Abstract
In the budding yeast Saccharomyces cerevisiae, the G1-specific cyclin-dependent kinases (Cdks) Cln1,2-Cdc28 and Pcl1,2-Pho85 are essential for ensuring that DNA replication and cell division are properly linked to cell polarity and bud morphogenesis. However, the redundancy of Cdks and cyclins means that identification of relevant Cdk substrates remains a significant challenge. We used array-based genetic screens (synthetic genetic array or SGA analysis) to dissect redundant pathways associated with G1 cyclins and identified Bni4 as a substrate of the Pcl1- and Pcl2-Pho85 kinases. BNI4 encodes an adaptor protein that targets several proteins to the bud neck. Deletion of BNI4 results in severe growth defects in the absence of the Cdc28 cyclins Cln1 and Cln2, and overexpression of BNI4 is toxic in yeast cells lacking the Pho85 cyclins Pcl1 and Pcl2. Phosphorylation of Bni4 by Pcl-Pho85 is necessary for its localization to the bud neck, and the bud neck structure can be disrupted by overexpressing BNI4 in pcl1Δpcl2Δ mutant cells. Our data suggest that misregulated Bni4 may bind in an uncontrolled manner to an essential component that resides at the bud neck, causing catastrophic morphogenesis defects.
INTRODUCTION
Cell cycle commitment in budding yeast is defined by a regulatory transition called START, after which cells initiate bud formation, spindle pole body duplication, and DNA replication (Tyers, 2000). Passage through START requires a buildup of cyclin-dependent-kinase (Cdk) activity, through activation of two sets of genes: CLN1 and CLN2, which encode cyclins that activate the Cdk Cdc28, and PCL1 and PCL2, which encode cyclins that activate the Cdk Pho85. The G1 cyclins are genetically redundant, because strains lacking any three of these cyclins still undergo cell cycle progression. However, a strain lacking all four G1 cyclins experiences a catastrophic morphogenesis (Moffat and Andrews, 2004), suggesting that Cln1 and Cln2, in association with Cdc28, work in parallel with Pcl1 and Pcl2, in association with Pho85, to regulate some essential morphogenesis event or events at the time of the G1/S transition.
As in other cells, polarized growth in budding yeast is a dynamic process linked to the cell division cycle. Early in the cell cycle, growth is polarized toward the bud tip and incipient bud site in an apical growth phase that permits bud emergence. Later, the growth orientation changes to isotropic, after which time the whole bud grows evenly in all directions. This allows the bud to enlarge and grow into an ellipsoidal cell. The switch from apical to isotropic growth correlates with changes in the structure of the septin ring (Faty et al., 2002). Septins are a family of conserved filament-forming proteins that organize the cortex at the mother-bud neck (Longtine et al., 1996; Gladfelter et al., 2001). The septins Cdc3, Cdc10, Cdc11, Cdc12, and Shs1 form a ring at the mother-bud neck, which acts as a barrier to prevent diffusion of integral and peripheral membrane proteins between the mother and the bud (Barral et al., 2000; Takizawa et al., 2000). The septin ring undergoes dynamic structural changes over the course of the cell cycle (Longtine and Bi, 2003; Gladfelter et al., 2005). The septin complex is essential for cell division; phenotypes of septin organization mutants include variations in septin localization, elongated buds, and wide necks (DeMarini et al., 1997; Cid et al., 2001; Gladfelter et al., 2005).
BNI4, along with other genes including CLN1, CLN2, BNI1, BUD3, BUD4, and BUD5, contributes to septin assembly and regulation (Gladfelter et al., 2005). Bni4 was originally identified as a scaffold protein that tethers the regulatory subunit of chitin synthase III, Chs4, to the bud neck through its interaction with septins (DeMarini et al., 1997; Kozubowski et al., 2003). Bni4 also targets the catalytic subunit of type 1 protein phosphatase (PP1, Glc7 in yeast), in which it is required to direct chitin synthase to the neck (Larson et al., 2008). Moreover, a genetic analysis identified several polarity genes that interact uniquely with BNI4, suggesting that Bni4 has other roles in bud neck organization beside its function in chitin regulation (Lesage et al., 2005). Bni4 is a phosphoprotein (Walsh et al., 2002; Kozubowski et al., 2003), and a mutant version of Bni4 that cannot bind to Glc7 exhibits hyperphosphorylation (Kozubowski et al., 2003), but the role of phosphorylation in Bni4 regulation and the relevant kinase remain unknown.
We took a genetic approach to identify effectors of G1 cyclins in their role of regulating cell polarity. One type of genetic interaction we chose to examine was synthetic lethality (SL). Two genes show an SL interaction if the combination of two mutations, neither by itself lethal, causes cell death (Boone et al., 2007). Synthetic lethal interactions may occur for genes within two distinct pathways if one process functionally compensates for or buffers the defects in the other (Boone et al., 2007). We reasoned that it should be possible to circumvent the genetic redundancy of the G1 cyclins by screening combinations of cyclin mutants for SL interactions. Information about genetic pathways also may also be revealed by systematic assessment of the effects of gene overexpression, which can perturb associated regulatory pathways. In particular, a synthetic dosage lethal (SDL) interaction occurs when overexpression of a gene is of little phenotypic consequence in a wild-type cell but causes a significant fitness defect when another gene is deleted (Kroll et al., 1996; Sopko et al., 2006). We have shown that systematic SDL screens are an effective way to identify downstream targets of protein kinases (Sopko et al., 2006; Sopko and Andrews, 2008). We reasoned that combining SL and SDL screens might be a particularly powerful genetic approach to identify substrates of redundant kinases. For example, because the Cln1, Cln2 pathway acts in parallel to the Pcl1, Pcl2 pathway, deletion of a gene encoding a specific target of the Pcl1, Pcl2 pathway could cause synthetic lethality in the absence of CLN1 and CLN2 and overexpression of the target gene could cause an SDL interaction when PCL1 and PCL2 are deleted (Figure 1C).
Figure 1.
Genetic interactions with G1 cyclin genes. (A) List of genes that have genetic interactions with G1 cyclin pairs identified from crosses with the deletion strain collection and with a collection of temperature-sensitive strains for synthetic lethal (SL/SS) interactions and with the pGAL-GST-open reading frame (ORF) overexpression collection for synthetic dosage lethal (SDL) interactions. Query genes are listed by row and genetic interactions by column. Synthetic sick interactions are shown in yellow. (B) Pie chart showing the genes that have genetic interactions (either SL/SS or SDL) with G1 cyclin pairs clustered by GO cellular role. Categories that contribute <8% of all genes are grouped as others. (C) Diagram showing one possible scenario for genetic interactions with G1 cyclin pairs. Genes that have SL/SS interaction with CLN1, CLN2 could be in a PCL1, PCL2 regulatory pathway, because quadruple deletion of CLN1, CLN2, PCL1, and PCL2 is lethal, as shown on the left. Genes downstream of PCL1, PCL2 will escape regulation when PCL1 and PCl2 are deleted; therefore, their overexpression may cause toxicity.
Using this approach, we identified BNI4 as a previously unappreciated target of the G1-specific Pho85 Cdks. We provide biochemical and genetic evidence that Bni4 is a substrate of Pcl1, Pcl2-Pho85 kinases and that phosphorylation of Bni4 regulates its subcellular localization. When an unphosphorylatable version of Bni4 is overexpressed, the septin ring structure is disrupted. We suggest that phosphorylation of Bni4 connects G1-specific cell cycle control to regulation of cell polarity at the bud neck.
