Abstract
Pax7 is a key regulator of skeletal muscle stem cells and is required along with Pax3 to generate skeletal muscle precursors. We have identified a collection of genes induced by either Pax3 or Pax7 in C2C12 muscle cells. Two notable Pax3/7 targets are the inhibitory helix-loop-helix (HLH) proteins inhibitor of DNA binding (Id) 2 and Id3, both of which are coordinately expressed with Pax7 in quiescent satellite cells and are induced in quiescent C2C12 myogenic cells after ectopic expression of either Pax3 or Pax7. Ectopic Pax7 activates expression of a luciferase reporter driven by the Id3 promoter, and maximal induction of this reporter requires a conserved Pax7 binding site located upstream of the Id3 gene. Chromatin immunoprecipitation indicated that Pax7 is bound upstream of the Id3 promoter in quiescent satellite cells. In addition, short hairpin RNA-mediated knockdown of Pax7 expression in cultured satellite cells coordinately decreased both Id2 and Id3 expression. Together, these findings indicate that Id3 is a direct transcriptional target for Pax7 in quiescent satellite cells, and they suggest that Pax7 acts to block premature differentiation of quiescent satellite cells by inducing the expression of Id2 and Id3, which in turn may act to block either the precocious induction of myogenic basic (b)HLH proteins, the activity of myogenic bHLH proteins, or both.
INTRODUCTION
Pax3 and Pax7 are two closely related transcription factors that are expressed in the dermomyotome, and they have been shown to be essential for generation of all fetal trunk musculature (Relaix et al., 2005). Pax3/7-expressing dermomyotomal cells have recently been shown to give rise to skeletal muscle progenitors, termed satellite cells, in the adult (Gros et al., 2005; Kassar-Duchossoy et al., 2005; Relaix et al., 2005; Schienda et al., 2006). Satellite cells are a small population of myogenic progenitors that reside between the sarcolemma and basal lamina of the muscle fiber, and they play a crucial role in postnatal muscle growth and regeneration (reviewed in Zammit et al., 2006). Following skeletal muscle injury or exercise-induced activation, the normally quiescent Pax7-expressing satellite cells proliferate extensively and up-regulate expression of MyoD (Smith et al., 2001; Yan et al., 2003; Montarras et al., 2005) and Myf-5 (Conboy and Rando, 2002; Yan et al., 2003), before differentiation into skeletal muscle.
Pax3 and Pax7 are members of the Pax transcription factor family and contain both paired (PD) and homeodomain (HD) DNA binding motifs. Pax3 and Pax7 can bind to DNA sequences containing either a consensus paired domain binding site (GTCAC A/G C/G A/T T/C) or a homeodomain binding site (ATTA) (Chalepakis et al., 1994; Chalepakis and Gruss, 1995). Pax3 and Pax7 activate MyoD expression (Maroto et al., 1997; Tajbakhsh et al., 1997; Relaix et al., 2003, 2004), and recently Pax3 has been shown to directly bind sequences that regulate the expression of either MyoD in C2C12 cells (Hu et al., 2008) or Myf-5 during development of hypaxial muscle (Bajard et al., 2006). Although it is clear that Pax3/7 can directly induce the expression of Myf5 (Bajard et al., 2006), MyoD (Hu et al., 2008), and fibroblast growth factor receptor 4 (Lagha et al., 2008), other relevant targets for Pax3/7 in satellite cells have yet to be identified. To identify potential targets for Pax3/7 in satellite cells, we examined the transcriptional profile of genes induced by these transcription factors in the C2C12 muscle cell line. Of the genes identified, we found that a subset were also expressed in quiescent satellite cells and therefore they could potentially be direct targets of Pax7 in these cells. In this report, we focus on two such putative Pax3/7 transcriptional targets, inhibitor of DNA binding (Id) 2 and Id3, which we found to be expressed in quiescent satellite cells. We report that Pax3/7 can drive expression of both Id2 and Id3 in C2C12 cells under low serum conditions, that the Id3 promoter contains a conserved Pax3/7 binding site, and that Pax3/7 can activate expression of a reporter construct driven by the Id3 promoter. In addition, we demonstrate that Pax7 is normally bound to the Id3 promoter in quiescent satellite cells and that short hairpin RNA (shRNA)-mediated knockdown of Pax7 expression in cultured satellite cells coordinately decreases both Id2 and Id3 expression. Together, these findings suggest that Id2 and Id3 are both transcriptional targets of Pax7 and that Id3 is a direct transcriptional target of Pax7 in satellite cells. Because the Id gene family contains key negative regulators of positively acting basic helix-loop-helix (bHLH) proteins (reviewed in Ruzinova and Benezra, 2003; Perk et al., 2005), our findings suggest that Pax7-induced expression of Id family members in quiescent satellite cells may act to block myogenic bHLH function in these cells.
MATERIALS AND METHODS
Antibodies
Anti-FLAG and anti-tubulin antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Anti-hemagglutinin (HA) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Pax3 and anti-Pax7 antibodies were obtained from Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). Anti-Id3 antibody was generously supplied by Robert Benezra (Memorial Sloan Kettering Cancer Center, New York, NY).
Cell Culture and Transient Transfection
CSM4B cells were isolated from 2-mo-old C57BL/6 mice as described previously (Cerletti et al., 2008). The cells were grown in F-12 media supplemented with 20% horse serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. C2C12 cells were grown in DMEM supplemented with 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. For differentiation, the media were changed to DMEM supplemented with 2% horse serum, 50 U/ml penicillin, and 50 μg/ml streptomycin. FuGENE 6 (Roche Diagnostics, Indianapolis, IN) was used for transient transfection per the manufacturer's directions.
