniv, 2001; Kadam and Emerson, 2003). Our data show that deficiency of Brg1, but not Brm, leads to the dissolution of discrete pericentromeric heterochromatin domains. This finding suggests that the Brg1 ATPase either represents a larger fraction of the total ATPase protein in the cell or that it has distinct functions from Brm. However, this effect on chromatin is readily apparent via multiple approaches and results in the structural dispersion of heterochromatin domains. These structures are known to be highly enriched in trimethylated histone H3 lysine 9 and histone H4 lysine 20. Correspondingly, Brg1 deletion results in a significant dissemination of these modifications. Interestingly, our data reveal that loss of SNF5, another core subunit of the SWI/SNF complex, does not elicit dissolution of heterochromatin domains, nor does it affect the localization of trimethylation on histone H4 lysine 20 or histone H3 lysine 9 (Supplemental Figure S2). It is well established that these histone markers are not required for the structural maintenance of the heterochromatin domains, because deletion of Suv39H1/H2 and retinoblastoma (RB)-related family members results in the loss of trimethylation of histone H3 lysine 9 and histone H4 lysine 20 at heterochromatin domains, respectively, without compromising the integrity of the overall chromatin domain structure (Peters et al., 2001; Gonzalo et al., 2005). Thus, the maintenance of pericentromeric heterochromatin domains is hierarchical, with an underlying Brg1-dependent function that is critical for structural integrity.
The regulation of transcription and modulation of chromatin structure are critically involved in cellular proliferation, and aberrations associated with these processes are implicated in tumorigenesis. The effect of SWI/SNF ATPase deficiency on cellular proliferation remains the subject of controversy, as there are tumor cell lines which harbor discrete loss of both ATPases (Strobeck et al., 2002; Reisman et al., 2003). These cell lines actively proliferate and are, in fact, compromised for the appropriate response to growth inhibitory signals as elicited through the RB pathway. Although such tumor cell models are important for interrogating pathways, it is not possible to determine the cellular requirement for ATPases because other genetic events could obviate their necessity. Thus, the analyses of cultured cells from gene-targeted animals afford an opportunity to define their intrinsic role in proliferation. Loss of Brm has minimal effect on proliferation, and primary and immortalized lines lacking Brm function can be readily propagated. However, Brg1 deletion resulted in a substantial reduction in cellular proliferation and BrdU incorporation and was selected against during culture. Moreover, this effect was observed in both primary cell culture and cultures specifically deficient in canonical p53 function. Thus, loss of Brg1 is not tolerated even in the context of rapidly proliferating immortalized populations. This finding is supportive of previous analyses in embryonal carcinoma cells (Sumi-Ichinose et al., 1997). Cell cycle analyses strongly suggest that the principle negative impact of Brg1 deficiency on proliferation is manifest during mitotic progression. Our data support the notion that Brg1 deficiency can be overcome by virtue of additional stochastic events; however, this process was highly sporadic even in the context of immortalized 3T3 populations and selection for Brg1 deletion. Thus, tumor cell lines and potentially other Brg1-deficient cell types, presumably use compensatory mechanisms to bypass the requirement that we observed.
Findings from multiple laboratories have suggested that deletion/loss of Brg1 may contribute to the genesis of cancer (Murphy et al., 1999; Lee et al., 2002); however, the underlying mechanism for this process is unclear. The data presented here indicate that loss of Brg1 results in aberrant mitotic progression and provides evidence of genomic instability. These findings are supported by previous studies that show both the localization of SWI/SNF to mitotic chromosomes (Xue et al., 2000), and the requirement for related complexes for proper mitosis and chromosome maintenance (Baetz et al., 2004; Campsteijn et al., 2007). This phenomenon is also similar to that observed with the knockout of a critical centromeric protein, inner centromere protein, which disrupts chromatin structure and leads to genomic instability (Cutts et al., 1999). Importantly, normal mitotic progression is dependent upon proper centromeric function, and the loss of Brg1 seems to result in disassembly of these pericentromeric heterochromatin domains. This suggests a mechanism for the observed mitotic abnormalities, because disruption of such domains compromises mitotic fidelity. It has been recently reported that tumors arising in Brg1+/− mice, although not mimicking specific pathways, are best characterized by genomic instability (Bultman et al., 2008). Importantly, this study also concludes that tumor formation in Brg1+/− mice occurs due to haploinsufficiency rather than loss of heterozygosity (Bultman et al., 2008), suggesting a lack of selection or proliferative advantage with the complete ablation of Brg1. Thus, in the context of Brg1 deficiency, resultant dispersion of pericentric heterochromatin domains and mitotic dysfunction could potentially represent the underlying key etiological feature relevant to tumorigenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Erik and Karen Knudsen's laboratories for critical review of the manuscript and insightful discussion. We also thank Drs. Christian Muchardt and Moshe Yaniv for providing Brm nullizygous mice, Drs. Daniel Metzger and Pierre Chambon for providing Brg1f/f mice, and to Dr. Charles Roberts for providing SNF5f/f mouse embryonic fibroblasts and the pBABE-Puro and pBABE-Puro-Cre plasmids. Finally, we thank Dr. A. Kathleen McClendon for providing the p53DD retrovirus. E.S.K. is supported by National Cancer Institute grant CA 104213.