MATERIALS AND METHODS
Yeast Strains, Plasmids, Growth Media, and Genetic Methods
Yeast strains were derived from BY4741 (Table 1; Brachmann et al., 1998). All yeast gene disruptions were achieved by homologous recombination using standard polymerase chain reaction (PCR)-based methods, and verified by PCR and phenotypic analyses (Longtine et al., 1998). Plasmids are listed in Table 2. Point mutations were made using PCR-based site-directed mutagenesis (Sopko et al., 2006) and confirmed by sequencing. Standard media, genetic and molecular biological methods were used except where noted.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4033 | Y2454 MATα cln1ΔNAT cln2ΔHPH | This study |
| BY1491 | Y2454 MATα pcl1ΔNAT pcl2ΔHPH | This study |
| BY4741 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 | Brachmann et al. (1998) |
| Y2454 | MATα ura3Δ0 leu2Δ0 can1ΔMFA1pr-HIS3 his3Δ1 lys2Δ0 | Tong et al. (2001) |
| BY4214 | MATaBNI4-TAP::HIS3 pGAL-CDC20::KAN ura3Δ | This study |
| BY4241 | MAhaBNI4-TAP::HIS3 pcl1ΔNAT pcl2ΔHPH GAL-CDC20 | This study |
| BY4226 | MATa BNI4-TAP::HIS3 | This study |
| BY4228 | MATaBNI4-GFP::HIS3 pcl1ΔNAT pcl2ΔHPH | This study |
| BY3059 | Y2454 MATα pho85ΔNAT | Sopko et al. (2006) |
| BY2131 | PCL1-13myc in BY263 background | Moffat et al. (2000) |
| BY4427 | MATaBNI4-GFP::HIS3 | Huh et al. (2003) |
| BY4470 | MATabni4-10A-GFP::HIS3 | This study |
| JRL265 | MATα leu2 his3 ura3 BNI4-mCitrine::SpHIS5 | This study |
| JRL646 | MATaleu2 his3 ura3 cln2Δ::KanMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| JRL648 | MATaleu2 his3 ura3 pcl2Δ::KanMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| JRL651 | MATα leu2 his3 ura3 pcl1Δ::NatMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| JRL652 | MATa leu2 his3 ura3 pho85Δ::NatMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| JRL685 | MATα leu2 his3 ura3 pcl1Δ::NatMX4 pcl2Δ::KanMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| JRL686 | MATaleu2 his3 ura3 cln1Δ::NatMX4 cln2Δ::KanMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| JRL689 | MATα leu2 his3 ura3 cln1Δ::NatMX4 BNI4-yEmCitrine::SpHIS5 | This study |
| BY4312 | Y2454 CDC11-mCherry | This study |
| BY4313 | Y2454 pcl1ΔNAT pcl2ΔHPH CDC11-mCherry | This study |
Table 2.
Plasmids used in this study
| Plasmid | Description | Reference or source | |
|---|---|---|---|
| pEGH | pGAL-GST-6xHis URA3 2-μm (vector) | Zhu et al. (2000) | |
| pGST-BNI4 | pEGH + BNI4 | Zhu et al. (2000) | |
| BA2046 | pDEST24-BNI4-GST | C-terminal GST-tagged BNI4 under T7 promoter for expression in E. coli | This study |
| BA2055 | pMET-BNI4 | BNI4 under control of MET promoter, URA3 CEN | This study |
| BA2220 | pGAL-BNI4-GFP | BNI4-GFP under GAL promoter, URA3 2-μm | This study |
Screening for SL and SDL Interactions
SGA analysis (Tong et al., 2001) was used to identify genes required for the optimal growth of strains deleted for either CLN1 and CLN2 or PCL1 and PCL2. To select triple deletion mutants, multiple selection steps were used in SGA analysis. Followed by standard steps of mating, diploid selection, sporulation, and MATa-type haploid selection, cells were sequentially selected on haploid selection media plus G418 once, and haploid selection media plus both G418, nourseothricin, and hygromycin twice. From triplicate SGA screens for each query strain, potential interactions were identified and confirmed first by random spore analysis and then by tetrad dissection as described previously (Tong et al., 2004).
A modified SGA method was used to introduce an array of plasmids, each overexpressing a different gene, into strains deleted for either CLN1, CLN2 or PCL1, PCL2 as described previously (Sopko et al., 2006). From triplicate screens, potential interactions were confirmed by serial dilution spot assays, comparing the growth of cells with overexpression of a given gene induced on SGal-ura plates (synthetic medium containing galactose and lacking uracil to select for plasmids) to that of cells not overexpressing the gene on SD-ura plates (synthetic medium containing glucose and lacking uracil). In addition, growth of strains overexpressing a given gene was compared with growth of the strain containing vector on SGal-ura.
Microscopy
Cells were imaged using two systems: 1) a CoolSNAP HQ high-speed digital camera (Roper Scientific, Trenton, NJ) mounted on a DM-LB microscope (Leica Microsystems, Deerfield, IL) and 2) a DMI 6000B fluorescence microscope (Leica Microsystems) equipped with a spinning-disk head and an argon laser (458, 488, and 514 nm; Quorum Technologies, Guelph, ON, Canada). Images from the confocal microscope were captured using an ImagEM-charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) and analyzed using Volocity software (Improvision, Coventry, United Kingdom). For Figure 4, cells were placed onto a pad of 2% agarose in synthetic medium containing 2% glucose and imaged for yellow fluorescent protein as described previously (Kozubowski et al., 2003). Fluorescence images in different z-axis planes (0.5 μm apart) were acquired using SlideBook software (Olympus, Center Valley, PA). Fluorescence levels were quantified as described previously (Kozubowski et al., 2003) by using the average of four adjacent pixels and subtracting background fluorescence from the cytoplasm.
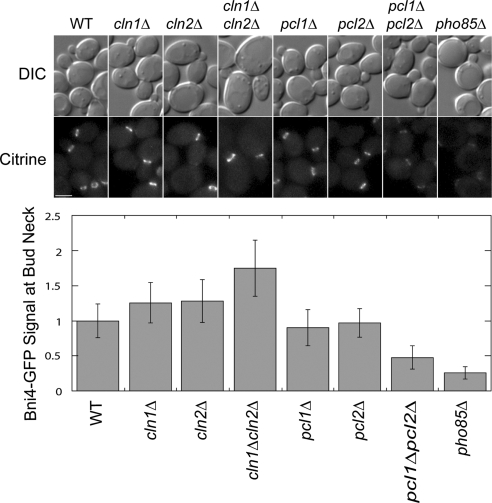
Figure 4.
Bni4 localization to the bud neck in Cdk mutant strains. DIC and fluorescence microscopy analysis of Bni4-mCitrine levels at the bud neck in strains of WT, cln1Δ, cln2Δ, cln1Δcln2Δ, pcl1Δ, pcl2Δ, pcl1Δpcl2Δ, and pho85Δ. Cultures were grown to mid-log phase and imaged by fluorescence microscopy. Fluorescence levels at the bud neck were quantified and normalized to wild-type levels. n ≥ 55. Error bars represent SD. Bar, 3 μm.
High-Throughput Microscopy
Ninety-four strains containing green fluorescent protein (GFP)-tagged proteins that localize to the bud neck or bud tip (BNT-GFP strains) were identified from the Yeast GFP Fusion Localization Database (http://yeastgfp.ucsf.edu/; Huh et al., 2003) and arrayed on a plate. Using the SGA method, the GFP-tagged bud neck genes were introduced into a pho85ΔNAT strain containing pGAL-GST-BNI4. As controls, the 94 GFP-tagged bud neck genes were introduced into a wild-type (WT) strain containing pGAL-GST-BNI4 and a pho85Δ strain containing vector. The final strains were inoculated into YPR (rich medium containing raffinose) in a 96-well microtiter dish, and grown at 30°C to an OD600 of ∼0.6. Galactose was added to 2% to induce the overexpression of BNI4 and cells were grown for 4 h at 30°C. Differential interference contrast (DIC) and GFP images were captured using a high-throughput microplate imaging system (ImageXpress 5000A; Axon Instruments, Foster City, CA).
Cell Cycle Western Analysis
WT and pcl1Δpcl2Δ strains expressing BNI4-TAP or bni4-10A-TAP were synchronized using a GAL-CDC20 allele as described previously (Shirayama et al., 1999). Cultures were first grown in YP + raffinose to log phase, and then glucose was added to 2% and cells incubated at 30°C for 2 h to arrest cells in metaphase due to the depletion of Cdc20. Metaphase arrest of cultures was confirmed by microscopic examination. Cells were washed and the medium was replaced by prewarmed YPGal to induce expression of CDC20 and allow resumption of the cell cycle. Samples were collected every 10 min for Western blot, fluorescence-activated cell sorting (FACS) analysis, and assessment of budding index.