Plasmids
Mouse Pax3 (Maroto et al., 1997) or Pax7d (Seale et al., 2004) cDNAs were polymerase chain reaction (PCR) amplified to remove stop codons and cloned in a retroviral vector (pOZ-FH-C-puro). pOZ-FH-C-puro vector was made by modifying the pOZ-FH-C vector (Nakatani and Ogryzko, 2003). A PCR-amplified puromycin resistance gene (isolated from pBabe-puro) was cloned in place of interleukin-2Rα using NcoI and BamHI site. pGL3-Id3(−934)-firefly luciferase and pGL3-Id3(−517)-firefly luciferase were made by PCR amplification of mouse Id3 proximal promoter fragments −934 to +13 and −517 to +13. These fragments were then ligated to KpnI–BglII cut pGL3-basic vector (Promega, Madison, WI). The HD and PD mutant Id3 reporters were made by PCR mutagenesis. The HD site (AATTAA) was mutated to an XhoI site (CTCGAG), whereas PD site (GTCACAAGAT) was mutated to create a NheI site (TTGCTAGCCC). The lentiviral vector expressing green fluorescent protein (GFP) shRNA was obtained from Addgene (Cambridge, MA) (deposited by Robert Weinberg, Massachusetts Institute of Technology, Cambridge, MA). The lentivirus packaging vector (psPAX2) and envelope vector (pMD2.G) were also obtained from Addgene (deposited by Didier Trono, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). The lentiviral vector expressing Pax7 shRNA was purchased from Open Biosystems (Huntsville, AL). The sequence of the Pax7 shRNA target site was GCTGTTGATTACCTGGCCAAA.
Generation of Pax3- or Pax7-expressing C2C12 Cells
Murine retroviruses expressing either Pax3 or Pax7 were made as described in Nakatani and Ogryzko (2003). C2C12 cells were infected with either pOZ-FH-C-puro or with pOZ-Pax3-Flag/HA-puro or pOZ-Pax7-Flag/HA-puro. Twenty-four hours after infection, stable polyclones were selected using puromycin.
Reverse Transcriptase (RT)-PCR
RNA was harvested from samples using the RNeasy mini kit (QIAGEN, Valencia, CA) per the manufacturer's instructions. For semiquantitative reverse transcriptase-PCR, reverse transcription and PCR analysis were carried out as described previously (Munsterberg et al., 1995). The primers used for PCR are described in Supplemental Table 2. The RT-quantitative (q)PCR was carried out using ABI Prism 7700 and SYBR Premix Ex Taq kit (Takara Bio USA, Madison, WI) per the manufacturer's instructions.
Immunostaining
Single myofibers were isolated from intact limb muscle and immunostained as described in Cerletti et al. (2008). In brief, myofibers were permeabilized with 0.2% Triton X-100 for 20 min, washed with phosphate-buffered saline, and then blocked for 1 h with M.O.M. Ig blocking reagent (Vector Laboratories, Burlingame, CA)/milk and 2% goat serum. Subsequently, myofibers were blocked with an avidin/biotin blocking kit (Vector Laboratories). The myofibers were then incubated overnight at 4°C with primary antibodies against Pax7 (mouse anti-Pax7) and Id3 (rabbit anti-Id3). After washing, the myofibers were incubated with goat anti-mouse Alexa 594 (Invitrogen, Carlsbad, CA) for Pax7 and biotinylated anti-rabbit IgG (Vector Laboratories) followed by streptavidin-Alexa 488 (Invitrogen) for Id3. Nuclei were stained with 4,6-diamidino-2-phenylindole (Vector Laboratories). Fluorescence images were acquired using an BX60 microscope with DPManager software (Olympus Optical, Center Valley, PA).
Electrophoretic Mobility Shift Assay (EMSA)
C2C12 nuclear extract was prepared using nuclear extract preparation kit (Active Motif, Carlsbad, CA) per the manufacturer's instructions. To make the probe, two complementary oligonucleotides, 5′-GCTTCACCGCAATTAATTTTTTCCCCCTCTGGTCACAAGATAATTCCTGA-3′ and 5′-TCAGGAATTATCTTGTGACCAGAGGGGGAAAAAATTAATTGCGGTGAAGC-3′, containing the HD and PD binding element of the Id3 promoter were annealed and radiolabeled using [α-32P]dATP. The nuclear extracts were incubated with radiolabeled DNA probe for 30 min at 25°C in a reaction mixture containing 10 mM Tris, pH 7.9, 50 mM NaCl, 2 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 5% glycerol (vol/vol), and 100 ng/ml poly(dI-dC). DNA–protein complexes were fractionated in a 6% nondenaturing polyacrylamide gel. For antibody interaction studies, nuclear extract was preincubated with antibody for 15 min.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP with C212 cells was carried out using the ChIP assay kit (Millipore, Billerica, MA) per the manufacturer's directions. ChIP on isolated CSM4B cells was performed as described in Attema et al. (2007). The immunoprecipitated DNA was recovered using a PCR purification kit (QIAGEN) and was used as template in PCR with specific primers spanning the HD and PD binding site of Id3 promoter. PCR products were run on 1% agarose gel and visualized by ethidium bromide staining. Primers used for amplification of mouse Id3 promoter were 5′-CCGGGCATACATTTAGTTCCT-3′ and 5′-TCTCTCTCTCCTCTCTCTCTCTCAA-3′.
