Abstract
PvdA catalyzes the hydroxylation of the sidechain primary amine of ornithine in the initial step of the biosynthesis of the Pseudomonas aeruginosa siderophore pyoverdin. The reaction requires FAD, NADPH, and O2. PvdA uses the same co-substrates as several flavin-dependent hydroxylases that differ one from another in the kinetic mechanisms of their oxidative and reductive half-reactions. Therefore, the mechanism of PvdA was determined by absorption stopped-flow experiments. By contrast to some flavin-dependent hydroxylases (notably, p-hydroxybenzoate hydroxylase), binding of the hydroxylation target is not required to trigger reduction of the flavin by NADPH: the reductive half-reaction is equally facile in the presence and absence of ornithine. Reaction of O2 with FADH2 in the oxidative half-reaction is accelerated by ornithine 80-fold, providing a mechanism by which PvdA can ensure coupling of NADPH and ornithine oxidation. In the presence of ornithine, the expected C(4a)-hydroperoxyflavin intermediate with 390-nm absorption accumulates and decays to the C(4a)-hydroxyflavin in a kinetically competent fashion. The slower oxidative half-reaction that occurs in the absence of ornithine involves accumulation of an oxygenated flavin species and two subsequent states that are tentatively assigned as C(4a)-peroxy- and -hydroperoxyflavin intermediates and the oxidized flavin. The enzyme generates stoichiometric hydrogen peroxide in lieu of hydroxyornithine. The data suggest that PvdA employs a kinetic mechanism that is a hybrid of those previously documented for other flavin-dependent hydroxylases.
The opportunistic human pathogen Pseudomonas aeruginosa produces pyoverdin, a hydroxamate siderophore, to scavenge iron in the iron-limiting conditions of the host, a process that has been linked to virulence (1–3). Pyoverdin contains a chromophore and a peptide backbone of 8–12 amino acids, formed by nonribosomal peptide synthetase enzymes (NRPS) (2, 4, 5). The amino acids of the peptide backbone are both proteinogenic and non-proteinogenic and include two formyl-hydroxyornithines. These two unusual amino acids provide hydroxamate ligands for iron chelation. Two accessory enzymes to the NRPS, the ornithine hydroxylase (PvdA) (6, 7) and the hydroxyornithine transformylase (PvdF) (8, 9), prepare ornithine for incorporation into the siderophore by the NRPS assembly system.
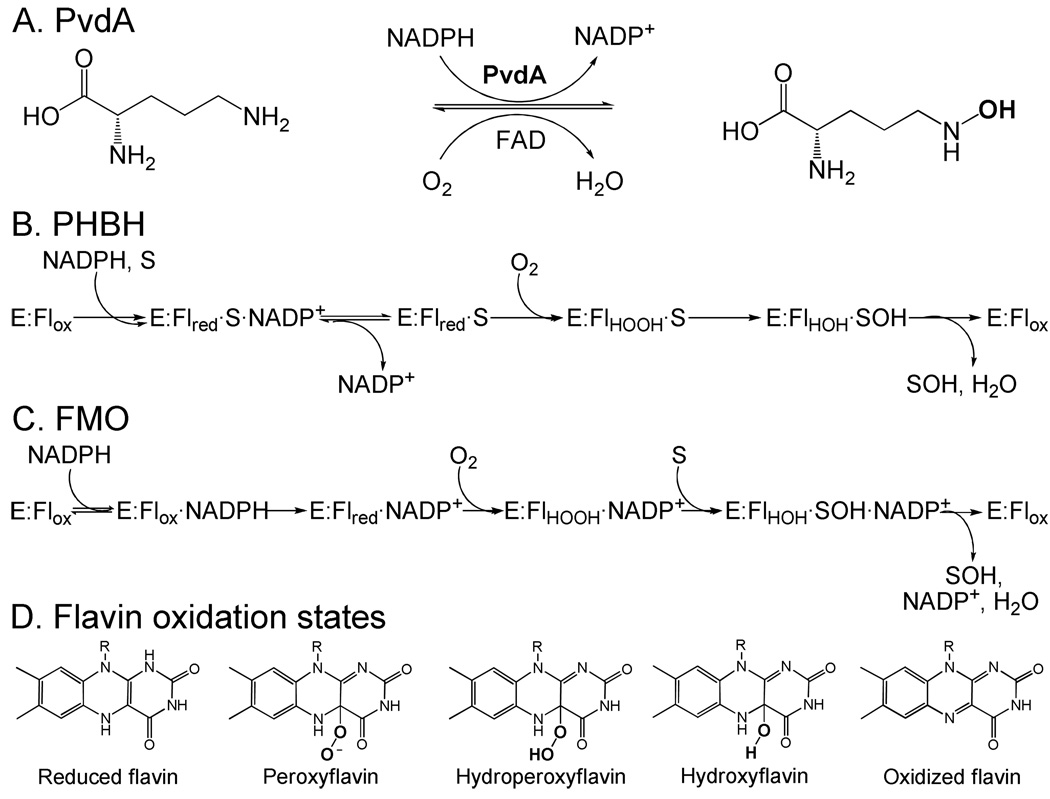
PvdA was recently characterized by steady-state kinetics and found to require both FAD and NADPH and to have high specificity for ornithine (Figure 1A) (10). The enzyme was tested for activity by two methods: the formation of hydroxylated product, which showed inhibition at high ornithine concentrations, and the oxidation of NADPH, which demonstrated simple saturation kinetics. PvdA was inhibited by chloride ions, and lysine served as a non-substrate effector, activating NADPH oxidation without the formation of hydroxylysine. It was noted that, in several respects, PvdA is functionally homologous to para-hydroxybenzoate hydroxylase (PHBH) and flavin-containing monooxygenase (FMO), two monooxygenases to which PvdA is comparably similar (∼ 35% at the level of primary structure). In both PHBH and FMO, addition of O2 to the reduced flavin generates a C(4a)-hydroperoxyflavin intermediate, which decays by substrate hydroxylation to the C(4a)-hydroxyflavin (11–15). This flavin intermediate sequence has been demonstrated by stopped-flow absorption and fluorescence experiments, in which both the hydroperoxy- and hydroxyflavins have absorbance maxima at 375–395 nm, whereas the hydroxyflavin is also highly fluorescent (excitation ∼400 nm and emission > 500 nm) (16–19). Despite the similarity of their chemical mechanisms, the two enzymes (PHBH and FMO) employ different kinetic mechanisms. PHBH requires the presence of substrate for the reduction of the flavin by NADPH (Figure 1B), which is hypothesized to be a regulatory mechanism to prevent uncoupled NADPH oxidation (12, 13, 18). In contrast, the flavin of FMO can be reduced by NADPH regardless of whether the target substrate is present (Figure 1C). The C(4a)-hydroperoxyflavin intermediate produced by subsequent addition of O2 to the reduced cofactor is remarkably stable (half-life ∼ 2 hrs) and awaits binding of the substrate. The protection of the intermediate ensures that, once NADPH has been oxidized, substrate oxidation ensues (11, 14, 18, 20). Thus, PHBH and FMO embody two distinct mechanistic strategies to ensure coupling of NADPH and substrate oxidation.