For Western blot analysis, 5 ml of culture was pelleted and resuspended in 375 μl of prechilled pronase buffer (1.4 M sorbitol, 25 mM Tris-HCl, pH 7.5, 20 mM NaN3, and 2 mM MgCl2), and cold trichloroacetic acid (TCA) was added to 10%. Samples were then stored at −80°C. To lyse the cells, an equal volume of glass beads was added, and samples were vortexed for 8 × 1 min at 4°C, with 1-min rests. To separate the lysate from the glass beads, a hole was made in the bottom of the tube with a hot needle, and the lysate was centrifuged at 1000 rpm for 2 min. The beads were then washed twice with 5% TCA. After precipitating the proteins on ice for 10 min, the lysate was centrifuged at 4°C for 10 min at 12,000 rpm. Pellets were resuspended in Thorner buffer (8 M urea, 5% SDS, 40 mM Tris-HCl, pH 6.8, 0.1 mM EDTA, 0.4 mg/ml bromphenol blue, and 1% β-mercaptoethanol). Samples were heated at 90°C for 5 min before loading on 6% polyacrylamide gels. After electrophoresis, protein was transferred to polyvinylidene difluoride (PVDF) membranes following standard procedures. Peroxidase anti-peroxidase complex (PAP) (P3039; Sigma-Aldrich, St. Louis, MO) was used at 1:2000 dilution to detect TAP-tagged proteins.
For FACS analysis, 2 ml of culture was collected, washed with phosphate-buffered saline/0.1% Tween (PBST), and resuspended in 2 ml of PBST and 5 ml of ethanol. Samples were prepared for FACS analysis with Sytox Green (Invitrogen) and sonicated. Analysis was done using flow cytometry with FACSCalibur (BD Biosciences, San Jose, CA) and CellQuest ProSoftware (Verity Software House, Topsham, ME). For budding index, 0.8-ml samples were added to 88 μl of formaldehyde to fix the cells, and then >100 cells were counted to determine the percentage of budded cells.
Kinase Assay
Recombinant Pcl2-Pho85, Pho85, and Cln2-Cdc28-Cks1 kinases were expressed and purified from insect cells and Pcl1 from Escherichia coli as described (Huang et al., 1999). Bni4 was expressed as a C-terminal glutathione transferase (GST)-tagged fusion protein in Escherichia coli and purified using glutathione-Sepharose. Kinase assays were performed as described previously (Friesen et al., 2003) and included 10 nM Bni4 and increasing concentration of kinase. Kinase activity was assessed using previously defined substrates: the GPA-SH3 domain of Rvs167 for Pcl1-Pho85 and Pcl2-Pho85 (Friesen et al., 2003) and histone H1 for Cln2-Cdc28 (Wittenberg et al., 1990).
Immunoblotting and Pulldown Assay
Western blots were probed with monoclonal α-myc 9E10 (sc-40; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal α-GST (sc-138; Santa Cruz Biotechnology), α-GFP (BD Living Color JL-8; BD Biosciences) and polyclonal α-Rvs167 antibodies (Lee et al., 1998). For the pulldown assay, cells were disrupted in lysis buffer (100 mM Tris-HCl pH 7.0, 200 mM NaCl, 1 mM EDTA, 5% glycerol, and 0.5% Triton X-100) and clarified by centrifugation at 13,000 rpm for 10 min. Crude extract was incubated with glutathione-Sepharose 4B beads (GE Healthcare, Chalfont St. Giles, United Kingdom) to purify GST-tagged and -associated proteins. Samples were separated on SDS 8 and 12% polyacrylamide gel and transferred to a PVDF membrane for detection.
RESULTS
Screens for Genes That Have Synthetic Lethal or Synthetic Dosage Lethal Interactions with G1 Cyclin Pairs
Because CLN1 and CLN2 function in parallel to PCL1 and PCL2, a genetic screen to identify genes that have synthetic lethal and synthetic sick (SL/SS) interactions with one set of G1 cyclins is likely to identify components in the parallel regulatory pathway regulated by the other set of G1 cyclins. More specifically, among the genes that have SL interactions with CLN1, CLN2, we expected to identify either upstream regulators or downstream targets of Pcl1 and Pcl2, and vice versa. We used the SGA method to conduct screens for SL/SS by using both a cln1Δcln2Δ strain and a pcl1Δpcl2Δ strain as queries. We identified 36 genes that caused an SL/SS phenotype in the absence of CLN1 and CLN2 (Figure 1A), including 11 previously known and 25 novel interactions. Screens for genes SL with PCL1, PCL2 identified no new interactions, but we confirmed three known interactions (Figure 1A). To home in on potential downstream targets of the cyclin pairs, we performed parallel screens for genes that have an SDL interaction with CLN1, CLN2 or PCL1, PCL2. The SDL screens identified 45 genes that caused a specific SDL phenotype in a cln1Δcln2Δ strain and three genes that caused SDL in a pcl1Δpcl2Δ strain (Figure 1A).
The roster of genes that have SL/SS and SDL interactions with G1 cyclins include genes involved in regulation of the actin cytoskeleton, chitin metabolism, vesicle trafficking, cell integrity, general transcription, and cell cycle. When we grouped these genes into categories defined by Gene Ontology (GO) cellular role (Dwight et al., 2002), almost one third of the identified genes involved regulation of various cell polarity events (Figure 1B), confirming a significant and redundant role for G1 Cdks in controlling cell polarity (Moffat et al., 2004). We reasoned that bona fide in vivo targets of G1 Cdks may show reciprocal patterns of SL and SDL interactions (Figure 1C). We identified BNI4 as a gene that showed an SL interaction with CLN1, CLN2 and caused SDL in the absence of PCL1 and PCL2, potentially placing Bni4 in a Pcl1, Pcl2-associated regulatory pathway, an idea we chose to explore further. In addition BNI4 had an SDL interaction with CLN1, CLN2 (Figure 2A), suggesting that its regulation may be complex (see Discussion).
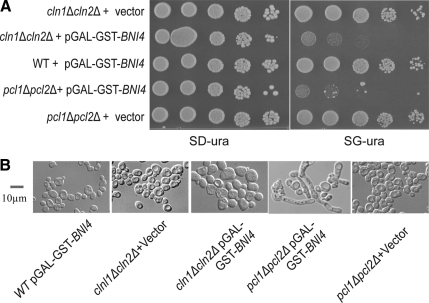
Figure 2.
Synthetic dosage phenotype caused by overexpression of BNI4 in the absence of G1 Cdks. (A) Cultures of WT cells carrying pGAL-GST-BNI4, cln1Δcln2Δ carrying vector (pEGH), cln1Δcln2Δ carrying pGAL-GST-BNI4, pcl1Δpcl2Δ carrying vector (pEGH), and pcl1Δpcl2Δ carrying pGAL-GST-BNI4 were serially diluted and spotted on both SD-ura and SG-ura plates. Plates were incubated 2 d for both SD-ura and SGal-ura plates at 30°C before being photographed. (B) Cultures were grown to log phase in SD-ura, and then transferred to SR-ura and incubated for another 5 h. Galactose was then added to 2%, and cultures were incubated at 30°C for an additional 4.5 h and examined by microscope. Bar, 10 μm.
Phenotypes Caused by Overexpression of BNI4 in G1 Cdk Mutant Backgrounds
To confirm the SDL phenotype associated with overexpression of BNI4 in the absence of G1 Cdks, cultures of WT, cln1Δcln2Δ, or pcl1Δpcl2Δ cells carrying pGAL-GST-BNI4, or vector control were tested by spot dilution on control medium (glucose) and plates containing galactose to induce BNI4 expression. BNI4 overexpression in either cln1Δcln2Δ or pcl1Δpcl2Δ led to a significant growth defect, whereas a wild-type strain overexpressing GST-BNI4, and a cln1Δcln2Δ and a pcl1Δpcl2Δ strain carrying vector grew normally (Figure 2A). The growth defect caused by overexpression of BNI4 in the pcl1Δpcl2Δ mutant strain was associated with a dramatic morphological defect, with most cells having extremely elongated buds (Figure 2B). In contrast, overexpression of BNI4 in cln1Δcln2Δ strain did not cause elongated buds (Figure 2B). These results implicate Pcl1, Pcl2-associated Pho85 kinase activity in the regulation of Bni4 function, perhaps during the switch from apical to isotropic growth.