RESULTS
Identification of Pax3/7 Transcriptional Targets in C2C12 Cells
To identify the transcriptional targets of Pax3 and Pax7, we infected C2C12 cells with either a retrovirus (pOZ-FH-C-puro) that encodes puromycin resistance, or with retroviruses programmed to encode both puromycin resistance and either HA/FLAG-tagged Pax3 or HA/FLAG-tagged Pax7 (pOZ-Pax3-FLAG/HA-puro or pOZ-Pax7-FLAG/HA-puro, respectively). Puromycin-resistant polyclones of C2C12 cells that stably expressed either Pax3 or Pax7 were selected. Because the parental C2C12 cells used in our laboratory express either no or only trace levels of endogenous Pax3 and Pax7 (Figure 2A), these cells are a good cellular context to evaluate the effects of exogenous Pax3/7 expression in a cellular background that is devoid of these proteins. We performed DNA microarray profiling by using the Mouse Expression Array 430_2.0 (Affymetrix, Santa Clara, CA) to identify genes whose expression was induced by either or both of these Pax genes in C2C12 cells cultured under low serum conditions. Of a total of 39,000 genes analyzed, we identified 156 whose expression was induced threefold or greater by either Pax3 or Pax7 (Supplemental Table 1). Eleven of these Pax3/7-inducible genes have been observed previously to be induced by Pax7 in C2C12 cells (McKinnell et al., 2008) (Supplemental Table 1). In addition, 28 of the genes we identified were shown previously to be expressed at higher levels in quiescent versus activated satellite cells (Fukada et al., 2007) (Supplemental Table 1), correlating with the greater level of expression of Pax7 in quiescent versus activated satellite cells (Figure 1A). The HLH inhibitor Id3 was one of the genes most highly induced by Pax7 in C2C12 cells cultured in low serum, showing a 33-fold induction and a high absolute level of expression (3168 U). Although Id2 was also induced by Pax7, Id2 displayed both a lower -fold induction (5-fold) and a lower absolute level of expression (542 U). Both these genes had been observed previously to be induced by Pax7 in C2C12 cells (McKinnell et al., 2008).
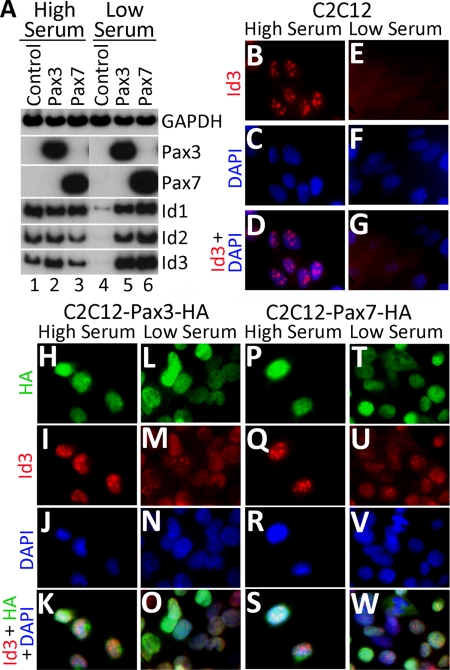
Figure 2.
Id2 and Id3 are induced in serum-starved C2C12 cells by either Pax3 or Pax7. (A) RT-PCR analysis of gene expression in either parental C2C12 cells (control) or in C2C12 polyclones expressing exogenous Pax3-HA or Pax7-HA. Cells were either cultured in 10% fetal calf serum (high serum) or in 2% horse serum (low serum) for 3 d. (B–W) Immunostaining for Id3 or Pax3/7-HA expression in either parental C2C12 cells or C2C12 polyclones expressing exogenous Pax3-HA or Pax7-HA, grown in either high or low serum conditions.
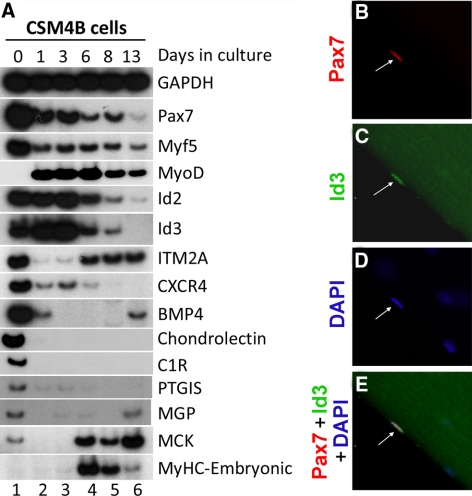
Figure 1.
Id2, Id3, and other putative Pax7 target genes are expressed in quiescent satellite cells. (A) CSM4B cells were isolated from the skeletal muscle of 2-mo-old mice. RT-PCR analysis of gene expression in either freshly isolated CSM4B cells or in cells cultured for designated days in vitro is displayed. (B–E) Id3 protein is apparent in satellite cells. Myofibers (and associated satellite cells) were isolated and immunostained with anti-Pax7 and anti-Id3. Pax7+, Id3+ satellite cell nucleus is indicated by the arrow. Although Id3 staining in the nucleus is specific (and confined to the Pax7+ nucleus; arrow), apparent Id3 staining in the myotube is due to autofluorescence.