Figure 1.
Reaction catalyzed by (A) ornithine hydroxylase (PvdA) from P. aerugionsa, (B) p-hydroxybenzoate hydroxylase (PHBH) from P. fluorescens, and (C) flavin-containing monooxygenase (FMO) from hog liver microsomes. The substrate for PHBH (p-hydroxybenzoate (33)) and FMO (variety of hydrophobic nitrogen and sulfur containing compounds (34–36)) is labeled as S, SOH represents the hydroxylated substrate (product), and the flavin states are oxidized (Flox), reduced (Flred), C4a-hydroperoxyflavin intermediate (Flhooh) and hydroxyflavin intermediate (Flhoh). (D) Flavin oxidation states.
The previous steady-state kinetic experiments on PvdA suggested that FAD reduction does not require ornithine. However, the upper limit that could be set for the half-life of the presumptive hydroxylating intermediate was much shorter than would seem to be required for PvdA to use the FMO coupling strategy (10). This work thus raised the question of whether PvdA ensures coupling of NADPH oxidation and ornithine hydroxylation, and how the enzyme might accomplish that coupling. Here, we have used stopped-flow absorption measurements to elucidate the PvdA mechanism. The results show that, whereas the enzyme follows the canonical chemical mechanism involving the key C(4a)-hydroperoxyflavin intermediate, PvdA coupling is regulated by a mechanism that is distinct from those employed by PHBH and FMO. This novel mechanism involves substrate triggering of O2 addition to the reduced flavin.
Materials and Methods
PvdA Purification
PvdA protein was over-expressed and purified as described previously (10). The purified protein in 100 mM potassium phosphate, pH 8.0 with 100 mM sodium citrate was concentrated to 260 – 290 µM using an Amicon stirred-cell.
Stopped-Flow Absorption
Stopped-flow experiments were performed at 22 °C using an Applied Photophysics SX.18MV stopped-flow apparatus (Surrey, U.K.) equipped with a photomultiplier detector housed in an anoxic chamber (MBraun). Absorbance traces were recorded with an optical path length of 1 cm. For the experiments determining the dependence of the flavin intermediate formation on [O2], the O2-free PvdA-flavin-NADPH complex was mixed in the stopped-flow apparatus with buffer containing varying [O2], which were prepared by mixing appropriate volumes of O2-saturated and O2-free buffer. The O2-saturated buffer was equilibrated at 22 °C, resulting in a solution with an oxygen concentration of 1.3 mM (21). These experiments were conducted with and without ornithine. The stopped-flow mixing ratio was 1:1. For multi-wavelength detection, a photodiode array detector was used in place of the photomultiplier. Experimental details are given in the figure legends. Curve-fitting analysis was performed with KaleidaGraph 4 (Synergy Software) to determine experimental rate constants from best-fit curves.
Hydrogen peroxide formation in the absence of substrate
The amount of NADPH oxidation and hydrogen peroxide formed were measured as follows. PvdA (5 µM) was incubated with 0.03 mM FAD and 0.15 mM NADPH in 100 mM potassium phosphate, pH 8.0 in a 1 ml final volume. NADPH oxidation was measured continuously for 4 minutes by monitoring the decrease in absorbance at 366 nm (ɛ = 2,850 M−1 cm−1) with a BioMate 3 spectrometer (Thermo Spectronics) at 24 °C (10). The amount of hydrogen peroxide produced was determined independently with an Amplex Red (Invitrogen) assay measuring the increase in absorbance at 572 nm (ɛ = 72,000 M−1 cm−1) at 24 °C. At 4 minutes, 250 µl of the assay was diluted into 750 µl of 100 mM potassium phosphate, pH 8.0 containing 0.02 mg/ml horseradish peroxidase and 0.05 mM Amplex Red. The change in absorbance was measured in triplicate with the BioMate 3 spectrometer.
Results
Rapid reaction kinetics
As with all flavin-dependent monooxygenases, the flavin chemistry by which PvdA effects hydroxylation of ornithine (Orn) can be studied as two half-reactions. First, FAD bound to the enzyme is reduced by NADPH in the reductive half-reaction. This is followed by the oxidative half-reaction, in which O2 adds to the enzyme-bound FADH2 and hydroxyornithine (OrnOH) is produced. FADH2 oxidation is expected also to occur in the absence of substrate. In this case, production of hydrogen peroxide accounts for the two oxidizing equivalents that are not transferred to ornithine. Using stopped-flow experiments monitoring absorbance, the flavin reaction intermediates were detected and rate constants determined for each catalytic step.