PCL1 and PCL2 Are Required for Phosphorylation of Bni4 In Vivo
To further explore the relationship between Pcl1, Pcl2-Pho85 and Bni4 in vivo, we used Western blot analysis to ask whether phosphorylation of Bni4 depends on Pcl1, Pcl2-associated Cdk activity. If phosphorylation of Bni4 depends on Pcl1, Pcl2-Pho85, we would expect an increase in Bni4 phosphorylation levels in a WT strain during the G1/S transition when Pcl1 and Pcl2 activities are at their peak (Measday et al., 1994, 1997), but not in a pcl1Δpcl2Δ strain. To test this, we assayed Bni4 phosphorylation over the course of a cell cycle. We synchronized pcl1Δpcl2Δ or WT strains carrying a tandem affinity purification (TAP)-tagged allele of BNI4 in metaphase by manipulating CDC20 expression (Shirayama et al., 1999). Samples were taken every 10 min after release, and protein was analyzed by Western blot (Figure 3A, top). Analysis of budding index and by FACS confirmed that the WT and pcl1Δpcl2Δ cultures had a similar rate of progression through the cell cycle (Figure 3A). Although total Bni4 protein levels remained fairly constant throughout the cell cycle (Figure 3A, top), a slower-migrating form of Bni4-TAP began to occur in extracts from a WT strain 40–50 min after release, when cells are transiting from G1 to S phase (Figure 3A), peaking by 60 min. Previous work established that the slow migrating isoform occurs due to Bni4 phosphorylation (Walsh et al., 2002; Kozubowski et al., 2003). Phosphorylated Bni4 was significantly reduced in the pcl1Δpcl2Δ strain (Figure 3A), revealing a substantial dependence of Bni4 phosphorylation on Pcl1, Pcl2-Pho85 in vivo.
Figure 3.
Phosphorylation of Bni4 by Pcl-Pho85 Cdks in vivo and in vitro. (A) Cell cycle Western blot of cultures of BNI4-TAP GAL-CDC20 and pcl1Δpcl2Δ BNI4-TAP GAL-CDC20. Samples were taken from cells at 10-min intervals after release from metaphase arrest for Western blot, FACS analysis, and budding index count. The panel on the top shows Bni4-TAP as detected by PAP. The panel in the middle is the bottom part of the same polyacrylamide gel used for Bni4-TAP detection, but it was probed with anti-Swi6 antibody for a loading control. The bottom panel shows FACS analysis results and budding index counts (% budded cells) for each time point. (B) In vitro phosphorylation of Bni4. C-terminal-GST-tagged Bni4, purified using an E. coli expression system, was mixed with kinase complex consisting of Pcl1 and Pho85; Pcl2 and Pho85; or Cln2, Cdc28, and Cks1; and [γ-32P]ATP in kinase reaction buffer, and incubated at 30°C for 30 min. Reaction products were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography (left). (C) Pulldown of GST-Bni4 and associated proteins from yeast extracts. Extracts were prepared from strain BY2131 PCL1-myc carrying pGAL-GST-BNI4 in which BNI4 expression had been induced for 3.5 h. Glutathione-Sepharose beads were used to pull down GST-Bni4, and associated proteins were analyzed by Western blot with anti-GST antibody to detect GST-Bni4 and with anti-myc antibody to detect Pcl1-myc. Western blot to detect GST in pull-down samples is not shown.
Bni4 Is Phosphorylated by Purified Pho85 Complexes and Associates with Pcl1 In Vitro
We next tested whether Bni4 can be directly phosphorylated by G1 Cdks using purified Pcl1-Pho85, Pcl2-Pho85, and Cln2-Cdc28 kinases in an in vitro kinase assay using [γ-32P]ATP with Bni4-GST as a substrate. The Bni4-GST preparation used in these assays included both full-length fusion protein and a number of degradation products (Figure 3B). We confirmed that all three kinase preparations were active by assaying phosphorylation of previously characterized substrates: Rvs167 for Pcl1-Pho85 and Pcl2-Pho85 and histone H1 for Cln2-Cdc28 (data not shown). Bni4-GST was phosphorylated by both Pcl1-Pho85 and Pcl2-Pho85 and the extent of incorporation of γ-32P into Bni4-GST was proportional to the concentration of kinase (Figure 3B). We also detected some phosphorylation of Bni4-GST by Cln2-Cdc28, but the extent of phosphorylation was unrelated to the concentration of kinase. These results are consistent with our in vivo assays suggesting Bni4 may be a significant substrate of G1-specific Pho85 kinases.
We further explored the biochemical relationship between Bni4 and Pcl-Pho85 kinases by assaying their association in vitro. To examine the physical interaction between Bni4 and Pho85 cyclins, we focused on Pcl1, which is most easily purified from yeast extracts. We purified GST-Bni4 from extracts derived from a yeast strain expressing PCL1-myc and carrying pGAL-GST-BNI4. We found that Pcl1-myc copurifies with GST-Bni4, but not with GST alone (Figure 3C). Together, our in vitro and in vivo assays suggest that Bni4 is a direct substrate of Pcl1, Pcl2-Pho85.
Bni4 Localization to the Bud Neck Is Reduced in pcl1Δpcl2Δ and pho85Δ Strains
Next, we sought to discover the functional role of Bni4 phosphorylation by G1 Cdks. Because Bni4 is thought to act as a scaffold protein at the bud neck (DeMarini et al., 1997), we reasoned that localization of Bni4 may be regulated by phosphorylation. To test this idea, we assayed localization of Bni4-mCitrine in various G1 cyclin/kinase deletion mutant strains: pcl1Δpcl2Δ, pcl1Δ, pcl2Δ, cln1Δcln2Δ, cln1Δ, cln2Δ, and pho85Δ. The C-terminal GFP tag has been previously shown not to disturb Bni4 function (Kozubowski et al., 2003).
Localization of Bni4 was clearly reduced in the pcl1Δpcl2Δ and pho85Δ strains relative to either single cyclin mutant or wild-type strains, consistent with the genetic redundancy of the PCL cyclins (Figure 4; Moffat and Andrews, 2004). Because BNI4 had an SDL interaction with CLN1 and CLN2, we also examined Bni4 localization in cln1Δcln2Δ, cln1Δ, and cln2Δ strains. As for the PCLs, deletion of either CLN1 or CLN2 had no obvious effect on Bni4 localization (Figure 4). However, the Bni4-mCitrine signal at the bud neck was reproducibly increased in the cln1Δcln2Δ strain (Figure 4), consistent with a direct or indirect role for Cln-Cdc28 kinases in Bni4 regulation. We favor an indirect model because BNI4 has an SL interaction with CLN1, CLN2, suggesting that it is more likely working in parallel with Cln1, Cln2. Regardless, these data establish a requirement for Pcl1, Pcl2-associated kinase activity for proper localization of Bni4 to the bud neck.
Wild-Type Strains Overexpressing BNI4 Mutated at Putative Phosphorylation Sites Have Similar Morphology Defects to pcl1Δpcl2Δ Mutants
Our in vivo and in vitro assays demonstrate that phosphorylation of Bni4 depends on Pcl1, Pcl2-associated kinase activity. To further substantiate the view that G1 Cdks phosphorylate Bni4 to regulate its localization, we analyzed the phenotype associated with overproduction of versions of Bni4 with alanine substitutions at broadly defined Cdk consensus phosphorylation sites (S/TP). We predicted that overproduction of a hypophosphorylated derivative of Bni4 might mimic the effect we saw with overproduction of wild-type Bni4 in pho85Δ or pcl mutant strains. Ten of 19 S/TP sites in Bni4 are conserved among closely related fungal species (Kellis et al., 2003, Figure 5A). We reasoned that these conserved residues were more likely required for function, and we focused our mutagenesis effort on this subset of sites. First, we asked whether overexpression of a putative phosphorylation site mutant of BNI4 would recapitulate the SDL phenotype caused by overexpression of wild-type BNI4 in pho85Δ mutant strains. Cultures of wild-type strains containing GAL-driven plasmids with single or various combinations of S/TP site mutations, along with appropriate control strains, were serially diluted and plated on galactose-containing and glucose-containing plates. Wild-type strains carrying plasmids with single mutations in putative Bni4 phosphorylation sites or combinations of two or three mutations showed no obvious growth defect when the overexpression of BNI4 was induced (data not shown; Figure 5B). However, strains overexpressing versions of BNI4 with all conserved serines or threonines in S/TP sites substituted with alanines, bni4-10A, showed a moderate but reproducible growth defect (Figure 5B). This result suggests that phosphorylation of Bni4 at the S/TP sites we substituted has a role in preventing toxicity upon overexpression.