Abbreviations used:
- Brg1
brahma-related gene 1
- Brm
brahma
- RB
retinoblastoma
- SNF
sucrose nonfermentable
- SNF5
SNF5/BAF47/INI1
- SWI
switch.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1224) on May 20, 2009.
REFERENCES
- Baetz K. K., Krogan N. J., Emili A., Greenblatt J., Hieter P. The ctf13–30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 2004;24:1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel J. A., Zhou J. Y., Rorke L. B., Stenstrom C., Wainwright L. M., Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- Bosco E. E., Wang Y., Xu H., Zilfou J. T., Knudsen K. E., Aronow B. J., Lowe S. W., Knudsen E. S. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J. Clin. Invest. 2007;117:218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S., et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman S. J., Herschkowitz J. I., Godfrey V., Gebuhr T. C., Yaniv M., Perou C. M., Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Campsteijn C., Wijnands-Collin A. M., Logie C. Reverse genetic analysis of the yeast RSC chromatin remodeler reveals a role for RSC3 and SNF5 homolog 1 in ploidy maintenance. PLoS Genet. 2007;3:e92. doi: 10.1371/journal.pgen.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coisy-Quivy M., Disson O., Roure V., Muchardt C., Blanchard J. M., Dantonel J. C. Role for Brm in cell growth control. Cancer Res. 2006;66:5069–5076. doi: 10.1158/0008-5472.CAN-05-0596. [DOI] [PubMed] [Google Scholar]
- Cutts S. M., Fowler K. J., Kile B. T., Hii L. L., O'Dowd R. A., Hudson D. F., Saffery R., Kalitsis P., Earle E., Choo K. H. Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum. Mol. Genet. 1999;8:1145–1155. doi: 10.1093/hmg/8.7.1145. [DOI] [PubMed] [Google Scholar]
- David G., Dannenberg J. H., Simpson N., Finnerty P. M., Miao L., Turner G. M., Ding Z., Carrasco R., Depinho R. A. Haploinsufficiency of the mSds3 chromatin regulator promotes chromosomal instability and cancer only upon complete neutralization of p53. Oncogene. 2006;25:7354–7360. doi: 10.1038/sj.onc.1209734. [DOI] [PubMed] [Google Scholar]
- David G., Turner G. M., Yao Y., Protopopov A., DePinho R. A. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 2003;17:2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroo B. J., Archer T. K. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene. 2001;20:3039–3046. doi: 10.1038/sj.onc.1204328. [DOI] [PubMed] [Google Scholar]
- Gonzalo S., et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Gregory R. I., Shiekhattar R. Chromatin modifiers and carcinogenesis. Trends Cell Biol. 2004;14:695–702. doi: 10.1016/j.tcb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Gunawardena R. W., Fox S. R., Siddiqui H., Knudsen E. S. SWI/SNF activity is required for the repression of deoxyribonucleotide triphosphate metabolic enzymes via the recruitment of mSin3B. J. Biol. Chem. 2007;282:20116–20123. doi: 10.1074/jbc.M701406200. [DOI] [PubMed] [Google Scholar]
- Gunawardena R. W., Siddiqui H., Solomon D. A., Mayhew C. N., Held J., Angus S. P., Knudsen E. S. Hierarchical requirement of SWI/SNF in retinoblastoma tumor suppressor-mediated repression of Plk1. J. Biol. Chem. 2004;279:29278–29285. doi: 10.1074/jbc.M400395200. [DOI] [PubMed] [Google Scholar]
- Hassan A. H., Neely K. E., Vignali M., Reese J. C., Workman J. L. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 2001;6:D1054–D1064. doi: 10.2741/hassan. [DOI] [PubMed] [Google Scholar]
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Kadam S., Emerson B. M. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treich I., Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Lee D., Kim J. W., Seo T., Hwang S. G., Choi E. J., Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. When the SWI/SNF complex remodels the cell cycle. Oncogene. 2001;20:3067–3075. doi: 10.1038/sj.onc.1204331. [DOI] [PubMed] [Google Scholar]
- Murphy D. J., Hardy S., Engel D. A. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Phelan M. L., Sif S., Narlikar G. J., Kingston R. E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Reisman D. N., Sciarrotta J., Wang W., Funkhouser W. K., Weissman B. E. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- Roberts C. W., Leroux M. M., Fleming M. D., Orkin S. H. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- Roberts C. W., Orkin S. H. The SWI/SNF complex–chromatin and cancer. Nat. Rev. Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Sevenet N., Sheridan E., Amram D., Schneider P., Handgretinger R., Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J. Hum. Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck M. W., Reisman D. N., Gunawardena R. W., Betz B. L., Angus S. P., Knudsen K. E., Kowalik T. F., Weissman B. E., Knudsen E. S. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 2002;277:4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer V. R., Brown P. O., Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi-Ichinose C., Ichinose H., Metzger D., Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell. Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Canman J. C., Lee C. S., Nie Z., Yang D., Moreno G. T., Young M. K., Salmon E. D., Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.