Conditions for reduction of PvdA-bound FAD
The previously determined dissociation constant for PvdA•FAD (26 µM (10); 10 µM (22)) is indicative of relatively weak binding. To limit the complications of free FAD in solution, all experiments were conducted such that FAD was predominantly bound to enzyme, and the PvdA•FAD complex was saturated with NADPH. Concentrations amenable to spectroscopic observation were (post-mixing): PvdA (145 µM), FAD (30 µM) and NADPH (180 µM).
Reduction of flavin is ornithine independent
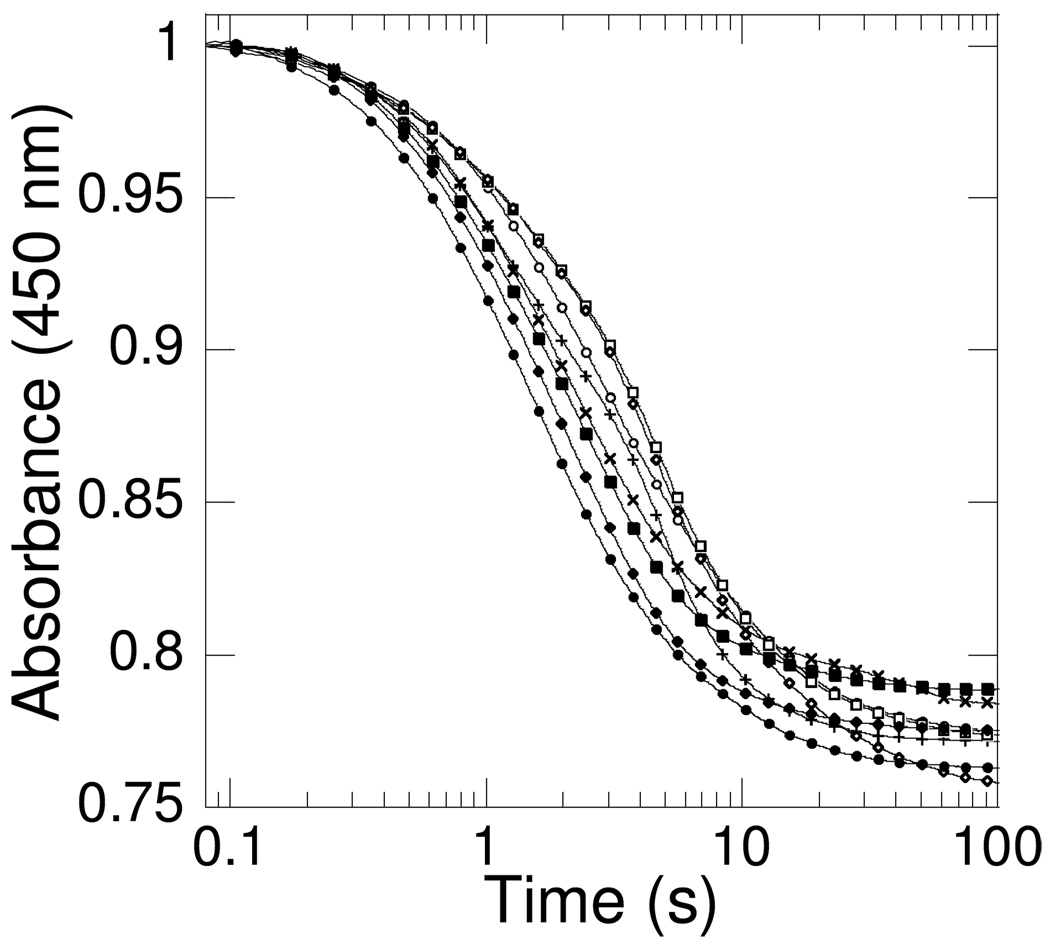
Several well-studied flavin-dependent monooxygenases require that the substrate be bound for NADH or NADPH to reduce the flavin (FAD or FMN), whereas others have no such requirement. To determine if reduction is dependent on ornithine for PvdA, the kinetics of the reduction of FAD by NADPH were examined (Figure 2). Enzyme that had been pre-mixed with no, one or two substrates was mixed in the stopped-flow apparatus with the other substrate(s). The effect of ornithine on the reduction kinetics was examined by separately including or omitting the substrate in the stopped-flow solutions. Pre-incubation of enzyme with ornithine, with or without flavin, and mixing with the remaining co-substrate(s) was also examined. A family of bi-phasic curves, presumably representing FAD reduction followed by NADP+ release, was generated. Flavin reduction occurs in the absence of ornithine, and the presence of the hydroxylation target accelerates the reaction by at most 2-fold. By comparison, p-hydroxybenzoate accelerates flavin reduction in PHBH by 104−105-fold (12, 13). The data in Figure 2 do not resolve the order of addition of all components, and further experiments monitoring the rate of reduction as a function of NADPH and ornithine concentrations are warranted to establish binding order. The important point is that FAD reduction by NADPH is essentially equally facile in the presence and absence of ornithine, inconsistent with the possibility that PvdA ensures coupling by the same mechanism employed by PHBH.
Figure 2.
Reductive half-reaction kinetics of PvdA. Absorbance-versus-time traces under different mixing conditions. Eight different mixing conditions were tested in oxygen-free 100 mM potassium phosphate pH 8.0 buffer containing PvdA (290 µM) at 22 °C. The coenzyme and cosubstrate concentrations were: FAD (60 µM) and NADPH (360 µM) in the same buffer and the spectra were recorded using a stopped-flow spectrometer equipped with a photomultiplier tube. Ornithine (10 mM) was included in some of the reaction mixtures. The curves were fit to a double exponential equation presumably for FAD reduction and NADP+ release (KaleidaGraph 4). E v FN (●) where E = enzyme, F = FAD, N = NADPH, and v = versus (mixed with), kobs = 0.63 and 0.11 s−1; ES v FN (♦) where S = ornithine, kobs = 0.50 and 0.08 s−1; E v FNS (■), kobs = 0.47 and 0.07s−1; EN v FS (+), kobs = 0.55 and 0.21 s−1; EN v F (×), kobs = 0.45 and 0.04 s−1; EF v N (○), kobs = 0.39 and 0.08 s−1; EF v NS (□), kobs = 0.33 and 0.11 s−1; EFS v N (◇), kobs = 0.24 and 0.04 s−1. The data points for each mixture were measured in duplicate and averaged. Every 20th data point is displayed for each fit curve.