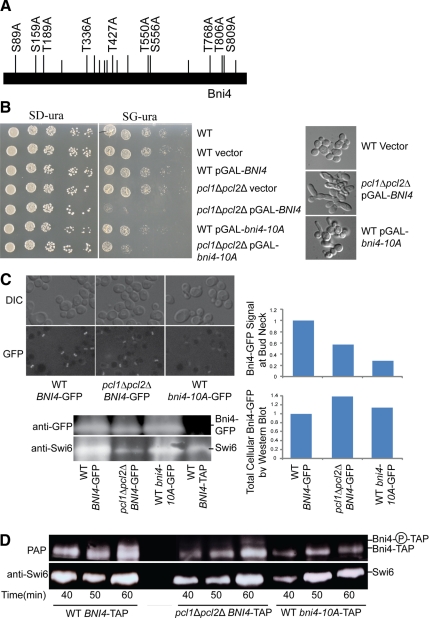
Figure 5.
Phenotype associated with Bni4 phosphorylation site mutants. (A) Diagram of Bni4 protein showing all S/TP sites (vertical lines above bar); S/TP sites conserved among members of the Saccharomyces sensu stricto group are shown as raised lines. The S/TP sites substituted with S/TA in bni4-10A are labeled with their amino acid position. (B) Cultures of wild-type strain, wild-type strain containing vector, or GAL-driven plasmid with BNI4, pcl1Δpcl2Δ strain containing vector or a GAL-driven plasmid with BNI4, a wild-type strain containing a GAL-driven plasmid with bni4-10A, and a pcl1Δpcl2Δ strain containing a GAL-driven plasmid with bni4-10A were serially diluted and plated on SD-ura (left) and SG-ura (right, inducing the overexpression of BNI4) and incubated for 2 d at 30°C. To the right are DIC images showing morphology changes in cells in which BNI4 or bni4-10A overexpression has been induced for 4 h. (C) Strains expressing WT BNI4-GFP, pcl1Δpcl2Δ BNI4-GFP, or WT bni4-10A-GFP, with BNI4 under the control of its own promoter, replacing the endogenous gene, were grown in SD medium and Bni4 localization to the bud neck was examined using DIC and fluorescence microscopy. Cultures were grown to mid-log phase. Fluorescence levels at the bud neck were imaged by fluorescence microscopy and quantified by Velocity software, n ≥ 55. Western blot analysis of these cultures is shown at the bottom, along with extract from a BNI4-TAP strain, as a negative control. The top panel was probed with anti-GFP antibody. The bottom panel shows a portion of the same polyacrylamide gel used for Bni4-GFP detection, but was probed with anti-Swi6 antibody as a loading control. The top graph shows relative Bni4-GFP at the bud neck quantified from micrographs as described in Figure 4. The bottom graph shows relative total cellular Bni4-GFP quantified by western blot and normalized to Swi6 levels. (D) Cell-cycle western blot of cultures of pGAL-CDC20 WT BNI4-TAP, pGAL-CDC20 pcl1Δpcl2Δ BNI4-TAP, or pGAL-CDC20 WT bni4-10A-TAP. Samples were analyzed from cultures taken at 10-min intervals from 40 min to 60 min after release from metaphase arrest. The top panel shows Bni4-TAP as detected by PAP complex (PAP). The label Bni4-P-TAP, to the right of the blot, refers to Bni4 phosphoforms, which are undetectable in the bni4-10A-TAP strain. The bottom panel shows a portion of the same polyacrylamide gel used for Bni4-TAP and bni4-10A-TAP detection but was probed with anti-Swi6 antibody as a loading control.
If the SDL phenotype of BNI4 in a pcl1Δpcl2Δ mutant were a consequence of not phosphorylating the 10 residues that were substituted with alanines, we would expect bni4-10A overexpression to generate the same level of toxicity in WT and in a pcl1Δpcl2Δ strain. As seen in Figure 5B, the pcl1Δpcl2Δ strain overexpressing bni4-10A was only slightly more defective in growth than WT overexpressing bni4-10A, suggesting that the SDL phenotype in a pcl1Δpcl2Δ results mostly from an inability to phosphorylate the 10 conserved S/TPs. Interestingly, the pcl1Δpcl2Δ mutant overexpressing WT BNI4 grew more slowly than the pcl1Δpcl2Δ mutant overexpressing bni4-10-A (Figure 5B). This suggests that another kinase, perhaps acting in opposition to Pcl1, Pcl2-Pho85, may phosphorylate Bni4 at one or more of the substituted residues.
Because the severity of the phenotype associated with overexpression of the alanine mutant in WT is less than that seen in the pcl1Δpcl2Δ strain overexpressing wild-type BNI4, it is possible that Pcl1, Pcl2-Pho85 phosphorylates Bni4 at additional residues or that Pcl1, Pcl2-Pho85 regulates other proteins that contribute to the toxicity of overexpressed BNI4. To confirm that the growth defect was a consequence of defective morphogenesis, we examined the morphology of these strains. Wild-type cells expressing WT BNI4 had normal bud morphology. However, pcl1Δpcl2Δ cells overexpressing WT BNI4 and WT cells overexpressing bni4-10A exhibited elongated bud phenotypes (Figure 5B) similar to that observed in pcl1Δpcl2Δ strains expressing pGAL-GST-BNI4 (Figures 2B and 5B, bottom). Our finding that the wild-type strain overexpressing bni4-10A partially phenocopies the pcl1Δpcl2Δ strain overexpressing wild-type BNI4, suggests that Pcl1, Pcl2-Pho85 phosphorylates Bni4 at some of the residues tested.
Because Bni4 localization to the bud neck was reduced in pcl1Δpcl2Δ and pho85Δ strains, we next asked whether bni4-10A had the same localization defect by examining the localization of bni4-10A fused to GFP under the control of its endogenous promoter. Like WT Bni4-GFP in the pcl1Δpcl2Δ strain, the GFP signal associated with bni4-10A was only weakly detectable in the bud neck compared with the Bni4-GFP signal we observed in WT cells, although cells still maintained a normal morphology (Figure 5C). Even less GFP signal was present at the bud neck in the WT strain expressing bni4-10A-GFP than was seen in pcl1Δpcl2Δ BNI4-GFP, suggesting that phosphorylation at these S/TP sites, by one or more kinases in addition to Pcl1, Pcl2-Pho85, may contribute to Bni4 localization. The disappearance of Bni4 from the bud neck was due to mislocalization rather than degradation because all the strains had the same amount of full-length Bni4-GFP as assayed by anti-GFP Western blot (Figure 5C). A partially redundant role for another kinase is also consistent with our cell cycle Western analysis of Bni4, where we still saw evidence of residual Bni4 phosphorylation at the G1/S transition in a pcl1Δpcl2Δ strain (Figure 3A).