Because it was not clear whether the Pax3/7-inducible genes that we had identified in C2C12 cells were indeed physiological targets of Pax3/7, we decided to focus our analysis on the putative Pax3/7 target genes that were also expressed in quiescent satellite cells, which constitutively express endogenous Pax7. We purified CD45−, Sca-1−, Mac-1−, CXCR4+, and β-1 integrin+ (CSM4B) myofiber-associated satellite cells by fluorescence-activated cell sorting (FACS) (Cerletti et al., 2008) from the skeletal muscle of 2-mo-old mice. Greater than 90% of CSM4B cells have been shown to express Pax7 after immediate isolation, and they efficiently differentiate into skeletal muscle both in vitro and in vivo (Cerletti et al., 2008). We assayed by RT-PCR the expression of Id2, Id3, and some other putative Pax7 targets that we had identified in the sorted CSM4B population. This analysis identified several putative Pax7 target genes that are specifically expressed in quiescent Pax7+ CSM4B cells, including Id2 and Id3, integral membrane protein 2A (ITM2A), the chemokine receptor CXCR4, Bone Morphogenic Protein 4 (BMP4), the sugar binding protein chondrolectin, complement component 1 R subunit (C1R), prostaglandin I2 (prostacyclin) synthase (PTGIS), and matrix Gla protein (MGP). RT-PCR analysis of gene expression in immediately harvested versus cultured CSM4B cells indicated that all these putative Pax7 targets are expressed in freshly isolated CSM4B cells (which express high levels of Pax7), and that in many cases their expression declines during culture of these cells (Figure 1A), in parallel with reduced Pax7 expression, induced proliferation, and eventual differentiation of these cells into myotubes (Cerletti et al., 2008). Id2 and Id3 were robustly expressed in both freshly isolated Pax7+, MyoD− CSM4B cells and in proliferating Pax7+, MyoD+ CSM4B cultures; expression of Id2 and Id3 declined and was eventually lost in differentiated cultures of Pax7−, MyoD+ CSM4B-derived myoblasts (Figure 1A, lanes 1–6). In contrast to Id2 and Id3, Id1 expression was not detected in either freshly isolated or cultured CSM4B cells (data not shown). Although the Id genes have been linked previously to cellular proliferation in at least some cellular contexts (Benezra et al., 1990), in CSM4B-derived myoblasts Id2 and Id3 are expressed in both quiescent and proliferating cells, indicating that their expression can be uncoupled from the cell cycle in this cell type.
Pax3 or Pax7 Induces the Expression of Id Family Members in C2C12 Cells and Id3 Protein Is Detectable in Quiescent Satellite Cells
To further analyze the ability of Pax3 or Pax7 to induce expression of Id proteins, we evaluated expression of Id1, Id2, and Id3 by RT-PCR in either parental C2C12 cells or in polyclones of C2C12 cells programmed to express exogenous Pax3 or Pax7. Both the parental C2C12 cells and the Pax3/Pax7-expressing C2C12 polyclones expressed approximately equivalent levels of Id1, Id2, and Id3 transcripts when the cells were cultured under high serum conditions (i.e., 10% fetal bovine serum) (Figure 2A, lanes 1–3). In striking contrast, when the cells were cultured under low serum conditions for 3 d (i.e., 2% horse serum), parental C2C12 cells down-regulated expression of Id1, Id2, and Id3, whereas the expression of these genes was maintained in the C2C12 polyclones programmed to express either Pax3 or Pax7 (Figure 2A, lanes 4–6).
Consistent with our analysis of Id3 transcript levels, we found that Id3 protein is apparent in discrete nuclear inclusions in proliferating C2C12 cells (Figure 2B), but it is not observed in quiescent C2C12 cells cultured in low serum conditions (Figure 2E). These findings are consistent with prior work indicating that Id family members are specifically expressed in proliferating cells and are down-regulated during the process of skeletal muscle differentiation and cell cycle withdrawal (Benezra et al., 1990). In contrast to the parental C2C12 cells, where Id3 only accumulates in dividing cells, Id3 protein is apparent both throughout the nucleus and in nuclear inclusions in C2C12 polyclones expressing either exogenous Pax3 or Pax7 in high serum and at slightly attenuated levels in low serum conditions (Figure 2, I, M, Q, and U). These findings indicate that Pax3 or Pax7 can induce the expression of Id2 and Id3 specifically in cells that are cultured under low serum conditions. To evaluate whether Id3 protein accumulates in quiescent satellite cells, we immunostained freshly dissected myofibers with their associated satellite cells for both Pax7 and Id3. Id3 protein was specifically detected in Pax7-expressing quiescent satellite cells located on the periphery of the myofiber (Figure 1, B–E). These data are consistent with the high level of expression of Id3 transcripts that are detectable in CSM4B cells isolated by FACS (Figure 1A, lane 1).