Formation of the flavin intermediates in the presence of ornithine
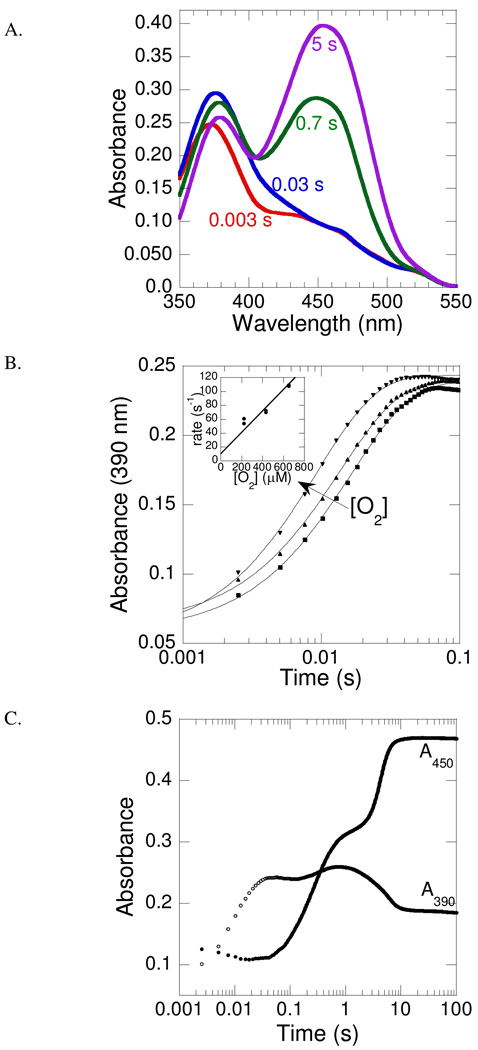
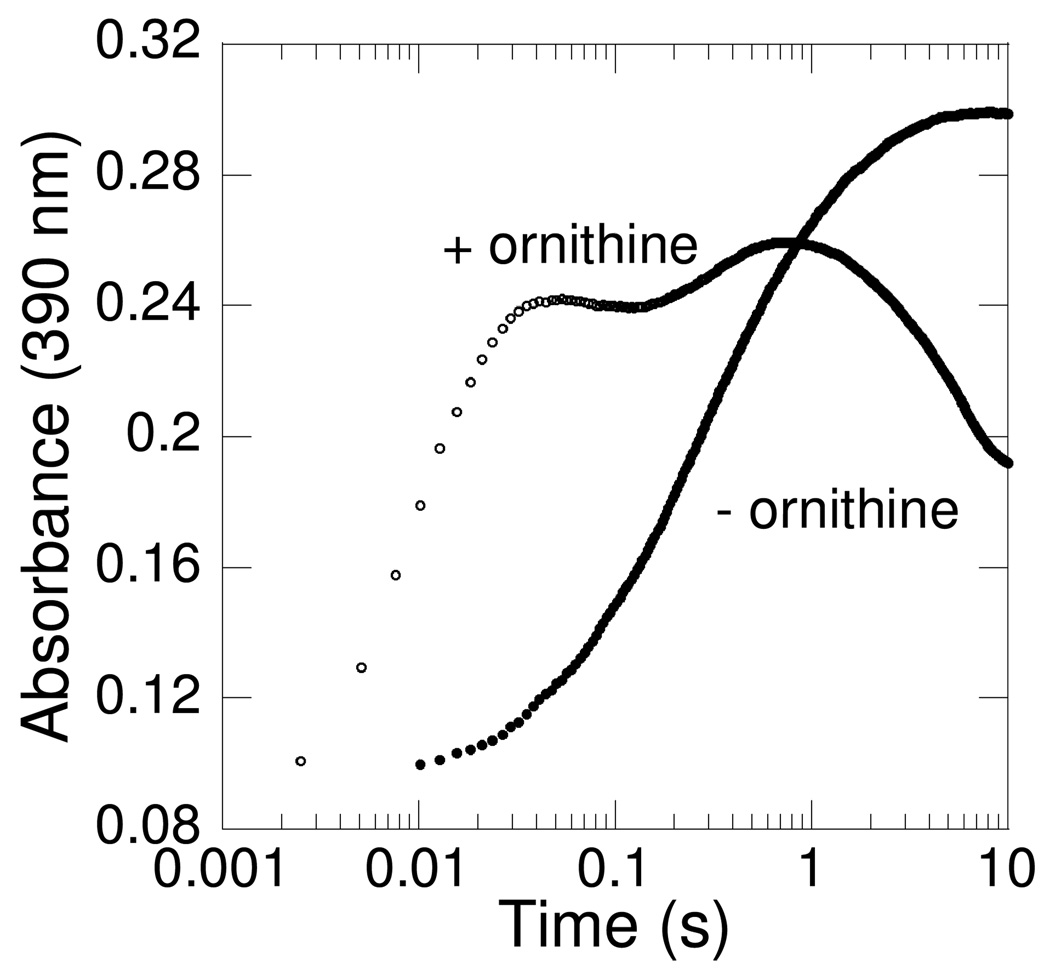
The oxidative half-reaction was investigated by mixing a solution containing PvdA (290 µM), FAD (60 µM), NADPH (360 µM) and ornithine (10 mM) with O2-containing buffer (1.3 mM) (Figure 3A). The hydroperoxyflavin intermediate accumulates rapidly under these reaction conditions, as evidenced by the characteristic absorption feature at 380 nm, which is similar to those from C(4a)-hydroperoxyflavin intermediates in related monooxygenases (23, 24). Measuring at 390 nm, the observed first-order rate constant for formation of the hydroperoxyflavin intermediate is linearly dependent on the oxygen concentration (Figure 3B). From this plot, the second-order rate constant for oxygen addition can be estimated as 150 ± 30 mM−1s−1. In Figure 3C, the increase in absorbance at 390 nm without an increase in 450 nm is also evident, correlating to this formation of hydroperoxyflavin. The oxygen is transferred from the hydroperoxyflavin to the substrate ornithine to produce the C(4a)-hydroxyflavin and the hydroxylated product, detected as an increase in absorbance at both 390 and 450 nm. The rate constant determined from the absorbance trace at 450 nm is 3.5 ± 0.7 s−1. The final species formed is the oxidized flavin, as a result of the dehydration of the hydroxyflavin. This species is characterized by an increase in absorbance at 450 nm and decrease in absorbance at 390 nm. The rate constant determined from the absorbance trace at 450 nm is 0.6 ± 0.1 s−1. The rates reported here are consistent with the steady-state kcat previously reported to be 0.4 s−1, which was determined with enzyme pre-equilibrated with excess FAD and NADPH (10).