We next examined the phosphorylation level of the bni4-10A mutant by Western blot, looking at points in the cell cycle when the Bni4 phosphoshift had been detected previously (Figure 5D). As observed previously, in wild-type cells, phosphoforms of Bni4 were apparent 50–60 min after release from a CDC20 block (Figure 5D, left). In contrast, in the pcl1Δpcl2Δ strain with wild-type Bni4 and in the wild-type strain with bni4-10A, little or no phosphorylated Bni4 was detectable (Figure 5D, middle and right). Because bni4-10A gives no detectable phosphoshift compared with wild-type Bni4, it is clear that some of the sites of phosphorylation of Bni4 by Pcl1, Pcl2-Pho85 must be located within the ten conserved residues. However, because many phosphorylation events do not lead to a phosphoshift, it is possible that Bni4 is additionally phosphorylated at residues that were not substituted in the 10A mutant. Indeed this possibility is supported by the finding that overexpressed bni4-10A is more toxic in pcl1, pcl2 than in wild type, which suggests that Pcl-Pho85 phosphorylates additional sites in Bni4.
Overexpression of BNI4 in pho85Δ Disrupts the Septin Ring Structure at the Bud Neck
Our genetic analysis revealed that in the absence of PCL1, PCL2, or PHO85, there was decreased Bni4 localization to its native site at the bud neck (Figures 4 and 5C) and overexpression of BNI4 caused cell polarity problems (Figure 2B). Because Bni4 is not essential, the defect in pcl1Δpcl2Δ cells overexpressing BNI4 is not due to the absence of Bni4 at the bud neck. Rather, we predicted that Bni4 may be titrating some essential component away from the bud neck with a consequent disastrous effect on cell morphology (Hartwell, 1971; Barral et al., 1999; Longtine et al., 2000).
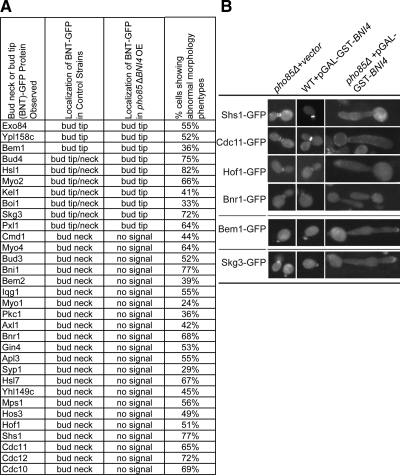
To test this hypothesis, we designed a screen to examine the localization of GFP-tagged bud neck proteins in cells lacking PHO85 and overexpressing BNI4. First, we assembled an array of 94 strains carrying GFP-tagged proteins reported to localize to the bud neck or bud tip (Huh et al., 2003), which we refer to as the BNT-GFP array. Then, we used the SGA method to introduce both a pho85Δ allele and a pGAL-GST-BNI4 plasmid into the BNT-GFP array. We also created two control arrays: 1) pho85Δ BNT-GFP carrying vector and 2) WT BNT-GFP carrying pGAL-GST-BNI4. After inducing BNI4 overexpression for 4 h, we examined the localization of BNT-GFP by using a high-throughput microscope. We then focused on the analysis of 32 proteins that maintained their proper localization in the two control arrays (data not shown).
Localization of the 32 proteins in control strains can be divided into three groups: I) bud neck only, II) bud tip only, and III) both bud tip and bud neck. We included bud tip proteins because polarization cues are required for both bud neck and bud tip localization, and we wanted to ask whether overexpression of mislocalized BNI4 had more general effects on proteins that respond to polarity signals. Because overexpressing BNI4 in the absence of PHO85 leads to elongated buds and abnormal morphology, we looked at localization of BNT-GFP proteins only in cells with abnormal morphology. We found that all 22 bud-neck–localized proteins, including all septin proteins (e.g., Shs1-GFP, Cdc11-GFP), lost their bud neck signal in the pho85Δ strain overexpressing BNI4. The GFP signals of these proteins either became undetectable (e.g., Bnr1-GFP) or improperly localized (e.g., Cdc11-GFP; Figure 6B, top). In addition, we noticed that three proteins that were localized to the bud tip in a WT strain, e.g., Bem1-GFP, still remained at the bud tip even though the morphology of the cells was abnormal with extremely elongated buds (Figure 6B, middle). In a WT strain background, some proteins localized to the bud tip initially and then migrated to the bud neck later in the cell cycle (class III; e.g., Skg3-GFP). These proteins remained at the bud tip at all times and were never found at the bud neck in a pho85Δ strain with BNI4 overexpressed (Figure 6B, bottom). In summary, each of the 29 group I or group III proteins, which localized to the bud neck in our control strains, was no longer found at the bud neck in a pho85Δ strain with overexpressed BNI4. This result suggests that the essential structure of the bud neck must be severely perturbed by expression of misregulated BNI4.
Figure 6.
Bud neck protein localization in strains overexpressing Bni4. (A) Localization of BNT-(bud neck or bud tip)-GFP fusion proteins observed in control strains, wild-type strains with overexpression of BNI4 or pho85Δ strains carrying vector (second column), or in pho85Δ strains overexpressing BNI4 (third column). Column 4 indicates the percentage of pho85Δ BNT-GFP pGAL-GST-BNI4 cells showing abnormal morphology (i.e., grossly elongated buds and polarity defects). (B) Representative images of cells showing abnormal morphology and mislocalization of BNT-GFP proteins. Overexpression of BNI4 was induced by adding galactose to a log phase culture and incubating at 30°C for 4 h. The GFP signal in these cultures was examined by high-throughput microscopy.
Septins Mislocalize Together with Bni4
To determine whether it was the septins that were titrated away from the bud neck by Bni4 in the absence of Pcl1, Pcl2-Pho85, we constructed a pcl1Δpcl2Δ CDC11-RFP strain carrying a pGAL-BNI4-GFP plasmid, which allowed us to observe the Cdc11-RFP and Bni4-GFP signal at the same time. As controls, we also constructed strains of WT CDC11-RFP cells carrying pGAL-BNI4-GFP and pcl1Δpcl2Δ CDC11-RFP with BNI4-GFP under control of its endogenous promoter. After inducing BNI4-GFP overexpression for 4 h, we examined these cells by using confocal microscopy.
We saw the expected cell cycle-dependent changes in the septin ring structure in the two control strains (WT CDC11-RFP with BNI4-GFP overexpression and pcl1Δpcl2Δ CDC11-RFP with endogenous level of Bni4-GFP; Figure 7). As expected, the Bni4-GFP ring structure occurred in the early cell cycle, before budding and Bni4 was localized to the mother side of the cell using the septin ring as a reference (Figure 7, top, GFP channel), consistent with previous findings (Kozubowski et al., 2005). Bni4-GFP formed a double ring structure at later cell cycle stages (Figure 7). Persistent localization of Bni4-GFP at the bud neck suggests that Bni4 may have a role in bud neck function throughout the cell cycle.
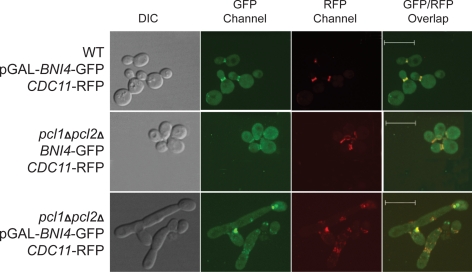
Figure 7.
Colocalization of Bni4 and septin proteins. WT CDC11-RFP and pcl1Δpcl2Δ CDC11-RFP strains carrying pGAL-BNI4-GFP or vector were grown in medium containing galactose for 4 h and observed using confocal microscopy. Cells were photographed using the DIC, GFP channel, RFP channel, and GFP/RFP overlaid. The RFP signal was switched to red using Volocity software to assist in viewing. Bars, 10 μm.
In the pcl1Δpcl2Δ CDC11-RFP strain carrying pGAL-BNI4-GFP, we observed proper septin ring and Bni4 ring structures at the bud neck at early cell cycle stages after shift to galactose-containing medium to induce BNI4 expression (data not shown). However, as expected from our earlier experiments, when BNI4 overexpression was induced for 5 h, abnormal morphology was observed in most cells. Among those cells with abnormal morphology, normal septin ring structures were not seen. Rather, the Cdc11-RFP signal was mislocalized to various places along the bud neck and in some cells, it seemed to localize in blobs (but not rings) where the bud neck should be (Figure 7, bottom, red fluorescent protein [RFP] channel). More importantly, we noticed that almost all of the mislocalized septin colocalized with the Bni4-GFP signal (shown as yellow in Figure 7 when GFP and RFP signals overlap). If the disruption of the septin ring structure were an indirect consequence of changes in the stability of the bud neck structure due to BNI4 overexpression, we might expect to see septin-related RFP signal distinct from Bni4-GFP signal (red only on overlaid image). However, all of the Cdc11-RFP signal overlapped with Bni4-GFP, suggesting that all mislocalized septin is accompanied by Bni4. This suggests that overproduced Bni4 is interacting with the septins directly, consistent with a role for Bni4 phosphorylation in preventing inappropriate interactions with the septin ring.