Pax7/Pax3 Can Bind to the Id3 Promoter and Induce the Transcriptional Activity of an Id3 Luciferase Reporter
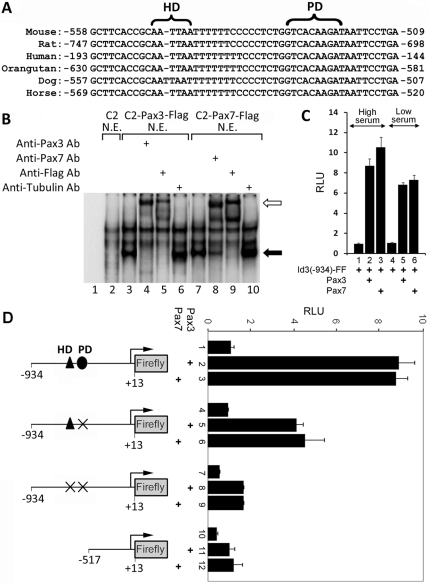
To examine whether Id3 is directly regulated by Pax3 or Pax7, we analyzed the proximal promoter of the mouse Id3 gene for putative PD and HD binding sites. Interestingly, we identified putative PD and HD binding sites upstream of the Id3 proximal promoter in several mammalian species, including mouse, rat, human, orangutan, dog, and horse (Figure 3A). To determine whether Pax3/7 bind to Id3 regulatory sequences, we monitored whether Pax3/7 present in nuclear extracts made from C2C12 cells programmed to express these proteins would interact in vitro with an oligonucleotide containing the putative Pax3/7 binding site located upstream of the Id3 promoter. Nuclear extracts derived from either parental C2C12 cells or such cells infected with a retrovirus encoding either Pax3-FLAG or Pax7-FLAG were incubated with a radiolabeled oligomer containing the putative Pax3/7 binding site in the Id3 promoter (Figure 3B). The Pax3/7-DNA protein complexes were visualized by EMSA. Nuclear extracts derived from C2C12 cells programmed to express either FLAG-tagged Pax3 or FLAG-tagged Pax7 gave rise to a new DNA–protein complex on the Id3 oligo (Figure 3B, lanes 3 and 7, black arrow designates new gel shift) that was not present in extracts derived from parental C2C12 cells (Figure 3B, lane 2). Importantly, these DNA–protein complexes were shifted to a lower electrophoretic mobility by inclusion of either anti-Pax3, anti-Pax7, or anti-FLAG antibodies (Figure 3B, lanes 4, 5, 8, and 9; hollow arrow designates antibody supershifted complex). In contrast, a control anti-tubulin antibody did not affect the mobility of the Pax3/7-DNA complexes (Figure 3B, lanes 6 and 10). These findings indicate that Pax3 and Pax7 can bind to a conserved sequence located just upstream of the Id3 promoter in vitro.
Figure 3.
A conserved Pax3/7 binding site is present upstream of the Id3 promoter. (A) Conserved HD and PD binding sites are present upstream of the Id3 promoter in several organisms. (B) EMSA using an oligo encoding the HD/PD binding sites upstream of the mouse Id3 gene with nuclear extracts made from parental C2C12 cells or polyclones expressing Pax3-FLAG or Pax7-FLAG. Anti-Pax3, anti-Pax7, anti-FLAG, or anti-tubulin were added to the EMSA as indicated. Pax3/7 gel shift is indicated by the black arrow; antibody supershift is indicated by the white arrow. (C) Serum factors affect Pax3 or Pax7 mediated induction of the Id3 reporter. C2C12 cells were cotransfected with pGL3-Id3(−934)-firefly luciferase and pSV40-Renilla-luciferase. In addition, the cells were cotransfected with either Pax3 or Pax7 expression vectors, as indicated, and grown in presence of either high or low serum. The relative luciferase units (RLU) of pGL3-Id3-firefly luciferase and pSV40-Renilla-luciferase are displayed. (D) Pax3 and Pax7 induction of an Id3 reporter requires both HD and PD binding sites in the Id3 promoter. C2C12 cells were cotransfected with either pGL3-Id3(−934)-firefly luciferase (containing the wild-type Id3 promoter) or mutated versions of this reporter lacking HD and PD binding sites as shown, plus pSV40-Renilla-luciferase. In addition, the cells were cotransfected with expression vehicles encoding either Pax3 or Pax7, as indicated. The RLUs of pGL3-Id3-firefly luciferase and pSV40-Renilla-luciferase is displayed.
To evaluate the importance of the putative Pax3/7 binding site located upstream of the Id3 promoter, we constructed a reporter vehicle containing 934 base pairs upstream of the Id3 transcriptional initiation site (−934 to +13) driving expression of firefly luciferase (Yeh and Lim, 2000). Forced expression of either Pax3 or Pax7 in C2C12 cells cotransfected with this Id3-luciferase reporter, resulted in an 8- to 10-fold increase in luciferase activity, indicating that both Pax3 and Pax7 can induce expression of a reporter driven by the Id3 promoter (Figure 3C). Because we noted that Id3 protein was decreased in C2C12 cells programmed to express either Pax3 or Pax7 in low serum conditions (Figure 2, 1, M, Q, and U), we evaluated whether serum factors would influence the ability of either Pax3 or Pax7 to induce expression of the Id3-luciferase reporter. Induction of the Id3 reporter by either Pax3 or Pax7 was greatest in high serum conditions (Figure 3C), suggesting that serum factors may indeed enhance the ability of these transcription factors to induce maximal Id3 expression. To investigate whether either the PD or HD binding sites were necessary for Pax3/7 to induce the expression of the Id3 reporter, we mutated these binding sites individually or in combination. Mutation of only the PD binding site resulted in about a 50% reduction in the expression of the Id3 reporter in response to either cotransfected Pax3 or Pax7, whereas mutation or deletion of both the PD and HD binding sites resulted in approximately an 80% reduction in the induction of this reporter by Pax3/Pax7 (Figure 3D).