Figure 3.
PvdA flavin oxidation in the presence of ornithine. A. An oxygen-free solution containing 290 µM PvdA, 60 µM FAD, 360 µM NADPH, 10 mM ornithine in 100 mM potassium phosphate pH 8.0 was mixed with an equal volume of the same buffer containing 1.3 mM oxygen at 22 °C. The spectra were recorded with a photodiode array. B. Representative absorbance-versus-time traces recorded at 390 nm using a photomultiplier tube. An oxygen-free solution containing 290 µM PvdA, 60 µM FAD, 360 µM NADPH, 10 mM ornithine in 100 mM potassium phosphate pH 8.0 was mixed with an equal volume of the same buffer containing oxygen concentrations such that the final O2 concentrations were 220 µM (■), 430 µM (▲), and 650 µM (▼). Each oxygen concentration was measured in duplicate and fit independently (KaleidaGraph 4) to produce the points for the inset plot. Inset: kobs versus oxygen concentration plot, indicating a second order rate constant of 150 ± 30 mM−1 s−1. Residual oxygen was detected in the system as evidenced by the non-zero rates at 0 µM O2, and therefore the line has a positive y-intercept. C. Representative absorbance-versus-time traces for absorbance at 390 nm (○) and 450 nm (●) were recorded using a photomultiplier tube. Enzyme, NADPH, FAD, ornithine and O2 mixing scheme as described in part A.
Formation of the flavin intermediates in the absence of ornithine
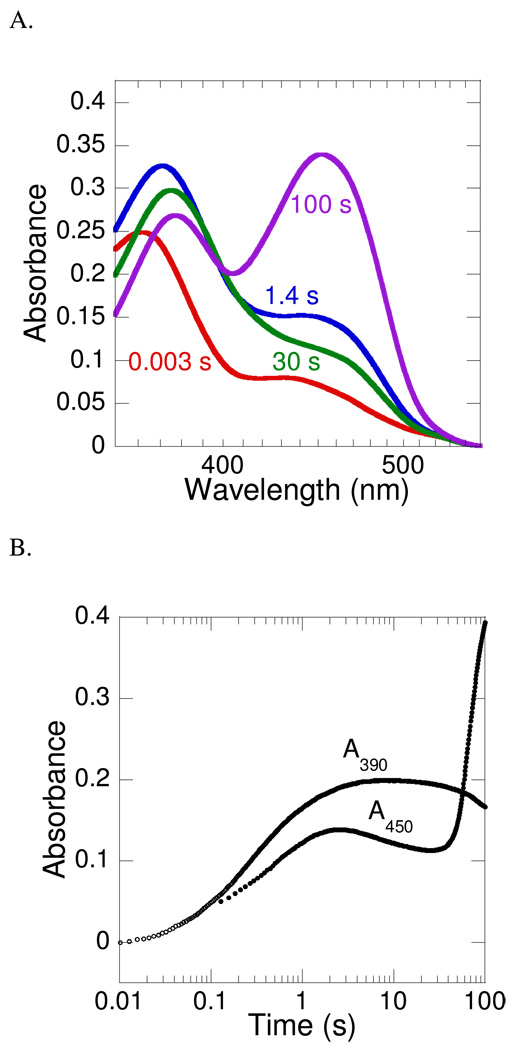
The oxidative half-reaction was also investigated in the absence of ornithine by mixing a solution containing PvdA (290 µM), FAD (60 µM), and NADPH (360 µM) with O2-containing buffer (1.3 mM). Close inspection of Figure 4A indicates that, in the first reliable spectrum (0.003 s), the absorbance maximum is at 361 nm, which shifts to 376 nm at later timepoints. In the reactions of both cyclohexanone monooxygenase (CHMO) (25) and phenol hydroxylase (PHHY) (26–28), C(4a)-peroxyflavin intermediates have been detected. For CHMO, the intermediate is characterized by a wavelength of maximum absorbance (λmax) of 366 nm, blue-shifted from the 383-nm λmax for the hydroperoxyflavin intermediate. Therefore, we have assigned the first flavin intermediate as a peroxyflavin (kobs = 1.8 ± 0.4 mM−1 s−1). Interestingly, this species shows an increase in absorbance at 450 nm, which may also be attributed to oxidized flavin in these turnover experiments (Figure 4B). After the formation of the peroxyflavin, the next step is protonation to form the hydroperoxyflavin. This second step is characterized by the spectral shift documented above and a decrease in absorbance at 450 nm, whereas the absorbance at 390 remains unchanged, characteristic of the hydroperoxyflavin detected in the presence of ornithine (Figure 4B). Finally, oxidized flavin begins to form by 100 s, as the increasing absorbance at 450 nm and decreasing absorbance at 390 nm indicate. This phase presumably represents the slow decay of the hydroperoxyflavin to the oxidized flavin with production of hydrogen peroxide rather than hydroxylated product.