DISCUSSION
The accurate spatial and temporal coordination of cell polarization with DNA replication and segregation guarantees the fidelity of genetic transmission. A previous study from our laboratory revealed that a buildup or burst of G1 Cdk activity, through activation of the cyclin genes CLN1, CLN2 and PCL1, PCL2, is essential for cell morphogenesis (Moffat and Andrews, 2004). Although a few substrates of G1 cyclins that regulate cell polarity have been identified, the specific morphological events that require G1 Cdks are still not clear. Previous studies have connected G1 Cdks to activation of the Cdc42 GTPase module that is required to establish cell polarity (Lenburg and O'Shea, 2001; Moffat and Andrews, 2004; Sopko et al., 2006). One of the septins, Shs1, also has been identified as a substrate of G1 cyclins (Egelhofer et al., 2008) and its phosphorylation is required for proper formation of the bud neck (Egelhofer et al., 2008). Previous studies also found other substrates of G1 cyclins, such as Rga2 and Gin4, that contribute to the regulation of cell polarity (Gladfelter et al., 2004; Sopko et al., 2007).
To discover new targets of G1 Cdks related to their role in cell polarity, we devised an approach that makes use of complementary functional genomic screens to place potential substrates in the Cln-Cdc28 or Pcl-Pho85 pathways. We used the SGA method to screen pairwise deletions of the G1 cyclins for both synthetic lethal or synthetic dosage lethal interactions. Systematic analysis of SL interactions by using SGA has proven an effective method to identify genetic interactions between parallel regulatory pathways (Tong et al., 2001, 2004), whereas SDL interactions tend to occur among components of the same pathway. Specifically, from our previous studies, we predicted that genes whose overexpression causes lethality in the absence of a kinase may encode substrates of that kinase (Sopko et al., 2006). We therefore reasoned that targets of either Cln1, Cln2-Cdc28 or Pcl1, Pcl2-Pho85 would be enriched among genes that showed a specific pattern of genetic interaction: SDL with deletion of the cognate kinase genes (for example, pcl1Δpcl2Δ) and SL with genes encoding kinases in the redundant pathway (cln1Δcln2Δ in this example). We note that this approach is conceptually extendable to any set of genetically redundant regulators.
BNI4 showed the pattern of genetic interaction we predicted for substrates of Pcl1, Pcl2-Pho85, a possibility we explored further with a variety of biochemical and cell biological assays. Bni4 has been reported previously to be localized to the bud neck (DeMarini et al., 1997; Kozubowski et al., 2005) and is thought to function as a scaffold protein because a strain deleted for BNI4 exhibits defects in tethering Chs4, the regulatory subunit of chitin synthase III, to the site of bud emergence (DeMarini et al., 1997; Kozubowski et al., 2003). Deletion of BNI4 leads to a chitin deposition defect (DeMarini et al., 1997; Kozubowski et al., 2003; Sanz et al., 2004), consistent with its role in chitin metabolism regulation; however, cells lacking BNI4 still have a proper septin ring structure throughout the cell cycle (Longtine et al., 1996), with almost normal morphology. Therefore, the presence of Bni4 is not essential for septin ring structure and cell cycle progression.
Our observation, that overexpression of BNI4 in the absence of PCL1, PCL2 causes severely abnormal morphology, reveals that Bni4 function needs to be tightly regulated by G1 cyclins. Overexpression of BNI4 has been reported to cause an elongated bud phenotype even in a WT strain (DeMarini et al., 1997; Kozubowski et al., 2003); however, in our experiments overproducing Bni4 with no tag, an N-terminal GST tag, or a C-terminal GFP tag (Figures 5 and 7), we observed little or no morphology defect. This variability could reflect the strain background used in our assays, which differs from that used in previous studies. Regardless, we observed a severe abnormal morphology when Bni4 was overproduced in a pc1lΔpcl2Δ background, reminiscent of the morphology of cells with defects in septin organization observed by other groups (Longtine et al., 1996; DeMarini et al., 1997; Mino et al., 1998; Richman et al., 1999; Gladfelter et al., 2001), suggesting that Bni4 helps to maintain the septin ring structure at the bud neck.
To investigate the mechanism of the morphology change we observed in pc1lΔpcl2Δ cells overexpressing BNI4, we first studied whether the phosphorylation of Bni4 is regulated by G1 cyclins, and then we examined how phosphorylation of Bni4 affects polarity regulation. Bni4 has been identified previously as a phosphoprotein (Kozubowski et al., 2003); however, the kinase responsible for the phosphorylation of Bni4, and its associated biological function was unknown. We have produced several lines of evidence that Pcl1, Pcl2-Pho85 phosphorylates Bni4. First, hyperphosphorylation of Bni4 occurs around the time of the G1/S transition, when Pcl1, Pcl2-Pho85 activity is at its peak, and this hyperphosphorylation is dependent on PCL1, PCL2 (Figure 3A). Second, BNI4 has an SL interaction with CLN1, CLN2 and an SDL interaction with PCL1, PCL2 (Figure 1), suggesting that Bni4 may be a downstream target of Pcl1, Pcl2-Pho85. Third, Pcl1- and Pcl2-Pho85 are able to phosphorylate Bni4 in vitro (Figure 3B). Fourth, Pcl1 and Pcl2 and Bni4 localize to the bud neck (Huh et al., 2003; Gladfelter et al., 2005; Sopko et al., 2007), and Pcl1 associates with Bni4 in vivo in a pulldown assay (Figure 3C).
The proper localization of Bni4 is regulated by Pcl1, Pcl2-Pho85, suggesting positive regulation by the kinase. SDL interactions can result from several different scenarios. One possible mechanism for SDL involves overexpression of a gene in the absence of its upstream negative regulator, as has been shown with some Pho85 substrates (Sopko et al., 2006). Our experiments provide an example of an additional mechanism that could cause SDL. As shown in our study of Bni4 regulation, phosphorylation of Bni4 serves as a positive signal to maintain its proper localization at the bud neck. In the absence of this regulation, overproduced Bni4 disrupts the structure of septins by titrating septins away from the bud neck, causing an SDL phenotype.
We predict that at least one additional kinase phosphorylates Bni4, because the defect we observed in bud neck localization of Bni4 in the pcl1Δpcl2Δ and pho85Δ strains was less severe than the localization defect of bni4-10A mutant in a WT strain. Furthermore, although it was considerably reduced, we observed a residual Bni4 phosphoshift in the pcl1Δpcl2Δ strain. The bud neck kinase Hsl1 plays a role in negatively regulating the localization of Bni4 to the bud neck. Regulation of Bni4 by Hsl1 is likely indirect because an hsl1Δ strain does not show reduced phosphorylation of Bni4 (Kozubowski et al., 2003). Another candidate G1-specific kinase is Cln1, Cln2-Cdc28. However, we think the interaction between CLN1, CLN2 and BNI4 is also likely to be indirect for the following reasons. First, the phosphoshift of Bni4 in G1 was observed in a cln1Δcln2Δ GAL-CLN3 strain (Kozubowski et al., 2003), suggesting that CLN1 and CLN2 are not required for Bni4 phosphorylation. Second, we found that in a cln1Δcln2Δ strain Bni4 localization to the bud neck was not reduced, and even showed some increase over WT (Figure 4). Third, Bni4 was not efficiently phosphorylated in vitro by Cln2-Cdc28 (Figure 3B). More work will be required to discover kinases in addition to Pcl-Pho85 that phosphorylate and regulate Bni4.