Pax7 Binds to the Id3 Promoter in Quiescent Satellite Cells
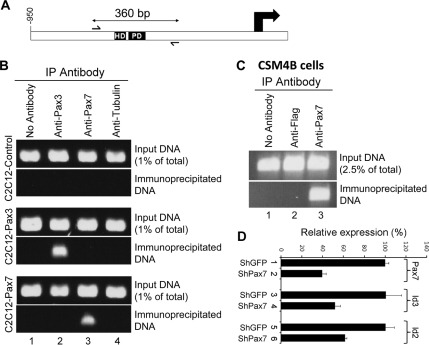
To evaluate whether Pax3/7 can bind to the Id3 promoter in vivo, we performed chromatin immunoprecipitation (IP) analysis with either parental C2C12 cells or C2C12 polyclones programmed to express either exogenous Pax3 or Pax7. Cells were cultured under low serum conditions, to stimulate Pax3/7-dependent induction of Id3 expression. After treatment of the cells with formaldehyde, chromatin was isolated, sheared by sonication, and regions of the genome bound by either Pax3 or Pax7 were immunoprecipitated. Both anti-Pax3 and anti-Pax7 antibodies precipitated chromatin encoding the Pax binding site located upstream of the Id3 transcription start site in C2C12 cells programmed to express either exogenous Pax3 or Pax7, respectively (Figure 4B). In contrast these antibodies did not precipitate the Id3 promoter in parental C2C12 cells (Figure 4B), which do not express detectable levels of Pax3 or Pax7 (Figure 2A).
Figure 4.
Pax7 is bound to the Id3 promoter in quiescent satellite cells. (A) Diagram of the Id3 promoter showing location of conserved HD and PD binding sites. (B) Chromatin IP with either parental C2C12 cells or C2C12 polyclones expressing either Pax3 or Pax7, indicate that exogenous Pax3 or Pax7 is bound to the Id3 promoter in cells cultured in low serum conditions. (C) Chromatin IP with freshly isolated CSM4B cells indicates that Pax7 is bound to the Id3 promoter in quiescent satellite cells. (D) Pax7 knockdown in CSM4B cells significantly decreases Id2 and Id3 expression. CSM4B cells were infected with lentivirus expressing either GFP shRNA or Pax7 shRNA. Pax7, Id2, and Id3 expression was analyzed by RT-qPCR.
To investigate whether endogenous Pax7 was similarly bound upstream of the Id3 promoter in quiescent satellite cells, we used FACS to isolate CSM4B cells fresh from the skeletal muscle of 2-mo-old mice. As stated, these cells have been documented to robustly differentiate into skeletal muscle both in vitro and in vivo, and to engraft into the satellite cell niche after intramuscular transplant (Cerletti et al., 2008). Coupled with the fact that >90% of sorted CSM4B cells express high levels of Pax7, whereas <5% express MyoD (Cerletti et al., 2008), it seems likely that this population of cells consists predominantly of quiescent satellite cells. Freshly isolated CSM4B cells, which express high levels of Id3 (Figure 1A), were purified and immediately cross-linked for chromatin IP analysis. Notably anti-Pax7 was able to immunoprecipitate chromatin encoding the Pax binding site located upstream of the Id3 promoter in these freshly isolated Pax7+ satellite cells (Figure 4C).
shRNA-mediated Knockdown of Pax7 Expression in Satellite Cells Significantly Reduces the Expression of Id2 and Id3
To examine whether Pax7 was necessary to maintain the expression of either Id2 or Id3 in satellite cells, we infected CSM4B cells, isolated from skeletal muscle tissue, with lentiviruses programmed to express shRNAs directed against either green fluorescent protein (shGFP; as a control) or Pax7 (shPax7). Four days after infection, RNA was isolated from the infected CSM4B cells, and RT-qPCR was used to assay gene expression. Expression of Pax7 was reduced to 40% of control levels in cells expressing Pax7 shRNA (Figure 4D). Notably, the expression of Id2 and Id3 was similarly reduced to 61 and 52% of control levels, respectively, in cells expressing Pax7 shRNA (Figure 4D). The reduction in expression of both Id2 and Id3 following knockdown of Pax7 expression suggests that Pax7 is necessary to maintain high level expression of both Id2 and Id3 in satellite cells.
DISCUSSION
Ectopic expression of either Pax3 or Pax7 in C2C12 cells led to the induction of Id1, Id2, and Id3, specifically when these cells were cultured under low serum conditions. Although these genes are both expressed in proliferating C2C12 cells grown in 10% fetal bovine serum, their expression is significantly attenuated when these cells are cultured in low serum conditions (2% horse serum) for 3 d. In striking contrast, in C2C12 cells programmed to express either exogenous Pax3 or Pax7, expression of these genes is maintained in low serum-containing medium. Although the Id genes have previously been linked to cellular proliferation in at least some cellular contexts (Benezra et al., 1990), in satellite cells Id2 and Id3 are expressed in both quiescent and proliferating cells, indicating that their expression can be uncoupled from the cell cycle in this cell type. shRNA-mediated knockdown of Pax7 expression in purified satellite cells leads to a significant loss in the expression of both Id2 and Id3 in activated satellite cells in vitro, suggesting that Pax7 is necessary to maintain high level expression of these Id family members in satellite cells. Because there are conserved paired domain and homeodomain binding sites located upstream of the mouse, rat, human, orangutan, dog, and horse Id3 genes, which are necessary for efficient induction of an Id3-luciferase reporter by either Pax3 or Pax7, we propose that Id3 is a direct transcriptional target of Pax3/7. Consistent with this notion, we have documented that Pax3 and Pax7 bind to chromatin containing this region of the Id3 gene in C2C12 cells programmed to express either Pax3 or Pax7 and that endogenous Pax7 is similarly bound to this region of the Id3 gene in quiescent satellite cells. Together, our findings suggest that Id2 and Id3 are transcriptional targets of Pax7 in satellite cells.