Figure 4.
PvdA flavin oxidation in the absence of ornithine. A. An oxygen-free solution containing 290 µM PvdA, 60 µM FAD, 360 µM NADPH in 100 mM potassium phosphate, pH 8.0 was mixed with an equal volume of the same buffer containing 1.3 mM oxygen at 22 °C. The spectra were recorded with a photodiode array. B. Representative absorbance-versus-time traces for absorbance at 390 nm (○) and 450 nm (●) were recorded using a photomultiplier tube. Enzyme, FAD, NADPH, and O2 mixing scheme as described in part A.
Hydrogen peroxide detection when ornithine is absent
To confirm the formation of hydrogen peroxide in the reaction without ornithine, and to correlate H2O2 production to NADPH oxidation, the amount of NADPH oxidation and hydrogen peroxide formed in the absence of substrate were measured. At a four-minute timepoint, 7 ± 2 nmol of NADPH were oxidized while 6.6 ± 0.2 nmol of hydrogen peroxide were formed. Therefore, in the absence of the substrate ornithine, the hydroperoxyflavin decays to form oxidized flavin, and hydrogen peroxide is released.
Discussion
Flavin monooxygenases use a variety of methods to prevent wasteful NADPH consumption. In the case of PHBH (12, 13) and PHHY (29), the flavin reduction is greatly accelerated by the addition of substrate. Other flavin-containing enzymes are reduced quickly regardless of the presence or absence of substrate. In the case of FMO (11, 14, 20), addition of oxygen then forms a long-lived hyroperoxyflavin as the enzyme awaits substrate, whereas CHMO (25) forms a peroxyflavin. Excess substrate causes uncoupling of NADPH oxidation and ornithine hydroxylation in PvdA (10). At physiological ornithine concentrations, we hypothesize that PvdA uses a hybrid of the methods for reaction coupling already demonstrated for flavin monooxygenases. Reduction of the flavin by NADPH is independent of the presence of ornithine, and addition of the oxygen to the flavin is accelerated by the presence of ornithine. In Figure 5 we have redrawn the A390 traces from Figure 3B and Figure 4B to emphasize the delay in oxygen addition to the flavin due to the absence of ornithine.
Figure 5.
Representative absorbance-versus-time traces for absorbance at 390 nm in the presence (○) and absence (●) of ornithine replotted from Figure 3B and Figure 4B for direct comparison.
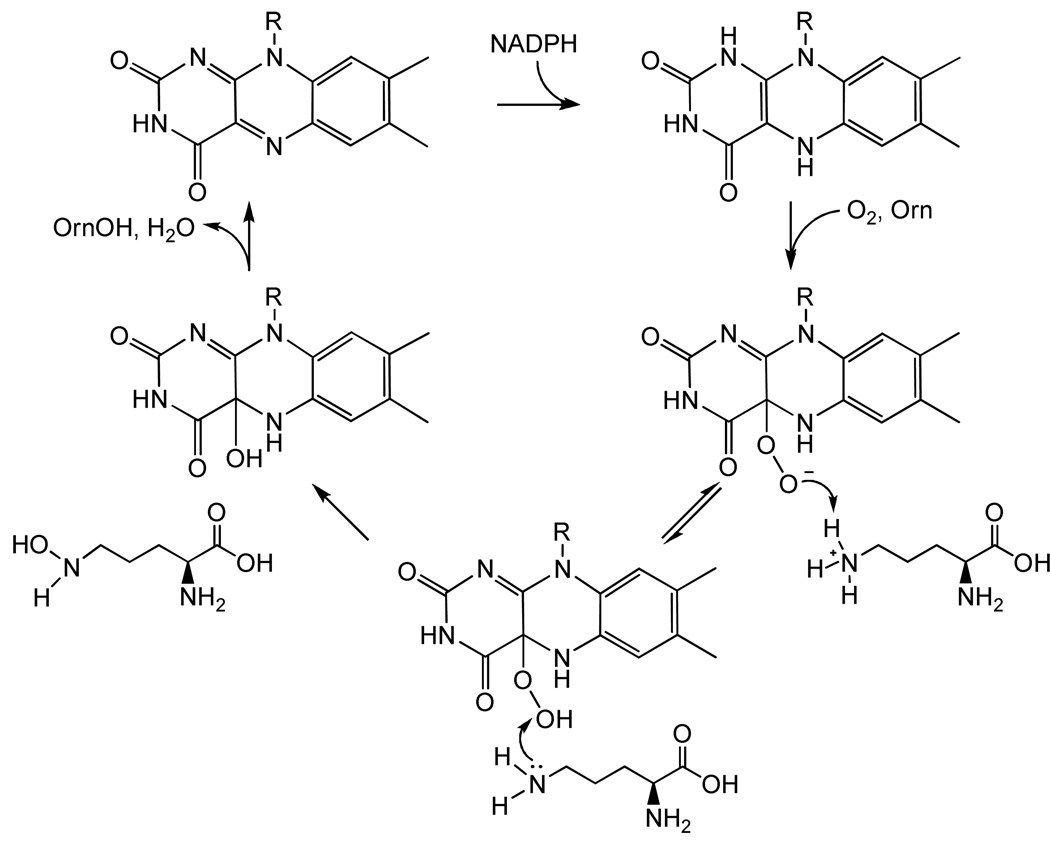
We propose that PvdA produces a peroxyflavin intermediate, also seen previously in cyclohexanone monooxygenase (25) and phenol hydroxylase (26–29). For CHMO, the peroxyflavin (λmax = 366 nm) is the reactive intermediate (the hydroperoxyflavin, λmax = 383 nm, is not reactive) to perform a Baeyer-Villiger rearrangement in which the cyclic ketone is converted to ɛ-caprolactone (25). In addition to their absorption signatures, the crucial evidence for the assignment of the two intermediates as protonated and unprotonated peroxide complexes is that they can be inter-converted by varying pH. In PvdA, the spectral shift is clearly evident without ornithine (Figure 4A) and is also evident with ornithine at very early timepoints (Figure 3A), suggesting that an unprotonated peroxide may also form early in the PvdA reaction and be rapidly converted to the C(4a)-hydroperoxyflavin by protonation. In the absence of ornthine, the formation of the peroxyflavin is 80-fold slower than in the presence of ornithine. The peroxy-and hydroperoxyflavin intermediates of CHMO are in equilibrium, and the reported pKa for this proton is 8.4 (25). In PvdA, it is tempting to hypothesize that the protonation of the peroxyflavin to make the hydroperoxyflavin is linked to deprotonation of the ornithine (pKa = 8.7, (30, 31)) making the ornithine reactive, ready to accept a hydroxyl group (Scheme 1). The similarity of the ornithine and hydroperoxyflavin pKas bolsters our hypothesis that the ornithine deprotonation and peroxyflavin protonation may be linked.