Our cell biological and genetic work supports a functionally significant role for Bni4 phosphorylation. First, phosphorylation of Bni4 by Pcl-Pho85 is required for its bud neck localization. Second, in the absence of phosphorylation, either from deleting the kinase or expressing a phosphorylation site mutant, overproduction of Bni4 results in disruption of the bud neck structure. In addition, the phosphorylation status of Bni4 corresponds well with its localization to the bud neck throughout the cell cycle. Bni4 is hypophosphorylated in early G1 when it is only weakly detectable at the bud neck, and becomes hyperphosphorylated in late G1 just before bud emergence, at which time it first localizes to the incipient bud site. Bni4-GFP levels at the bud neck fall at cytokinesis, at which time it is hypophosphorylated (Kozubowski et al., 2003, Figure 5). Previous studies found that the phosphatase Glc7 binds to Bni4 and that the Bni4–Glc7 complex is required for efficient bud neck localization of both proteins (DeMarini et al., 1997; Kozubowski et al., 2003). Bni4 acts as a targeting subunit for Glc7 to direct chitin synthesis to the bud neck (Larson et al., 2008), but there is no evidence that association with Glc7 regulates Bni4 phosphorylation in a cell cycle-dependent manner because Bni4 seems to be hyperphosphorylated during the time it is associated with Glc7 (Kozubowski et al., 2003).
Although Bni4 is not essential, its failure to localize to the bud neck is detrimental, raising the question of what important cellular function is affected by reducing proper Bni4 localization. The abnormal cell biological phenotype associated with the SDL interaction between BNI4 and PCL1, PCL2 reveals some clues. Specifically, our survey of bud neck-localized proteins revealed a general defect in localization of proteins to the bud neck when BNI4 was overexpressed in a pho85Δ background (Figure 6), suggesting that the essential structure of the bud neck has been perturbed. One possibility is that Bni4 physically interacts with some bud neck protein to help establish or maintain the proper bud neck structure. When Bni4 is overexpressed and inappropriately regulated (in a pcl1Δpcl2Δ strain), Bni4 fails to localize to the bud neck and may titrate away an important bud neck component, leading to the severe phenotype we observed. We observed colocalization between overproduced Bni4-GFP and Cdc11-RFP in a pcl1Δpcl2Δ strain suggesting that the primary “titrated” component may be the septins, consistent with a known two-hybrid interaction between Bni4 and the septin Cdc10 (DeMarini et al., 1997).
CONCLUSION
It has been shown previously that Bni4 is required for Chs4 localization to the bud neck (DeMarini et al., 1997) and for regulation of the localization of the phosphatase Glc7. In this work, we show that Bni4 also has a role in regulating the dynamic changes of the septin ring. Phosphorylation of Bni4 by G1 Cdks, and possibly other kinases, regulates the localization of Bni4 and may influence its binding affinity for other proteins.
ACKNOWLEDGMENTS
We thank Wei Ye and Wendy Liang for technical assistance. This work was supported by an operating grant from the National Cancer Institute of Canada with funds from the Canadian Cancer Society. Our SGA laboratory is supported Canadian Institutes of Health Research grant GSP-41567 and by funds from Genome Canada through the Ontario Genomics Institute.
Abbreviations used:
- Cdk
cyclin-dependent kinase
- SDL
synthetic dosage lethal
- SL
synthetic lethal
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1255) on May 20, 2009.
REFERENCES
- Barral Y., Mermall V., Mooseker M. S., Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Barral Y., Parra M., Bidlingmaier S., Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C., Bussey H., Andrews B. J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Cid V. J., Adamikova L., Sanchez M., Molina M., Nombela C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 2001;147:1437–1450. doi: 10.1099/00221287-147-6-1437. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Adams A. E., Fares H., De Virgilio C., Valle G., Chuang J. S., Pringle J. R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight S. S., et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer T. A., Villen J., McCusker D., Gygi S. P., Kellogg D. R. The septins function in G1 pathways that influence the pattern of cell growth in budding yeast. PLoS ONE. 2008;3:e2022. doi: 10.1371/journal.pone.0002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faty M., Fink M., Barral Y. Septins: a ring to part mother and daughter. Curr. Genet. 2002;41:123–131. doi: 10.1007/s00294-002-0304-0. [DOI] [PubMed] [Google Scholar]
- Friesen H., Murphy K., Breitkreutz A., Tyers M., Andrews B. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Mol. Biol. Cell. 2003;14:3027–3040. doi: 10.1091/mbc.E02-09-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Kozubowski L., Zyla T. R., Lew D. J. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 2005;118:1617–1628. doi: 10.1242/jcs.02286. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Zyla T. R., Lew D. J. Genetic interactions among regulators of septin organization. Eukaryot. Cell. 2004;3:847–854. doi: 10.1128/EC.3.4.847-854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Huang D., Patrick G., Moffat J., Tsai L. H., Andrews B. Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1999;96:14445–14450. doi: 10.1073/pnas.96.25.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Larson J. R., Tatchell K. Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:3455–3466. doi: 10.1091/mbc.E04-09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L., Panek H., Rosenthal A., Bloecher A., DeMarini D. J., Tatchell K. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell. 2003;14:26–39. doi: 10.1091/mbc.E02-06-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll E. S., Hyland K. M., Hieter P., Li J. J. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J. R., Bharucha J. P., Ceaser S., Salamon J., Richardson C. J., Rivera S. M., Tatchell K. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:3040–3051. doi: 10.1091/mbc.E08-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Colwill K., Aneliunas V., Tennyson C., Moore L., Ho Y., Andrews B. Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 1998;8:1310–1321. doi: 10.1016/s0960-9822(07)00561-1. [DOI] [PubMed] [Google Scholar]
- Lenburg M. E., O'Shea E. K. Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics. 2001;157:39–51. doi: 10.1093/genetics/157.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Shapiro J., Specht C. A., Sdicu A. M., Menard P., Hussein S., Tong A. H., Boone C., Bussey H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., DeMarini D. J., Valencik M. L., Al-Awar O. S., Fares H., De Virgilio C., Pringle J. R. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Theesfeld C. L., McMillan J. N., Weaver E., Pringle J. R., Lew D. J. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Moore L., Ogas J., Tyers M., Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- Measday V., Moore L., Retnakaran R., Lee J., Donoviel M., Neiman A. M., Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino A., Tanaka K., Kamei T., Umikawa M., Fujiwara T., Takai Y. Shs1p: a novel member of septin that interacts with spa2p, involved in polarized growth in saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1998;251:732–736. doi: 10.1006/bbrc.1998.9541. [DOI] [PubMed] [Google Scholar]
- Moffat J., Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat. Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- Moffat J., Huang D., Andrews B. Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog. Cell Cycle Res. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- Richman T. J., Sawyer M. M., Johnson D. I. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 1999;274:16861–16870. doi: 10.1074/jbc.274.24.16861. [DOI] [PubMed] [Google Scholar]
- Sanz M., Castrejon F., Duran A., Roncero C. Saccharomyces cerevisiae Bni4p directs the formation of the chitin ring and also participates in the correct assembly of the septum structure. Microbiology. 2004;150:3229–3241. doi: 10.1099/mic.0.27352-0. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Toth A., Galova M., Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Sopko R., Andrews B. J. Linking the kinome and phosphorylome—a comprehensive review of approaches to find kinase targets. Mol. Biosyst. 2008;4:920–933. doi: 10.1039/b801724g. [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., Andrews B. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Smith J. C., Figeys D., Andrews B. J. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 2007;26:4487–4500. doi: 10.1038/sj.emboj.7601847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P. A., DeRisi J. L., Wilhelm J. E., Vale R. D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tyers M. The Yeast Nucleus: Frontiers in Molecular Biology. Oxford, United Kingdom: Oxford University Press; 2000. [Google Scholar]
- Walsh E. P., Lamont D. J., Beattie K. A., Stark M. J. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry. 2002;41:2409–2420. doi: 10.1021/bi015815e. [DOI] [PubMed] [Google Scholar]
- Wittenberg C., Sugimoto K., Reed S. I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- Zhu H., Klemic J. F., Chang S., Bertone P., Casamayor A., Klemic K. G., Smith D., Gerstein M., Reed M. A., Snyder M. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]