Previous work has indicated that >90% of CSM4B cells isolated from skeletal muscle express Pax7 and that such cells exhibit the functional characteristics of quiescent skeletal muscle stem cells or satellite cells (Cerletti et al., 2008). When freshly isolated, this cell population displays robust expression of Pax7, Myf5, and both Id2 and Id3, yet it does not express appreciable levels of Id1. Culturing these cells in vitro leads to both their entry into the cell cycle (Cerletti et al., 2008), induction of MyoD expression, and subsequent loss of Pax7 expression (Figure 1A). The expression of Id2 and Id3 eventually declines in these cultures as well, in parallel with loss of Pax7 expression and induction of skeletal muscle differentiation. Previous work has indicated that expression of Id1 is high in proliferating C2C12 cells and significantly decreases when these cells are induced to differentiate in low serum-containing medium (Benezra et al., 1990). The induction of Id genes by Pax3/Pax7 in C2C12 cells cultured in low serum, or expression of Id2 and Id3 in quiescent satellite cells, suggests that the expression of Id factors can be divorced from cellular proliferation. Interestingly, however, maximal induction of the Id3-luciferase reporter by cotransfected Pax3/7 required the presence of high serum.
Chromatin IP analysis demonstrated that Pax7 binds to a conserved binding site adjacent to the promoter of the Id3 gene in freshly isolated, quiescent satellite cells, and mutagenesis of the Id3 promoter has indicated that this Pax7 binding site is necessary for Pax7-mediated induction of an Id3-promoter-luciferase construct in transfected C2C12 cells. Based upon these findings, we propose that Pax7 may act to maintain the expression of Id3 (and potentially Id2) specifically in quiescent satellite cells (outlined in Figure 5). Once satellite cells are induced to enter the cell cycle and express MyoD, expression of Id2/3 may be maintained either by signaling pathways downstream of serum factors and/or by Pax7 that is also expressed in proliferating satellite cells. Indeed knockdown of Pax7 attenuated the expression of both Id2 and Id3 in cultures of proliferating satellite cells. Consistent with the notion that Pax7 may be necessary to maintain the expression of Id2/3 in growth arrested cells, expression of both Pax7 and Id2/3 is lost in differentiated myotubes.
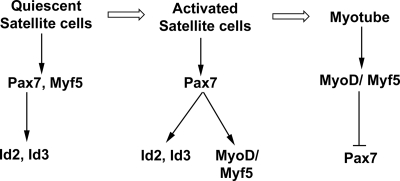
Figure 5.
Pax7 activates the expression of Id2 and Id3 in quiescent satellite cells. Based on our findings we speculate that Pax7 induces the expression of Id2/3 in quiescent satellite cells, where these dominant-negative HLH factors may act to block premature differentiation of these cells. In activated satellite cells, Pax7 may induce the expression of both Id2/3 and MyoD/Myf5 before differentiation. In differentiated myotubes, MyoD/Myf5 represses expression of Pax7 and as a consequence Id2/3 expression is lost.
The Id family of HLH proteins have been demonstrated to block the function of bHLH proteins such as MyoD and Myf5, by titrating their E protein binding partners (Benezra et al., 1990); and to also block negative autoregulation of the Notch effector Hes1 (Bai et al., 2007) and thereby maintain high level expression of Hes1 and potentially other members of the hairy/enhancer of split family. In this light, it is interesting that quiescent satellite cells express relatively robust levels of both Id2 and Id3 (this study) and several members of the hairy/enhancer of split family, including Heyl (Fukada et al., 2007), Hes1 and Hey1 (Wagers, unpublished observations). It is possible that high-level expression of Id2 and Id3 in quiescent satellite cells acts to either block the activity of Myf5 that is readily expressed (at least at the RNA level) in at least 90% of quiescent satellite cells (Kuang et al., 2007) or to maintain the expression of the Notch-induced transcriptional repressors Hes1, Hey1, and/or Heyl, which in turn may block expression of MyoD and Myf5. Thus, by inducing the expression of Id2 and Id3, Pax7 may act to block the premature differentiation of quiescent satellite cells. In addition, Pax7 may also block skeletal muscle differentiation by targeting myogenic bHLH proteins for degradation (Olguin et al., 2007). In contrast, in activated satellite cells, Pax7 may act to directly induce the expression of Myf5 (Bajard et al., 2006) and MyoD (Hu et al., 2008), which in turn will compete with Id proteins for E protein dimerization. In this scenario, Pax7 serves two roles in satellite cells: blocking their differentiation via Id2/3 induction during quiescence and promoting their differentiation via Myf5/MyoD induction during activation (Figure 5). Interestingly, Id family members have been implicated in both promoting self-renewal and blocking differentiation of hematopoietic (Jankovic et al., 2007) and neural (Robert Benezra, personal communication) stem cells, and our work suggests that they may play a similar role in satellite cells as well. Although mice deleted for Id3 are viable and show no discernible muscle phenotype (Pan et al., 1999), mice lacking both Id2 and Id3 are embryonic lethal (Robert Benezra, personal communication). Because quiescent satellite cells express both Id2 and Id3 (Figure 1), it is possible that the lack of a skeletal muscle phenotype in mice lacking only Id3 (Pan et al., 1999) is due to functional redundancy with Id2. Therefore, to determine whether Id2 and Id3 are indeed necessary to block differentiation of quiescent satellite cells, it will be necessary to conditionally delete both these genes in this cell type.