Scheme 1.
PvdA was previously hypothesized to be mechanistically similar to either PHBH or FMO (10). PvdA is comparably similar to both enzymes (∼35% sequence similarity) despite the lack of structural homology between PHBH and FMO. Based on this new mechanistic information, the sequence of PvdA and PHHY were aligned. With the removal of the C-terminal domain of unknown function of PHHY (32) and alignment of the proposed co-enzyme and co-substrate binding regions, the similarity reaches 50% (20% identical). We therefore hypothesize that PvdA may be most similar in structure to the two N-terminal domains of PHHY.
Conclusions
PvdA, the ornithine monooxygenase from Pseudomonas aeruginosa, demonstrates a unique kinetic mechanism. In the reductive half-reaction, substrate is not required for NADPH-reduction of FAD. The oxidative half-reaction includes three flavin intermediates: peroxy-, hydroperoxy-, and hydroxyflavins; of which the hydroperoxyflavin is the reactive intermediate. Therefore, ornithine accelerates oxygen addition to the flavin, suggesting a novel mechanism for the conservation of NADPH in the absence of substrate.
Acknowledgements
We thank T. C. Gamblin for the use of the Cary WinUV Microplate Spectrophotometer. We are indebted to B. Palfey for continued discussions about flavin intermediates.
Abbreviations and Full Textual Notes
- CHMO
cyclohexanone monooxygenase from Acinetobacter sp.
- FlHOO-
peroxyflavin
- FlHOOH
hydroperoxyflavin
- FlHOH
hydroxyflavin
- Flox
oxidized FAD
- Flred
reduced FAD
- FMO
flavin-containing monooxygenase
- NRPS
nonribosomal peptide synthetase
- Orn
ornithine
- OrnOH
N5-hydroxyornithine
- PvdA
ornithine hydroxylase or L-ornithine N5-oxygenase from Pseudomonas aeruginosa
- PvdF
hydroxyornithine transformylase from Pseudomonas aeruginosa
- PHBH
para-hydroxybenzoate hydroxylase from Pseudomonas fluorescens
- PHHY
phenol hydroxylase from Trichosporon cutaneum.
Footnotes
This publication was made possible by NIH Grant Number P20 RR-17708-05 from the National Center for Research Resources of the National Institute of Health. K.M.M. was a recipient of a National Institutes of Health Predoctoral Training Grant Fellowship (GM08545).
References
- 1.Crosa JH. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 2000;174:135–142. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- 3.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox CD, Adams P. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect. Immun. 1985;48:130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendenbaum S, Demange P, Dell A, Meyer JM, Abdallah MA. The Structure of Pyoverdine Pa, the Siderophore of Pseudomonas aeruginosa. Tetrahedron Letters. 1983;24:4877–4880. [Google Scholar]
- 6.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme L-ornithine N5-oxygenase in Pseudomonas aeruginosa. J. Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visca P, Serino L, Orsi N. Isolation and characterization of Pseudomonas aeruginosa mutants blocked in the synthesis of pyoverdin. J. Bacteriol. 1992;174:5727–5731. doi: 10.1128/jb.174.17.5727-5731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamont IL, Martin LW. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology. 2003;149:833–842. doi: 10.1099/mic.0.26085-0. [DOI] [PubMed] [Google Scholar]
- 9.McMorran BJ, Kumara HM, Sullivan K, Lamont IL. Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiology. 2001;147:1517–1524. doi: 10.1099/00221287-147-6-1517. [DOI] [PubMed] [Google Scholar]
- 10.Meneely KM, Lamb AL. Biochemical characterization of a flavin adenine dinucleotide-dependent monooxygenase, ornithine hydroxylase from Pseudomonas aeruginosa, suggests a novel reaction mechanism. Biochemistry. 2007;46:11930–11937. doi: 10.1021/bi700932q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beaty NB, Ballou DP. The oxidative half-reaction of liver microsomal FAD-containing monooxygenase. J. Biol. Chem. 1981;256:4619–4625. [PubMed] [Google Scholar]
- 12.Howell LG, Spector T, Massey V. Purification and Properties of p-Hydroxybenzoate Hydroxylase from Pseudomonas fluorescens. J. Biol. Chem. 1972;247:4340–4350. [PubMed] [Google Scholar]
- 13.Husain M, Massey V. Kinetic studies on the reaction of p-hydroxybenzoate hydroxylase. Agreement of steady state and rapid reaction data. J. Biol. Chem. 1979;254:6657–6666. [PubMed] [Google Scholar]
- 14.Jones KC, Ballou DP. Reactions of the 4a-hydroperoxide of liver microsomal flavin-containing monooxygenase with nucleophilic and electrophilic substrates. J. Biol. Chem. 1986;261:2553–2559. [PubMed] [Google Scholar]
- 15.Spector T, Massey V. p-Hydroxybenzoate Hydroxylase from Pseudomonas fluorescens, Evidence for an Oxygenated Flavin Intermediate. J. Biol. Chem. 1972;247:5632–5636. [PubMed] [Google Scholar]
- 16.Moran GR, Entsch B, Palfey BA, Ballou DP. Electrostatic effects on substrate activation in para-hydroxybenzoate hydroxylase: studies of the mutant lysine 297 methionine. Biochemistry. 1997;36:7548–7556. doi: 10.1021/bi9706327. [DOI] [PubMed] [Google Scholar]
- 17.Moran GR, Entsch B, Palfey BA, Ballou DP. Mechanistic insights into p-hydroxybenzoate hydroxylase from studies of the mutant Ser212Ala. Biochemistry. 1999;38:6292–6299. doi: 10.1021/bi990021+. [DOI] [PubMed] [Google Scholar]
- 18.Palfey BA, Massey V. Flavin-Dependent Enzymes. In: Sinnott M, editor. Comprehensive Biological Catalysis. San Diego: Academic Press; 1998. pp. 83–154. [Google Scholar]
- 19.Yeh E, Cole LJ, Barr EW, Bollinger JM, Jr, Ballou DP, Walsh CT. Flavin redox chemistry precedes substrate chlorination during the reaction of the flavin-dependent halogenase RebH. Biochemistry. 2006;45:7904–7912. doi: 10.1021/bi060607d. [DOI] [PubMed] [Google Scholar]
- 20.Beaty NB, Ballou DP. The reductive half-reaction of liver microsomal FAD-containing monooxygenase. J. Biol. Chem. 1981;256:4611–4618. [PubMed] [Google Scholar]
- 21.Hitchman ML. Measurement of dissolved oxygen. New York: Wiley; 1978. [Google Scholar]
- 22.Ge L, Seah SY. Heterologous expression, purification, and characterization of an L-ornithine N(5)-hydroxylase involved in pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7205–7210. doi: 10.1128/JB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Entsch B, Ballou DP, Massey V. Flavin-oxygen derivatives involved in hydroxylation by p-hydroxybenzoate hydroxylase. J. Biol. Chem. 1976;251:2550–2563. [PubMed] [Google Scholar]
- 24.Ghisla S, Entsch B, Massey V, Husein M. On the structure of flavin-oxygen intermediates involved in enzymatic reactions. Eur. J. Biochem. 1977;76:139–148. doi: 10.1111/j.1432-1033.1977.tb11579.x. [DOI] [PubMed] [Google Scholar]
- 25.Sheng D, Ballou DP, Massey V. Mechanistic studies of cyclohexanone monooxygenase: chemical properties of intermediates involved in catalysis. Biochemistry. 2001;40:11156–11167. doi: 10.1021/bi011153h. [DOI] [PubMed] [Google Scholar]
- 26.Maeda-Yorita K, Massey V. On the reaction mechanism of phenol hydroxylase. New information obtained by correlation of fluorescence and absorbance stopped flow studies. J. Biol. Chem. 1993;268:4134–4144. [PubMed] [Google Scholar]
- 27.Xu D, Ballou DP, Massey V. Studies of the mechanism of phenol hydroxylase: mutants Tyr289Phe, Asp54Asn, and Arg281Met. Biochemistry. 2001;40:12369–12378. doi: 10.1021/bi010962y. [DOI] [PubMed] [Google Scholar]
- 28.Xu D, Enroth C, Lindqvist Y, Ballou DP, Massey V. Studies of the mechanism of phenol hydroxylase: effect of mutation of proline 364 to serine. Biochemistry. 2002;41:13627–13636. doi: 10.1021/bi020446n. [DOI] [PubMed] [Google Scholar]
- 29.Detmer K, Massey V. Effect of monovalent anions on the mechanism of phenol hydroxylase. J. Biol. Chem. 1984;259:11265–11272. [PubMed] [Google Scholar]
- 30.Batchelder AC, Schmidt CLA. The effects of certain salts on the dissociation of aspartic acid, arginine and ornithine. J. Phys. Chem. 1940;44:893–909. [Google Scholar]
- 31.Kuo LC, Herzberg W, Lipscomb WN. Substrate specificity and protonation state of ornithine transcarbamoylase as determined by pH studies. Biochemistry. 1985;24:4754–4761. doi: 10.1021/bi00339a007. [DOI] [PubMed] [Google Scholar]
- 32.Enroth C, Neujahr H, Schneider G, Lindqvist Y. The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis. Structure. 1998;6:605–617. doi: 10.1016/s0969-2126(98)00062-8. [DOI] [PubMed] [Google Scholar]
- 33.Hosokawa K, Stanier RY. Crystallization and properties of p-hydroxybenzoate hydroxylase from Pseudomonas putida. J. Biol. Chem. 1966;241:2453–2460. [PubMed] [Google Scholar]
- 34.Poulsen LL, Hyslop RM, Ziegler DM. S-oxidation of thioureylenes catalyzed by a microsomal flavoprotein mixed-function oxidase. Biochem. Pharm. 1974;23:3431–3440. doi: 10.1016/0006-2952(74)90346-3. [DOI] [PubMed] [Google Scholar]
- 35.Prough RA. The N-oxidation of alkylhydrazines catalyzed by the microsomal mixed-function amine oxidase. Arch. Biochem. Biophys. 1973;158:442–444. doi: 10.1016/0003-9861(73)90641-3. [DOI] [PubMed] [Google Scholar]
- 36.Ziegler DM, Jollow D, Cook DE. In: The Third International Symposium on Flavins and Flavoproteins. Kamin H, editor. Baltimore: University Park Press; 1971. pp. 475–497. [Google Scholar]