In addition to Id2 and Id3, we noted that several other genes induced by Pax7 in C2C12 cells were also specifically expressed in quiescent satellite cells. These genes include ITM2A, the chemokine receptor CXCR4, BMP4, the sugar binding protein chondrolectin, C1R, PTGIS, and MGP. Interestingly, the expression of many of these Pax7 inducible genes was greatest in quiescent satellite cells and expression of these genes was extinguished during either proliferation or differentiation of these cells. CXCR4 has already been demonstrated to play a crucial role in migration of skeletal muscle cells during embryogenesis (Vasyutina et al., 2005). It will be interesting to determine the role of both Id2/Id3 and these other putative Pax7 transcriptional targets in quiescent satellite cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Lim for generously supplying the Id3 promoter; Michael Rudnicki for the Pax7 cDNA; Hyung-song Nam, Yvette Chin, and Robert Benezra for characterizing and supplying the anti-Id3 rabbit monoclonal antibody (now available from CalBioreagents, San Mateo, CA); and Robert Benezra for thoughtful comments on our work. This work was funded by National Institutes of Health grant GM-054879 (to A.B.L.) and grants from the Harvard Stem Cell Institute, Jain Foundation, and Beckman Foundation (to A.J.W.). D. K. was funded by a fellowship from the Muscular Dystrophy Association.
Abbreviations used:
- C1R
complement component 1 R subunit
- ChIP
chromatin immunoprecipitation
- CSM4B
CD45− Sca-1− Mac-1− CXCR4+ β-1 integrin+
- EMSA
electrophoretic mobility shift assay
- HD
homeo domain
- Id
inhibitor of DNA binding
- ITM2A
integral membrane protein 2A
- MGP
matrix gla protein
- PD
paired domain
- PTGIS
prostaglandin I2 (prostacyclin) synthase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1185) on May 20, 2009.
REFERENCES
- Attema J. L., Papathanasiou P., Forsberg E. C., Xu J., Smale S. T., Weissman I. L. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl. Acad. Sci. USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F., Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Bajard L., Relaix F., Lagha M., Rocancourt D., Daubas P., Buckingham M. E. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Cerletti M., Jurga S., Witczak C. A., Hirshman M. F., Shadrach J. L., Goodyear L. J., Wagers A. J. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G., Goulding M., Read A., Strachan T., Gruss P. Molecular basis of splotch and Waardenburg Pax-3 mutations. Proc. Natl. Acad. Sci. USA. 1994;91:3685–3689. doi: 10.1073/pnas.91.9.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G., Gruss P. Identification of DNA recognition sequences for the Pax3 paired domain. Gene. 1995;162:267–270. doi: 10.1016/0378-1119(95)00345-7. [DOI] [PubMed] [Google Scholar]
- Conboy I. M., Rando T. A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Fukada S., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gros J., Manceau M., Thome V., Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Hu P., Geles K. G., Paik J. H., DePinho R. A., Tjian R. Codependent activators direct myoblast-specific MyoD transcription. Dev. Cell. 2008;15:534–546. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic V., Ciarrocchi A., Boccuni P., DeBlasio T., Benezra R., Nimer S. D. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L., Giacone E., Gayraud-Morel B., Jory A., Gomes D., Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M. A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M., Kormish J. D., Rocancourt D., Manceau M., Epstein J. A., Zaret K. S., Relaix F., Buckingham M. E. Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev. 2008;22:1828–1837. doi: 10.1101/gad.477908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M., Reshef R., Munsterberg A. E., Koester S., Goulding M., Lassar A. B. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- McKinnell I. W., Ishibashi J., Le Grand F., Punch V. G., Addicks G. C., Greenblatt J. F., Dilworth F. J., Rudnicki M. A. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Munsterberg A. E., Kitajewski J., Bumcrot D. A., McMahon A. P., Lassar A. B. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9:2911–2922. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Olguin H. C., Yang Z., Tapscott S. J., Olwin B. B. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J. Cell Biol. 2007;177:769–779. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Sato S., Frederick J. P., Sun X. H., Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol. Cell Biol. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perk J., Iavarone A., Benezra R. Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Relaix F., Polimeni M., Rocancourt D., Ponzetto C., Schafer B. W., Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004;18:1088–1105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F., Rocancourt D., Mansouri A., Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Ruzinova M. B., Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Schienda J., Engleka K. A., Jun S., Hansen M. S., Epstein J. A., Tabin C. J., Kunkel L. M., Kardon G. Somitic origin of limb muscle satellite and side population cells. Proc. Natl. Acad. Sci. USA. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Ishibashi J., Scime A., Rudnicki M. A. Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS Biol. 2004;2:E130. doi: 10.1371/journal.pbio.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. K., Maxwell L., Rodgers C. D., McKee N. H., Plyley M. J. Exercise-enhanced satellite cell proliferation and new myonuclear accretion in rat skeletal muscle. J. Appl. Physiol. 2001;90:1407–1414. doi: 10.1152/jappl.2001.90.4.1407. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Vasyutina E., Stebler J., Brand-Saberi B., Schulz S., Raz E., Birchmeier C. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 2005;19:2187–2198. doi: 10.1101/gad.346205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Choi S., Liu X., Zhang M., Schageman J. J., Lee S. Y., Hart R., Lin L., Thurmond F. A., Williams R. S. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 2003;278:8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- Yeh K., Lim R. W. Genomic organization and promoter analysis of the murine Id3 gene. Gene. 2000;254:163–171. doi: 10.1016/s0378-1119(00)00274-2. [DOI] [PubMed] [Google Scholar]
- Zammit P. S., Partridge T. A., Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J. Histochem. Cytochem. 2006;54:1177–1191. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.